Abstract
Rising ocean temperatures and extreme temperature events have precipitated declines and local extinctions in many marine species globally, but patterns of loss are often uneven across species ranges for reasons that are poorly understood. Knowledge of the extent of local adaptation and gene flow may explain such patterns and help predict future trajectories under scenarios of climate change. We test the extent to which local differentiation in thermal tolerance is influenced by gene flow and local adaptation using a widely distributed intertidal seaweed (Hormosira banksii) from temperate Australia. Population surveys across ~2,000 km of the species range revealed strong genetic structuring at regional and local scales (global F ST = 0.243) reflecting extremely limited gene flow, while common garden experiments (14‐day exposures to 15, 18, 21°C) revealed strong site differences in early development and mortality in response to elevated temperature. Embryos from many sites spanning a longitudinal thermal gradient showed suppressed development and increased mortality to elevated water temperatures, but populations originating from warmer and more variable thermal environments tended to be less susceptible to warming. Notably, there was significant local‐scale variation in the thermal responses of embryos within regions which was corroborated by the finding of small‐scale genetic differences. We expect the observed genetic and phenotypic differentiation to lead to uneven responses to warming sea surface temperatures in this important marine foundation species. The study highlights the challenges of predicting species responses to thermal stress and the importance of management strategies that incorporate evolutionary potential for “climate‐proofing” marine ecosystems.
Keywords: climate change, evolutionary potential, gene flow, local adaptation, marine ecosystem engineer
1. INTRODUCTION
The speed and magnitude of biotic shifts being triggered by climate change pose a major challenge for marine biodiversity conservation. Rising ocean temperatures, increasing acidification, and changing ocean currents are contributing to fundamental and irreversible ecological transformations in marine ecosystems at a global scale (Babcock et al., 2019; Harris et al., 2018; Hoegh‐Guldberg & Bruno, 2010). Species living close to their physiological limits are of particular concern and will become increasingly dependent on their ability to overcome environmental change via dispersal, physical tolerance, and evolutionary adaptation (Hoffmann & Sgro, 2011; Portner & Gutt, 2016). Understanding the environmental resilience of ecosystem engineers (i.e., keystone and foundation species) is of particular importance, given their response to environmental change will have major impacts on community structure and ecosystem function (Babcock et al., 2019; Hoegh‐Guldberg & Bruno, 2010; Smale et al., 2019; Thomson et al., 2015). Such information is necessary to inform marine conservation planning directed at minimizing biodiversity loss, and preserving environmental, economic, and cultural values (Magris, Pressey, Weeks, & Ban, 2014).
Macrophytes are key foundation species in temperate oceans underpinning reef ecosystem services (Dayton, 1972; Jones, Lawton, & Shachak, 1994) but are at increasing threat from environmental change with declines seen as a result of warming (Vergés et al., 2016, 2014), heatwaves (Thomson et al., 2015; Wernberg, Bennett, et al., 2016), over‐harvesting (Steneck et al., 2002), and urbanization (Airoldi & Beck, 2007; Coleman, Kelaher, Steinberg, & Millar, 2008; Connell et al., 2008). Critically, such declines appear to be long term or even permanent, highlighting a need to understand the adaptability and vulnerability of marine macrophytes to future change. This is pertinent in areas where marine macrophyte communities are showing signs of acute and chronic climate stress, such as climate change “hot spots” that are particularly prone to extreme temperature events, and that are experiencing sea surface temperature orders of magnitude above the global average (Babcock et al., 2019; Hobday & Pecl, 2014; Wernberg et al., 2011). This is a major concern as thermal stress is known to impair photosynthetic, respiratory, and cellular function in macrophyte species, inhibiting growth, inducing mortality and disease, and resulting in range contractions and local extirpations (Flukes, Wright, & Johnson, 2015; Mathieson & Dawes, 1986; Pineiro‐Corbeira, Barreiro, Cremades, & Arenas, 2018; Smale & Wernberg, 2013; Wernberg, Bettignies, Joy, & Finnegan, 2016). Furthermore, thermal stress can suppress macrophyte resilience to environmental stressors associated with other anthropogenic and natural perturbations (Wernberg et al., 2010) as well as alter key ecological interactions that impact macrophyte populations, such as grazing and predation (Miranda et al., 2019; Provost et al., 2017; Vergés et al., 2014). Coupled climate–ecosystem models suggest that declines are likely to intensify in coming decades, with projections of major range contractions of temperate seaweeds, and potential risks of extinction for many species (Martinez et al., 2018).
The resilience of marine macrophyte populations to environmental change will be largely determined by levels of standing adaptive genetic variation and patterns of gene flow among populations. Traditionally, insights into climate resilience have been gained by characterizing patterns of gene flow among populations distributed across thermal gradients. While gene flow can be an impediment to local adaptation, it can also assist the adaptation process by making thermally adapted genotypes available for selection (Hoffmann & Sgro, 2011; Hoffmann & Willi, 2008). Also, quantitative approaches (e.g., common garden and transplant experiments) are used to assess the extent to which species and populations have adapted to different environments, by testing the genetic basis of trait variation spanning environmental gradients (Hoffmann & Willi, 2008). Such studies have been particularly useful for assessing historical responses to environmental change and predicting the evolvability of species from both marine and terrestrial systems (Sgro, Lowe, & Hoffmann, 2011; Sherman & Ayre, 2008), but they are limited for marine macrophytes.
Predicting species responses to warming environments requires an understanding of intraspecific variation in thermal responses and gene flow across species ranges (King, McKeown, Smale, & Moore, 2018). Predictive tools such as species distribution and climate niche models do not directly consider physiological variation unless this variation contributes to extant distributions (Razgour et al., 2019; Willis et al., 2015). Such models may therefore be limited in predicting range shifts, particularly at edges of a species’ distribution, where temperatures are expected to exceed theoretical species “thermal niche” limits or safety margins (Bennett, Wernberg, Joy, Bettignies, & Campbell, 2015), which may be countered by evolutionary shifts (Bush et al., 2016). Variability in thermal physiology between populations has been established for several marine macrophytes (reviewed in King et al., 2018), which may contribute to uneven population responses to thermal stress (Bennett et al., 2015; Carballo, Olabarria, & Osuna, 2002; Filbee‐Dexter, Feehan, & Scheibling, 2016; Starko et al., 2019; Tegner & Dayton, 1987; Thomsen et al., 2019). This variation often coincides with fine‐scale genetic structuring (King et al., 2018), but empirical tests of this are lacking. Information on the genetic structure of populations and phenotypic differences among populations is needed to help understand the uneven thermal response of populations, identify those most vulnerable to thermal stress, and indicate where genotypes resilient to future climate changes might reside. These are all important components of climate change adaptation and restoration programs (Foden et al., 2019; Jordan, Hoffmann, Dillon, & Prober, 2017; Willis et al., 2015; Wood et al., 2019).
Hormosira banksii (Turner) Decaisne is the dominant intertidal macrophyte across temperate Australasia and is an autogenic ecosystem engineer with no functional equivalents (Bishop et al., 2009; Pocklington, Keough, O'Hara, & Bellgrove, 2019; Schiel, 2006). This species is highly sensitive to environmental disturbance associated with coastal development and urbanization (Doblin & Clayton, 1995; Keough & Quinn, 1998; Kevekordes, 2000), and thermal stress (Alestra & Schiel, 2015; Tait & Schiel, 2013). H. banksii embryo development is particularly sensitive to upwards shifts in temperature, with water 3°C higher than ambient levels causing significant mortality (Alestra & Schiel, 2015; Clark, Poore, Ralph, & Doblin, 2013). Yet significant genotype × environment interactions for embryo growth have been reported, as well as growth and photosystem traits that show heritable genetic variation relating to temperature (Clark, Poore, & Doblin, 2018; Clark et al., 2013). These findings suggest there is potential for selection to result in the evolution of more thermally tolerant populations. However, despite potential standing genetic variation in some populations for thermal adaptation, gene flow may be limited in H. banksii, restricting the adaptive potential of this species. Previous studies indicate strong genetic structuring among populations from the central region of Australia's east coast (Coleman et al., 2011; Coleman, Clark, Doblin, Bishop, & Kelaher, 2019) suggesting that gene flow is unlikely to facilitate the natural movement of genetic variants across temperature gradients to aid adaptation and recovery of depleted populations. Such genetic structuring could also contribute to local adaptation and variation in thermal tolerances among populations spanning thermal gradients in Australian temperate waters. However, the genetic structure across the full range of this species, and levels of population variability in thermal tolerance, is currently unknown.
In this study, we assessed the potential adaptability of H. banksii to rising ocean temperatures across much of its distribution. First, we use microsatellite markers to investigate patterns of genetic diversity, gene flow, and population connectivity from sites spanning ~2,000 km of the distribution of this species around mainland Australia and covering a wide temperature gradient (mean summer sea surface temperatures ranging from 16 to 24°C). Second, we undertake a common garden experiment to explore spatial variation in thermal responses among populations with different thermal histories by comparing embryo development at different experimental temperatures. Outcomes from these analyses will help determine the role of gene flow and local adaptation in affecting responses of populations to thermal stress. We expected populations under thermal extremes in the sampling range to be phenotypically adapted to those temperature environments, particularly in the presence of low levels of gene flow. Any adaptive changes would produce uneven thermal responses among populations across the species range. We discuss the implications of our results in the context of the future evolutionary potential of H. banksii, and conservation/ restoration approaches that might be considered to catalyze adaptation and recovery processes in light of climate change.
2. METHODS
2.1. Study species
Hormosira banksii is a perennial, dioecious, habitat‐forming fucoid macroalga with a distribution extending >2,000 km from Albany in Western Australia to Lennox Head in New South Wales on mainland Australia, around Tasmania, the North and South Islands of New Zealand, and some of the smaller offshore islands in southern Australasia (Clark et al., 2018; Osborn, 1948). H. banksii is fertile throughout the year, releasing gametes on emersion during low tide (Levring, 1949). Once fertilized, eggs are thought to attach rapidly, thereby limiting dispersal (Bellgrove, Clayton, & Quinn, 1997, 2004). The thallus of H. banksii consists of multiple chains of hollow, water‐filled, vesicles that arise from a diffuse holdfast (Osborn, 1948) capable of vegetative regeneration (Keough & Quinn, 1998; Schiel & Taylor, 1999), with individual plants surviving >11 years (Kain, 2015). H. banksii shows significant morphological variability among populations and across geographic regions, and between marine and estuarine environments shown to have functional consequences (Clark et al., 2018). This is thought to be correlated with local environmental conditions (Macinnis‐Ng, Morrison, & Ralph, 2005; Ralph, Morrison, & Addison, 1998) and may be linked to genetic differentiation (Coleman et al., 2019).
2.2. Population genetic analysis
2.2.1. Sampling
Tissue samples of H. banksii were collected between December 2006 and April 2007 at 3 spatial scales across the geographic distribution of this species, with nesting present within each scale: large scale (1,000s km separation between states and across the Tasman Sea to New Zealand; hereafter referred to as states), intermediate scale (regions separated by 100s km), and small scale (sites separated by 10s km). We sampled from two rocky‐shore sites within each of nine regions within three states (a total of 18 sample locations): three regions in each of South Australia, Victoria, and New South Wales (Table 1; Figure 1a,b). At each site, 1–2 vesicles from each of 32 individual clumps (defined as an individual with one holdfast and multiple fronds) of H. banksii were collected, washed in freshwater, blotted dry, and desiccated in silica gel for later genetic analysis. Genomic DNA was extracted from 10 mg sample of dried material tissue for individual specimens using the NucleoSpin® 96 Plant II protocol (Macherey‐Nagel Inc., Düren, KO, GER).
Table 1.
Location details and corresponding codes for 18 collection sites of Hormosira banksii from southern, southeastern, Western Australia, and New Zealand which were used for genetic analyses
| Region | Site | Code | Latitude | Longitude |
|---|---|---|---|---|
| New South Wales | ||||
| Central Coast | McMasters | MCM | −33.5011 | 151.4261 |
| Terrigal | TER | −33.4565 | 151.4385 | |
| Sydney Region | Long Reef | LOR | −33.7378 | 151.3131 |
| Sutherlands Point | SUT | −34.0035 | 151.2282 | |
| South Coast | Dalmeny | DAL | −36.1628 | 150.1297 |
| Mystery Bay | MYS | −36.3036 | 150.1389 | |
| Victoria | ||||
| Wilsons Promontory | Cape Patterson | CPA | −38.6667 | 145.6167 |
| Walkerville | WKV | −38.8333 | 146.0000 | |
| Mornington Peninsula | Flinders West | FLW | −38.4667 | 145.1000 |
| London Bridge | LOB | −38.3167 | 144.6833 | |
| Southwest Coast | Backyards | BAK | −38.4333 | 142.5833 |
| Oigles | OIG | −38.3833 | 142.2167 | |
| South Australia | ||||
| Fleurieu Peninsula | Aldinga | ALD | −35.2833 | 138.4403 |
| Willunga | WIL | −35.2703 | 138.4431 | |
| Yorke Peninsula | Gleeson Landing | GLE | −35.0089 | 136.9506 |
| Stenhouse | STN | −35.2794 | 136.9422 | |
| Eyre Peninsula | Porter Bay | POR | −34.1278 | 135.2664 |
| Point Avoid | PTA | −34.6781 | 135.3267 | |
Figure 1.
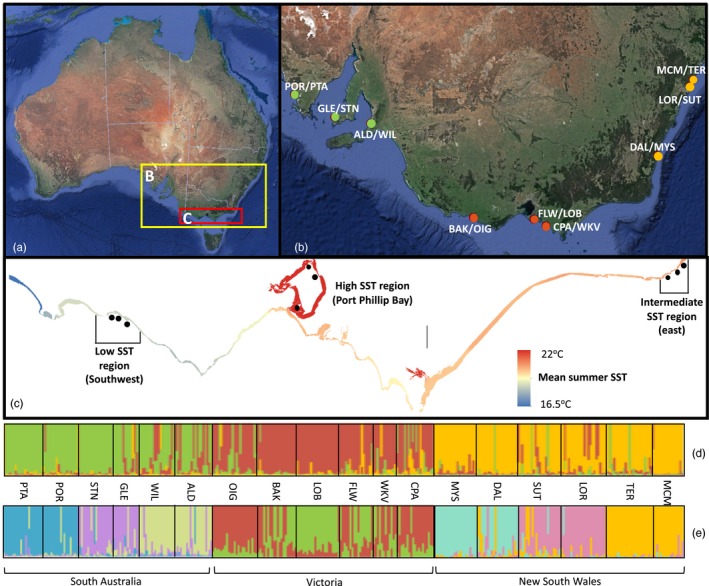
Maps showing Hormosira banksii sampling locations for the population genetic analysis (map “B”) and common garden experiments (map “C,” modified from Ierodiaconou et al., 2018). Map c shows the nesting of collection sites within three distinctive temperature zones (low, intermediate, and high), with the coastline color coded according to local mean summer sea surface temperatures (SST). Summary plots of the population subdivision based on STRUCTURE analyses of Hormosira banksii multilocus genotypes are also included (“d” and “e”). Single vertical lines broken into segments reflect each individual in the STRUCTURE summary plot, where segments are proportional to the membership coefficient for each of the population clusters. Individuals are arranged into sites from which they were sampled following the order given in Table 1 (site codes are also derived from Table 1). STRUCTURE plots are provided for the first hierarchical level of structure (all states included in the analysis; plot “d”) and the second hierarchical level of structure (independent analysis of sites from New South Wales, Victoria, and South Australia; plot “e”)
2.2.2. Microsatellite analysis
A total of 373 H. banksii individuals from 26 sites spanning New South Wales, Victoria, and South Australia were genotyped at 10 microsatellite loci previously developed by Bellgrove et al. (2017). Loci were amplified by multiplex PCR using Eppendorf Mastercycler S gradient units following the protocol described by Blacket, Robin, Good, Lee, and Miller (2012). Genotyping was performed using an Applied Biosystems 3730 capillary analyzer (AGRF, Melbourne Australia), and microsatellite profiles were examined and scored manually using GENEMAPPER version 4.0 (Applied Biosystems). Independence of loci (absence of linkage) was assessed using the software GENEPOP, version 3.4 (Raymond & Rousset, 1995) and the probability test function, which found no significant association among loci. The software MICRO‐CHECKER (Van Oosterhout, Hutchinson, Wills, & Shipley, 2004) was used to assess microsatellite loci for null alleles and scoring errors using formula 1 outlined by Brookfield (1996) and found no evidence of null alleles across sites and loci. Descriptive statistics were calculated for the microsatellite data using FSTAT version 2.9.3 (Goudet, 1995), including allelic richness per population averaged over loci, Weir and Cockerham's inbreeding coefficient (F IS: the deficiency of heterozygotes relative to the level expected with random mating), and a global estimate of population differentiation (F ST) with 95% confidence limits (Weir & Cockerham, 1984). In addition, population pairwise measures of F ST and their significance were determined using permutation (10,000 permutations). In order to overcome potential limitations of F ST calculations using multiallelic loci (Jost, 2008), additional estimates of population differentiation, global D est, and population pairwise measures of D est (significance determined using 10,000 permutations) were generated using GenAlEx 6.5 (Peakall & Smouse, 2006). The false discovery rate (FDR) procedure (Benjamini & Hochberg, 1995) was used to adjust significance levels when performing multiple simultaneous comparisons.
Estimates of observed (Ho) and expected (He) heterozygosity were determined using the Excel Microsatellite Toolkit (Park, 2001) and deviations from Hardy–Weinberg equilibrium (HWE) were determined using Genepop version 3.4 (Raymond & Rousset, 1995). An analysis of molecular variation (AMOVA) was performed in GenAlEx 6.5 (Peakall & Smouse, 2006) using pairwise F ST as the distance measure, with 10 000 permutations and missing data for loci set at 10%. The model for analysis partitioned variation among regions, among sample sites within regions, and within sample sites. A discriminant analysis of principal components (DAPC), implemented in the adegenet package for R (Jombart, 2008; Jombart & Ahmed, 2011), summarized patterns of genetic differentiation between sample sites.
Bayesian analyses implemented in STRUCTURE (Pritchard, Stephens, & Donnelly, 2000) were conducted to estimate the number of populations within the sample data. STRUCTURE identifies the number of distinct population clusters, assigns individuals to clusters, and identifies migrants and admixed individuals using genetic data only. To determine the number of populations (K), five independent simulations for K = 1–18 with 100,000 burn‐in and 1,000,000 data iterations were run. Analyses were performed using the admixture model of population structure (i.e., each individual draws some fraction of their genome from each of K populations) with no prior population information included, and allele frequencies set as independent among populations. The most likely K was estimated using Evanno's ΔK (Evanno, Regnaut, & Goudet, 2005) in Structure Harvester (Earl & Vonholdt, 2012).
Rates of recent migration were estimated among each of the sampling locations using a Bayesian algorithm implemented in BAYESASS v.3.0.3 (Wilson & Rannala, 2003). The program estimates migration among populations within the last three generations. To identify movements among populations, five independent runs of 107 Markov chain Monte Carlo (MCMC) iterations were used following a burn‐in period of 107 and a sampling interval of 500 steps. Chains were compared with a stationary posterior distribution for convergence by performing multiple runs with dispersed starting values. The proportion of individuals that were assigned as migrants (migration rates), and associated 95% confidence intervals (CIs) were estimated among each of the sampling locations.
2.3. Testing for local adaptation
2.3.1. Site selection and sampling
We tested for genetically based variation in thermal responses by performing common garden experiments on H. banksii sampled from across a longitudinal temperature gradient in south eastern Australia. Sampling was conducted according to a spatially hierarchical design, with H. banksii sampled from three sites nested within three distinct temperature regions of the Victorian coastline (Table 2; Figure 1c). Processed sea surface temperature (SST) datasets for the years 1995–2015 were evaluated from the Integrated Marine Observing System (IMOS; Ierodiaconou et al., 2018) and used to identify the three most variable SST zones, including southwestern Victoria (low temperature and variability), eastern Victoria/southern NSW (intermediate temperature and variability), and Port Phillip Bay (high temperature and variability) (hereafter Southwest, East, and PPB, respectively; Figure 1c). Sea surface temperature rather than air temperature was recognized as the most important variable for early life stage development, as fertilization and early embryonic development occurs during submersion after gamete release during low tide (Bellgrove, McKenzie, McKenzie, & Sfiligoj, 2010), and intertidal seaweeds are most physiologically competent during periods of submersion (Hurd, Harrison, Bischof, & Lobban, 2014). Sampling was performed in October 2018 (austral spring), with simultaneous collections from all three regions. Approximately 2 kg of mature H. banksii fronds was randomly collected by hand at low tide from a minimum of 50 individuals distributed over an area of 50–100 m2 at each site. Samples were transported to the laboratory on ice and stored in labeled plastic bags at 4°C in the dark for less than 3 days prior to experimentation. Sites from the southwestern and eastern Victorian regions were included in both the population genetic and common garden experiments, while sites from Port Phillip Bay were used only for the common garden experiment.
Table 2.
Location details for 9 collection sites of Hormosira banksii from southeastern Australia used for the common garden experiment, including mean summer and mean seasonal ranges in sea surface temperature in decimal degrees (°C)
| Region | Location/Site | Latitude | Longitude | Sea surface temperature (°C) | |
|---|---|---|---|---|---|
| Mean summer | Mean seasonal range | ||||
| Southwest | Warrnambool | −38.4044 | 142.4760 | 18.12 | 4.11 |
| Backyards | −38.4384 | 142.5877 | 18.04 | 3.89 | |
| Peterborough | −38.6061 | 142.8696 | 17.86 | 3.84 | |
| Port Phillip Bay | Queenscliff | −38.2779 | 144.6401 | 19.91 | 7.04 |
| Williamstown | −37.8694 | 144.8901 | 21.57 | 9.92 | |
| Black Rock/Half Moon Bay | −37.9942 | 145.0509 | 21.61 | 9.85 | |
| East | Mallacoota | −37.5684 | 149.7644 | 19.08 | 4.85 |
| Merimbula | −36.8997 | 149.9152 | 20.33 | 4.81 | |
| Eden | −37.0567 | 149.9131 | 20.28 | 4.80 | |
2.3.2. Thermal response experiments
H. banksii zygotes from each of the three defined thermo‐geographic regions (Southwest, East, and PPB) were reared under common culture conditions for experimental purposes following the collections outlined in Section 2.3.1. Thermal response experiments were performed by exposing zygotes from each site to three controlled temperature treatments, 15, 18, and 21℃. These temperatures were based on 15 and 18℃ falling within the bounds of annual sea surface temperatures (SST) across all regions, while only sites from the PPB region are exposed to average summer SSTs as high as 21℃ (Table 2). Initially, gamete release for all sample sites was stimulated in accordance with Kevekordes and Clayton (1996), whereby thalli were rinsed in freshwater, patted dry using paper towel, and exposed to ambient temperature and light for up to 1 hr until gametes were exuded. Eggs were collected prior to sperm to maximize fertilization success, as sperm viability is known to deteriorate quickly upon release into seawater (Doblin & Clayton, 1995; Levring, 1949). Male and female gamete solutions were made for each of the nine sample locations by submerging 10 male or 10 female fronds in 150 ml of 15℃, 0.22 µm filtered seawater (Sterivac Millipore Corporation, Bedford MA). Relative gamete concentrations were determined using UV spectrophotometry and subsequently standardized using 15℃ microfiltered seawater to achieve female:male gamete concentrations of approximately 1:50 to avoid polyspermy (Kevekordes & Clayton, 1996). Once standardized, gamete solutions from each source population were mixed to initiate fertilization (within population crosses only), with 700 µl from each zygote solution immediately pipetted onto 6 individual glass coverslips placed inside each of 18 replicate Petri dishes (6 for each temperature treatment) and left to settle for approximately 5 min (Kevekordes & Clayton, 1996). Twenty mL of 15℃ microfiltered seawater was gently added to each Petri dish to submerge the glass coverslips, after which six Petri dishes from each sample location were randomly assigned to one of three temperature‐control cabinets set at 15, 18, and 21℃, fitted with full spectrum (T5 Fluorescent Kit 4 × 54 Watt Hydro 44, 35 μmolm‐2s‐1) lights on 12:12‐hr light:dark cycle. Petri dishes representing the different sample locations were also randomized within the cabinets.
Embryonic development for each sample location under each temperature treatment was assessed microscopically (100× magnification, ZEISS—Axiostar Plus Microscope), with developmental milestones being recorded for 100 individuals per replicate, adapted from Kevekordes and Clayton (1996): fertilization at 1–2 hr, >4 cell divisions and rhizoid length at 7 days, and mortality and meristematic activity (evidenced by presence of apical hairs) at 14 days. Fertilization success was scored from one randomly selected replicate glass coverslip in each of three replicate Petri dishes for each sample location and temperature treatment prior to manipulation, whereas all other dependent variables were scored from one randomly selected replicate glass coverslip from each of the six replicate Petri dishes for each sample location and temperature treatment. If insufficient zygotes had settled onto a single coverslip for scoring, a second one was sampled. At day 7, photographs of embryos were taken with a mounted camera (ZEISS—Axiocam ERc 5s) at ×100 magnification and subsequently 5 embryos randomly selected from each replicate for measurement of the rhizoid lengths using the ImageJ analysis package (version: 2.0.0‐rc‐43/1.51p, https://imagej.net).
2.3.3. Statistical analyses for common garden experiment
Fertilization success prior to temperature manipulation was analyzed by a two‐factor nested analysis of variance (ANOVA) with region (three levels: Southwest, East, and PPB; fixed) and sites nested within regions (nine levels; random). Data from the common garden experiment were then analyzed by partially nested, mixed model (split plot) design ANOVAs with temperature (three levels: 15, 18, and 21°C; fixed), region (three levels: Southwest, East, and PPB; fixed), and sites nested within regions (nine levels; random). Assumptions of normality and homogeneity of variances were visually checked with box plots and residual plots (Quinn & Keough, 2002). Where significant temperature × region interactions occurred, simple main effects (SEM) analyses examined the effects of temperature for each region separately with a reduced 2‐factor ANOVA design (temperature and site) using the error term from the full model to test the effect of temperature (Quinn & Keough, 2002). Tukey's HSD pairwise comparisons compared temperature treatments after ANOVAs. All analyses were performed on untransformed data in SYSTAT v13.2 and tested at α = 0.05.
Relationships between home site temperature environments and thermal response were estimated by regressing site‐level mean summer sea surface temperatures and mean seasonal variation in sea surface temperatures (evaluated from the Integrated Marine Observing System (IMOS) for the years 1995–2015) against mean values for each thermal response variable from each site in the 21°C treatment of the common garden experiment.
Q ST–F ST comparisons were also performed to determine whether the degree of phenotypic differentiation observed among sites departs from neutral expectations, where Q ST > F ST suggests that quantitative traits show a higher level of differentiation than what would have been expected under the influence of genetic drift, indicative of selection. The R package Pstat (Da Silva & Da Silva, 2018) was used to estimate PST (analogous to Q ST) for each thermal response variable for each site in the 21°C treatment (the highest experimental temperature), using P ST = c/h 2σ2 b/(c/h 2σ2 b + 2σ2 w), where σ2 b and σ2 w are the respective phenotypic variances between and within groups of populations, c is an estimate of the proportion of the total variance due to additive genetic effects across populations, and h 2 is narrow‐sense heritability, the proportion of phenotypic variance due to additive genetic effects (Brommer, 2011). The c/h 2 default level of 1 was used; 95% confidence intervals were calculated with 1,000 bootstrapping data frames. As values of c/h 2 are often not known for wild populations and can be variable among populations, the robustness of this ratio on phenotypic divergence was evaluated using additional c/h 2 values of 0.5 and 0.1. As microsatellite genotypes were not available for sites included in the common garden experiment, we contrasted P ST against global F ST, and against estimates of genetic divergence between and within regions.
3. RESULTS
3.1. Population genetic analysis
Across the 10 microsatellite loci and 18 sample locations, we detected a total of fifty‐one alleles (one of which was exclusive to South Australian sites), with a mean of 6.0 alleles per locus (range 4–8). Levels of genetic diversity (allelic richness and heterozygosity) were largely consistent across sites over all loci and heterozygosity, with a mean allelic richness of 2.59 (range 2.16–3.00) and a mean observed heterozygosity of 0.41 (range 0.31–0.48; Table S1). Most sites were found to conform to HWE (Table S1), with the exceptions of sites FLW, LOB, and POR which all showed heterozygote deficits. This resulted in large and significant positive inbreeding coefficients (F IS) for these sites. Closer inspection indicated that a single locus influenced these deviations for site LOB, while several different loci influenced deviations for sites FLW and POR, suggesting caution when interpreting estimates of differentiation involving these sites.
We detected strong genetic structure among sites with global estimates of F ST and D est across all loci being significantly different from zero [F ST = 0.243; 95% confidence interval (CI) = 0.20–0.29; D est = 0.29; 95% CI = 0.17–0.31] indicating limited gene flow between sampling sites. Out of 153 pairwise population comparisons of F ST, only two did not differ significantly from zero, reflecting restricted gene flow among regions and most sites within regions (Table 3). Similarly, AMOVA analyses indicated that a significant proportion of the genetic variance (9%) could be attributed to difference among regions (p < .01, F ST = 0.092), and 16% of the variance due to differences among sites within regions (p < .01, F ST = 0.166). The relationship between genetic and geographic distance (Figure S1) suggested moderate isolation by distance (Mantel r = .35, p < .05). These analyses were repeated for New South Wales (NSW), Victoria (VIC), and South Australia (SA) separately, with only Victorian sites exhibiting significant isolation by distance (NSW: r = .07, p > .05; VIC: r = .35, p < .05; SA: r = .265, p > .05).
Table 3.
Pairwise estimates of F ST (lower diagonal) and D est (upper diagonal) between 26 Hormosira banksii collection sites
| MCM | TER | LOR | SUT | DAL | MYS | CPA | WKV | FLW | LOB | BAK | OIG | ALD | WIL | GLE | STN | POR | PTA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCM | 0.14 | 0.24 | 0.23 | 0.12 | 0.30 | 0.25 | 0.25 | 0.28 | 0.39 | 0.41 | 0.39 | 0.26 | 0.23 | 0.35 | 0.29 | 0.37 | 0.23 | |
| TER | 0.23 | 0.33 | 0.17 | 0.16 | 0.19 | 0.21 | 0.23 | 0.21 | 0.28 | 0.28 | 0.28 | 0.15 | 0.12 | 0.24 | 0.23 | 0.28 | 0.21 | |
| LOR | 0.29 | 0.31 | 0.18 | 0.21 | 0.37 | 0.20 | 0.28 | 0.22 | 0.33 | 0.35 | 0.37 | 0.31 | 0.29 | 0.40 | 0.37 | 0.33 | 0.24 | |
| SUT | 0.34 | 0.35 | 0.31 | 0.17 | 0.26 | 0.15 | 0.27 | 0.21 | 0.42 | 0.34 | 0.26 | 0.28 | 0.24 | 0.35 | 0.27 | 0.27 | 0.15 | |
| DAL | 0.17 | 0.19 | 0.17 | 0.29 | 0.12 | 0.17 | 0.20 | 0.21 | 0.36 | 0.35 | 0.31 | 0.20 | 0.17 | 0.26 | 0.38 | 0.26 | 0.26 | |
| MYS | 0.35 | 0.25 | 0.31 | 0.39 | 0.12 | 0.17 | 0.15 | 0.18 | 0.27 | 0.23 | 0.21 | 0.20 | 0.20 | 0.26 | 0.39 | 0.37 | 0.28 | |
| CPA | 0.22 | 0.25 | 0.23 | 0.22 | 0.20 | 0.24 | 0.02 | 0.04 | 0.20 | 0.10 | 0.08 | 0.07 | 0.07 | 0.20 | 0.21 | 0.23 | 0.25 | |
| WKV | 0.27 | 0.28 | 0.28 | 0.33 | 0.21 | 0.23 | 0.06 | 0.03 | 0.13 | 0.09 | 0.10 | 0.10 | 0.11 | 0.25 | 0.26 | 0.33 | 0.28 | |
| FLW | 0.26 | 0.26 | 0.25 | 0.25 | 0.22 | 0.25 | 0.03 | 0.06 | 0.06 | 0.07 | 0.08 | 0.08 | 0.10 | 0.28 | 0.30 | 0.26 | 0.27 | |
| LOB | 0.30 | 0.27 | 0.27 | 0.36 | 0.27 | 0.27 | 0.16 | 0.12 | 0.06 | 0.08 | 0.20 | 0.17 | 0.20 | 0.37 | 0.39 | 0.42 | 0.42 | |
| BAK | 0.36 | 0.31 | 0.31 | 0.37 | 0.28 | 0.26 | 0.13 | 0.13 | 0.10 | 0.08 | 0.08 | 0.11 | 0.12 | 0.29 | 0.32 | 0.39 | 0.41 | |
| OIG | 0.40 | 0.34 | 0.31 | 0.36 | 0.26 | 0.25 | 0.17 | 0.19 | 0.17 | 0.22 | 0.12 | 0.13 | 0.14 | 0.31 | 0.25 | 0.35 | 0.39 | |
| ALD | 0.24 | 0.20 | 0.25 | 0.30 | 0.17 | 0.22 | 0.10 | 0.12 | 0.10 | 0.13 | 0.10 | 0.14 | 0.00 | 0.11 | 0.25 | 0.15 | 0.34 | |
| WIL | 0.23 | 0.18 | 0.23 | 0.29 | 0.15 | 0.20 | 0.11 | 0.14 | 0.13 | 0.16 | 0.12 | 0.14 | 0.00 | 0.12 | 0.25 | 0.16 | 0.31 | |
| GLE | 0.33 | 0.32 | 0.34 | 0.35 | 0.24 | 0.30 | 0.18 | 0.25 | 0.24 | 0.28 | 0.26 | 0.29 | 0.11 | 0.13 | 0.33 | 0.11 | 0.34 | |
| STN | 0.32 | 0.28 | 0.32 | 0.33 | 0.31 | 0.37 | 0.21 | 0.27 | 0.27 | 0.30 | 0.28 | 0.26 | 0.22 | 0.22 | 0.29 | 0.43 | 0.27 | |
| POR | 0.36 | 0.34 | 0.34 | 0.30 | 0.29 | 0.39 | 0.21 | 0.30 | 0.24 | 0.33 | 0.36 | 0.37 | 0.20 | 0.21 | 0.17 | 0.37 | 0.36 | |
| PTA | 0.27 | 0.30 | 0.30 | 0.27 | 0.27 | 0.34 | 0.24 | 0.28 | 0.25 | 0.31 | 0.33 | 0.37 | 0.26 | 0.26 | 0.30 | 0.29 | 0.34 |
Values shown in bold are nonsignificant (p > .001) after 10,000 permutations and correction for multiple comparisons. Site codes are derived from Table 1.
Genetic differentiation between regions and sites within regions is depicted by the discriminant analysis of principal components (DAPC) of the microsatellite variation (Figure S2). When all sites were included in the analysis, clear regional differences were evident, with sites from NSW, VIC, and SA separating across the axes and representing three distinct clusters (estimated using the find cluster function; Figure S2a). DAPCs performed on sites from each state show regional genetic structuring with sites from common regions typically clustering more closely, but with centroids of population clusters rarely overlapping (Figure S2a–d). STRUCTURE Bayesian clustering analyses confirmed significant genetic differentiation between the NSW, VIC, and SA sites, with analyses indicating assignment of individuals from the respective states to three distinct population clusters (ΔK = 3; Figure 1d). STRUCTURE analysis of each region suggested significant structuring with each of the regions, consistent with the DAPC analysis. Further structuring at the state scale was evident when STRUCTURE analyses were performed on sites from each region independently (Figure 1e). Analyses of NSW sample locations revealed three distinct population clusters (ΔK = 3) corresponding with the Central Coast, Sydney, and South Coast regions. Similarly, significant regional differences were observed in SA where individuals were assigned to three population clusters (ΔK = 3) corresponding with the Fleurieu Peninsula, Yorke Peninsula, and Eyre Peninsula regions. Regional genetic structuring was less evident in Victoria where two population clusters were determined (ΔK = 2), but not reflecting an obvious geographic pattern. We expect this is likely due to comparatively lower level of genetic differentiation among Victorian sample locations, and the limited ability of STRUCTURE to resolve fine‐scale patterns of genetic structure (Evanno et al., 2005).
Results from the Bayesian migration analyses indicated limited migration among sites within the last three generations (Table S1). Estimates of the strength and directionality of migration demonstrated that each site has been largely dependent on recruitment from local sources, with minimal migration from nonlocal sources. Evidence of weak recent migration (average 8.3% migrants per generation) was recorded between three pairs of neighboring sites. In the Sydney region, evidence of unidirectional migration of 5% was found between LOR and SUT, while bidirectional migration of 9%–11% between sites WIL and ALD on the Fleurieu Peninsula separated by <2 km was also detected.
3.2. Testing for local adaptation
3.2.1. Fertilization success
Fertilization success was high for all sites (mean ± SE: 92.7 ± 0.98%, N = 81 pooling across sites; Figure S3), and there was a marginally nonsignificant difference in fertilization success among regions (2‐factor nested ANOVA: F (2,6) = 5.08; p = .051) and no significant difference among sites within regions (F (6,72) = 1.09; p = .377) at the beginning of the experiment. This was expected as all gamete mixes were initially prepared under common temperatures (15°C), where fertilization was expected to be largely complete within minutes (Kevekordes & Clayton, 1996), prior to exposure to experimental temperatures.
3.2.2. Embryo development and rhizoid growth (Day 7)
The response of developing embryos to different temperature treatments was complex and variable among sites and regions. The percentage of mature embryos at day 7 (>4 cell divisions and presence of rhizoids) among temperature treatments were consistent across sites within regions (ANOVA temperature × site(region); F (12,135) = 0.77; p = .685) but not among regions (ANOVA temperature × region; F (4,12) = 8.58; p = .002). There was no significant difference in the percentage of mature embryos among temperature treatments for sites from the PPB region (SME temperature F (2,12) = 2.352; p = .137; Figure 2a), although this region did have the lowest proportion of mature embryos compared with sites from the Southwest and East regions. In contrast, the percentage of mature embryos differed between temperatures for the Southwest and East regions, with significantly lower development in the warmest treatment compared with 15 and 18°C (Tukey's HSD, p < .05; Figure 2a). Rhizoid lengths increased significantly with temperature, with length being greatest at 21°C (ANOVA temperature; F (2,4) = 13.91; p = .001; Tukey's HSD p < .05 for all pairwise comparisons); this pattern was consistent across sites within regions (ANOVA temperature × site(region); F (12,134) = 1.66; p = .082) and among regions (ANOVA temperature × region interaction; F (4,12) = 2.35; p = .114; Figure 2b).
Figure 2.
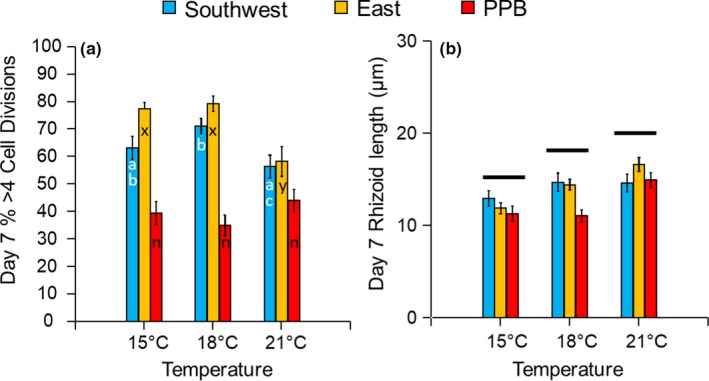
Development of embryos of Hormosira banksii from three source regions (southwest Victoria, eastern Victoria/southern NSW, and Port Phillip Bay, pooling three sites in each region) after 7 days of culture at 15, 18, and 21°C showing a) mean (±SE) percentage of embryos with greater than four cell divisions and well‐developed rhizoids, and (b) mean (±SE) rhizoid length. Broken horizontal lines above bars in (b) indicate significant differences among temperature treatments based on Tukey's test after the full ANOVA model, whereas letters on bars in (a) indicate Tukey's test results among treatments for each region individually after simple main effects analysis (because of significant temperature × region interaction in the full model)
3.2.3. Embryo mortality and meristematic activity (Day 14)
Embryo mortality by day 14 averaged 12.3 ± 0.84% (N = 162) but varied among sites within regions with temperature (ANOVA temperature × site(region) interaction; F (12,135) = 3.12; p = .001; Figure 3), which was largely driven by significantly higher mortality at 21°C than either 15 or 18°C for one site (Backyards) in the Southwest (Tukey's HSD, Bonferroni‐p = .017 & .004, respectively; Figure 3). Of the embryos that survived to day 14, meristematic activity (the presence of apical hairs) among temperature treatments was not consistent among sites within regions (ANOVA temperature × site(region); F (12,135) = 2.66; p = .003; Figure 5), but temperature effects on meristematic activity were consistent among regions (ANOVA temperature × region; F (4,12) = 2.55; p = .093). Overall, there were significantly more embryos with apical hairs at day 14 in the 21°C treatment compared with both 18 and 15°C treatments (Tukey's HSD, p < .001 for 21 versus 18°C and 21 versus 15°C), but this pattern was only significant for 2 sites in each of the PPB and East regions (Tukey's HSD after SME temperature; Figure 4).
Figure 3.
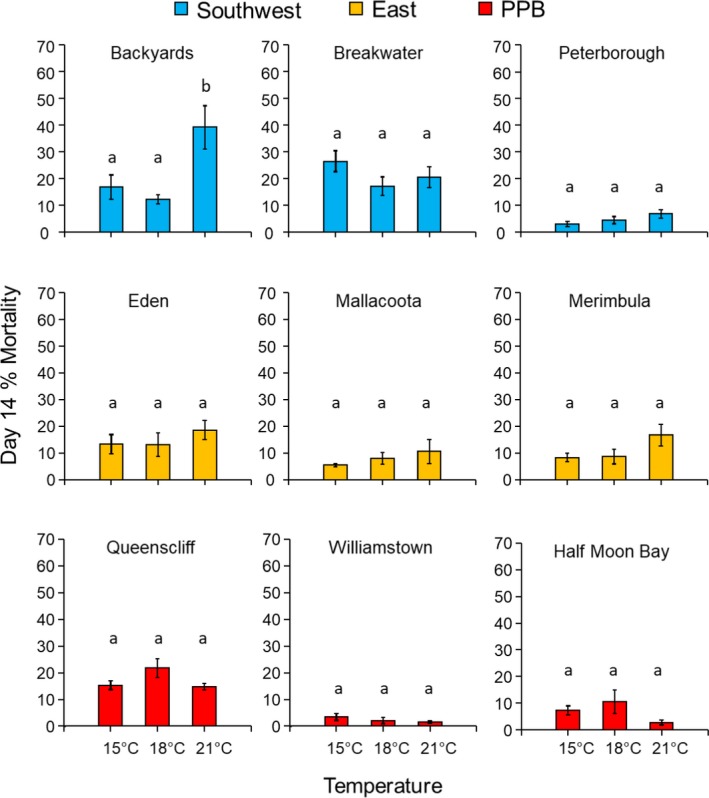
Mean (±SE) percentage mortality of embryos of Hormosira banksii from three random sites in each of three source regions (southwest Victoria, eastern Victoria/southern NSW, and Port Phillip Bay) after 14 days of culture at 15, 18, and 21°C. Letters above bars indicate Tukey's test results among temperature treatments for each site individually after simple main effects analysis (because of significant temperature × site(region) interaction in the full model)
Figure 4.
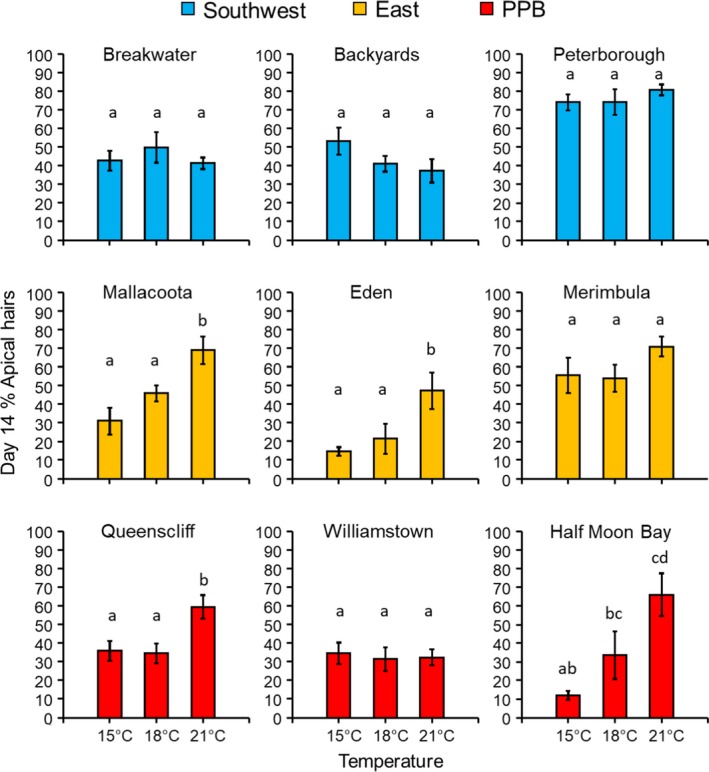
Development of embryos of Hormosira banksii from three random sites in each of three source regions (southwest Victoria, eastern Victoria/southern NSW, and Port Phillip Bay) after 14 days of culture at 15, 18, and 21°C showing mean (±SE) percentage of embryos with meristematic activity evidenced by apical hair development. Letters above bars indicate Tukey's test results among temperature treatments for each site individually after simple main effects analysis (because of significant temperature × site(region) interaction in the full model)
3.2.4. Regression analyses and P ST–F ST comparisons
Regression analyses suggest an association between home site temperature environments and thermal response in the common garden experiment. We found a significant negative relationship between home site mean seasonal variation in sea surface temperature and 7‐day rhizoid development (r = −.64, p = .007) and 14‐day mortality (r = −.65, p < .001) at 21°C (Figure 5). Nonsignificant negative trends were also noted when home site mean summer SST was regressed against 7‐day rhizoid development (r = −.45, p = .225) and 14‐day mortality (r = −.58, p = .104) at 21°C (Figure 5). There was no association between home site mean seasonal variation in sea surface temperature or site mean summer temperature with 7‐day rhizoid length (r = .05, p = .905; r = .05, p = .908; respectively), and 14‐day apical hair development (r = .18, p = .647; r = .10, p = .801; respectively) at 21°C.
Figure 5.
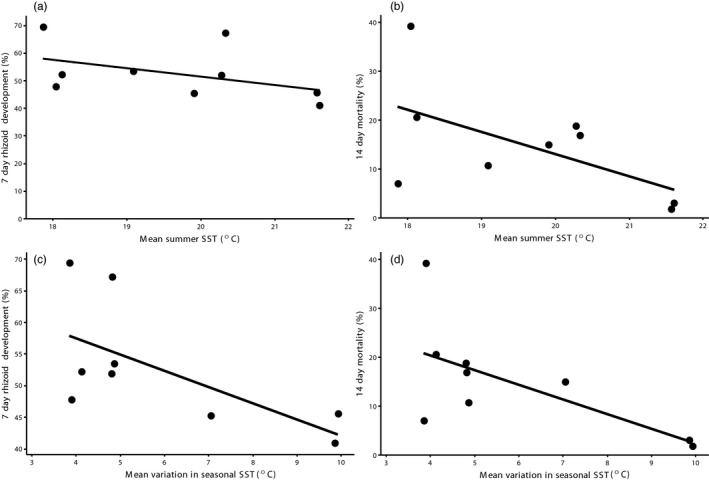
Regression analyses of home site temperature environments and thermal response at 21°C in the common garden experiment. Nonsignificant negative relationships for home site mean summer temperature regressed against (a) 7‐day rhizoid development (r = −.45, p = .225), and (b) 14‐day mortality (r = −.58, p = .104). Significant negative relationships between home site mean seasonal variation in sea surface temperature and (c) 7‐day rhizoid development (r = −.64, p = .007), and (d) 14‐day mortality (r = −.65, p < .001)
Phenotypic variance (P ST) estimates were highest for 14‐day mortality (P ST = 0.85; 95% CI = 0.77–0.95), intermediate for 14‐day apical hair development (P ST = 0.79; 95% CI = 0.71–0.92) and 7‐day rhizoid length (P ST = 0.70; 95% CI = 0.61–0.88), and lowest for percentage of mature embryos after 7 days (embryos with >4 cell divisions and rhizoids present; P ST = 0.44; 95% CI = 0.29–0.81). While P ST for percentage of mature embryos was similar to F ST, P ST measures for the remaining three traits were significantly higher than F ST globally (F ST = 0.24; 95% CI = 0.20–0.29), between regions (F ST = 0.09), and within regions (F ST = 0.17).
4. DISCUSSION
4.1. Population connectivity
Genetic and phenotypic differences among populations may contribute to uneven species responses to climate change; such unevenness may result from the genetic history of populations as well as selection associated with prior thermal history. Understanding the interplay of these factors helps predict thermal responses of species to increasing incidences of acute and chronic thermal stress. Within this context, our study on the dominant intertidal macrophyte in Australasia, H. banksii, indicates highly restricted gene flow across much of its contemporary distribution with evidence of strong genetic structuring on all spatial scales examined. These findings are consistent with those reported previously for H. banksii over smaller geographic ranges (Bellgrove et al., 2017; Coleman et al., 2011, 2019) and reports of fine‐scale genetic structuring in other marine macrophytes including fucoids (Coleman & Brawley, 2005; Williams & Difiori, 1996). Common garden experiments revealed a heterogeneous spatial distribution of thermally tolerant phenotypes, which may be related to local‐scale thermal adaptation. These findings suggest that the impacts on this important foundation species of warming ocean temperatures under climate change may be uneven. Opportunities for recovery of depleted populations and adaptation in populations vulnerable to thermal stress through natural gene flow may be limited, but interventions such as translocation and assisted migration could assist the recovery or bolster the resilience of existing populations given the existence of relatively tolerant populations across the species range (Marzinelli, Leong, Campbell, Steinberg, & Verges, 2016; Wood et al., 2019).
4.2. Heterogeneity in thermal response
Consistent with Clark et al. (2013) and Alestra et al. (2015), our study suggests the potential for selection to increase thermal tolerance across the species range. Our common garden experiments indicate strong differences in early development between sites sampled from three geographical regions in response to differences in temperature. High water temperatures (21°C) often led to suppressed embryonic development and increased mortality in individuals from sites from cooler regions, while others from warmer and more variable temperature regions were relatively unaffected. Our correlation analyses indicated negative linear relationships between homesite, sea surface temperatures (mean summer temperature and seasonal variation), and thermal response at 21°C. These findings suggest genetically determined variation in thermal tolerance across broad thermal gradients in H. banksii, consistent with results reported for other fucoid and laminarian macrophytes (Bennett et al., 2015; King et al., 2018; Mohring, Wernberg, Wright, Connell, & Russell, 2014; Staehr & Wernberg, 2009).
Our results also indicate site variation within regions in the thermal responses of embryos to temperature stress, reflecting potential adaptation on local spatial scales. Although there are few studies on micro‐scale variation in marine macrophytes, signatures of salinity‐associated adaptation among fucoid (Fucus serratus) populations separated by as little as 12 km have been previously demonstrated (Coyer et al., 2011), and reciprocal transplants of genotypes from low to high intertidal zones (10s of meters scale) in another fucoid (Silvetia compressa) demonstrated clear home site advantages (Hays, 2007). These findings add to a growing literature supporting the notion that selection processes operate on finer spatial scales than currently assumed in marine ecosystems, with local habitat heterogeneity contributing to genetic adaptation in a range of marine organisms (Babin, Gagnaire, Pavey, & Bernatchez, 2017; Miller et al., 2019; Sherman & Ayre, 2008).
While the majority of studies reporting phenotypic variation across environmental gradients in marine macrophytes have not considered the relative contributions of local adaptation and plasticity (King et al., 2018), our study suggests genetic variation among H. banksii populations. This is further supported by Q ST–F ST comparisons which indicate departures of Q ST from neutral expectations in three of four thermal response traits. Thermal responses are often affected by environmental conditions (i.e., are largely plastic); for instance, macroalgae often show plastic responses in morphology and physiology to changes in temperature (Flukes et al., 2015; Hurd et al., 2014; Reusch, 2014). However, in our case we provide evidence for adaptive heritable variation that can contribute to phenotypic variation alongside this plasticity. The development of heritable differences among sites may reflect limited gene flow in the species. In the absence of gene flow, populations are expected to evolve phenotypes in response to local selection pressures, helping to produce the patterns observed here. Local adaptation over short distances has been shown to occur in terrestrial plant systems when gene flow patterns are conducive to adaptation (e.g., Buehler et al., 2013; Byars, Papst, & Hoffmann, 2007).
Although we have interpreted differences among sites as reflecting genetic variation, such comparisons should ideally involve additional generations because of the possibility of cross‐generation effects induced by environmental conditions (Schiffer, Hangartner, & Hoffmann, 2013). Such effects can lead to phenotypic changes spanning multiple generations (i.e., transgenerational plasticity), including those due to epigenetic mechanisms (Walsh et al., 2016), in the absence of fixed genetic differences that reflect changes in allele frequencies. Cross‐generation studies will help determine the relative contributions of genetic, environmental, and epigenetic effects to the temperature variation we observed among sites.
We have demonstrated substantial genetic and phenotypic differences among H. banksii populations, which may manifest in uneven responses to warming sea surface temperatures. Such differences may help to explain nonuniform responses to thermal stress in other macrophyte species. For example, variable responses to temperature stress have been reported at local and regional scales for a range of kelp species from southern California Bight, Nova Scotia, British Columbia, Australia, and New Zealand following extreme climatic events (Bennett et al., 2015; Carballo et al., 2002; Filbee‐Dexter et al., 2016; Starko et al., 2019; Tegner & Dayton, 1987; Thomsen et al., 2019). Such variation is often explained by heterogeneous habitat features (i.e., exposure, depth, bathymetry, and reef geomorphology) that contribute to different thermal environments at local and regional scales (Ierodiaconou et al., 2018). However, habitat differences are also expected to contribute to genetic and phenotypic differences influencing the climate niche of local populations due to selection, particularly for species with low levels of dispersal and strong genetic structuring (King et al., 2018; Miller et al., 2019). This highlights the need for improved evolutionary information on marine macrophytes at the population level to help understand what drives differential responses among populations to thermal stress. Also, this highlights the importance of species distribution and climate niche models that account for population differentiation in order to generate more reliable predictions of species responses to future climates (Martinez et al., 2018; Razgour et al., 2018; Valladares et al., 2014).
Genomic sequencing approaches (i.e., reduced genome representation, whole genome, and transcriptome sequencing) will help in further elucidating patterns of differentiation across sites. Such methods provide unprecedented sensitivity for resolving fine‐scale genetic structure, as well as identifying signatures of selection and putative candidate genes that underlie adaptive differences between species populations (Jordan et al., 2017; Miller et al., 2019). Unfortunately, this study was restricted to the use of microsatellite markers due to DNA not being of sufficient quality for high‐throughput sequencing methods. However, the 10 polymorphic loci used in this study sufficiently resolved patterns of population genetic structure, and microsatellite markers have proven effective for such analyses in other marine macrophyte systems (Reynolds et al., 2017; Wernberg et al., 2018).
4.3. Assisted migration and adaptation
Our findings have implications for predicting the recovery potential of depleted H. banksii in areas affected by coastal development, urbanization, and thermal stress (Bellgrove et al., 2010; Keough & Quinn, 1998). The apparent lack of gene flow among sites within regions suggests opportunities for natural recolonization following local mortality are low (Coleman et al., 2008). Instead, interventions such as translocation and reseeding may be needed to assist recovery (Bellgrove et al., 2010; Campbell, Marzinelli, Verges, Coleman, & Steinberg, 2014). Moreover, strong population genetic structuring in H. banksii also suggests that gene flow is unlikely to assist populations in adapting to warming sea surface temperatures via the migration of thermally adapted genotypes (Alestra & Schiel, 2015; Clark et al., 2013). Instead, a lack of gene flow will necessitate in situ adaptation. While empirical studies suggest that maladaptation can be mitigated in the presence of weak gene flow across environmental gradients (Hoffmann & Sgro, 2011; Sgro et al., 2011), our results suggest that little to no migration is occurring between populations separated by 10s of kilometers. Consequently, the future resilience of local populations to rising sea surface temperatures will depend on standing genetic variation and plasticity, in the absence of any intervention.
While our study suggests that thermally resistant genotypes can occur, selection may struggle to keep pace with a rapidly shifting climatic envelope and increasingly frequent extreme temperature events (Wernberg, Bettignies, et al., 2016). This is particularly pertinent for macrophyte species in climate change hot spots such as southwestern and southeastern Australia. For locally adapted species with limited dispersal ability such as H. banksii, adaptive management strategies might be needed. These could include assisted migration of thermally adapted genotypes to populations showing signs of climate stress. Such approaches are being widely advocated as a tool for “climate proofing” threatened marine and terrestrial animal and plant communities (Aitken & Whitlock, 2013; Prober et al., 2015; Sgro et al., 2011). For species showing genetically determined clinal variation in thermal response, such approaches might involve the translocation of genotypes from warm environments into cooler areas (Bansal, Harrington, Gould, St, & B., 2015; Schueler et al., 2013). However, alternative and more tailored approaches may be needed for species such as H. banksii where thermal adaptations across the species range are heterogeneous. In such cases, assisted migration strategies may require composite provenancing approaches, involving mixed genotypes from multiple source populations (Broadhurst et al., 2008; Prober et al., 2015; Weeks et al., 2011). This can help to broaden the genetic basis of introduced genotypes, providing at least some genetic variants that are preadapted to warmer oceanographic conditions.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
This project was conceived by A.D.M, A.B, M.A.C, and C.D.H.S, with the population genetic data collected by A.D.M, A.B, J.C, and M.A.C, and analysis led by A.D.M. Common garden experiments were led by A.D.M, A.B, and A.A.H; the collection of phenotypic data was assisted by R.C and Z.N, with A.D.M, A.B, A.A.H, M.A.C, and C.D.H.S contributing to data analyses. Writing of the manuscript was led by A.D.M. with assistance from all authors.
Supporting information
ACKNOWLEDGEMENTS
This work was partially funded by Deakin University funding and Wettenhall Environment Trust awarded to A Miller and A Bellgrove, an Australian Research Council Discovery Grant (DP0663550) awarded to M.A. Coleman, and UTS funding awarded to MA Doblin. We thank JB Pocklington, O Holland, E Cumming, and E Marks for assistance with sample collection for the common garden experiment; and BP Kelaher and MJ Bishop for assistance with sample collection for population genetics analysis. PF Francis (nee McKenzie) is acknowledged for early contributions to assessing the population genetics of H. banksii. GP Quinn is thanked for advice relating to statistical analyses, and M Young for assistance with geospatial aspects of the project.
Miller AD, Coleman MA, Clark J, et al. Local thermal adaptation and limited gene flow constrain future climate responses of a marine ecosystem engineer. Evol Appl. 2020;13:918–934. 10.1111/eva.12909
DATA AVAILABILITY STATEMENT
Genetic and phenotypic data will be made publicly available in the DRYAD: ://doi.org/10.5061/dryad.g4f4qrfmb (Miller, 2019).
REFERENCES
- Airoldi, L. , & Beck, M. W. (2007). Loss, status and trends for coastal marine habitats of Europe In Gibson R. N., Atkinson R. J. A., & J. D. M. Gordon (Eds.), Oceanography and Marine Biology (pp. 345–405). Boca Raton: CRC Press. [Google Scholar]
- Aitken, S. N. , & Whitlock, M. C. (2013). Assisted gene flow to facilitate local adaptation to climate change. Annual Review of Ecology, Evolution, and Systematics, 44, 367–388. 10.1146/annurev-ecolsys-110512-135747 [DOI] [Google Scholar]
- Alestra, T. , & Schiel, D. R. (2015). Impacts of local and global stressors in intertidal habitats: Influence of altered nutrient, sediment and temperature levels on the early life history of three habitat‐forming macroalgae. Journal of Experimental Marine Biology and Ecology, 468, 29–36. 10.1016/j.jembe.2015.03.017 [DOI] [Google Scholar]
- Babcock, R. C. , Bustamante, R. H. , Fulton, E. A. , Fulton, D. J. , Haywood, M. D. E. , Hobday, A. J. , … Vanderklift, M. A. (2019). Severe continental‐scale impacts of climate change are happening now: Extreme climate events impact marine habitat forming communities along 45% of Australia’s coast. Frontiers in Marine Science, 6 10.3389/fmars.2019.00411 [Epub ahead of print]. [DOI] [Google Scholar]
- Babin, C. , Gagnaire, P. A. , Pavey, S. A. , & Bernatchez, L. (2017). RAD‐seq reveals patterns of additive polygenic variation caused by spatially‐varying selection in the American Eel (Anguilla rostrata). Genome Biology and Evolution, 9, 2974–2986. 10.1093/gbe/evx226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, S. , Harrington, C. A. , Gould, P. J. , St, C. J. , & B., (2015). Climate‐related genetic variation in drought‐resistance of Douglas‐fir (Pseudotsuga menziesii). Global Change Biology, 21, 947–958. [DOI] [PubMed] [Google Scholar]
- Bellgrove, A. , Clayton, M. N. , & Quinn, G. P. (1997). Effects of secondarily treated sewage effluent on intertidal macroalgal recruitment processes. Marine and Freshwater Research, 48, 137–146. 10.1071/MF96011 [DOI] [Google Scholar]
- Bellgrove, A. , Clayton, M. N. , & Quinn, G. P. (2004). An integrated study of the temporal and spatial variation in the supply of propagules, recruitment and assemblages of intertidal macroalgae on a wave‐exposed rocky coast, Victoria, Australia. Journal of Experimental Marine Biology and Ecology, 310, 207–225. 10.1016/j.jembe.2004.04.011 [DOI] [Google Scholar]
- Bellgrove, A. , McKenzie, P. F. , McKenzie, J. L. , & Sfiligoj, B. J. (2010). Restoration of the habitat‐forming fucoid alga Hormosira banksii at effluent‐affected sites: Competitive exclusion by coralline turfs. Marine Ecology Progress Series, 419, 47–56. 10.3354/meps08843 [DOI] [Google Scholar]
- Bellgrove, A. , van Rooyen, A. , Weeks, A. R. , Clark, J. S. , Doblin, M. A. , & Miller, A. D. (2017). New resource for population genetics studies on the Australasian intertidal brown alga, Hormosira banksii: Isolation and characterization of 15 polymorphic microsatellite loci through next generation DNA sequencing. Journal of Applied Phycology, 29(3), 1721–1727. 10.1007/s10811-016-1015-0 [DOI] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate ‐ a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bennett, S. , Wernberg, T. , Joy, B. A. , De Bettignies, T. , & Campbell, A. H. (2015). Central and rear‐edge populations can be equally vulnerable to warming. Nature Communications, 6, 10280 10.1038/ncomms10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, M. J. , Morgan, T. , Coleman, M. A. , Kelaher, B. P. , Hardstaff, L. K. , & Evenden, R. W. (2009). Facilitation of molluscan assemblages in mangroves by the fucalean alga Hormosira banksii . Marine Ecology Progress Series, 392, 111–122. 10.3354/meps08247 [DOI] [Google Scholar]
- Blacket, M. J. , Robin, C. , Good, R. T. , Lee, S. F. , & Miller, A. D. (2012). Universal primers for fluorescent labelling of PCR fragments- an efficient and cost-effective approach to genotyping by fluorescence. Molecular Ecology Resources, 12, 456–463. [DOI] [PubMed] [Google Scholar]
- Broadhurst, L. M. , Lowe, A. , Coates, D. J. , Cunningham, S. A. , McDonald, M. , Vesk, P. A. , & Yates, C. (2008). Seed supply for broadscale restoration: Maximizing evolutionary potential. Evolutionary Applications, 1, 587–597. 10.1111/j.1752-4571.2008.00045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brommer, J. E. (2011). Whither Pst? The approximation of Qst by Pst in evolutionary and conservation biology. Journal of Evolutionary Biology, 24, 1160–1168. 10.1111/j.1420-9101.2011.02268.x [DOI] [PubMed] [Google Scholar]
- Brookfield, J. F. Y. (1996). A simple new method for estimating null allele frequency from heterozygote deficiency. Molecular Ecology, 5, 453–455. 10.1111/j.1365-294X.1996.tb00336.x [DOI] [PubMed] [Google Scholar]
- Buehler, D. , Poncet, B. N. , Holderegger, R. , Manel, S. , Taberlet, P. , & Gugerli, F. (2013). An outlier locus relevant in habitat‐mediated selection in an alpine plant across independent regional replicates. Evolutionary Ecology, 27, 285–300. 10.1007/s10682-012-9597-8 [DOI] [Google Scholar]
- Bush, A. , Mokany, K. , Catullo, R. , Hoffmann, A. , Kellermann, V. , Sgrò, C. , … Ferrier, S. (2016). Incorporating evolutionary adaptation in species distribution modelling reduces projected vulnerability to climate change. Ecology Letters, 19, 1468–1478. 10.1111/ele.12696. [DOI] [PubMed] [Google Scholar]
- Byars, S. G. , Papst, W. , & Hoffmann, A. A. (2007). Local adaptation and cogradient selection in the alpine plant, Poa hiemata, along a narrow altitudinal gradient. Evolution, 61, 2925–2941. 10.1111/j.1558-5646.2007.00248.x [DOI] [PubMed] [Google Scholar]
- Campbell, A. H. , Marzinelli, E. M. , Verges, A. , Coleman, M. A. , & Steinberg, P. D. (2014). Towards restoration of missing underwater forests. PLoS ONE, 9, e84106 10.1371/journal.pone.0084106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo, J. L. , Olabarria, C. , & Osuna, T. G. (2002). Analysis of four macroalgal assemblages along the Pacific Mexican coast during and after the 1997–98 El Nino. Ecosystems, 5, 749–760. [Google Scholar]
- Clark, J. S. , Poore, A. G. B. , & Doblin, M. A. (2018). Shaping up for stress: Physiological flexibility is key to survivorship in a habitat‐forming macroalga. Journal of Plant Physiology, 231, 346–355. 10.1016/j.jplph.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Clark, J. S. , Poore, A. G. B. , Ralph, P. J. , & Doblin, M. A. (2013). Potential for adaptation in response to thermal stress in an intertidal macroalga. Journal of Phycology, 49, 630–639. 10.1111/jpy.12067 [DOI] [PubMed] [Google Scholar]
- Coleman, M. A. , & Brawley, S. H. (2005). Spatial and temporal variability in dispersal and population genetic structure of a rockpool alga. Marine Ecology Progress Series, 300, 63–77. 10.3354/meps300063 [DOI] [Google Scholar]
- Coleman, M. A. , Chambers, J. , Knott, N. A. , Malcolm, H. A. , Harasti, D. , Jordan, A. , & Kelaher, B. P. (2011). Connectivity within and among a network of temperate marine reserves. PLoS ONE, 6, e20168 10.1371/journal.pone.0020168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, M. A. , Clark, J. S. , Doblin, M. A. , Bishop, M. J. , & Kelaher, B. P. (2019). Genetic differentiation between estuarine and open coast ecotypes of a dominant ecosystem engineer. Marine and Freshwater Research, 70, 977–985. 10.1071/MF17392 [DOI] [Google Scholar]
- Coleman, M. A. , Kelaher, B. P. , Steinberg, P. D. , & Millar, A. J. K. (2008). Absence of a large brown macroalga on urbanized rocky reefs around Sydney, Australia, and evidence for historical decline. Journal of Phycology, 44, 897–901. [DOI] [PubMed] [Google Scholar]
- Connell, S. D. , Russell, B. D. , Turner, D. J. , Shepherd, S. A. , Kildea, T. , Miller, D. , … Cheshire, A. (2008). Recovering a lost baseline: Missing kelp forests from a metropolitan coast. Marine Ecology Progress Series, 360, 63–72. 10.3354/meps07526 [DOI] [Google Scholar]
- Coyer, J. A. , Hoarau, G. , Pearson, G. , Mota, C. , Jüterbock, A. , Alpermann, T. , … Olsen, J. L. (2011). Genomic scans detect signatures of selection along a salinity gradient in populations of the intertidal seaweed Fucus serratus on a 12 km scale. Marine Genomics, 4, 41–49. 10.1016/j.margen.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Da Silva, S. B. , & Da Silva, A. (2018). Pstat: An R package to assess population differentiation in phenotypic traits. R Journal, 10, 447–454. 10.32614/RJ-2018-010 [DOI] [Google Scholar]
- Dayton, P. K. (1972). Toward an understanding of community resilience and the potential effects of enrichment to the benthos as McMurdo Sound, Antarctica. In Proceedings of the Colloquium on Conservation problems in Antarctica (pp. 81–96). Lawrence, Kansas: Allen Press. [Google Scholar]
- Doblin, M. A. , & Clayton, M. N. (1995). Effects of secondarily treated sewage effluent on the early life‐history stages of 2 species of brown macroalgae ‐ Hormosira banksii and Durvillaea potatorum . Marine Biology, 122, 689–698. [Google Scholar]
- Earl, D. A. , & Vonholdt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Filbee‐Dexter, K. , Feehan, C. J. , & Scheibling, R. E. (2016). Large‐scale degradation of a kelp ecosystem in an ocean warming hotspot. Marine Ecology Progress Series, 543, 141–152. 10.3354/meps11554 [DOI] [Google Scholar]
- Flukes, E. B. , Wright, J. T. , & Johnson, C. R. (2015). Phenotypic plasticity and biogeographic variation in physiology of habitat‐forming seaweed: response to temperature and nitrate. Journal of Phycology, 51, 896–909. 10.1111/jpy.12330 [DOI] [PubMed] [Google Scholar]
- Foden, W. B. , Young, B. E. , Akçakaya, H. R. , Garcia, R. A. , Hoffmann, A. A. , Stein, B. A. , … Huntley, B. (2019). Climate change vulnerability assessment of species. Wiley Interdisciplinary Reviews‐Climate Change, 10, e551 10.1002/wcc.1551 [DOI] [Google Scholar]
- Goudet, J. (1995). FSTAT (version 1.2): A computer program to calculate F‐statistics. Journal of Heredity, 86, 485–486. 10.1093/oxfordjournals.jhered.a111627 [DOI] [Google Scholar]
- Harris, R. M. B. , Beaumont, L. J. , Vance, T. R. , Tozer, C. R. , Remenyi, T. A. , Perkins‐Kirkpatrick, S. E. , … Bowman, D. M. J. S. (2018). Biological responses to the press and pulse of climate trends and extreme events. Nature Climate Change, 8, 579–587. 10.1038/s41558-018-0187-9 [DOI] [Google Scholar]
- Hays, C. G. (2007). Adaptive phenotypic differentiation across the intertidal gradient in the alga Silvetia compressa. Ecology, 88, 149–157. [DOI] [PubMed] [Google Scholar]
- Hobday, A. J. , & Pecl, G. T. (2014). Identification of global marine hotspots: Sentinels for change and vanguards for adaptation action. Reviews in Fish Biology and Fisheries, 24, 415–425. 10.1007/s11160-013-9326-6 [DOI] [Google Scholar]
- Hoegh‐Guldberg, O. , & Bruno, J. F. (2010). The impact of climate change on the world's marine ecosystems. Science, 328, 1523–1528. 10.1126/science.1189930 [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A. , & Sgro, C. M. (2011). Climate change and evolutionary adaptation. Nature, 470, 479–485. 10.1038/nature09670 [DOI] [PubMed] [Google Scholar]
- Hoffmann, A. A. , & Willi, Y. (2008). Detecting genetic responses to environmental change. Nature Reviews Genetics, 9, 421–432. 10.1038/nrg2339 [DOI] [PubMed] [Google Scholar]
- Hurd, C. L. , Harrison, P. J. , Bischof, K. , & Lobban, C. S. (2014). Seaweed Ecology and Physiology. Cambridge: Cambridge University Press. [Google Scholar]
- Ierodiaconou, D. A. , Young, M. , Miller, A. D. , Treml, E. , Swearer, S. , Sherman, C. D. H. , … Gorfine, H. (2018). Patterns of interaction between habitat and oceanographic variables affecting the connectivity and productivity of invertebrate fisheries. Fisheries Research and Development Corporation, Final Report project 2018-025.
- Jombart, T. (2008). adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Jombart, T. , & Ahmed, I. (2011). adegenet 1.3‐1: New tools for the analysis of genome‐wide SNP data. Bioinformatics, 27, 3070–3071. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C. G. , Lawton, J. H. , & Shachak, M. (1994). Organisms as ecosystem engineers. Oikos, 69, 373–386. 10.2307/3545850 [DOI] [Google Scholar]
- Jordan, R. , Hoffmann, A. A. , Dillon, S. K. , & Prober, S. M. (2017). Evidence of genomic adaptation to climate in Eucalyptus microcarpa: Implications for adaptive potential to projected climate change. Molecular Ecology, 26, 6002–6020. [DOI] [PubMed] [Google Scholar]
- Jost l., (2008). Gst and its relatives do not measure differentiation. Molecular Ecology, 17, 4015–4026. [DOI] [PubMed] [Google Scholar]
- Kain, J. M. (2015). Hormosira banksii (Phaeophyceae): A tough survivor in the harsh conditions of high intertidal pools in southeast Australia. European Journal of Phycology, 50, 408–421. [Google Scholar]
- Keough, M. J. , & Quinn, G. P. (1998). Effects of periodic disturbances from trampling on rocky intertidal algal beds. Ecological Applications, 8, 141–161. 10.1890/1051-0761(1998)008[0141:EOPDFT]2.0.CO;2 [DOI] [Google Scholar]
- Kevekordes, K. (2000). The effects of secondary‐treated sewage effluent and reduced salinity on specific events in the early life stages of Hormosira banskii (Phaeophyceae). European Journal of Phycology, 35, 365–371. [Google Scholar]
- Kevekordes, K. , & Clayton, M. N. (1996). Using developing embryos of Hormosira banksii (Phaeophyta) as a marine bioassay system. International Journal of Plant Sciences, 157, 582–585. [Google Scholar]
- King, N. G. , McKeown, N. J. , Smale, D. A. , & Moore, P. J. (2018). The importance of phenotypic plasticity and local adaptation in driving intraspecific variability in thermal niches of marine macrophytes. Ecography, 41, 1469–1484. 10.1111/ecog.03186 [DOI] [Google Scholar]
- Levring, T. (1949). Fertilization experiments with Horinosira Banksii (Tnrn.) Dcne. Physiologia Plantarum, 2, 45–55. 10.1111/j.1399-3054.1949.tb07647.x [DOI] [Google Scholar]
- Macinnis‐Ng, C. M. O. , Morrison, D. A. , & Ralph, P. J. (2005). Temporal and spatial variation in the morphology of the brown macroalga Hormosira banksii (Fucales, Phaeophyta). Botanica Marina, 48, 198–207. 10.1515/BOT.2005.031 [DOI] [Google Scholar]
- Magris, R. A. , Pressey, R. L. , Weeks, R. , & Ban, N. C. (2014). Integrating connectivity and climate change into marine conservation planning. Biological Conservation, 170, 207–221. 10.1016/j.biocon.2013.12.032 [DOI] [Google Scholar]
- Martínez, B. , Radford, B. , Thomsen, M. S. , Connell, S. D. , Carreño, F. , Bradshaw, C. J. A. , … Wernberg, T. (2018). Distribution models predict large contractions of habitat‐forming seaweeds in response to ocean warming. Diversity and Distributions, 24, 1350–1366. 10.1111/ddi.12767 [DOI] [Google Scholar]
- Marzinelli, E. M. , Leong, M. R. , Campbell, A. H. , Steinberg, P. D. , & Verges, A. (2016). Does restoration of a habitat‐forming seaweed restore associated faunal diversity? Restoration Ecology, 24, 81–90. 10.1111/rec.12292 [DOI] [Google Scholar]
- Mathieson, A. C. , & Dawes, C. J. (1986). Photosynthetic responses of florida seaweeds to light and temperature ‐ a physiological survey. Bulletin of Marine Science, 38, 512–524. [Google Scholar]
- Miller, A. (2019). Local thermal adaptation and limited gene flow constrain future climate responses of a marine ecosystem engineer Dryad Dataset, 10.5061/dryad.g4f4qrfmb [DOI] [PMC free article] [PubMed]
- Miller, A. D. , Hoffmann, A. A. , Tan, M. H. , Young, M. , Ahrens, C. , Cocomazzo, M. , … Sherman, C. D. H. (2019). Local and regional scale habitat heterogeneity contribute to genetic adaptation in a commercially important marine mollusc (Haliotis rubra) from southeastern Australia. Molecular Ecology, 28, 3053–3072. [DOI] [PubMed] [Google Scholar]
- Miranda, R. J. , Coleman, M. A. , Tagliafico, A. , Rangel, M. S. , Mamo, L. T. , Barros, F. , & Kelaher, B. P. (2019) Invasion mediated effects on marine trophic interactions under climate change: Positive feedbacks favour kelp persistence. Proceedings of the Royal Society B‐Biological Sciences, 286, 20182866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohring, M. B. , Wernberg, T. , Wright, J. T. , Connell, S. D. , & Russell, B. D. (2014). Biogeographic variation in temperature drives performance of kelp gametophytes during warming. Marine Ecology Progress Series, 513, 85–96. 10.3354/meps10916 [DOI] [Google Scholar]
- Osborn, J. E. M. (1948) The structure and life history of Hormosira banksii (Turner) Decaisne. Transactions and Proceedings of the Royal Society of New Zealand, 77, 47–71. [Google Scholar]
- Park, S. D. E. (2001). Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Dublin, Ireland: Genetic Department, University of Dublin. [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GENALEX 6: Genetic analysis in Excel. Population genetics software for teaching and research. Molecular Ecology Notes, 6, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro‐Corbeira, C. , Barreiro, R. , Cremades, J. , & Arenas, F. (2018). Seaweed assemblages under a climate change scenario: Functional responses to temperature of eight intertidal seaweeds match recent abundance shifts. Scientific Reports, 8, 12978 10.1038/s41598-018-31357-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocklington, J. B. , Keough, M. J. , O'Hara, T. D. , & Bellgrove, A. (2019). The influence of canopy cover on the ecological function of a key autogenic ecosystem engineer. Diversity‐Basel, 11, 79 10.3390/d11050079 [DOI] [Google Scholar]
- Portner, H. O. , & Gutt, J. (2016). Impacts of climate variability and change on (marine) animals: Physiological underpinnings and evolutionary consequences. Integrative and Comparative Biology, 56, 31–44. 10.1093/icb/icw019 [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober, S. M. , Byrne, M. , McLean, E. H. , Steane, D. A. , Potts, B. M. , Vaillancourt, R. E. , & Stock, W. D. (2015). Climate‐adjusted provenancing: A strategy for climate‐resilient ecological restoration. Frontiers in Ecology and Evolution, 3, 65 10.3389/fevo.2015.00065 [DOI] [Google Scholar]
- Provost, E. J. , Kelaher, B. P. , Dworjanyn, S. A. , Russell, B. D. , Connell, S. D. , Ghedini, G. , … Coleman, M. A. (2017). Climate‐driven disparities among ecological interactions threaten kelp forest persistence. Global Change Biology, 23, 353–361. 10.1111/gcb.13414 [DOI] [PubMed] [Google Scholar]
- Quinn, G. P. , & Keough, M. J. (2002). Experimental design and data analysis for biologists. Cambridge: Cambridge University Press. [Google Scholar]
- Ralph, P. J. , Morrison, D. A. , & Addison, A. (1998). A quantitative study of the patterns of morphological variation within Hormosira banksii (Turner) Decaisne (Fucales : Phaeophyta) in south‐eastern Australia. Journal of Experimental Marine Biology and Ecology, 225, 285–300. 10.1016/S0022-0981(97)00232-3 [DOI] [Google Scholar]
- Raymond, M. , & Rousset, F. (1995). An exact test for population differentiation. Evolution, 49, 1280–1283. 10.1111/j.1558-5646.1995.tb04456.x [DOI] [PubMed] [Google Scholar]
- Razgour, O. , Forester, B. , Taggart, J. B. , Bekaert, M. , Juste, J. , Ibáñez, C. , … Manel, S. (2019). Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proceedings of the National Academy of Sciences of the United States of America, 116, 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razgour, O. , Taggart, J. B. , Manel, S. , Juste, J. , Ibáñez, C. , Rebelo, H. , … Park, K. (2018). An integrated framework to identify wildlife populations under threat from climate change. Molecular Ecology Resources, 18, 18–31. 10.1111/1755-0998.12694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch, T. B. H. (2014). Climate change in the oceans: Evolutionary versus phenotypically plastic responses of marine animals and plants. Evolutionary Applications, 7, 104–122. 10.1111/eva.12109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, L. K. , Stachowicz, J. J. , Hughes, A. R. , Kamel, S. J. , Ort, B. S. , & Grosberg, R. K. (2017). Temporal stability in patterns of genetic diversity and structure of a marine foundation species (Zostera marina). Heredity, 118, 404–412. 10.1038/hdy.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel, D. R. (2006). Rivets or bolts? When single species count in the function of temperate rocky reef communities. Journal of Experimental Marine Biology and Ecology, 338, 233–252. 10.1016/j.jembe.2006.06.023 [DOI] [Google Scholar]
- Schiel, D. R. , & Taylor, D. I. (1999). Effects of trampling on a rocky intertidal algal assemblage in southern New Zealand. Journal of Experimental Marine Biology and Ecology, 235, 213–235. 10.1016/S0022-0981(98)00170-1 [DOI] [Google Scholar]
- Schiffer, M. , Hangartner, S. , & Hoffmann, A. A. (2013). Assessing the relative importance of environmental effects, carry‐over effects and species differences in thermal stress resistance: A comparison of Drosophilids across field and laboratory generations. Journal of Experimental Biology, 216, 3790–3798. 10.1242/jeb.085126 [DOI] [PubMed] [Google Scholar]
- Schueler, S. , Kapeller, S. , Konrad, H. , Geburek, T. , Mengl, M. , Bozzano, M. , … Olrik, D. C. (2013). Adaptive genetic diversity of trees for forest conservation in a future climate: A case study on Norway spruce in Austria. Biodiversity and Conservation, 22, 1151–1166. 10.1007/s10531-012-0313-3 [DOI] [Google Scholar]
- Sgro, C. M. , Lowe, A. J. , & Hoffmann, A. A. (2011). Building evolutionary resilience for conserving biodiversity under climate change. Evolutionary Applications, 4, 326–337. 10.1111/j.1752-4571.2010.00157.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, C. D. H. , & Ayre, D. J. (2008). Fine‐scale adaptation in a clonal sea anemone. Evolution, 62, 1373–1380. 10.1111/j.1558-5646.2008.00375.x [DOI] [PubMed] [Google Scholar]
- Smale, D. A. , & Wernberg, T. (2013). Extreme climatic event drives range contraction of a habitat‐forming species. Proceedings of the Royal Society B: Biological Sciences, 280(1754), 20122829 10.1098/rspb.2012.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale, D. A. , Wernberg, T. , Oliver, E. C. J. , Thomsen, M. , Harvey, B. P. , Straub, S. C. , … Moore, P. J. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nature . Climate Change, 9, 306–312. 10.1038/s41558-019-0412-1 [DOI] [Google Scholar]
- Staehr, P. A. , & Wernberg, T. (2009). Physiological responses of Ecklonia Radiata (Laminariales) to a latitudinal gradient in ocean temperature. Journal of Phycology, 45, 91–99. [DOI] [PubMed] [Google Scholar]
- Starko, S. , Bailey, L. A. , Creviston, E. , James, K. A. , Warren, A. , Brophy, M. K. , … Neufeld, C. J. (2019). Environmental heterogeneity mediates scale‐dependent declines in kelp diversity on intertidal rocky shores. PLoS ONE, 14(3), e0213191 10.1371/journal.pone.0213191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneck, R. S. , Graham, M. H. , Bourque, B. J. , Corbett, D. , Erlandson, J. M. , Estes, J. A. , & Tegner, M. J. (2002). Kelp forest ecosystems: Biodiversity, stability, resilience and future. Environmental Conservation, 29, 436–459. 10.1017/S0376892902000322 [DOI] [Google Scholar]
- Tait, L. W. , & Schiel, D. R. (2013). Impacts of temperature on primary productivity and respiration in naturally structured macroalgal assemblages. PLoS ONE, 8, e74413 10.1371/journal.pone.0074413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegner, M. J. , & Dayton, K. (1987). El Niño effects on southern California kelp forest communities. Advances in Ecological Research, 17, 243–279. [Google Scholar]
- Thomsen, M. S. , Mondardini, L. , Alestra, T. , Gerrity, S. , Tait, L. , South, P. M. ,… Schiel, D. R. (2019) Local extinction of bull kelp (Durvillaea spp.) due to a marine heatwave. Frontiers in Marine Science, 6 10.3389/fmars.2019.00084 [Epub ahead of print]. [DOI] [Google Scholar]
- Thomson, J. A. , Burkholder, D. A. , Heithaus, M. R. , Fourqurean, J. W. , Fraser, M. W. , Statton, J. , & Kendrick, G. A. (2015). Extreme temperatures, foundation species, and abrupt ecosystem change: An example from an iconic seagrass ecosystem. Global Change Biology, 21, 1463–1474. 10.1111/gcb.12694 [DOI] [PubMed] [Google Scholar]
- Valladares, F. , Matesanz, S. , Guilhaumon, F. , Araújo, M. B. , Balaguer, L. , Benito‐Garzón, M. , … Zavala, M. A. (2014). The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecology Letters, 17, 1351–1364. 10.1111/ele.12348 [DOI] [PubMed] [Google Scholar]
- Van Oosterhout, C. , Hutchinson, W. F. , Wills, D. P. M. , & Shipley, P. (2004). MICRO‐CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535–538. 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- Vergés, A. , Doropoulos, C. , Malcolm, H. A. , Skye, M. , Garcia‐Pizá, M. , Marzinelli, E. M. , … Steinberg, P. D. (2016). Long‐term empirical evidence of ocean warming leading to tropicalization of fish communities, increased herbivory, and loss of kelp. Proceedings of the National Academy of Sciences, 113(48), 13791–13796. 10.1073/pnas.1610725113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergés, A. , Steinberg, P. D. , Hay, M. E. , Poore, A. G. B. , Campbell, A. H. , Ballesteros, E. , … Wilson, S. K. (2014). The tropicalization of temperate marine ecosystems: climate‐mediated changes in herbivory and community phase shifts. Proceedings of the Royal Society B: Biological Sciences, 281, 20140846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, M. R. , Castoe, T. , Holmes, J. , Packer, M. , Biles, K. , Walsh, M. , … Post, D. M. (2016). Local adaptation in transgenerational responses to predators. Proceedings of the Royal Society B: Biological Sciences, 283, 20152271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, A. R. , Sgro, C. M. , Young, A. G. , Frankham, R. , Mitchell, N. J. , Miller, K. A. , … Hoffmann, A. A. (2011). Assessing the benefits and risks of translocations in changing environments: A genetic perspective. Evolutionary Applications, 4, 709–725. 10.1111/j.1752-4571.2011.00192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B. , & Cockerham, C. (1984). Estimating F‐statistics for the analysis of population structure. Evolution, 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wernberg, T. , Bennett, S. , Babcock, R. C. , de Bettignies, T. , Cure, K. , Depczynski, M. , … Wilson, S. (2016). Climate‐driven regime shift of a temperate marine ecosystem. Science, 353, 169–172. 10.1126/science.aad8745 [DOI] [PubMed] [Google Scholar]
- Wernberg, T. , Coleman, M. A. , Bennett, S. , Thomsen, M. S. , Tuya, F. , & Kelaher, B. P. (2018). Genetic diversity and kelp forest vulnerability to climatic stress. Scientific Reports, 8, 1851 10.1038/s41598-018-20009-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernberg, T. , de Bettignies, T. , Joy, B. A. , & Finnegan, P. M. (2016). Physiological responses of habitat‐forming seaweeds to increasing temperatures. Limnology and Oceanography, 61, 2180–2190. 10.1002/lno.10362 [DOI] [Google Scholar]
- Wernberg, T. , Russell, B. D. , Moore, P. J. , Ling, S. D. , Smale, D. A. , Campbell, A. , … Connell, S. D. (2011). Impacts of climate change in a global hotspot for temperate marine biodiversity and ocean warming. Journal of Experimental Marine Biology and Ecology, 400, 7–16. 10.1016/j.jembe.2011.02.021 [DOI] [Google Scholar]
- Wernberg, T. , Thomsen, M. S. , Tuya, F. , Kendrick, G. A. , Staehr, P. A. , & Toohey, B. D. (2010). Decreasing resilience of kelp beds along a latitudinal temperature gradient: Potential implications for a warmer future. Ecology Letters, 13, 685–694. 10.1111/j.1461-0248.2010.01466.x [DOI] [PubMed] [Google Scholar]
- Williams, S. L. , & Difiori, R. E. (1996). Genetic diversity and structure in Pelvetia fastigiata (Phaeophyta: Fucales): Does a small effective neighborhood size explain fine scale genetic structure? Marine Biology, 126, 371–382. 10.1007/BF00354619 [DOI] [Google Scholar]
- Willis, S. G. , Foden, W. , Baker, D. J. , Belle, E. , Burgess, N. D. , Carr, J. A. , … Butchart, S. (2015). Integrating climate change vulnerability assessments from species distribution models and trait‐based approaches. Biological Conservation, 190, 167–178. 10.1016/j.biocon.2015.05.001 [DOI] [Google Scholar]
- Wilson, G. A. , & Rannala, B. (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics, 163, 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, G. , Marzinelli, E. M. , Coleman, M. A. , Campbell, A. H. , Santini, N. S. , Kajlich, L. , … Vergés, A. (2019). Restoring subtidal marine macrophytes in the Anthropocene: Trajectories and future‐proofing. Marine and Freshwater Research, 70, 936–951. 10.1071/MF18226 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Miller, A. (2019). Local thermal adaptation and limited gene flow constrain future climate responses of a marine ecosystem engineer Dryad Dataset, 10.5061/dryad.g4f4qrfmb [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Genetic and phenotypic data will be made publicly available in the DRYAD: ://doi.org/10.5061/dryad.g4f4qrfmb (Miller, 2019).


