Abstract
Synaptic pathology is associated with several brain disorders, thus positron emission tomography (PET) imaging of synaptic vesicle glycoprotein 2A (SV2A) using the radioligand [11C]UCB-J may provide a tool to measure synaptic alterations. Given the pivotal role of mouse models in understanding neuropsychiatric and neurodegenerative disorders, this study aims to validate and characterize [11C]UCB-J in mice. We performed a blocking study to verify the specificity of the radiotracer to SV2A, examined kinetic models using an image-derived input function (IDIF) for quantification of the radiotracer, and investigated the in vivo metabolism. Regional TACs during baseline showed rapid uptake of [11C]UCB-J into the brain. Pretreatment with levetiracetam confirmed target engagement in a dose-dependent manner. VT (IDIF) values estimated with one- and two-tissue compartmental models (1TCM and 2TCM) were highly comparable (r=0.999, p < 0.0001), with 1TCM performing better than 2TCM for K1 (IDIF). A scan duration of 60 min was sufficient for reliable VT (IDIF) and K1 (IDIF) estimations. In vivo metabolism of [11C]UCB-J was relatively rapid, with a parent fraction of 22.5 ± 4.2% at 15 min p.i. In conclusion, our findings show that [11C]UCB-J selectively binds to SV2A with optimal kinetics in the mouse representing a promising tool to noninvasively quantify synaptic density in comparative or therapeutic studies in neuropsychiatric and neurodegenerative disorder models.
Keywords: Animal models, kinetic modeling, positron emission tomography, SV2A, synaptic density
Introduction
Synaptic vesicles play an essential role in neurotransmitter release from the presynaptic terminals following action potential stimulation.1 The synaptic vesicle glycoprotein 2 (SV2) is a highly conserved protein family in all vertebrates that displays a vital role in physiological synaptic functionality.2 SV2 proteins are divided into three isoforms, which show differential expression. SV2A is the predominant isoform, SV2B is also widely-expressed, although not as extensively as SV2A, and SV2C is only present in specific brain regions.3–5 Indeed, SV2A is expressed ubiquitously in virtually all synapses3 and its role has been shown to be vital as mice lacking SV2A develop spontaneous seizures and die postnatally due to destabilization of synaptic circuits.4,6 In addition, SV2A is the binding site of the antiepileptic drug levetiracetam (LEV)7 and dysfunction of SV2A has also been associated with the pathophysiology of epilepsy8 as well as other neurological disorders.9
Since SV2A is homogenously distributed in synaptic vesicles,10 quantifying SV2A offers the opportunity to monitor changes in synaptic density. Different radiotracers to visualize SV2A in vivo using positron emission tomography (PET) have been described, including [11C]UCB-J, [18F]UCB-H, and [11C]UCB-A.11–14 [11C]UCB-J appears the most promising due to its optimal pharmacokinetics and quantification properties as recently reported in both non-human primates11 and healthy humans.15,16 In addition, recent studies have demonstrated the feasibility of [11C]UCB-J PET in detecting pathological changes in synaptic density in patients with mesial temporal lobe epilepsy,16 Alzheimer's disease,17 and depression.18
However, although in vivo PET imaging using [11C]UCB-J has been published in humans and non-human primates, to date no study validating the binding nor characterizing the kinetics of [11C]UCB-J PET imaging in rodents has been described. Mouse models play a crucial role in biomedical research19 for the translation of basic research findings into the clinical applications. Given the invaluable importance of mouse models in studying neuropsychiatric and neurodegenerative disorders, and the broad spectrum of diseases characterized by synaptic dysfunction, application of [11C]UCB-J PET imaging in mice would provide significant advances in understanding the pathophysiology and diagnosis of neurological disorders.20 The aim of the present study was to validate the specific binding and to provide the methodology for noninvasive in vivo quantification of [11C]UCB-J PET imaging in mice in order to apply this radiotracer in preclinical studies. We performed a blocking study to verify the specificity of the radiotracer to SV2A, examined kinetic models and parameter estimations for quantification of the radiotracer, and investigated the in vivo metabolism of [11C]UCB-J in plasma of healthy and Huntington's Disease model (HD) mice.
Materials and methods
Animals
A total of 38 male mice (C57BL/6J) were obtained from Jackson Laboratories (Bar Harbour, Maine, USA) and included in the study. In vivo PET imaging was performed in eight-months-old male wild-type (C57BL/6J, WT, n = 10). For the generation of a population-based metabolite curve, six-months-old WT and heterozygous littermates (Q175DN mouse model of Huntington's disease, HD) were included (n = 15 per genotype). Animals were group-housed in individually ventilated cages under a 12-h light/dark cycle in a temperature- and humidity-controlled environment with food and water ad libitum. At least one week of acclimatization was allowed before the start of the procedures. All experiments were performed according to the European Committee Guidelines (decree 2010/63/CEE) and the Animal Welfare Act (7 USC 2131), and reported in compliance with the ARRIVE guidelines. Experiments were approved by the Ethical Committee for Animal Testing (ECD 2017-27) at the University of Antwerp (Belgium).
Radiotracer synthesis
[11C]UCB-J synthesis was performed on an automated synthesis module (Carbosynthon I, Comecer, The Netherlands) adapting the previously described procedure11 to our system. Briefly, [11C]UCB-J compound was synthetized via a single-step carbon-11 labelling starting with a mixture of trifluoroborate and boric acid precursors (ratio 95:5), which was prepared prior to the synthesis and reacted with [11C]MeI already trapped into DMF. Reaction occurred in the presence of palladium catalyst (Pd2(dba)3), P(o-tol)3, and a base (K2CO3) in DMF for 5 min at 100℃. Trapping of [11C]MeI in the reactor with DMF prior to the addition of the mixture was required for the reaction. Average radiochemical purity was greater than 99%, and the molar activity was 78.8 ± 23.0 GBq/µmol.
PET acquisition and reconstruction
Ninety-minute dynamic microPET/computed tomography (CT) images were acquired on two Siemens Inveon PET-CT scanners (Siemens Preclinical Solution, Knoxville, USA). Anaesthesia was induced and maintained using isoflurane (Forene, Belgium; 5% and 1.5%, respectively) in oxygen. Animals (n = 10) were catheterized in the tail vein for intravenous (i.v.) administration of the tracer and positioned onto the scanner. Physiological parameters (respiration and temperature) were continuously monitored and kept constant throughout the procedure using a Monitoring Acquisition Module (Minerve, France). At the onset of the dynamic PET scan, bolus injection was performed using an automated pump (Pump 11 Elite, Harvard Apparatus, USA) over a 12 s interval (1 ml/min). Following the microPET scan, a 10 min 80 kV/500 μA CT scan was performed for attenuation correction as well as for anatomical registration.
A blocking study to validate specificity of [11C]UCB-J binding was performed using the antiepileptic drug LEV (Merck, Germany), a high affinity ligand for SV2A,21 at two different doses that displayed antiepileptic effects in mouse models of epilepsy22,23 (50 and 200 mg/kg i.p.; n = 4 per dose) in the same animals scanned at baseline. LEV was dissolved in saline and injected 30 min before administration of [11C]UCB-J. Methodologically, no significant difference between baseline and blocking PET scans was observed in the injected activity (baseline = 4.5 ± 1.2 MBq, LEV50 =3.6 ± 0.7 MBq, LEV200 = 4.3 ± 0.3 MBq; p = 0.251), body weight (baseline = 31.2 ± 0.9 g, LEV50 =30.7 ± 0.1 g, LEV200 = 31.4 ± 1.5 g; p = 0.606), or injected mass (baseline = 1.0 ± 0.3 µg/kg, LEV50 =1.2 ± 0.4 µg/kg, LEV200 = 1.02 ± 0.3 µg/kg; p = 0.226).
Acquired PET data were histogrammed and reconstructed into 39 frames of increasing length (12 × 10 s, 3 × 20 s, 3 × 30 s, 3 × 60 s, 3 × 150 s and 15 × 300 s). For quantitative analysis, all images were reconstructed using a list-mode iterative reconstruction with proprietary spatially variant resolution modeling in 8 iterations and 16 subsets of the 3D ordered subset expectation maximization (OSEM 3D) algorithm. Normalization, dead time, and CT-based attenuation corrections were applied. PET image frames were reconstructed on a 128 × 128 × 159 grid with 0.776 × 0.776 × 0.776 mm3 voxels.
Image processing and kinetic modeling
Analysis and processing of the PET data were performed using PMOD 3.6 software (Pmod Technologies, Zurich, Switzerland). In order to achieve spatial normalization of the PET images, a [11C]UCB-J PET template was generated using the 90-min static PET images of the baseline scans, adapting the procedure we previously described24 to [11C]UCB-J. Individual static [11C]UCB-J PET images were spatially normalized to this [11C]UCB-J PET template through non rigid brain normalization. The spatial transformations were saved and applied to dynamic PET imaging frames. For the blocking experiment, the PET template approach could not be applied due to lack of specific signal. Therefore, for these data, individual CT images were spatially normalized to the CT image of the [11C]UCB-J PET template adapting the procedure we previously described.24 All images were visually checked for accuracy following spatial transformation.
Since the [11C]UCB-J PET template was generated in the same space of the Waxholm atlas,25 volumes-of-interest (VOIs) were adapted from that atlas. Regional time-activity curves (TACs) were calculated for the following regions: striatum (STR), thalamus (THAL), motor cortex (MC), hippocampus (HC), and cerebellum (CB).
Kinetic analysis was performed by fitting the regional TACs with the one-tissue compartmental model (1TCM), two-tissue compartmental model (2TCM) and Logan plot method.26 We estimated the total blood radioactivity using a noninvasive image-derived input function (IDIF) to obtain VT (IDIF) as a surrogate of VT. For the compartmental models, the blood volume fraction (VB) was fixed at 3.6%,27 while for the Logan plot the linear phase (t*) was determined from the curve fitting with t* ranging from 7.5 to 12.5 min. The aforementioned IDIF was extracted from the PET images measuring the whole blood activity in the lumen of the left ventricle of the heart. Such was achieved by generating a volume-of-interest on the early time frame exhibiting maximal activity in the heart with a threshold considering 50% of the max intensity as previously described.28 This approach allows to perform comparative studies in mice avoiding extensive blood (or plasma) sampling which is not achievable in longitudinal studies.
When performing full kinetic modeling, it is possible to estimate the microparameters K1 and k229 in addition to the macroparameter VT, which is the theoretical volume of tissue needed to contain the total amount of radiotracer at the concentration in metabolite-corrected plasma at equilibrium. K1 represents the delivery rate of the tracer and is hence an indication of the cerebral blood flow, while k2 depicts the transport of the tracer from the tissue back to the blood. K1 is dependent on perfusion, while k2 is coupled to K1.
During the blocking study, only the Logan plot method was used for kinetic modeling since assessment of the microparameters was not required. In addition, an estimation of the regional non-displaceable binding potential (BPND) was calculated as ratio of VT and VND minus 1 (DVR-1)30 using the individual regional VT (IDIF) values of baseline scans for VT and the group regional VT (IDIF) values during the blocking with highest dose (200 mg/kg) as an estimate for the non-displaceable volume of distribution (VND) (Supplemental Table 1).
Parametric images were generated with PMOD 3.6 software using the pixel-wise modeling tool (PXMOD) through voxel-based graphical analysis (Logan plot, VT (IDIF) images) and 1TCM31 (VT (IDIF) and K1 (IDIF) images) with the IDIF as input function, represented as group averages, and overlaid onto a 3D coronal/sagittal/transversal mouse brain template for anatomical reference.
To determine the relative performance of each model to fit the regional PET data, the goodness-to-fit was calculated using the Akaike Information Criterion (AIC),32 the Model Selection Criterion (MSC), a modified version of the AIC criterion used in the Scientist Software (MicroMath, Saint Louis, Missouri USA), and the F-test.33
To estimate the extent of target occupancy reached during the PET scan with LEV pretreatment, we employed the Lassen plot,34 which also provides an estimate of the VND,35 based on the regional changes in VT (IDIF) following drug administration.
To evaluate the time stability of the VT (IDIF) and K1 (IDIF) values, PET data were analysed by repeatedly excluding the last 5 min of PET acquisition from 90 min down to 20 min. The 90 min PET acquisition was considered the reference outcome to which values obtained from shorter acquisition were compared to. Quantification of PET data was considered acceptable when the average percentage difference compared to the 90 min PET acquisition was less than 10% with an inter-individual standard deviation lower than 5%.
Metabolite analysis and correction
In vivo plasma metabolite analysis was performed at 5, 15, 30, and 45 min post-injection (p.i.). The procedure was done in six-months-old wild-type (WT) mice as well as Q175DN heterozygouos mice (n = 3 per time point/genotype) with the aim to characterize the metabolism of [11C]UCB-J in mice and to investigate possible disease-associated differences in radiotracer metabolism as well as for generation of a population-based curve to correct for this radiometabolism in the quantification.
Mice were injected via the lateral tail vein with [11C]UCB-J (4.9 ± 1.6 MBq in 200 μl) and blood was withdrawn via cardiac puncture at 5, 15, 30, and 45 min p.i. Our procedure was previously described36 and currently adapted as follows: plasma samples (200 μl), obtained after centrifugation of blood at 4500 × r/min (2377 × rcf) for 5 min, were mixed with equal amounts of ice-cold acetonitrile. Then, 10 µl of cold reference were added (1 mg/ml), and subsequently centrifuged at 4500 × rpm (2377 × rcf) for 5 min to precipitate denatured proteins. Supernatant was separated from the precipitate and both fractions were counted in a gamma counter to calculate the extraction efficiency (percentage of recovery of radioactivity), which was 97.3 ± 4.8%. Next, 100 μl of supernatant were loaded onto a pre-conditioned reverse-phase (RP)-HPLC system (Kinetex, 150 × 4.6 mm, 5 μm HPLC column +Phenomenex security guard pre-column) and eluted with NaOAc 0.05M pH 5.5 and acetonitrile (68:32 v/v) buffer at a flow rate of 1.5 ml/min. RP-HPLC fractions were collected at 0.5 min intervals for 8 min and radioactivity was measured in a gamma counter. The radioactivity was expressed as a percentage of the total area of the peaks based on the radiochromatograms.
To determine the recovery of [11C]UCB-J as well as the stability of the tracer during the workup, control experiments were performed using blood spiked in vitro with 37 kBq of radiotracer. Sample workup was identical as described and confirmed that no degradation of the tracers occurred during procedural work-up (99.7 ± 0.25% of intact tracer).
To generate the population-based curve to account for peripheral radiometabolism, we used the parent fractions measured in WT mice at different times p.i. In PMOD 3.6 software, values were fitted using a sigmoid curve as this model provided the best fit compared to other models (i.e. Watabe, Hill, 1-Exponential, 2-Exponentials, and 3-Exponentials). Thus, each individual IDIF was corrected for the sigmoid curve of the parent fraction and the plasma-to-whole blood ratio, calculated based on the radiometabolite analysis data, in order to derive the metabolite-corrected input function to use for kinetic modeling.
Statistical analysis
Two-way ANOVA was applied to the blocking experiment to compare baseline and blockade scans in the different regions. Pearson's correlation tests as well as Bland-Altman plots were used to compare VT (IDIF) and K1 (IDIF) values determined with different kinetic models. In addition, Pearson's correlation tests were used to compare the effect of shortening the PET acquisition, to correlate the VT (IDIF) values either corrected or not for metabolism, and for the Lassen plots. The percentage of the intra-animal coefficient of variation (COV) within each kinetic model and brain region was calculated as follows
where N represents the number of measurements in the group, xi and SDi respectively the mean and standard deviation of the values for measurement i. All analyses were performed with GraphPad Prism (v 6.0) statistical software. The data are represented as mean ± standard deviation (SD). All tests were two-tailed and significance was set at p < 0.05.
Results
Validation of [11C]UCB-J binding in mice
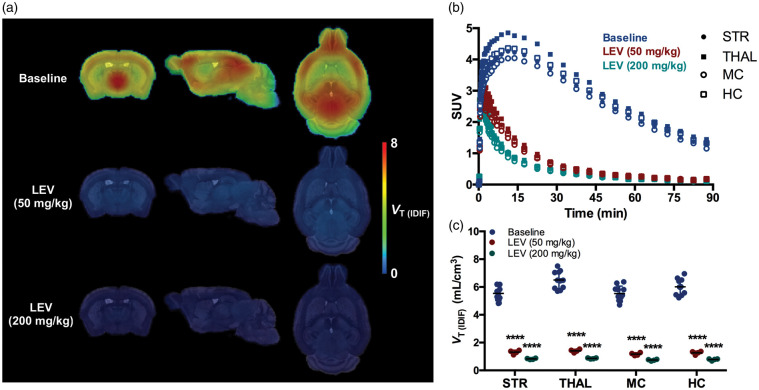
To validate the specificity of [11C]UCB-J binding to SV2A in the mouse, we performed a blocking study with LEV. Baseline and blockade scans are shown in Figure 1. The average SUV TACs during baseline showed the rapid brain uptake of [11C]UCB-J, peaking within the first 15 min in the different investigated brain regions, while the pretreatment with LEV at two different doses (50 and 200 mg/kg, i.p.) resulted in a manifest decline of [11C]UCB-J binding (Figure 1(b)). Accordingly, VT (IDIF) (Logan) values were significantly reduced in all investigated brain regions following pretreatment with LEV in a dose-dependent manner (e.g. in striatum: baseline = 5.17 ± 0.34 mL/cm3; LEV50 = 1.31 ± 0.14 mL/cm3, −74.6%, p < 0.0001; LEV200 = 0.82 ± 0.04 mL/cm3, −84.1%, p < 0.0001) (Figure 1(c)). The effect of pretreatment with LEV on [11C]UCB-J VT (IDIF) values for each individual region and the estimated BPND are reported in Table 1.
Figure 1.
Validation of [11C]UCB-J binding in the mouse brain. (a) Effect of blockade with levetiracetam (LEV) on [11C]UCB-J uptake in WT mice. Parametric microPET images are overlaid onto a MRI mouse brain template for anatomical localization. (b) SUV time-activity curves in four brain regions during baseline and pretreatment with levetiracetam for a 90-min acquisition. (c) VT (IDIF) (Logan) quantification during blockade resulted in a statistically significant reduced values in all investigated regions compared to baseline. ****p < 0.0001. Baseline: n = 10; LEV: n = 4 per dose. STR: striatum; THAL: thalamus; MC: motor cortex; HC: hippocampus; VT (IDIF): total volume of distribution using IDIF.
Table 1.
Effect of blockade of [11C]UCB-J binding using levetiracetam determined by Logan plot based on 90-min acquisition.
| Baseline |
Pretreatment with levetiracetam |
Estimated BPND |
||||
|---|---|---|---|---|---|---|
|
VT (IDIF)
(mL/cm3) |
50 mg/kg |
200 mg/kg |
DVR-1 |
|||
| Region | Mean ± SD | Mean ± SD | Diff (%) | Mean ± SD | Diff (%) | Mean ± SD |
| Striatum | 5.17 ± 0.44 | 1.31 ± 0.14 | −74.6§ | 0.82 ± 0.04 | −84.1§ | 5.26 ± 0.42 |
| Thalamus | 6.28 ± 0.53 | 1.38 ± 0.16 | −78.0§ | 0.86 ± 0.04 | −86.3§ | 6.17 ± 0.49 |
| Motor cortex | 5.46 ± 0.48 | 1.16 ± 0.10 | −78.7§ | 0.74 ± 0.05 | −86.4§ | 6.22 ± 0.51 |
| Hippocampus | 5.82 ± 0.47 | 1.26 ± 0.12 | −78.3§ | 0.77 ± 0.06 | −86.7§ | 6.38 ± 0.48 |
VT(IDIF): total volume of distribution using IDIF; BPND: non-displaceable binding potential; DVR: distribution volume ratio; Diff: difference.
§p < 0.0001.
To estimate the SV2A occupancy reached with the pretreatment with LEV at the 2 different doses, we applied the Lassen graphical analysis. A linear relationship was observed for both doses (50 and 200 mg/kg) with fitted occupancy estimates of 79.2% and 97.3%, respectively (Supplemental Figure 1), with the latter indicating an approximate VND (IDIF) of 0.60 mL/cm3.
Kinetic analysis of [11C]UCB-J binding in the mouse brain
Kinetic analysis was performed on the regional TACs by applying different models, namely Logan plot, 1TCM, and 2TCM, in order to estimate VT (IDIF). First, selection of the most appropriate model was performed. The relative performance of each model was evaluated in different brain regions. According to the AIC values, where the model with the lowest value is the preferred, the 1TCM appeared to be preferable compared to 2TCM or Logan plot (e.g. in striatum: 1TCM = 40.9, 2TCM = 44.5, and Logan = 90.4) (Supplemental table 2). The MSC values, where the preferred model has the highest value, were in agreement with the AIC values with 1TCM being slightly better than 2TCM (e.g. in striatum: 1TCM = 2.6, 2TCM = 2.5) (Supplemental Table 3). Consistently, the F-test indicated 1TCM as the preferred model since the F-statistic was lower than the tabulated F value (F = 3.3 for p = 0.05) in all investigated regions (striatum = 0.6, thalamus = 0.7, motor cortex = 0.3, hippocampus = 1.8, and cerebellum = 0.1).
The 1TCM microparameters and VT (IDIF) values as well as the 2TCM and Logan plot VT (IDIF) values are reported in Table 2. The microparameters for 2TCM are not reported due to the low reliability (relative standard errors were above 20%, especially for k3 and k4). The estimated K1 values based on 1TCM were high, ranging from 1.99 ± 0.33 mL/cm3/min in thalamus to 1.40 mL/cm3/min in cerebellum, while the k2 were ranging from 0.32 ± 0.05 per min in cerebellum to 0.26 ± 0.03 per min in motor cortex and hippocampus (Table 2). Notably, the relative standard errors for the K1 and k2 estimates based on 1TCM were low, ranging from 1.6% to 6.4% for K1 and from 2.3% to 8.8% for k2.
Table 2.
In vivo kinetic parameters of [11C]UCB-J determined using 1TCM, 2TCM, and Logan plot based on 90-min acquisition in the mouse brain.
| 1TCM |
2TCM |
Logan plot |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
K1(IDIF)
(mL/cm3/min) |
k2 (IDIF) (per
min) |
VT (IDIF)
(mL/cm3) |
VT (IDIF)
(mL/cm3) |
VT (IDIF)
(mL/cm3) |
||||||
| Region | Mean ± SD | COV (%) | Mean ± SD | COV (%) | Mean ± SD | COV (%) | Mean ± SD | COV (%) | Mean ± SD | COV (%) |
| Striatum | 1.52 ± 0.21 | 13.67 | 0.27 ± 0.029 | 10.85 | 5.63 ± 0.43 | 7.67 | 5.66 ± 0.44 | 7.91 | 5.17 ± 0.34 | 6.69 |
| Thalamus | 1.99 ± 0.33 | 16.91 | 0.28 ± 0.037 | 13.12 | 6.96 ± 0.55 | 7.94 | 6.98 ± 0.54 | 7.79 | 6.28 ± 0.43 | 6.85 |
| Motor Cortex | 1.59 ± 0.28 | 17.78 | 0.26 ± 0.03 | 14.13 | 6.00 ± 0.47 | 7.91 | 6.01 ± 0.48 | 7.94 | 5.46 ± 0.38 | 6.97 |
| Hippocampus | 1.65 ± 0.24 | 14.83 | 0.26 ± 0.03 | 11.35 | 6.28 ± 0.48 | 7.61 | 6.30 ± 0.48 | 7.56 | 5.82 ± 0.37 | 6.45 |
| Cerebellum | 1.40 ± 0.27 | 19.62 | 0.32 ± 0.05 | 17.35 | 4.41 ± 0.35 | 8.03 | 4.42 ± 0.36 | 8.07 | 3.82 ± 0.27 | 7.05 |
Note: The microparameters for 2TCM are not reported due to the low reliability (relative standard errors were above 20%).
1TCM: one-tissue compartmental model; 2TCM: two-tissue compartmental model; VT (IDIF): total volume of distribution using IDIF; COV: coefficient of variation.
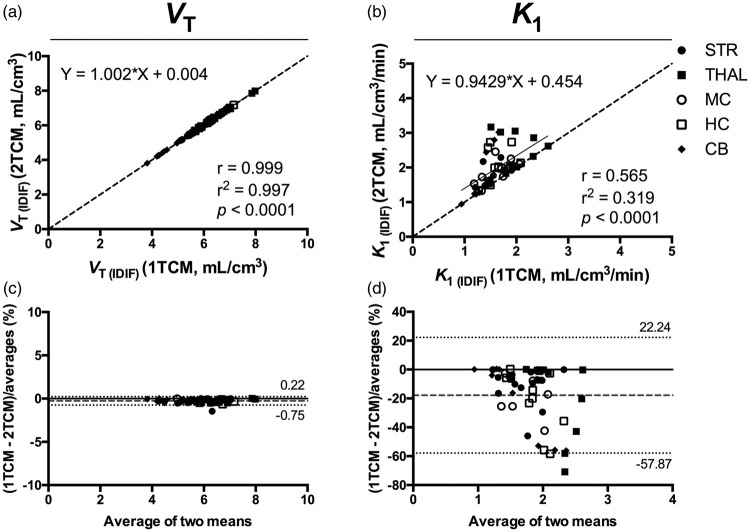
Comparison of VT (IDIF) values between 1TCM and 2TCM resulted in a nearly perfect correlation (r = 0.999, p < 0.0001), with no deviation from the identity line (slope = 1.002 and intercept = +0.004) (Figure 2). Bland–Altman plot between the two models confirmed the extremely high agreement (bias = −0.26%) (Figure 2). On the contrary, the K1 estimates based on 2TCM were unstable and affected by large relative standard errors (>20%) in one-third of the samples. Thus, K1 estimates calculated with 2TCM were overestimated for those samples compared to 1TCM, resulting in only a moderate correlation between the 2 models (r = 0.565, p < 0.0001) with deviation from the identity line (slope = 0.9429 and intercept = +0.454) and a significant bias (−17.82%) according to the Bland-Altman plot as shown in Figure 2.
Figure 2.
Comparison of the total volume of distribution (VT) and K1 values determined using 1TCM and 2TCM based on IDIF. Correlation of the VT (IDIF) (a) and K1 (IDIF) (b) values calculated using 1TCM and 2TCM based on 90-min acquisition. The solid line represents the linear regression, while the dashed line depicts the identity line. (c and d) Bland–Altman plot to compare 1TCM and 2TCM in estimation of VT (IDIF) and K1 (IDIF), respectively. Dashed line represents the bias between the two approaches, while the dotted lines denote the 95% limits of agreement. n = 10. Each symbol represents a different region. 1TCM: one-tissue compartmental model; 2TCM: two-tissue compartmental model; VT (IDIF): total volume of distribution using IDIF.
VT (IDIF) values determined using the Logan plot were consistently slightly underestimated when compared to 1TCM or 2TCM, nonetheless showing strong correlations (r = 0.973, p < 0.0001 and r = 0.974, p < 0.0001, respectively), with negligible deviation from the identity line (slope = 1.029 and intercept = +0.397; slope = 1.031 and intercept = +0.399, respectively) (Supplemental Figure 2). Bland–Altman plots confirmed the consistent bias towards underestimation of the VT (IDIF) values determined by Logan plot compared to 1TCM and 2TCM (bias = −10.1% and −10.28%, respectively) (Supplemental Figure 2).
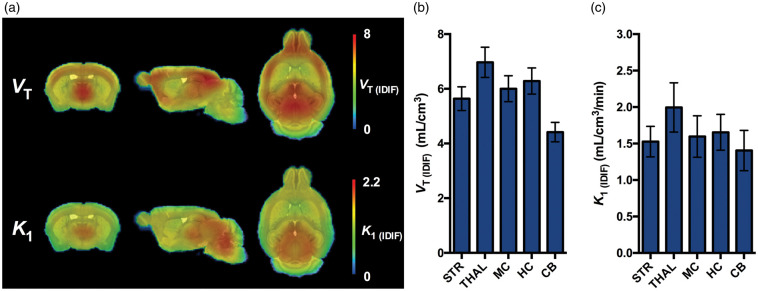
Overall, 1TCM resulted in the most reliable model to estimate both VT (IDIF) and K1 (IDIF) values in the mouse brain. The average paramteric VT (IDIF) and K1 (IDIF) images of [11C]UCB-J determined with 1TCM and 90-min acquisition in the mouse brain are shown in Figure 3(a), while the VT (IDIF) and K1 (IDIF) values based on regional TACs are reported in Figure 3(b) and (c).
Figure 3.
[11C]UCB-J PET imaging in the mouse brain. (a) Average microPET parametric images for VT (IDIF) and K1 (IDIF) of [11C]UCB-J in healthy WT mice using 1TCM over 90-min acquisition. Parametric microPET images are overlaid onto a MRI mouse brain template for anatomical localization. (b) Regional [11C]UCB-J VT (IDIF) values. (c) Regional [11C]UCB-J K1 (IDIF) values. n = 10. 1TCM: one-tissue compartmental model; VT (IDIF) : total volume of distribution using IDIF; STR: striatum; TH: thalamus; MC: motor cortex; HC: hippocampus; CB: cerebellum.
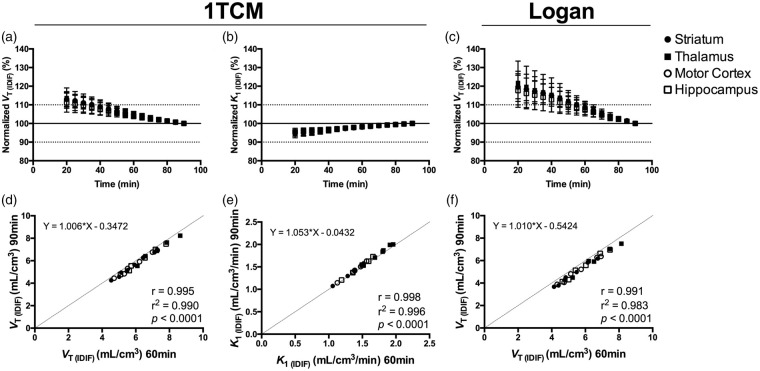
To assess the time stability of the parameter estimation, the effect of the scan duration on VT (IDIF) (1TCM and Logan) and K1 (1TCM) values was investigated. As shown in Figure 4, the VT (IDIF) and K1 (IDIF) values based on 1TCM were stable, with a deviation of only 4.8 ± 1.5% and 1.9 ± 0.6%, respectively, when considering 60-min acquisition. VT (IDIF) values based on Logan deviated 8.6 ± 2.8% when shortening the acquisition to 60 min (Figure 4(c)). Importantly, comparison of the VT (IDIF) and K1 (IDIF) values based on 90 and 60 min resulted in excellent correlations with both 1TCM (VT (IDIF): r = 0.995, p < 0.0001, slope = 1.006 and intercept = –0.3472; K1: r = 0.998, p < 0.0001, slope = 1.053 and intercept = –0.0432) and Logan plot (VT (IDIF): r = 0.991, p < 0.0001, slope = 1.010 and intercept = –0.5424) (Figure 4) indicating that a scan acquisition of 60 min is sufficient to obtain accurate estimates.
Figure 4.
Time stability of the outcome parameters in different brain regions. VT (IDIF) and K1 (IDIF) calculated using 1TCM (a and b) and VT (IDIF) based on Logan plot (c) normalized to the values obtained during 90-min acquisition. Correlations of the outcome parameters VT (IDIF) and K1 (IDIF) calculated using 1TCM (d and e) and VT (IDIF) based on Logan plot (f) when shortening the acquisition time from 90 to 60 min. n = 10. 1TCM = one-tissue compartmental model, VT (IDIF) = total volume of distribution using IDIF.
Plasma metabolism of [11C]UCB-J
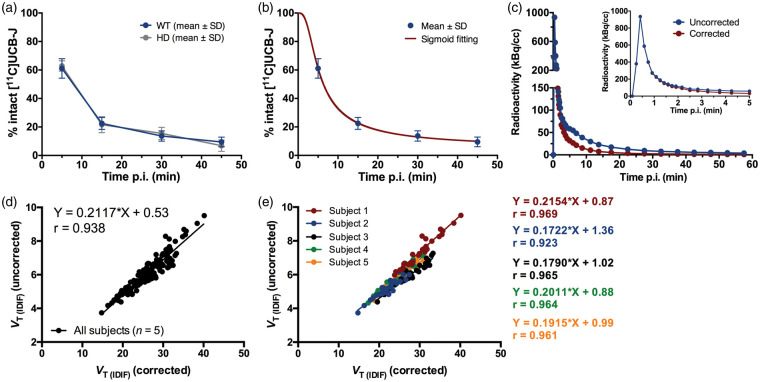
In vivo metabolism of [11C]UCB-J was relatively rapid. The parent fraction in plasma of WT mice accounted for 61.1 ± 6.8%, 22.5 ± 4.2%, 13.6 ± 3.7%, and 9.5 ± 3.4% of the total radioactivity at 5, 15, 30, and 45 min p.i., respectively (Figure 5(a) and Supplemental Table 4). Also, the percentage of unchanged [11C]UCB-J in plasma of diseased mice (heterozygous for HD), accounting for 62.8 ± 3.6%, 21.6 ± 5.6%, 15.5 ± 4.3%, and 6.9 ± 3.9% of the total radioactivity at 5, 15, 30, and 45 min p.i., respectively, was comparable to the values obtained in WT (Figure 5(a) and Supplemental Table 4). Radiochromatograms of both WT and diseased mice are shown in Supplemental Figure 3.
Figure 5.
Population-based plasma metabolism of [11C]UCB-J in mice. (a) Population-based curve of intact [11C]UCB-J in plasma of healthy wild-type (WT) and diseased (model of Huntington's disease, HD) mice. n = 3 per genotype at each time point. (b) Sigmoid fitting curve of the intact [11C]UCB-J in WT mice. (c) Comparison of representative image-derived input functions (IDIF) corrected and uncorrected for the population-based sigmoid fitting of intact [11C]UCB-J. Scatter plots between VT (IDIF) (Logan) corrected and uncorrected for the population-based sigmoid fitting of intact [11C]UCB-J based on a total of five WT mice (d) and correlations for each individual WT mouse (e) in different brain regions. Individual animals presented different regression line equations.
The values of parent fraction at the different times p.i. in WT mice were corrected for the plasma-to-whole blood ratio and fitted using a sigmoid curve in order to generate a population-based curve of intact [11C]UCB-J (Figure 5(b)), which was applied to the IDIF in order to correct for metabolism as shown in Figure 5(c). Comparison of the VT values based on Logan plot calculated using the corrected and uncorrected IDIF in all Waxholm atlas brain regions of five animals resulted in a high correlation (r = 0.938; p < 0.0001, slope = 0.2117 and intercept = +0.53) (Figure 5(d)). Noteworthy, the comparison of the VT values of each individual subject resulted in even higher correlations (average r = 0.956, p < 0.0001) (Figure 5(e)).
Discussion
Rodent models are vital to understanding pathophysiology, identifying novel biomarkers, and testing candidate therapies to translate basic research findings into clinical applications. In our present study, we aimed to characterize [11C]UCB-J PET imaging in mice by validating the specificity of [11C]UCB-J binding to SV2A, and to determine the optimal pharmacokinetic model for noninvasive in vivo quantification. First, we validated the specificity of [11C]UCB-J by pretreating mice with LEV and confirmed the dose-dependent blockade of [11C]UCB-J binding site in the whole brain as was expected given the ubiquitous distribution of SV2A in the grey matter.3 We reached a LEV SV2A occupancy of up to 97%, which corresponded to a decrease of 86% in [11C]UCB-J VT (IDIF). This suggests that only a small component of the total [11C]UCB-J brain uptake is related to VND, while the majority of [11C]UCB-J is specifically binding to the target (VS). This fairly low VND indicates [11C]UCB-J PET imaging might be applicable in target occupancy studies in mice as was recently shown in humans.37
In addition to [11C]UCB-J, [11C]UCB-A and [18F]UCB-H have been reported as radiotacers targeting SV2A.12,14 Both radiotracers have shown specific binding to SV2A; however, [11C]UCB-A was limited by its slow kinetics and metabolism in both rats and pigs.12 On the other hand, [18F]UCB-H had good kinetics, but direct comparison with [11C]UCB-J in non-human primates suggested the latter has higher specific binding.11 Consistently, Warnock et al.38 reported a decrease of approximately 44% in the whole-brain [18F]UCB-H binding in rats pretreated with 100 mg/kg of LEV, while in the present study, with a 50 mg/kg dose in mice, we could observe a decrease of 78% of the [11C]UCB-J binding to SV2A. Assuming a comparable VND across rodent species, this might be a further indication of the higher specific binding of [11C]UCB-J compared to [18F]UCB-H. However, comparative studies in the same animals are recommended for a conclusive assessment.
Previous studies reported the utility of white matter structures (e.g. centrum semiovale) as reference region for kinetic analysis in non-human primates and humans11,15,37; however, the much smaller size of the mouse brain did not allow the application of white matter tissue as reference region even though microPET scanners feature higher spatial resolution. Alternatively, to perform noninvasive quantification, it is possible to use an IDIF.39 Although this approach is not suitable for absolute quantification, since it often overestimates the blood activity, we previously validated it to be reliable for comparative studies.28
Kinetic analysis of the TACs indicated a near perfect agreement between VT (IDIF) values estimated using either 1TCM or 2TCM (r = 0.999). According to model selection criteria (AIC, MSC, and F-test), 1TCM was the preferred model, as also suggested by the more accurate K1 estimations in agreement with the previously published human data.15 Indeed, in one-third of the cases, K1 calculated using 2TCM was unstable resulting in overestimation of the values, hence the moderate correlation with K1 estimates determined with 1TCM (r = 0.565). This evidence suggests 2TCM could not adequately discriminate between K1 and k3 possibly due to [11C]UCB-J kinetic profile. Notably, the Logan plot seemed to be a valid alternative, despite showing a small but consistent underestimation of the VT (IDIF) values. Nonetheless, the main advantage of applying 1TCM is the ability to estimate K1 values as an indication of the cerebral blood flow. Importantly, reliable 1TCM-based parametric images of both VT (IDIF) and K1 (IDIF) could be generated for each subject, which was not the case when applying the 2TCM.
We evaluated the time stability of the estimated VT (IDIF) and K1 (IDIF) values to verify whether the 90-min acquisition could be shortened. Overall, 60-min acquisition resulted in <5% of deviation from the 90 min scan and nearly perfect correlations between the 2 measurements using 1TCM. Thus, [11C]UCB-J parameter estimation showed high stability, indicating the possibility to reduce scan acquisition down to 60 min as recently shown in humans.15
Plasma analysis indicated a relatively rapid metabolism of the tracer with generation of at least one radiometabolite that was more polar than [11C]UCB-J. This result as well as the calculated parent fraction curve were consistent with the literature.11,15 Nabulsi et al.11 reported the generation of at least two polar radiometabolites in plasma of rats. Similarly, three polar radiometabolites were found in humans.15 In the present study, only one radiometabolite peak could be determined. This could be related to the limited time resolution of our setup (0.5 min), which did not allow us to discriminate between radiometabolites with similar retention times. Concerning possible radiometabolites in the brain, previous work reported only a minor amount of radiometabolites (<1.5%) in the rat brain11; however, such negligible amount could be related to the blood volume fraction present in the brain, which is estimated to approximately account for 3.6 to 4.1% of the total rat brain volume.27 Altogether, the reported rat data and the clear blocking results from the present study suggest that no brain-penetrant radiometabolites contribute to the brain PET signal.
Arterial blood sampling in mice is very challenging, especially if applied to repeated scans (for instance the blocking study), due to the limited amount of blood that can be collected. Indeed, this often represents an end of life procedure when using standard techniques for metabolite analysis. A possible approach to circumvent this limitation and still be able to correct for plasma radiometabolites is the use of a population-based metabolite curve. An essential assumption in performing a population-based correction is that the inter-individual variation in the rate of metabolism is small enough to avoid under- or over-corrections of the plasma input curve and hence the VT. In addition, it is conceivable that disease-specific animal models might be characterized by different metabolism; in the present study, we used an animal model of Huntington's Disease that did not show altered metabolic rate of the radiotracer.
Even though we obtained strong correlations between VT values calculated using the uncorrected and population-based corrected IDIF, we observed relevant changes in the slopes of the correlations across animals. Given that the same correction was applied to all subjects, this finding might suggest that the assumption of low inter-individual variability was not met, resulting in the introduction of inter-individual variability between the two methods of analysis. Overall, it appeared that correcting the IDIF for population-based plasma metabolites did not represent an improvement. Therefore, when performing comparative studies in mice, the uncorrected IDIF curve can be used, as long as there is no difference in rate of metabolism between the investigated groups.
In conclusion, we showed that [11C]UCB-J selectively binds to SV2A in mice. The present study provides the methodology based on 1TCM with IDIF for noninvasive in vivo quantification of [11C]UCB-J PET imaging in mice. Similar to the previously published work on non-human primates and humans, [11C]UCB-J displayed optimal kinetics in mice, representing a promising tool for noninvasive quantification of synaptic density in comparative rodent studies.
Supplemental Material
Supplemental Material for Validation and noninvasive kinetic modeling of [11C]UCB-J PET imaging in mice by Daniele Bertoglio, Jeroen Verhaeghe, Alan Miranda, Istvan Kertesz, Klaudia Cybulska, Špela Korat, Leonie Wyffels, Sigrid Stroobants, Ladislav Mrzljak, Celia Dominguez, Longbin Liu, Mette Skinbjerg, Ignacio Munoz-Sanjuan and Steven Staelens in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
The authors thank Philippe Joye, Caroline Berghmans, Eleni Van der Hallen, and Annemie Van Eetveldt of the Molecular Imaging Center Antwerp (MICA) for their valuable assistance.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by CHDI Foundation, Inc., a nonprofit biomedical research organization exclusively dedicated to developing therapeutics that will substantially improve the lives of HD-affected individuals. DB is supported by a PhD fellowship from the Research Foundation Flanders (FWO, 11W2516N/11W2518N). Antwerp University, Belgium founded the work through a partial assistant professor position for JV and LW , and a full professor position for SStr and SSta. LW and SStr are also supported by Antwerp University Hospital, Belgium through a departmental position.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' contributions
DB, JV, LW, SStr, LM, CD, LL, MS, IMS, and SSta designed the study. DB performed the PET experiments. AM and JV reconstructed the PET data. DB, ŠK, and KC performed the metabolite analysis. IK, KC, ŠK, and LW produced the radiotracer. DB and JV performed data analysis. DB, JV, and SSta wrote the manuscript. All authors approved the final version of this manuscript.
Supplementary material
Supplemental material for this article is available online
References
- 1.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature 2012; 490: 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowack A, Yao J, Custer KL, et al. SV2 regulates neurotransmitter release via multiple mechanisms. Am J Physiol Cell Physiol 2010; 299: C960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajjalieh SM, Frantz GD, Weimann JM, et al. Differential expression of synaptic vesicle protein 2 (SV2) isoforms. J Neurosci 1994; 14: 5223–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janz R, Sudhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience 1999; 94: 1279–1290. [DOI] [PubMed] [Google Scholar]
- 5.Bajjalieh SM, Peterson K, Linial M, et al. Brain contains two forms of synaptic vesicle protein 2. Proc Natl Acad Sci U S A 1993; 90: 2150–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowder KM, Gunther JM, Jones TA, et al. Abnormal neurotransmission in mice lacking synaptic vesicle protein 2A (SV2A). Proc Natl Acad Sci U S A 1999; 96: 15268–15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch BA, Lambeng N, Nocka K, et al. The synaptic vesicle protein SV2A is the binding site for the antiepileptic drug levetiracetam. Proc Natl Acad Sci U S A 2004; 101: 9861–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loscher W, Gillard M, Sands ZA, et al. Synaptic vesicle glycoprotein 2A ligands in the treatment of epilepsy and beyond. CNS Drugs 2016; 30: 1055–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepeta K, Lourenco MV, Schweitzer BC, et al. Synaptopathies: synaptic dysfunction in neurological disorders – a review from students to students. J Neurochem 2016; 138: 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mutch SA, Kensel-Hammes P, Gadd JC, et al. Protein quantification at the single vesicle level reveals that a subset of synaptic vesicle proteins are trafficked with high precision. J Neurosci 2011; 31: 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabulsi NB, Mercier J, Holden D, et al. Synthesis and preclinical evaluation of 11C-UCB-J as a PET tracer for imaging the synaptic vesicle glycoprotein 2A in the brain. J Nucl Med 2016; 57: 777–784. [DOI] [PubMed] [Google Scholar]
- 12.Estrada S, Lubberink M, Thibblin A, et al. [(11)C]UCB-A, a novel PET tracer for synaptic vesicle protein 2A. Nucl Med Biol 2016; 43: 325–332. [DOI] [PubMed] [Google Scholar]
- 13.Cai H, Mangner TJ, Muzik O, et al. Radiosynthesis of (11)C-levetiracetam: a potential marker for PET imaging of SV2A expression. ACS Med Chem Lett 2014; 5: 1152–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bretin F, Warnock G, Bahri MA, et al. Preclinical radiation dosimetry for the novel SV2A radiotracer [18F]UCB-H. EJNMMI Res 2013; 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finnema SJ, Nabulsi NB, Mercier J, et al. Kinetic evaluation and test-retest reproducibility of [(11)C]UCB-J, a novel radioligand for positron emission tomography imaging of synaptic vesicle glycoprotein 2A in humans. J Cereb Blood Flow Metab 2017; 38: 2041–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnema SJ, Nabulsi NB, Eid T, et al. Imaging synaptic density in the living human brain. Sci Transl Med 2016; 8: 348ra396. [DOI] [PubMed] [Google Scholar]
- 17.Chen MK, Mecca AP, Naganawa M, et al. Assessing synaptic density in alzheimer disease with synaptic vesicle glycoprotein 2A positron emission tomographic imaging. JAMA Neurol 2018; 75: 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes SE, Scheinost D, Finnema SJ, et al. Lower synaptic density is associated with depression severity and network alterations. Nat Commun 2019; 10: 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox J, Barthold S, Davisson M, et al. The mouse in biomedical research, 2nd ed. Amsterdam: Elsevier, 2006, pp. 2192. [Google Scholar]
- 20.Cai Z, Li S, Matuskey D, et al. PET imaging of synaptic density: A new tool for investigation of neuropsychiatric diseases. Neurosci Lett 2018; 691: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillard M, Chatelain P, Fuks B. Binding characteristics of levetiracetam to synaptic vesicle protein 2A (SV2A) in human brain and in CHO cells expressing the human recombinant protein. Eur J Pharmacol 2006; 536: 102–108. [DOI] [PubMed] [Google Scholar]
- 22.Marrosu F, Bortolato M, Frau R, et al. Levetiracetam attenuates spontaneous spike-and-wave discharges in DBA/2J mice. Epilepsy Res 2007; 75: 224–227. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira AA, Almeida JP, Freitas RM, et al. Effects of levetiracetam in lipid peroxidation level, nitrite-nitrate formation and antioxidant enzymatic activity in mice brain after pilocarpine-induced seizures. Cell Mol Neurobiol 2007; 27: 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertoglio D, Verhaeghe J, Kosten L, et al. MR-based spatial normalization improves [18F]MNI-659 PET regional quantification and detectability of disease effect in the Q175 mouse model of Huntington's disease. PLoS One 2018; 13: e0206613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson GA, Badea A, Brandenburg J, et al. Waxholm space: an image-based reference for coordinating mouse brain research. Neuroimage 2010; 53: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logan J, Fowler JS, Volkow ND, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab 1990; 10: 740–747. [DOI] [PubMed] [Google Scholar]
- 27.Julien-Dolbec C, Tropres I, Montigon O, et al. Regional response of cerebral blood volume to graded hypoxic hypoxia in rat brain. Br J Anaesth 2002; 89: 287–293. [DOI] [PubMed] [Google Scholar]
- 28.Verhaeghe J, Bertoglio D, Kosten L, et al. Noninvasive relative quantification of [(11)C]ABP688 PET imaging in mice versus an input function measured over an arteriovenous shunt. Front Neurol 2018; 9: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koeppe RA, Holthoff VA, Frey KA, et al. Compartmental analysis of [11C]flumazenil kinetics for the estimation of ligand transport rate and receptor distribution using positron emission tomography. J Cereb Blood Flow Metab 1991; 11: 735–744. [DOI] [PubMed] [Google Scholar]
- 30.Innis RB, Cunningham VJ, Delforge J, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 2007; 27: 1533–1539. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Huang S-C, Bergsneider M. Linear ridge regression with spatial constraint for generation of parametric images in dynamic positron emission tomography studies. IEEE Transac Nucl Sci 2001; 48: 125–130. [Google Scholar]
- 32.Akaike H. A new look at the statistical model identification. IEEE Transac Autom Control 1974; 19: 716–723. [Google Scholar]
- 33.Cunningham VJ. Non-linear regression techniques in data analysis. Med Inform 1985; 10: 137–142. [DOI] [PubMed] [Google Scholar]
- 34.Lassen NA, Bartenstein PA, Lammertsma AA, et al. Benzodiazepine receptor quantification in vivo in humans using [11C]flumazenil and PET: application of the steady-state principle. J Cereb Blood Flow Metab 1995; 15: 152–165. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham VJ, Rabiner EA, Slifstein M, et al. Measuring drug occupancy in the absence of a reference region: the Lassen plot re-visited. J Cereb Blood Flow Metab 2010; 30: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldron AM, Wyffels L, Verhaeghe J, et al. Longitudinal characterization of [18F]-FDG and [18F]-AV45 uptake in the double transgenic TASTPM mouse model. J Alzheimers Dis 2017; 55: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koole M, van Aalst J, Devrome M, et al. Quantifying SV2A density and drug occupancy in the human brain using [(11)C]UCB-J PET imaging and subcortical white matter as reference tissue. Eur J Nucl Med Mol Imaging 2018; 46: 396–406. [DOI] [PubMed] [Google Scholar]
- 38.Warnock GI, Aerts J, Bahri MA, et al. Evaluation of 18F-UCB-H as a novel PET tracer for synaptic vesicle protein 2A in the brain. J Nucl Med 2014; 55: 1336–1341. [DOI] [PubMed] [Google Scholar]
- 39.Zanotti-Fregonara P, Chen K, Liow JS, et al. Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 2011; 31: 1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for Validation and noninvasive kinetic modeling of [11C]UCB-J PET imaging in mice by Daniele Bertoglio, Jeroen Verhaeghe, Alan Miranda, Istvan Kertesz, Klaudia Cybulska, Špela Korat, Leonie Wyffels, Sigrid Stroobants, Ladislav Mrzljak, Celia Dominguez, Longbin Liu, Mette Skinbjerg, Ignacio Munoz-Sanjuan and Steven Staelens in Journal of Cerebral Blood Flow & Metabolism