Abstract
A dramatic life history switch that has evolved numerous times in marine invertebrates is the transition from planktotrophic (feeding) to lecithotrophic (nonfeeding) larval development—an evolutionary tradeoff with many important developmental and ecological consequences. To attain a more comprehensive understanding of the molecular basis for this switch, we performed untargeted lipidomic and proteomic liquid chromatography-tandem mass spectrometry on eggs and larvae from three sea urchin species: the lecithotroph Heliocidaris erythrogramma, the closely related planktotroph Heliocidaris tuberculata, and the distantly related planktotroph Lytechinus variegatus. We identify numerous molecular-level changes possibly associated with the evolution of lecithotrophy in H. erythrogramma. We find the massive lipid stores of H. erythrogramma eggs are largely composed of low-density, diacylglycerol ether lipids that, contrary to expectations, appear to support postmetamorphic development and survivorship. Rapid premetamorphic development in this species may instead be powered by upregulated carbohydrate metabolism or triacylglycerol metabolism. We also find proteins involved in oxidative stress regulation are upregulated in H. erythrogramma eggs, and apoB-like lipid transfer proteins may be important for echinoid oogenic nutrient provisioning. These results demonstrate how mass spectrometry can enrich our understanding of life history evolution and organismal diversity by identifying specific molecules associated with distinct life history strategies and prompt new hypotheses about how and why these adaptations evolve.
Keywords: development, echinoderms, life history evolution
1 |. INTRODUCTION
Life history evolution is driven by tradeoffs in reproduction, development, and resource utilization. The interplay between these traits and the environment has created numerous behavioral and morphological adaptations that contribute enormously to organismal diversity (Flatt & Heyland, 2011; Stearns, 1992). A mature body of theory and extensive field data provide powerful insights into the fitness consequences of shifts in life history, including fecundity, offspring size, longevity, and mortality rate but much less is known about the molecular-level differences underlying alternative life history strategies. Analysis of the specific molecules important to contrasting life history strategies has the potential to provide a deeper understanding of organismal diversity by revealing the molecular basis for life history shifts.
Marine invertebrates provide an outstanding system for studying life history evolution as a huge diversity of larval forms and life history modes are frequently present in closely related species (Strathmann, 1985). In these animals, life cycles are often biphasic, including a pelagic (living in the water column) larva followed by metamorphosis into a benthic (sea floor-dwelling) adult. Transitions in developmental mode between planktotrophic (feeding) and lecithotrophic (nonfeeding) larvae can have profound ecological consequences on marine invertebrate populations via gene flow, local adaptation, and speciation (Jablonski & Lutz, 1983; Palumbi, 1994; Wray & Raff, 1991). A recent survey of marine invertebrate larval life history and biogeography reports ocean productivity and temperature are strongly associated with developmental mode and offspring size such that planktotrophic species are more common in areas of high temperature and food availability that promote rapid larval development, and in turn, reduce larval mortality (Marshall, Krug, Kupriyanova, Byrne, & Emlet, 2012; Morgan, 1995). Furthermore, planktotrophic larvae possess a much greater dispersal potential relative to lecithotrophs due to their prolonged larval phase. However, because of relatively large, maternally derived energetic stores, lecithotrophic larvae can settle normally unfavorable, low-nutrient habitats more readily (Strathmann, 1985). Although researchers have investigated the ecology of this life history switch for many decades, less is known of the molecular differences underlying changes in maternal investment and development.
The evolution of lecithotrophy is thought to generally occur in two steps: increase in egg size and maternal provisioning of nutrients followed by loss of larval feeding structures and accelerated premetamorphic development (Wray & Raff, 1991). Therefore, both physiological and developmental adaptations are essential to this recurrent life history switch. In echinoids, changes associated with the evolution of lecithotrophic larvae include first transitioning from an obligately to facultatively feeding larva that can reach metamorphosis without feeding, followed by loss of feeding ability and reduced larval morphology until finally a highly simplified, nonfeeding larval form is reached (Wray, 1996). Importantly, this switch has occurred numerous times within many marine invertebrate phyla (Haszprunar, von Salvini-Plawen, & Rieger, 1995; Strathmann, 1985), and in Echinodermata alone, lecithotrophy has evolved at least 20 times from the inferred ancestral planktotrophic state (Hart, 2002; McEdward & Miner, 2001; Raff, 1987; Wray, 1996; Wray & Raff, 1991). As a result, comparative analyses of egg provisioning strategies between planktotrophs and lecithotrophs provide insight into the initial evolutionary steps from feeding to nonfeeding larval development in marine invertebrates.
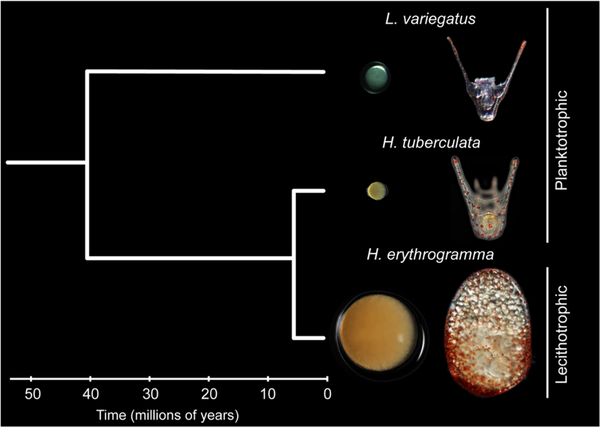
Perhaps the most extensively studied case of this life history switch is found in the echinoid genus Heliocidaris. The planktotroph Heliocidaris tuberculata produces relatively small eggs (~95 μm in diameter) and its larvae feed in the plankton for 3–5 weeks before metamorphosing into juveniles (Emlet, McEdward, & Strathmann, 1987; Laegdsgaard, Byrne, & Anderson, 1991; Williams & Anderson, 1975; Figure 1). The lecithotroph Heliocidaris erythrogramma, in contrast, produces large, positively buoyant eggs (~430 μm in diameter) that develop into simplified, nonfeeding larvae that undergo metamorphosis in just 4 days (Figure 1; Laegdsgaard et al., 1991; Williams & Anderson, 1975). These are classic life history trade-offs: less time spent in the plankton results in lower larval mortality but at the expense of reduced fecundity and dispersal (McEdward, 1995, Strathmann, 1985, Thorson, 1950, Vance, 1973). Embryological investigations of H. erythrogramma have uncovered dramatic temporal and spatial modifications to developmental mechanisms accompanying the transition to lecithotrophy over ~5 million years (Zigler, Raff, Popodi, Raff, & Lessios, 2003) of divergence (for reviews see Raff, 1992, Raff & Byrne, 2006). In addition, a battery of oogenic and metabolic adaptations fuse with these genetic and developmental alterations to bolster development beyond metamorphosis including a 100-fold increase in egg volume and hypertrophy of oocyte lipid provisioning (Byrne et al., 1999; Emlet & Hoegh-Guldberg, 1997; Hoegh-Guldberg & Emlet, 1997; Villinski, Villinski, Byrne, & Raff, 2002; Williams & Anderson, 1975), reported previously to be composed primarily of wax esters (Villinski et al., 2002).
FIGURE 1.
Eggs and larvae of the three study species: Heliocidaris erythrogramma, Heliocidaris tuberculata, and Lytechinus variegatus. Small eggs of the two distantly related planktotrophs (L. variegatus and H. tuberculata) develop into morphologically similar feeding larvae. Large eggs of the lecithotroph (H. erythrogramma) develop into simplified, nonfeeding larvae with an abundance of vesicle-bound lipid stores
To more precisely and comprehensively identify the developmental and energetic drivers underlying larval life history evolution in Heliocidaris, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to generate lipidomic and proteomic profiles of eggs and early stage larvae from three sea urchin species: H. erythrogramma, H. tuberculata, and Lytechinus variegatus, a planktotrophic species that diverged ~40 million years ago from the lineage leading to Heliocidaris (Smith, 1989; Smith, Littlewood, & Wray, 1995; Figure 1). By including L. variegatus as an outgroup comparison, we can more confidently assign differences in egg provisioning as being associated with phylogeny (Heliocidaris vs. Lytechinus) or life history evolution (planktotrophy vs. lecithotrophy). These untargeted, high-resolution analyses of lipid and protein content compare relative differences in lipid and protein abundance and, along with transcriptomic approaches, allow for a more detailed and complete interpretation of the biological correlates of life history evolution. This approach provides an innovative window into the evolution of life history, revealing predicted as well as unexpected maternal provisioning strategies and adaptations in developmental physiology associated with lecithotrophy in H. erythrogramma.
2 |. MATERIALS AND METHODS
2.1 |. Sample collection
H. erythrogramma and H. tuberculata adults were collected near Sydney, Australia, and L. variegatus adults were collected at the Duke University Marine Lab in Beaufort, NC. H. erythrogramma and H. tuberculata were held in aquaria filled with natural seawater in a 22°C temperature-controlled room at the University of Sydney for 3–5 days before spawning. L. variegatus was kept in artificial seawater for 2 weeks at the Duke University main campus in Durham, NC. For all three species, adults were not fed while in captivity. A unique female–male pair of adult sea urchins was included in each cross, resulting in three biological replicates for each species. Spawning was induced by 0.5M potassium chloride injection. H. erythrogramma and H. tuberculata embryos were reared in glass culture dishes at 23°C with gentle bubbling from a glass pipette to maintain circulation. L. variegatus embryos were reared at 23°C. in large glass culture dishes, and seawater changes were performed every 8 hr to minimize deleterious effects from hypoxia. Approximately 100 unfertilized eggs and unfed, early stage larvae were collected from each cross for mass spectrometric analysis. In this study, “early stage larvae” denotes the two-armed pluteus stage for L. variegatus (42 hr postfertilization [hpf]) and H. tuberculata (52 hpf), and the developmental stage immediately following gastrula in H. erythrogramma at which the embryo acquires an elongated morphology and an early rudiment has started developing (48 hpf; see Figure 1). Unfertilized eggs and arrested embryos were removed from cultures before sample collection of the larval stage to minimize confounding lipid and protein measurements from earlier developmental stages. In total, 36 samples were collected for lipidomic and proteomic profiling.
2.2 |. Lipid sample preparation
Lipid samples were solubilized in ammonium bicarbonate with sonication and 500 μl of each sample was used for lipid profiling. The Bradford assay (BioRad Inc., Hercules, CA) was used to normalize the samples to total protein biomass and two samples (one L. variegatus egg and one L. variegatus larvae) were excluded from analysis due to insufficient sample quantity, leaving a total of 16 lipid samples. Two hundred microliters of methanol was added to normalized 50 μl volumes of sample lysate and mixed at 25°C for 5 min. Six hundred microliters of methyl tert-butyl ether was added to each sample and mixed for 60 cycles of 0.5 ml automated aspiration with a Vioflo96 pipettor (Integra Biosciences, Hudson, NH). After mixing at 37°C for 10 min and centrifugation at 2000g for 10 min at room temperature, 400 μl of the organic layer was isolated from each sample, and extracts were dried under N2 evaporation and resuspended in 200 μl of 4/3/1 (v/v/v) isopropyl alcohol/acetonitrile/ water. After 10 min of shaking at 37°C, samples were immediately processed for mass spectrometry analysis.
2.3 |. Lipid mass spectrometry
Samples were analyzed using ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry (UPLC-ESI-MS/MS) allowing chromatographic resolution by lipid class, total carbons, and degree of saturation. Block design of the mass spectrometry analysis is described in Table S1 and quality control pools were prepared by mixing equal volumes of all samples; these quality control pools were analyzed five times throughout the study to determine technical variance for each lipid feature. Two microliter injection was used in ESI + ionization mode and 6 μl was used in ESI − ionization mode. UPLC lipid separation was performed in a nanoACQUITY UPLC system (Waters, Milford, MA) with a 2.1 mm × 100 mm 1.7 μl CSH C18 column. Mobile phase A was 10 mM ammonium formate and 0.1% formic acid in 40/60 (v/v) water/acetonitrile. Mobile phase B was 1% formic acid in 10/90 (v/v) acetonitrile/2-propanol. Samples were introduced to a Synapt G2 HDMS mass spectrometer (Waters, Milford, MA) and analyzed in acquisition sensitivity mode (~15,000 Rs), with 0.4 s MS full scan followed by two 0.2 s data-dependent MS/MS scans.
Raw data were imported and analyzed in Rosetta Elucidator v3.3 (Rosetta Biosoftware, Seattle, WA) for peak-picking and alignment between samples. Raw peak intensities were reported for 6,223 electrospray-positive features and 3,210 electrospray-negative features. Intensity scaling was performed by scaling each sample to the median across all samples excluding the top and bottom 10% features when calculating the median. This was performed independently in each ionization mode and these normalized data sets used for statistical analysis are available in Supporting Information Data 1.
2.4 |. Lipid identification
Lipids were identified by selecting the minimum difference between the input mass/charge (m/z) ratio of features from the mass spectra analysis and reference lipid m/z ratios in the Lipid Maps virtual database (Fahy et al., 2009). An initial search using the Lipid Maps structure database revealed a large number of high abundance features without any definitive identification; these were subsequently putatively identified (using Lipid Maps virtual database) as diacylglycerol ethers, which are not annotated in the Lipid Maps structure database. For every input m/z from the mass spectra analysis, the minimum difference between the input and possible database matches with a maximum difference of ±0.01 m/z allowed was selected. Next, features with greater than 30% coefficient of variation from the pooled quality control samples were filtered out and spectra intensities of m/z ratios matching to the same lipid (class: total carbons: double bonds) were summed for each sample including different ionization forms of the same lipid feature.
In total, 1,667 unique lipid molecules were identified at the level of lipid class (triacylglycerol, diacylglycerol, diacylglycerol ether, monoacylglycerol, wax ester, fatty acid, acyl-carnitine, cholesterol ester, glycerophospholipid, and lyso-glycerophospholipid), fatty acid chain length, and degree of saturation (Supporting Information Data 1). During the classification process, manual annotation and use of limited MS/MS spectra are required when a single m/z ratio from the mass spectra analysis has tied minimal differences to multiple reference m/z ratios in the Lipid Maps database. For this reason, glycerophospholipids and lyso-glycerophospholipids were classified as two lipid classes instead of broken down as specific types of glycerophospholipids (i.e., phosphatidylcholine and phosphatidylserine). Also, diacylglycerol ethers were typically annotated as matched ties between triacylglycerols and diacylglycerols of the same carbon number and degree of saturation.
2.5 |. Protein sample preparation
Two hundred microliters of each protein sample was resuspended in 50 mM ammonium bicarbonate (pH 8) containing Roche cOmplete™ (Roche, Basel, Switzerland) protease inhibitors followed by sonication and centrifugation at 15,000g for 10 min. Protein concentrations from the supernatant were quantified by the Bradford assay and each sample was normalized to 0.25 μg/μl with ammonium bicarbonate containing a 0.2% final concentration of acid labile surfactant (ALS-1) for each sample. Dithiothreitol was added to a final concentration of 10 mM, and samples were denatured at 80°C for 10 min followed by alkylation with 25 mM iodoacetamide for 30 min in the dark. Proteins were digested at 37°C with sequencinggrade modified trypsin (Promega, Madison, WI; 1:50 w/w trypsin/protein) overnight. Then, samples were acidified with 1% trifluoroacetic acid and 2% acetonitrile, followed by heating at 60°C for 2 hr to inactivate the trypsin and degrade the ALS-1. Waters trypsinized MassPREP yeast alcohol dehydrogenase 1 was added at 50 fmol/μg as a surrogate standard. Following centrifugation, supernatants were transferred to Maximum Recovery LC vials (Waters, Milford, MA) and quality control pools were prepared by mixing equal volumes of egg and larva samples.
2.6 |. Protein mass spectrometry
LC-MS/MS was performed using a nanoACQUITY UPLC system (Waters) coupled to a Q-Exactive Plus high-resolution tandem mass spectrometer (Thermo Fisher Scientific, Waltham, MA) via a nanoelectrospray ionization source as previously described. Approximately 333 ng of peptide digests of each sample was analyzed in a random order and interspersed with quality pools as described in Table S2.
Label-free quantitation used Rosetta Elucidator v 4.0 (Rosetta Biosoftware, Seattle, WA) as previously described in Bauernfeind et al. (2015). Briefly, all samples were aligned using accurate mass and retention time and relative peptide abundance was calculated based on areaunder-the-curve of aligned features across all runs. MS/MS data were searched using Mascot Server v2.5 (Matrix Sciences, Mount Prospect, IL) with a database containing H. erythrogramma, H. tuberculata, and L. variegatus developmental transcriptomes (Israel et al., 2016) and an equal number of reverse entries for false discovery rate (FDR) determination (901,770 total entries). Database search parameters included fixed modification on Cys (carbamidomethyl) and variable modifications on Met (oxidation) and Asn/Gln (deamidation). Data were annotated at a 1% peptide FDR using the PeptideTeller algorithm in Elucidator and scaled to the robust median intensity across all samples.
Initially, 13,290 peptides with hits to the database generated from each of the three sea urchin species developmental transcriptomes were quantified. Because the spectrum intensity of a peptide is sensitive to its primary amino acid sequence, only peptides with shared primary amino acid sequence hits to all three sea urchins’ transcriptomes (see “Primary Protein Name” and “Shared Proteins” columns of Supporting Information Data 2) were included for relative abundance comparisons between species (2,900 peptides encompassing 993 unique proteins). For comparison, of these 993 shared proteins, 674 were also quantified in a recent phosphoproteomic mass spectrometry analysis of Strongylocentrotus purpuratus eggs (Guo et al., 2015). Normalized spectra of peptides with hits to the same protein were summed together for downstream statistical analysis (Supporting Information Data 2). In addition, identified peptides with significant hits to the sea urchin transcriptomes were also annotated with S. purpuratus gene models using consensus alignments of assembled transcripts from each study species BLASTed (Altschul, Gish, Miller, Myers, & Lipman, 1990) at the protein level to the S. purpuratus gene models (e-value ≤ 1e-10; Sea Urchin Genome Sequencing et al., 2006) as described in Israel et al. (2016).
2.7 |. Statistical analysis and data visualization
Results of the lipid and protein mass spectrometry analyses were imported into R, and the “dpylr” v. 0.7.5 (Wickham & Francois, 2016) and “purrr” v. 0.2.5 (Henry & Wickham, 2017) packages were utilized for data organization and analysis. Normalized average spectra intensities for eggs and larvae samples of each sea urchin species were calculated, as well as fold-change (FC) for each comparison of interest. For comparisons between the three urchin sea species, one-way analysis of variance (ANOVA) followed by Tukey’s honest significant difference (HSD) test was performed. Only protein molecules that were quantified in at least two of the three biological replicates were included in comparisons between species, which resulted in 955 and 984 proteins for comparison between eggs and larvae, respectively. Because our study utilizes a discovery-based methodology, this criterion was implemented to maximize the number of proteins included for comparative analysis while controlling for false-positives arising from experimental groups with proteins represented by only one biological replicate. In this study, we considered differentially abundant lipids or proteins to have a relative abundance FC ≥ 2 between sample sets and a Tukey’s HSD corrected p ≤ 10%. Lipids and proteins assigned as potentially associated with lecithotrophic or planktotrophic development were significantly differentially abundant between H. erythrogramma and H. tuberculata but not significantly differentially abundant between the two planktotroph species, H. tuberculata and L. variegatus. Sets of differentially abundant lipids and proteins are available in Supporting Information Data 3 and 4, respectively.
The principal component analysis was carried out with the prcomp function in R on normalized lipidomic and proteomic spectra intensities. Messenger RNA (mRNA) read counts were retrieved from the transcriptomic data sets produced in Israel et al. (2016). Gene Ontology (GO; Ashburner et al., 2000) enrichment analyses of differentially abundant proteins on a background set of all S. purpuratus gene identifiers were carried out with the “piano” v 1.16.4 R package (Varemo, Nielsen, & Nookaew, 2013) using the runGSAhyper function with Benjamini–Hochberg correction (Benjamini, Drai, Elmer, Kafkafi, & Golani, 2001; Supporting Information Data 4). Adobe Photoshop was utilized to darken the background and brighten specimens of sea urchin egg and larva micrographs for visualization purposes. Figures were generated with the “ggplot2” v. 2.2.1 R package (Wickham, 2016) and Adobe Illustrator.
2.8 |. Data accessibility
The mass spectrometry lipidomics data have been deposited on the MassIVE public data repository at the University of California-San Diego with the data set identifier MSV000082501. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the data set identifier PXD007065. Normalized spectra intensities used for statistical analysis of lipidomic and proteomic data sets are available in Supporting Information Data 1 and 2, respectively.
3 |. RESULTS
3.1 |. Lipidomic mass spectrometry
We collected lipid samples from three biological replicates of unfertilized eggs and unfed, early stage larvae for each sea urchin species: H. erythrogramma, H. tuberculata, and L. variegatus (Figure 1). Two L. variegatus samples (one egg and one larva) were not processed due to insufficient sample amount, so 16 samples were analyzed in total. Samples were analyzed by label-free quantitative lipidomics using accurate mass and retention time alignment followed by area-under-the-curve measurements of identified lipids. Lipidomic LC-MS/MS data were aligned in Rosetta Elucidator for quantification and searched against the Lipid Maps virtual database (Fahy et al., 2009) for putative lipid identifications, which resulted in 1,667 specific lipid molecules for comparative analysis. Lipids were identified at the resolution of chain length, degree of saturation (number of double bonds), and lipid class including tri-, di-, and monoacylglycerol, diacylglycerol ether, wax ester, fatty acid, acyl-carnitine, cholesterol ester, glycerophospholipid, and lyso-glycerophospholipid.
3.2 |. Life history drives developmental lipidome divergence
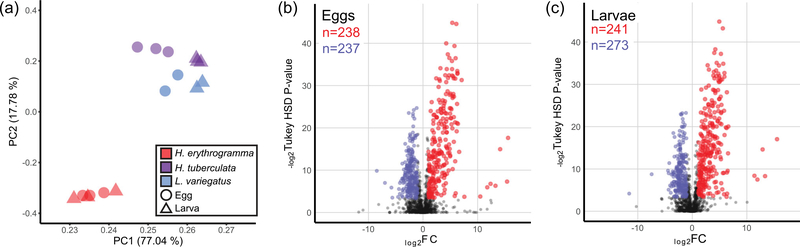
We first aimed to parse the relative contributions of phylogenetic divergence, life history, and developmental stage to differences in the quantitative lipidomic profiles among samples. Principal component analysis (PCA) demonstrates life history (lecithotrophy or planktotrophy) dominates the variation in lipid content, with the first two principal components explaining 94.82% of the variation (Figure 2a). In addition, PC 1 distinguishes egg and larva samples in H. tuberculata and L. variegatus, but not in H. erythrogramma, which suggests global lipidomic changes during embryogenesis may be more extensive in the planktotrophs relative to the lecithotroph. Differential abundance analyses between planktotroph and lecithotroph samples identify numerous molecular shifts in lipid composition in egg and larva samples (Figure 2b,c). Specifically, 475 distinct lipid molecules were differentially abundant between H. erythrogramma and H. tuberculata eggs, and not differentially abundant between either planktotroph species, which accounts for 28.5% of all lipid molecules identified. Of these, 238 molecules were more abundant in H. erythrogramma whereas 237 were more abundant in the planktotrophs (Figure 2b). However, the median FC of lipids significantly more abundant in H. erythrogramma was 8.4, but only 4.5 for lipids significantly more abundant in H. tuberculata, which suggests an especially pronounced shift in the lipidome has evolved in H. erythrogramma. A comparable number of lipids were differentially abundant between the larval stages of these species (Figure 2c; 514 lipids).
FIGURE 2.
Variation in lipid content of sea urchin eggs and larvae is driven by life history evolution. (a) PC analysis of normalized lipidomic mass spectrum intensities measured from egg and larva samples from each sea urchin species. PC 1 explains 77.04% of the variation and distinguishes Heliocidaris erythrogramma samples from those of the two planktotrophs. (b,c) Volcano plots illustrating differential relative abundance of 1,667 unique lipid molecules between (b) eggs and (c) larvae in H. erythrogramma (red) and Heliocidaris tuberculata (blue). Only lipids that are differentially abundant between H. erythrogramma and H. tuberculata and not differentially abundant between H. tuberculata and Lytechinus variegatus are colored to mark a high confidence set of lipids potentially associated with either life history strategy. Significantly differentially abundant lipids (n) are called as having an FC ≥ 2 and a Tukey’s honest significant difference corrected p ≤ 10%. FC: fold-change; PC: principle component
3.3 |. Lipid storage strategy is concordant with a life history strategy
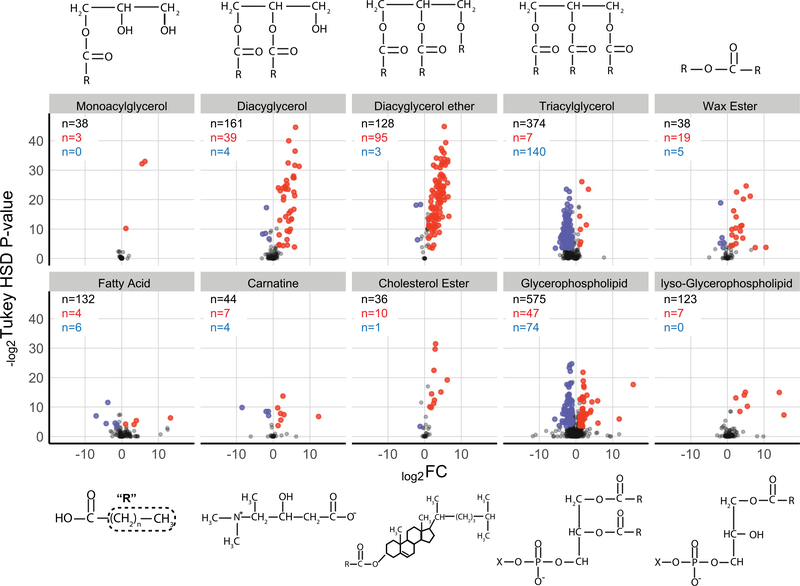
Breaking down differentially abundant lipid molecules by lipid class demonstrates strong compositional alterations to egg provisioning associated with developmental mode (Figure 3). Relative to the two planktotrophic species, eggs and larvae of H. erythrogramma are dominated by diacylglycerol ether (DAGE) lipids, which are low-density ether analogues of triacylglycerols (TAGs). At the egg and larval stages, 95 and 94 distinct DAGE molecules were more abundant in H. erythrogramma, respectively, which accounts for nearly 40% of all differentially abundant lipid molecules in H. erythrogramma and over 70% of all DAGE molecules identified in this analysis (Figure 3; Supporting Information Data 3). DAGEs are characterized by slower metabolic turnover rates relative to TAGs in marine animals, which suggests they play an important role in long-term energy storage (Lee, Hagen, & Kattner., 2006). In contrast, TAGs constitute most of the energetic lipid content in the planktotrophic species. Specifically, 140 and 123 TAGs were more abundant in H. tuberculata egg and larva samples, accounting for ~60% and ~45%, respectively, of all differentially abundant lipids relative to H. erythrogramma (Figure 3; Supporting Information Data 3). Compared to long-term storage lipids like DAGEs and wax esters, TAGs serve as a short-term energy storage lipid in many marine zooplankton taxa (Lee et al., 2006). This energy storage strategy is commensurate with the metabolic requirements of planktotrophic development in which larvae spend weeks feeding and obtaining energy in the plankton rather than relying on maternally deposited energetic stores.
FIGURE 3.
Arranging differentially abundant lipids between Heliocidaris erythrogramma (red) and Heliocidaris tuberculata (blue) eggs (from Figure 2b) according to lipid class reveals qualitative differences in the energetic composition of oocyte provisioning between lecithotrophic and planktotrophic development. Long-term, low-density diacylglycerol ether lipids comprise many of the differentially abundant lipid molecules in H. erythrogramma eggs relative to H. tuberculata, whereas energetic lipid content in H. tuberculata eggs is dominated by triacylglycerols. Significantly differentially abundant lipids (n) are called as having an FC ≥ 2 and a Tukey’s honest significant difference corrected p ≤ 10%. FC: fold-change; R: hydrocarbon chain
3.4 |. Diacylglycerol ether lipids are not significantly catabolized pre-metamorphosis in H. erythrogramma
Comparing H. erythrogramma egg and larval stages, we measured 84 differentially abundant lipid molecules (Supporting Information Data 3). Surprisingly, DAGEs show minimal change in relative abundance between these developmental stages (average FC per lipid: 0.046), indicating that the huge lipid stores of H. erythrogramma may not be significantly catabolized before metamorphosis. The ether linkage of DAGE lipids is thought to be cleaved by only one enzyme, alkylglycerol mono-oxygenase (Watschinger & Werner, 2013), also known as transmembrane protein 195. Our proteomic analyses did not quantify expression of alkylglycerol mono-oxygenase in the eggs or larvae of H. erythrogramma, likely because it is an integral membrane protein; interestingly, however, examination of previously published transcriptional profiling of the same three urchins and developmental time span (Israel et al., 2016) uncovers ~10-fold higher normalized mRNA expression of a gene (SPU_003820) encoding alkylglycerol mono-oxygenase in H. erythrogramma larvae samples and a robust increase in expression of this gene not found in either planktotroph species over the same developmental time period (Figure S1). These results suggest translation and utilization of this enzyme, and thus digestion of DAGEs, occurs after the larval stage examined here and perhaps primarily following metamorphosis. It is likely that a different set of metabolites and physiological pathways serve as the primary energetic source for rapid premetamorphic development in H. erythrogramma. Metabolism of short-term energetic lipids like TAGs offer one possible premetamorphic energy supply. For example, we quantified a subtle reduction (FC ≥ 1.25) on an average abundance of 25 TAG molecules from egg to larvae stage in H. erythrogramma, and of these TAGs, 22 and 17 molecules also decreased in abundance through development in H. tuberculata and L. variegatus, respectively.
3.5 |. Mass spectrometry highlights lipid molecular diversity across sea urchin species
The experimental approach carried out in this study, UPLC-ESI-MS/MS, permits high-resolution identification of distinct lipid molecules. The most diverse lipid classes in terms of fatty acid chain length and saturation were glycerophospholipids (GPLs; 575 lipids), the primary lipid constituent of biological membranes, and TAGs (374 lipids), energetic storage lipids. Closer examination of structural differences within these lipid classes reveals species-specific trends in the molecular composition of their developmental lipidomes.
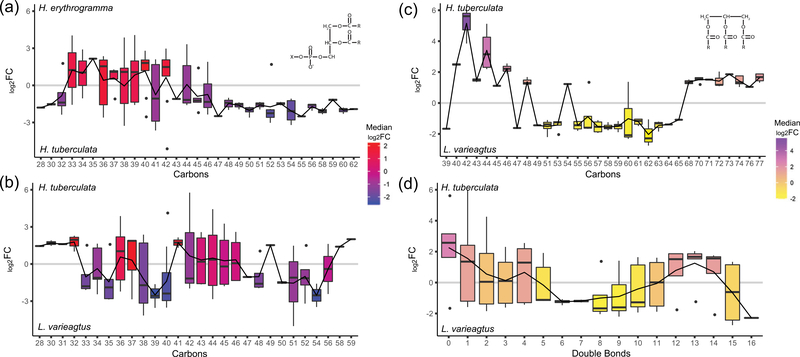
Differentially abundant GPLs in the eggs of the lecithotroph H. erythrogramma relative to its planktotrophic congener H. tuberculata are enriched for GPLs with short chain fatty acids (33–40 carbons), whereas GPLs more abundant in H. tuberculata are characterized by very short (28–32 carbons) and long chain fatty acids (46–62 carbons; Figure 4a). Similarly, GPLs with long chain fatty acids (43–56 carbons) are also relatively more abundant in L. variegatus compared with H. erythrogramma (Figure S2a). However, differentially abundant GPLs between the two planktotrophic species, H. tuberculata and L. variegatus, do not show a clear association with fatty acid chain length (Figure 4b), and our analyses did not detect a relationship with differential GPL abundance and degree of saturation in any species comparison (Figure S2b–d). This result suggests compositional remodeling of cell membrane in the large eggs of the lecithotroph H. erythrogramma may exist as a shift in fatty acid chain length of GPL molecules.
FIGURE 4.
Species-specific structural differences within lipid classes highlights lipidomic molecular diversity across sea urchin species. (a) Among differentially abundant lipids, very short (28–32 carbons) and long (46+ carbons) chain length GPLs are relatively more abundant in Heliocidaris tuberculata than Heliocidaris erythrogramma eggs. GPLs more abundant in H. erythrogramma were enriched for fatty acid chain lengths between 33 and 40 carbons long. (b) A similar relationship was not quantified between H. tuberculata and Lytechinus variegatus eggs, so structural differences in GPL composition may reflect molecular remodeling of the large eggs of H. erythrogramma. Differentially abundant TAG between the two planktotroph species, H. tuberculata and L. variegatus, show species-specific differences in (c) fatty acid chain length and (d) degree of saturation. Box color indicates median log2 FC in GPL or TAG abundance between species for each chain length or number of double bonds. Black line traces the mean log2 FC in GPL or TAG abundance between species for each fatty acid chain length or number of double bonds. FC: fold-change; GPL: glycerophospholipid; TAG: triacylglycerols
Although there is a clear difference in TAG abundance in the two planktotrophic species relative to H. erythrogramma (see above), a number of distinct TAG molecules were also differentially abundant between the eggs (103) and larvae (108) of either planktotrophic species (Supporting Information Data 1). A sharp disparity in chain length distinguishes differentially abundant TAGs between H. tuberculata and L. variegatus eggs: TAGs with short (40–48 carbons) and long (68–77 carbons) chain fatty acids comprise relatively more abundant molecules in H. tuberculata, and TAGs with intermediate chain length (49–65 carbons) fatty acids are relatively more abundant in L. variegatus (Figure 4c). These differentially abundant TAGs also appear to differ in degree of saturation (number of double bonds) between each species, such that highly saturated TAGs (0–1 double bonds) are relatively more abundant in H. tuberculata eggs and more unsaturated TAGs (6–8 double bonds) are relatively more abundant in L. variegatus eggs (Figure 4d). These results highlight the astounding molecular diversity among the lipidomes of these sea urchin species and exemplify the widespread compositional divergence between and within different types of lipids in marine organisms (Lee et al., 2006, Parrish, 2013).
3.6 |. Proteomic mass spectrometry
We also performed label-free, quantitative proteomic mass spectrometry on three biological replicates of egg and early stage larvae for each sea urchin species. Peptide precursor (MS1) data was aligned in Rosetta Elucidator and corresponding product ion (MS/MS) data were searched against a single reverse decoy database containing developmental transcriptomes of each of the three sea urchin species (Israel et al., 2016). After annotation and quantitation of identified peptides, relative protein abundances were calculated as the aggregate sum of peptide intensities as described in Reidel et al. (2011). In addition, we annotated peptides with pre-existing gene models of the well-studied sea urchin species S. purpuratus (e-value ≤ 1e-10; Sea Urchin Genome Sequencing et al., 2006). However, because peptide spectra intensity can vary with species-specific amino acid sequence differences, only peptides with identical amino acid sequences among the three species were included for comparative protein abundance analysis (2,901 peptides encompassing 993 proteins). We still draw from the much larger set of all peptides for examples of highly expressed proteins of interest potentially associated with lecithotrophic or planktotrophic development in these sea urchins (13,290 peptides encompassing 1,959 proteins; Supporting Information Data 2).
3.7 |. Phylogeny and life history differentially affect variation in developmental proteomes
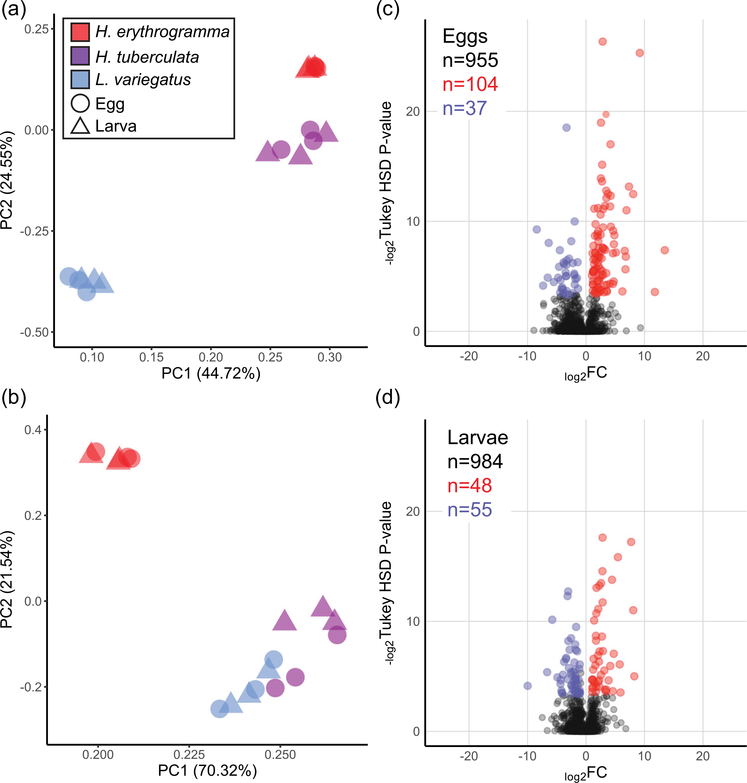
When normalized spectra intensity of all 13,290 peptides from the analysis are considered, PCA distinguishes most of the variation in egg and larva protein content by phylogeny (i.e., Heliocidaris vs. Lytechinus; Figure 5a). However, when only the set of peptides with sequence identity across all three species are included in the PCA (2,901 peptides), developmental life history explains most of the variation between protein samples (i.e., lecithotrophy vs. planktotrophy; Figure 5b). Therefore, there is a significant phylogenetic contribution to divergence in maternal provisioning of protein content, which may be partially attributed to bias in spectra intensity from higher peptide sequence-similarity between the two Heliocidaris species relative to L. variegatus. Nonetheless, when only peptides with identical sequences are considered, life history strategy dominates quantitative differences in the developmental proteomes between these three species.
FIGURE 5.
Variation in peptide abundance is dependent on phylogeny and developmental life history. (a) When all peptides are considered (13,290 peptides encompassing 1,959 unique proteins), PC analysis of normalized spectra intensities distinguishes samples by their phylogenetic relationships (Heliocidaris vs. Lytechinus). (b) If only peptides with sequence identity across all three species are included (2,901 peptides encompassing 993 unique proteins), then most of the variation in protein content is explained by developmental life history (lecithotrophy vs. planktotrophy). This shared set of peptides was included in the differential abundance analyses between species. (c,d) Volcano plots illustrating differential relative abundance of protein molecules between (c) eggs and (d) larvae of Heliocidaris erythrogramma (red) and Heliocidaris tuberculata (blue). Only proteins that are differentially abundant between H. erythrogramma and H. tuberculata and not differentially abundant between H. tuberculata and Lytechinus variegatus are colored to mark a high confidence set of proteins potentially associated with either life history strategy. Significantly differentially abundant proteins are called as having an FC ≥ 2 and a Tukey’s honest significant difference corrected p ≤ 10%. FC: fold-change; PC: principle component
To identify which peptides may be driving the variation between the developmental proteomes of H. erythrogramma relative to H. tuberculata and L. variegatus, we applied the same differential expression criteria as in the lipidomic comparisons to the shared set of 993 proteins and required that for any experimental group (species, developmental stage), the protein must be quantified in at least two of the three biological replicates (Supporting Information Data 2). Comparison of planktotroph to lecithotroph eggs identified 141 differentially expressed proteins, of which 104 were elevated in H. erythrogramma egg samples compared to 37 elevated in H. tuberculata (Figure 5c). At the larval stage, the total number of differentially expressed proteins decreased to 103 (Figure 5d).
3.8 |. Adaptations to lecithotrophic development revealed by comparative proteomics
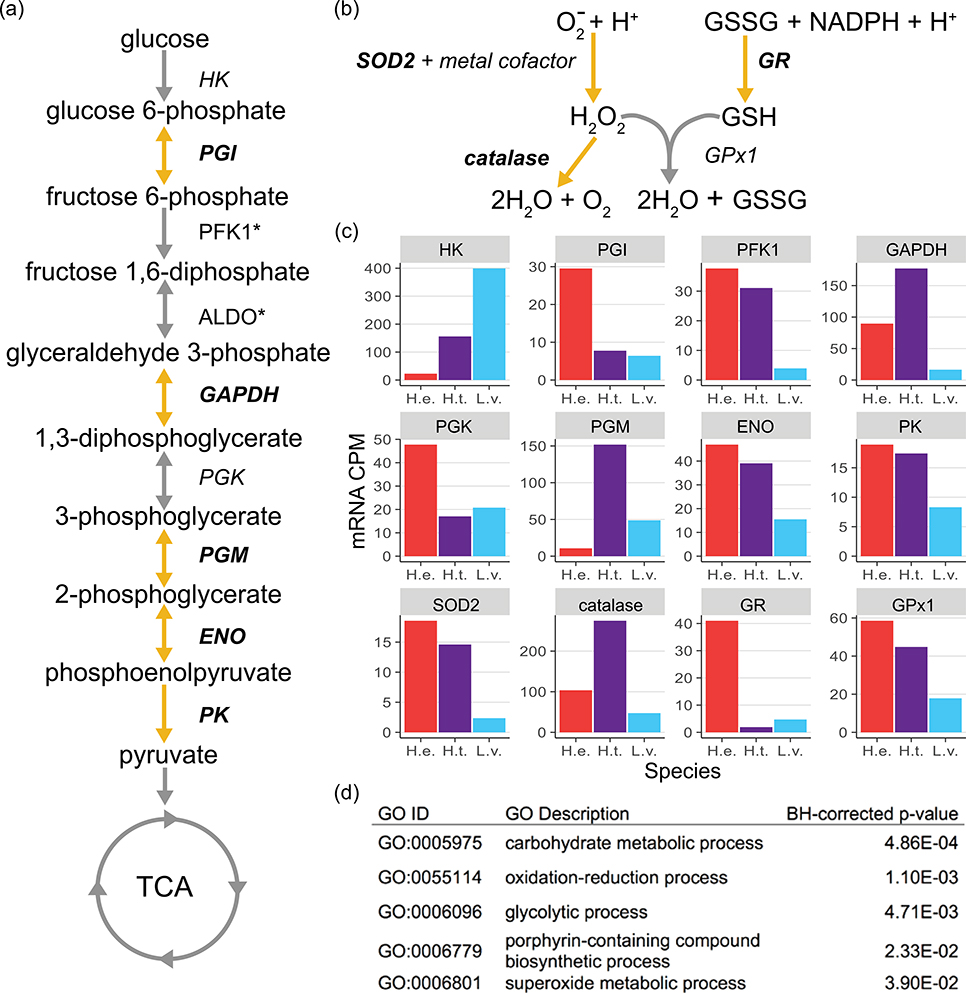
Many of the 104 differentially expressed proteins in H. erythrogramma eggs relative to those of the planktotrophs may be associated with the evolution of lecithotrophy in this species. First, a suite of proteins involved in carbohydrate metabolism are relatively more abundant in H. erythrogramma eggs (Supporting Information Data 4). Specifically, five of the nine enzymes involved in glycolysis were more highly expressed in this species (Figure 6a), as well as isocitrate dehydrogenase (SPU_000236, FC: + 26.5), the rate-limiting enzyme of the TCA cycle. Several other enzymes involved in sugar metabolism are also expressed at higher levels in H. erythrogramma eggs including β-galactosidase (SPU_027889, FC: + 111.9), α-mannosidase (SPU_006585, FC: +34.7), fucosidase (SPU_026835, FC: +8.3), fructose-1,6-bisphosphatase (SPU_016377, FC: +7.2), and neuraminidase 1-1 (SPU_006441, FC: +6.6). Many of these enzymes break down complex carbohydrates, potentially providing glycolysis with an increased input of metabolites. Analysis of previous published transcriptomic comparisons of these three sea urchin species (Israel et al., 2016) demonstrates mRNA abundance of these glycolytic genes can be associated with developmental life history (planktotrophy or lecithotrophy) or lineage specificity (Figure 6c). Furthermore, the top three most enriched GO Biological Processes among differentially expressed proteins in H. erythrogramma are “carbohydrate metabolic process,” “oxidation–reduction process,” and “glycolytic process” (Benjamini–Hochberg corrected p value: 4.86 × 10−4, 1.10 × 10−3, and 4.71 × 10−3, respectively; Figure 6d). Collectively, these data suggest elevated carbohydrate metabolism may be critical to sustaining the high metabolic rate and energetic demands of rapid premetamorphic development in H. erythrogramma.
FIGURE 6.
Biological pathways enriched for differentially expressed proteins between lecithotroph and planktotroph egg samples: (a) glycolysis and (b) regulation of oxidative stress. Significantly upregulated proteins in Heliocidaris erythrogramma eggs are bolded and the reaction catalyzed is highlighted in orange. (c) Normalized egg mRNA abundance (CPM) of proteins from panels (a) and (b) retrieved from Israel et al. (2016). (d) Five most enriched GO “Biological Process” categories among significantly differentially abundant proteins in H. erythrogramma eggs. *Protein was not included in differential abundance analyses due to lack of peptide sequence-similarity or lacking quantification in two or more biological replicates in a sample group (species, developmental stage). CPM: counts-per-million; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; GO: Gene Ontology; GPx: glutathione peroxidase; GR; glutathione reductase; mRNA: messenger RNA
In addition, several proteins involved in oxidative stress response (detoxifying radical oxygen species) are relatively more abundant in H. erythrogramma egg samples, including superoxide dismutase 2 (SPU_024657, FC: +3.1), catalase (SPU_000281, FC: +11.0), and glutathione reductase (SPU_025989, FC: +11.2; Figure 6b; Supporting Information Data 4). Similar to mRNA levels of the glycolytic enzymes, mRNA abundance of these oxidative stress enzymes is associated with life history differences (glutathione reductase) or phylogeny (Heliocidaris: superoxide dismutase 2, glutathione peroxidase 1; H. tuberculata: catalase; Figure 6c). Furthermore, the fifth-most enriched GO Biological Process among proteins more abundant in H. erythrogramma eggs is “superoxide metabolic process” (BH corrected p = 3.90 × 10−2; Figure 6d).
Apolipoprotein B (apoB) is a large lipid transfer protein that functions in lipid transport and metabolism across many invertebrate and vertebrate animal groups (Smolenaars, Madsen, Rodenbrug, and van der Horst, 2007). Although we did not quantify any peptides representing apoB with 100% sequence identity across all three species (thus it was not included in our original comparative abundance analyses), we noted a proteincoding for apoB is among of the most differentially abundant proteins in H. erythrogramma egg and larva samples when all peptides are considered (SPU_028684; 10 peptides; egg = FC: +418.1, larva = FC: +102.6; Supporting Information Data 2). Elevated expression levels of apoB in H. erythrogramma eggs is likely associated with altered maternal provisioning toward high lipid content during oogenesis in this species. In contrast, one of the most differentially expressed proteins in planktotroph eggs compared with lecithotroph eggs is the sea urchin major yolk protein (MYP; SPU_013301), which is relatively much more abundant in eggs of each of the planktotrophic species than those of H. erythrogramma (H. tuberculata = FC: +45.2; L. variegatus = FC: +54.5; Supporting Information Data 2). MYP is a transferrin-like, iron-binding glycoprotein thought to potentially play a role in gamete nutrient transfer to the egg during oogenesis (Brooks & Wessel, 2002). Large differences in apoB and MYP abundance between H. erythrogramma and the two planktotrophic species suggests shifts in egg energetic content necessitates accompanying changes in the provisioning of nutrient transfer proteins.
4 |. DISCUSSION
In this study, high-resolution, comparative analyses of metabolites and proteins have significantly enhanced our understanding of planktotrophic and lecithotrophic development by identifying the molecular basis for possible adaptive changes in physiology and development in H. erythrogramma. We demonstrate the utility and feasibility of mass spectrometry-based lipidomics and proteomics for studying life history evolution by way of egg provisioning in three sea urchin species. This discovery-based approach identifies specific molecules potentially important to each sea urchin species or life history strategy, and notably, our findings prompt new hypotheses about the evolution of lecithotrophy, a life history transition that has evolved numerous times in phylogenetically diverse groups of marine invertebrates (Haszprunar et al., 1995; Strathmann, 1985).
4.1 |. DAGE lipids are maternally provisioned for postmetamorphic survivorship
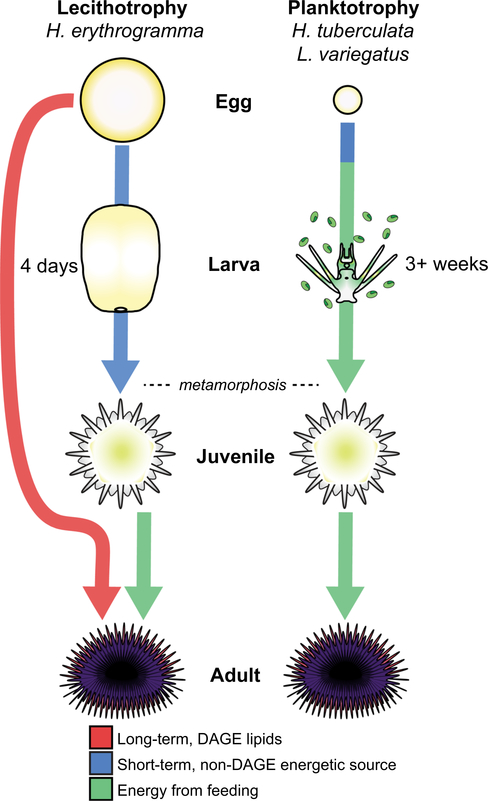
The eggs of planktotrophs by definition lack sufficient energy reserves to complete development through metamorphosis, while those of lecithotrophs are generally overprovisioned with excess energy reserves. Here we show that distinct sets of energy-rich molecules deposited into the eggs of lecithotrophs are likely differentially utilized during pre- and postmetamorphic development. Specifically, our results suggest that maternally deposited DAGEs, which account for the bulk of energetic material in H. erythrogramma eggs, serve as a long-term energy source for juvenile growth and postmetamorphic survivorship in H. erythrogramma rather than fueling rapid embryogenesis and larval development (Figure 7). The hypothesis that DAGE lipids are provisioned for postmetamorphic survivorship in H. erythrogramma is consistent with previous work in which experimental removal of embryonic lipid stores did not affect larval viability, developmental metabolic rate, or time to metamorphosis, but instead resulted in loss of buoyancy and smaller, shorter-lived postmetamorphic juveniles (Emlet & Hoegh-Guldberg, 1997; Villinski et al., 2002). Furthermore, recent work using a targeted, thin-layer chromatographic approach to measure absolute DAGE abundance in this species demonstrates, in agreement with the results of this study, no reduction in DAGE levels between fertilization and 3-day old larvae. However, in 6-day old unfed juveniles (2 days postmetamorphosis), 70% of DAGE levels remain, followed by 49% remaining in 14-day old juveniles (Byrne & Sewell, 2019). These results support the idea that elevated energy store deposition in lecithotrophic marine invertebrates may be more important for the development of a high-quality juvenile than for supporting embryogenesis and construction of the larval body form, as previously suggested for other sea urchins (Alcorn & Allen, 2009), sea stars (Moreno & Hoegh-Guldberg, 1999), and polychaetes (Pernet, Amiel, & Seaver, 2012). Taken together, these findings highlight the importance of considering the linkage between temporally disparate life history phases when investigating the impacts and constraints of adaptations associated with life history evolution (Marshall & Morgan, 2011).
FIGURE 7.
Working model of developmental nutritive strategies of lecithotrophic and planktotrophic sea urchin species. Left: Heliocidaris erythrogramma primarily relies on maternally provisioned, non-diacylglycerol ether, energetic sources (blue) for embryogenesis and larval development. Upregulated carbohydrate metabolism may be associated with rapid premetamorphic development (~4 days) in this species. Abundant diacylglycerol ether lipid stores are instead provisioned for postmetamorphic utilization and survivorship (red), which promotes larval settlement and juvenile development in normally unfavorable environments with low food availability. Right: Heliocidaris tuberculata and Lytechinus variegatus rely on maternal provisions for embryogenesis and early larval development (blue). However, small eggs lacking overprovisioned energetic stores in these species requires that they acquire nutrition from external sources by feeding in the plankton to complete larval development, metamorphosis, and juvenile growth (green)
Previous histological comparisons of H. erythrogramma and H. tuberculata gonadal tissue describe hypertrophy of oocyte lipid deposition as a derived, secondary oogenic phase during which oocytes in H. erythrogramma acquire most of their mass and size (Byrne et al., 1999). The present study detected DAGE molecules in samples from all three sea urchin species examined, suggesting that upregulation of a pre-existing physiological pathway, rather than the evolution of a novel one, underlies increased deposition of DAGEs during H. erythrogramma oogenesis. High levels of DAGE deposition in eggs have also been reported in lecithotrophic species of two other echinoderm classes, Asteroidea (Prowse, Falkner, Sewell, & Byrne, 2009) and Ophiuroidea (Falkner, Sewell, & Byrne, 2015). Thus, maternal provisioning of this specific class of lipids appears to represent a convergent evolutionary modification to lecithotrophy in echinoderms and potentially other marine invertebrate taxa.
4.2 |. Support for novel and existing hypotheses on marine invertebrate larval life history evolution
Our results prompt three new hypotheses about the molecular evolution of lecithotrophy in H. erythrogramma. One is that upregulated carbohydrate metabolism may be critical to fueling rapid embryonic and larval development in this species. Higher relative abundance of enzymes involved in glycolysis and aerobic respiration in H. erythrogramma suggest a greater flux of molecules through the core energy-producing catabolic pathway exists in this species, consistent with measurements of higher metabolic rates during the first 4 days postfertilization relative to H. tuberculata (Hoegh-Guldberg & Emlet, 1997). However, reduced abundance of several TAG molecules through development in this species may indicate this class of lipids is utilized for embryonic and larval morphogenesis. Previous work has demonstrated TAGs are metabolized for premetamorphic development in other echinoids (Meyer, Green, Moore, & Manahan, 2007; Prowse, Sewell, & Byrne, 2017; Sewell, 2005; Whitehill & Moran, 2012), so it is reasonable to hypothesize that TAGs may serve as an energetic source for early development in H. erythrogramma as well.
In addition, especially high expression levels of apoB were quantified in H. erythrogramma eggs and larvae samples relative to the two planktotrophic species. Given that large lipid transfer proteins including apoB bind and assist with lipid storage across metazoans (Smolenaars et al., 2007), we propose this protein may play a crucial role in transporting and provisioning DAGEs and other lipids during oogenesis in H. erythrogramma (see Figures 1 and 3). Vitellogenin typically serves as a precursor to MYPs that assist with oogenic lipid transfer in many metazoan taxa (Smolenaars et al., 2007) including some echinoderms like the Asteroidea (e.g., sea stars; Prowse & Byrne, 2012). Interestingly, the MYP of Echinozoa (e.g., sea urchins and sea cucumber) transports iron and zinc and is encoded by a transferrin-like gene (Brooks & Wessel, 2002; Unuma, Yamamoto, Akiyama, Shiraishi & Ohta, 2003). The high abundance of apoB quantified in this study suggests these species and other echinoids may instead utilize an apoB-like LLTP for primary lipid transport during oogenesis. In this scenario, greater relative expression of apoB in H. erythrogramma eggs compared with the planktotrophic species would reflect hypertrophic oogenic lipid provisioning in this species. Future studies directly aimed at investigating the significance of apoB in oogenic lipid transfer in sea urchins will be crucial for testing this hypothesis.
Finally, our comparative proteomics results suggest defense against oxidative stress is especially crucial for H. erythrogramma embryos. Proteins involved in these physiological pathways may be important for managing high oxidative stress during the initial respiratory burst following fertilization (Shapiro, 1991) or from elevated developmental metabolic rates in H. erythrogramma. Another possible explanation is ultraviolet radiation (UVR)-induced oxidative stress, which is known to cause significant physiological and developmental stress on marine invertebrates (Lesser, 2006), including sea urchin larvae (Adams, Campanale, & Foltz, 2012). Many of the same oxidative stress proteins reported in this study have greater activity in sea urchin larvae exposed to increased experimental (Campanale, Tomanek, & Adams, 2011) and natural UVR (Lister, Lamare, & Burritt, 2010), and the degree of this UVR-induced oxidative stress is dependent on depth in the water column (Lesser, 2010). Since UVR is more intense for the positively buoyant eggs of H. erythrogramma which float closer to the ocean surface (as opposed to the negatively buoyant eggs of either planktotroph species), a higher abundance of proteins involved in regulating radical oxygen species may be adaptive to UVR-induced oxidative stress in H. erythrogramma embryos and larvae.
The adaptations implicated in this study to be associated with the evolution lecithotrophy in H. erythrogramma are consistent with evolutionary tradeoffs found across many examples of this developmental life history transition. In general, greater maternal investment per offspring results in a lower mortality rate in lecithotrophic developers, but at the expense of reduced fecundity (Strathmann, 1985; Vance, 1973). Here, we show that DAGE lipids are exceedingly more abundant in the egg and larval lipidomes of H. erythrogramma than in the planktotrophs H. tuberculata and L. variegatus, and these lipids constitute the bulk of lipid provisioning in H. erythrogramma (Byrne & Sewell, 2019). DAGEs are likely utilized for postmetamorphic survivorship in a resource-poor environment rather than fueling accelerated larval development in this species. Oogenic upregulation of maternally deposited lipid transfer proteins like apoB may be crucial to provisioning lipids and thus lecithotrophy as a whole in H. erythrogramma. Longer larval periods spent in the plankton lead to higher offspring die-off rates due to predation, starvation, or chance mortality (Wray & Raff, 1991), so accelerated premetamorphic development driven instead by upregulated carbohydrate metabolism may be a key adaptation acquired during this life history transition. However, a shorter larval period comes at the cost of reduced dispersal potential in lecithotrophs (Emlet, 1995; Paulay & Meyer, 2006). Lastly, increased regulation of oxidative stress may serve as an auxiliary trait to physical and metabolic modifications to oogenesis and developmental physiology to further increase offspring chance of survival before metamorphosis.
Our results also provide additional support for existing hypotheses regarding maternal provisioning strategies of planktotrophic developers. We find conservation of a shortterm, triacylglycerol-based lipid storage system in both planktotrophic sea urchin species that corroborates previous reports of an ancestral, TAG-based maternal lipid provisioning program in planktotrophic echinoderms (Byrne et al., 2008; Falkner et al., 2015; Prowse et al., 2009). Also, a higher relative abundance of yolk protein in the eggs of both planktotroph species compared to those of H. erythrogramma supports previous reports of the potential importance of yolk protein for nutrition during planktotrophic development in sea urchins and other marine invertebrates (Jaeckle, 1995; Prowse, Sewell, & Byrne, 2008). These findings are concordant with the larval development strategies of H. tuberculata, L. variegatus, and many other planktotrophic marine invertebrates that rely briefly on maternally derived egg provisions during embryogenesis followed by the development of a larva that feeds in the water column for weeks to months before metamorphosis (Figure 7). In contrast to lecithotrophy, this developmental strategy is advantageous in environments where food availability is abundant and seasonally reliable (Marshall et al., 2012, Morgan, 1995), as more offspring will have a higher chance at survivorship despite less nutritional content per egg.
5 |. CONCLUSION
This study highlights the advantages of applying high-resolution LC-MS/MS to exploring molecular adaptations potentially associated with life history evolution and organismal diversity. Future comparative studies that identify molecular differences between planktotrophic and lecithotrophic species in other clades are needed to test and refine our hypotheses of the evolution of lecithotrophy in H. erythrogramma. These results will shed light on an additional novel and convergent evolutionary innovations associated with this recurrent life history switch.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. B. Emlet for helpful discussion; E. C. Raff for Heliocidaris egg and larva micrographs; A. N. George for assistance acquiring Lytechinus variegatus egg and larva micrographs; members of Wray Lab for helpful comments on the manuscript; and J. W. Israel for providing L. variegatus samples. P. L. D. was supported by the Developmental and Stem Cell Biology Program at Duke University, funded by National Institute of Health Training (grant no. 2T32HD040372-16). This project was funded by National Science Foundation (grant no. IOS-1457305).
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Number: 2T32HD040372-16; Division of Integrative Organismal Systems, Grant/Award Number: 1457305; National Institute of Health; National Science Foundation
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Adams NL, Campanale JP, & Foltz KR (2012). Proteomic responses of sea urchin embryos to stressful ultraviolet radiation. Integrative and Comparative Biology, 52, 665–680. [DOI] [PubMed] [Google Scholar]
- Alcorn NJ, & Allen JD (2009). How do changes in parental investment influence development in echinoid echinoderms? Evolution and Development, 11, 719–727. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, & Lipman DJ (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, … Sherlock G (2000). Gene ontology: Tool for the unification of biology. Nature Genetics, 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind AL, Soderblom EJ, Turner ME, Moseley MA, Ely JJ, Hof PR, … Babbitt CC (2015). Evolutionary divergence of gene and protein expression in the brains of humans and chimpanzees. Genome Biology and Evolution, 7, 2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, & Golani I (2001). Controlling the false discovery rate in behavior genetics research. Behavioural Brain Research, 125, 279–284. [DOI] [PubMed] [Google Scholar]
- Brooks JM, & Wessel GM (2002). The major yolk protein on sea urchins is a transferrin-like, iron binding protein. Developmental Biology, 245, 1–12. [DOI] [PubMed] [Google Scholar]
- Byrne M, Prowse TAA, Sewell MA, Dworjanyn S, Williamson JE, & Vaitilingon D (2008). Maternal provisioning for larvae and larval provisioning for juveniles in the toxopneustid sea urchin Tripneustes gratilla. Marine Biology, 155, 473–482. [Google Scholar]
- Byrne M, Villinski JT, Cisternas P, Siegel RK, Popodi E, & Raff RA (1999). Maternal factors and the evolution of developmental mode: Evolution of oogenesis in Heliocidaris erythrogramma. Development Genes and Evolution, 209, 275–283. [DOI] [PubMed] [Google Scholar]
- Byrne M, & Sewell M (2019). Evolution of maternal provisioning strategies in echinoids – selection for high quality juveniles. Marine Ecology Progress Series. Accepted, in press. 10.3354/meps12938 [DOI] [Google Scholar]
- Campanale JP, Tomanek L, & Adams NL (2011). Exposure to ultraviolet radiation causes proteomic changes in embryos of the purple sea urchin, Strongylocentrotus purpuratus. Journal of Experimental Marine Biology and Ecology, 397, 106–120. [Google Scholar]
- Emlet RB (1995). Developmental mode and species geographic range in regular sea urchins (Echinodermata: Echinoidea). Evolution, 49, 476–489. [DOI] [PubMed] [Google Scholar]
- Emlet RB, & Hoegh-Guldberg O (1997). Effects of egg size on postlarval performance: Experimental evidence from a sea urchin. Evolution, 51, 141–152. [DOI] [PubMed] [Google Scholar]
- Emlet RB, McEdward LR, & Strathmann RR (1987). Echinoderm larval ecology viewed from the egg In Lawrence JM (Ed.), Echinoderm studies (1st ed., pp. 55–136). Rotterdam, the Netherlands: Balkema. [Google Scholar]
- Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, … Dennis EA (2009). Update of the LIPID MAPS comprehensive classification system for lipids. Journal of Lipid Research, 50(Suppl), S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner I, Sewell MA, & Byrne M (2015). Evolution of maternal provisioning in ophiuroid echinoderms: Characterisation of egg composition in planktotrophic and lecithotrophic developers. Marine Ecology Progress Series, 525, 1–13. [Google Scholar]
- Flatt T, & Heyland A (2011). Mechanisms of life history evolution: The genetics and physiology of life history traits and trade-offs (1st ed.). London, UK: Oxford University Press. [Google Scholar]
- Guo H, Garcia-Vedrenne AE, Isserlin R, Lugowski A, Morada A, Sun A, … Emili A (2015). Phosphoproteomic network analysis in the sea urchin Strongylocentrotus purpuratus reveals new candidates in egg activation. Proteomics, 15, 4080–4095. [DOI] [PubMed] [Google Scholar]
- Hart MW (2002). Life history evolution and comparative developmental biology of echinoderms. Evolution and Development, 4, 62–71. [DOI] [PubMed] [Google Scholar]
- Haszprunar G, von Salvini-Plawen L, & Rieger RM (1995). Larval planktotrophy—A primitive trait in the bilateria. Acta Zoological-Stockholm, 76, 141–154. [Google Scholar]
- Henry L and Wickham H (2017) purrr: Functional Programming Tools. R package version 0.2.5. Retrieved from https://CRAN.Rproject.org/package=purrr [Google Scholar]
- Hoegh-Guldberg O, & Emlet RB (1997). Energy use during the development of a lecithotrophic and a planktotrophic echinoid. Biological Bulletin, 192, 27–40. [DOI] [PubMed] [Google Scholar]
- Israel JW, Martik ML, Byrne M, Raff EC, Raff RA, McClay DR, & Wray GA (2016). Comparative developmental transcriptomics reveals rewiring of a highly conserved gene regulatory network during a major life history switch in the sea urchin genus Heliocidaris. PLOS Biology, 14, e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski D, & Lutz RA (1983). Larval ecology of marine benthic invertebrates: Paleobiological implications. Biological Reviews, 58, 21–89. [Google Scholar]
- Jaeckle WB (1995). Variations in the size, energy content and biochemical composition of invertebrate eggs: Correlates to the mode of larval development In McEdward LR (Ed.), Ecology of marine invertebrate larvae (1st ed., pp. 49–78). Boca Raton, FL: CRC Press. [Google Scholar]
- Laegdsgaard P, Byrne M, & Anderson DT (1991). Reproduction of sympatric populations of Heliocidaris erythrogramma and H. tuberculata (Echinoidea) in New South Wales. Marine Biology, 110, 359–374. [Google Scholar]
- Lee RF, Hagen W, & Kattner G (2006). Lipid storage in marine zooplankton. Marine Ecology Progress Series, 307, 273–306. [Google Scholar]
- Lesser MP (2006). Oxidative stress in marine environments: Biochemistry and physiological ecology. Annual Review of Physiology, 68, 253–278. [DOI] [PubMed] [Google Scholar]
- Lesser MP (2010). Depth-dependent effects of ultraviolet radiation on survivorship, oxidative stress and dna damage in sea urchin (Strongylocentrotus droebachiensis) embryos from the Gulf of Maine. Photochemistry and Photobiology, 86, 382–388. [DOI] [PubMed] [Google Scholar]
- Lister KN, Lamare MD, & Burritt DJ (2010). Oxidative damage in response to natural levels of UV-B radiation in larvae of the tropical sea urchin Tripneustes gratilla. Photochemistry and Photobiology, 86, 1091–1098. [DOI] [PubMed] [Google Scholar]
- Marshall DJ, Krug PJ, Kupriyanova EK, Byrne M, & Emlet RB (2012). The biogeography of marine invertebrate life histories. Annual Review of Ecology, Evolution, and Systematics, 43, 97–114. [Google Scholar]
- Marshall DJ, & Morgan SG (2011). Ecological and evolutionary consequences of linked life history stages in the sea. Current Biology, 21, R718–R725. [DOI] [PubMed] [Google Scholar]
- McEdward LR (1995). Ecology of Marine Invertebrate Larvae (1st ed.). Boca Raton, FL: CRC Press. [Google Scholar]
- McEdward LR, & Miner BG (2001). Larval and life-cycle patterns in echinoderms. Canadian Journal of Zoology, 79, 1125–1170. [Google Scholar]
- Meyer E, Green AJ, Moore M, & Manahan DT (2007). Food availability and physiological state of sea urchin larvae (Strongylocentrotus purpuratus). Marine Biology, 152, 179–191. [Google Scholar]
- Moreno G, & Hoegh-Guldberg O (1999). The energetics of development of three congeneric seastars (Patiriella Verrill, 1913) with different types of development. Journal of Experimental Marine Biology and Ecology, 235, 1–20. [Google Scholar]
- Morgan SG (1995). Life and death in the plankton: Larval mortality and adaptation In McEdward LR (Ed.), Ecology of marine invertebrate larvae (1st ed., pp. 279–322). Boca Raton, FL: CRC Press. [Google Scholar]
- Palumbi SR (1994). Genetic divergence, reproductive isolation, and marine speciation. Annual Review of Ecology and Systymatics, 25, 546–572. [Google Scholar]
- Parrish CC (2013). Lipids in marine ecosystems. ISRN Oceanography, 2013, 16–16. [Google Scholar]
- Paulay G, & Meyer C (2006). Dispersal and divergence across the greatest ocean region: Do larvae matter? Integrative and Comparative Biology, 46, 269–281. [DOI] [PubMed] [Google Scholar]
- Pernet B, Amiel A, & Seaver EC (2012). Effects of maternal investment on larvae and juveniles of the annelid Capitella teleta determined by experimental reduction of embryo energy content. Invertebrate Biology, 131, 82–95. [Google Scholar]
- Prowse TAA, & Byrne M (2012). Evolution of yolk protein genes in the Echinodermata. Evolution and Development, 14, 139–151. [DOI] [PubMed] [Google Scholar]
- Prowse TAA, Falkner I, Sewell MA, & Byrne M (2009). Long-term storage lipids and developmental evolution in echinoderms. Evolutionary Ecology Research, 11, 1069–1083. [Google Scholar]
- Prowse TAA, Sewell MA, & Byrne M (2008). Fuels for development: Evolution of maternal provisioning in asterinid sea stars. Marine Biology, 153, 337–349. [Google Scholar]
- Prowse TAA, Sewell MA, & Byrne M (2017). Three-stage lipid dynamics during development of planktotrophic echinoderm larvae. Marine Ecology Progress Series, 583, 149–161. [Google Scholar]
- Raff RA (1987). Constraint, flexibility, and phylogenetic history in the evolution of direct development in sea urchins. Developmental Biology, 119, 6–19. [DOI] [PubMed] [Google Scholar]
- Raff RA (1992). Direct-developing sea urchins and the evolutionary reorganization of early development. BioEssays, 14, 211–218. [DOI] [PubMed] [Google Scholar]
- Raff RA, & Byrne M (2006). The active evolutionary lives of echinoderm larvae. Heredity, 97, 244–252. [DOI] [PubMed] [Google Scholar]
- Reidel B, Thompson JW, Farsiu S, Moseley MA, Skiba NP, & Arshavsky VY (2011). Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Molecular and Cellular Proteomics, 10, M110.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium, Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA., … Burgess DR. (2006). The genome of the sea urchin Strongylocentrotus purpuratus. Science, 314, 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell MA (2005). Utilization of lipids during early development of the sea urchin Evechinus chloroticus. Marine Ecology Progress Series, 304, 133–142. [Google Scholar]
- Shapiro BM (1991). The control of oxidant stress at fertilization. Science, 252, 533–536. [DOI] [PubMed] [Google Scholar]
- Smith AB (1989). RNA sequence data in phylogenetic reconstruction: Testing the limits of its resolution. Cladistics, 5, 321–344. [DOI] [PubMed] [Google Scholar]
- Smith AB, Littlewood DTJ, & Wray GA (1995). Comparing patterns of evolution: Larval and adult life history stages and ribosomal-RNA of post-Paleozoic echinoids. Philiosophical Transactions of the Royal Society B, 349, 11–18. [Google Scholar]
- Smolenaars MMW, Madsen O, Rodenburg KW, & van der Horst DJ (2007). Molecular diversity and evolution of the large lipid transfer protein superfamily. Journal of Lipid Research, 48, 489–502. [DOI] [PubMed] [Google Scholar]
- Stearns SC (1992). The evolution of life histories (1st ed.). London: Oxford University Press. [Google Scholar]
- Strathmann RR (1985). Feeding and nonfeeding larval development and life-history evolution in marine invertebrates. Annual Review of Ecology and Systematics, 16, 339–361. [Google Scholar]
- Thorson G (1950). Reproductive and larval ecology of marine bottom invertebrates. Biological Reviews, 25, 1–45. [DOI] [PubMed] [Google Scholar]
- Unuma T, Yamamoto T, Akiyama T, Shiraishi M, & Ohta H (2003). Quantitative changes in yolk protein and other components in the ovary and testis of the sea urchin Pseudocentrotus depressus. Journal of Experimental Biology, 206, 365–372. [DOI] [PubMed] [Google Scholar]
- Vance RR (1973). On the reproductive strategies of marine benthic invertebrates. The American Naturalist, 107, 339–352. [DOI] [PubMed] [Google Scholar]
- Varemo L, Nielsen J, & Nookaew I (2013). Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Research, 41, 4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villinski JT, Villinski JC, Byrne M, & Raff RA (2002). Convergent maternal provisioning and life-history evolution in echinoderms. Evolution, 56, 1764–1775. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, … Hermjakob H (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Research, 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watschinger K, & Werner ER (2013). Alkylglycerol monooxygenase. IUBMB Life, 65, 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehill EAG, & Moran AL (2012). Comparative larval energetics of an ophiuroid and an echinoid echinoderm. Invertebrate Biology, 131, 345–354. [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis, New York, NY: Springer-Verlag; 10.1007/978-0387-98141-3 [DOI] [Google Scholar]
- Wickham H and Francois R (2016) dplyr: A Grammar of Data Manipulation. R package version 0.7.5. Retrieved from https://CRAN.R-project.org/package=dplyr [Google Scholar]
- Williams DHC, & Anderson DT (1975). Reproductive system, embryonic development, larval development and metamorphosis of sea urchin Heliocidaris erythrogramma (Val.) (Echinoidea: Echinometridae). Australian Journal of Zoology, 23, 371–403. [Google Scholar]
- Wray GA (1996). Parallel evolution of nonfeeding larvae in echinoids. Systematic Biology, 45, 308–322. [Google Scholar]
- Wray GA, & Raff RA (1991). The evolution of developmental strategy in marine invertebrates. Trends in Ecology and Evolution, 6, 45–50. [DOI] [PubMed] [Google Scholar]
- Zigler KS, Raff EC, Popodi E, Raff RA, & Lessios HA (2003). Adaptive evolution of bindin in the genus Heliocidaris is correlated with the shift to direct development. Evolution, 57, 2293–2302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry lipidomics data have been deposited on the MassIVE public data repository at the University of California-San Diego with the data set identifier MSV000082501. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner repository with the data set identifier PXD007065. Normalized spectra intensities used for statistical analysis of lipidomic and proteomic data sets are available in Supporting Information Data 1 and 2, respectively.