Abstract
Pharmacogenomic drug labels in the Summary of Product Characteristics (SmPC) provide an instrument for clinical implementation of pharmacogenomics. We compared pharmacogenomic guidance by Clinical Pharmacogenetics Implementation Consortium (CPIC), Dutch Pharmacogenetics Working Group (DPWG), the US Food and Drug Administration (FDA), and by the European agencies the European Medicines Agency (EMA), College ter Beoordeling van Geneesmiddelen Medicines Evaluation Board (CBG‐MEB), and Federal Institute for Drugs and Medical Devices (FIDMD), collectively assigned as EMA/FIDMD+MEB shortened as EMA/FM. Of 54 drugs with an actionable gene–drug interaction in the CPIC and DPWG guidelines, only 50% had actionable pharmacogenomic information in the SmPCs and the agencies were in agreement in only 18% of the cases. We further compared 450 additional drugs, lacking CPIC or DPWG guidance, and found 126 actionable gene–drug labels by the FDA and/or the EMA/FM. Based on these 126 drugs in addition to the 54 above, the consensus of actionable pharmacogenomic labeling between the FDA and the EMA/FM was only 54%. In conclusion, guidelines provided by CPIC/DPWG are only partly implemented into the SmPCs and the implementation of pharmacogenomic drug labels into the clinics would strongly gain from a higher extent of consensus between agencies.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ The Clinical Pharmacogenetics Implementation Consortium (CPIC), the Dutch Pharmacogenetics Working Group (DPWG), the US Food and Drug Administration (FDA), the European Medicines Agency (EMA), as well as the Dutch College ter Beoordeling van Geneesmiddelen Medicines Evaluation Board (CBG‐MEB) and German Federal Institute for Drugs and Medical Devices (BfArM) medical product agencies all provide pharmacogenomic guidelines and Summary of Product Characteristics (SmPC) labels for the use of genomic biomarkers in clinical practice.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ To what extent is the provided pharmacogenomic information in concordance between the different consortia and regulatory agencies?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ We find that there is a huge discrepancy between information given by the CPIC and DPWG and the pharmacogenomic information in drugs labels in the corresponding SmPC. We also find important differences between the US and European regulatory bodies.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The results indicate the necessity to re‐review and validate the current pharmacogenomic drug labels in order to obtain a more coherent and efficient instrument for implementation of pharmacogenomics into the clinics.
Interindividual differences in drug metabolism, response, and toxicity are important. These are inherent to differences in physiological factors like age, sex, body mass index, and lifestyle, but also by drug interactions at the enzyme, transporter, or target levels as well as by genetic factors. Overall estimations have been made, identifying 20–30% of this variability to be attributable to genetic factors, although an exact figure is difficult to define.1, 2 Twin studies do indicate that the genetic influence for the pharmacokinetics of certain drugs is very high.3 Indeed, in some cases, the genetic background for such variability still has to be identified, including the exact role of rare genetic variants.4, 5
In the past, much work has been focused on the definition of genetic variants of importance for interindividual variations in drug pharmacokinetics, toxicity, and response.4 This work has resulted in the characterization of genetic markers in a set of 15–20 genes that can already be used to inform drug prescribing.6 Furthermore, several different initiatives have been taken to support clinical application of pharmacogenetics. The Clinical Pharmacogenetics Implementation Consortium (CPIC)7 and the Dutch Pharmacogenetics Working Group (DPWG)8 independently created guidelines that focus on how genetic test results should be translated into specific prescribing actions. Regulatory authorities (e.g., the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA)) also provide pharmacogenomic information in their labels.9, 10 Obviously, a recommendation to perform a pharmacogenomic test presents a strong stimulus for clinical implementation.11
In order to analyze the actual pharmacogenomic information given in different guidelines in comparison to the actual SmPC in the United States and Europe, we have scrutinized the pharmacogenomic labeling information regarding 505 different drugs as provided by the CPIC, DPWG, the FDA, the EMA, and, for drugs approved prior to the founding of the EMA in 1995, the corresponding information in the SmPC from the Dutch College ter Beoordeling van Geneesmiddelen Medicines Evaluation Board (CBG‐MEB) and German Federal Institute for Drugs and Medical Devices (FIDMD) medical product agencies.
The results indicate severe discrepancies in the manner by which the regulatory agencies and the academic guidelines present recommendations for clinical implications of pharmacogenomic biomarkers and emphasize the need for action to reach consensus on how the pharmacogenomic label information should be interpreted and presented to the clinicians.
Methods
Data sources
The Summary of Product Characteristics (SmPC) of the EMA‐approved drugs were screened for pharmacogenomic labeling. Drugs registered prior to the founding of the EMA (1995) were represented by SmPC from the Dutch (CBG‐MEB, which will be referred to as Medicines Evaluation Board (MEB)) and German (BfArM, which will be referred to as the FIDMD agencies), which were retrieved from two databases,12, 13 respectively. The SmPCs retrieved from BfArM are written in the German language and were screened and translated (only pharmacogenomics, if applicable) by a native German speaker (L.S.). Thus, the European labels are provided by the EMA or FIDMD+MEB, defined as the EMA/FM. The FDA approved drugs were screened in the “Table of Pharmacogenomic Biomarkers in Drug Labeling” (last updated December 2018), available on the FDA website.14 Nonhuman genotypes are excluded in the table on the FDA website, but were included in our screening and, therefore, the SmPCs of the FDA approved antiinfectives were screened manually after retrieval from the Drugs@FDA database.15 The FDA SmPC of inotersen was also checked manually, as the indication was not included in the aforementioned FDA resource. In those drugs that are registered under multiple trade names, one SmPC was checked as a representative. All information was accessed on January 1, 2019, except for novel drug approvals, which were checked until June 15, 2019. During screening of the information, pharmacogenomic labels were extracted and collected in a separate file. Subsequently, they were assigned a label category by two of the authors (R.S. and L.S.), independently. The categorization was then compared and in case of disagreement was discussed in order to reach consensus.
Definitions
In case of no registration, awaiting approval, or unavailable SmPC, no label was obtained, and it was indicated with not available in the tables. Labels were further classified as (i) no information on any gene–drug interaction (no information), (ii) information on any gene–drug interaction (pharmacogenomic information), (iii) stating a recommended action (recommendation), or (iv) a specific recommended action for any gene–drug interaction (strong recommendation), (v) contraindicated in specific genetic backgrounds (recommendation dealing with contraindication), (vi) indicated in specific genetic backgrounds (indication), and (vii) mandatory testing or adjustments (mandatory). The categories mandatory, indication, recommendation dealing with contraindication, strong recommendation, and recommendation (category > 3) are considered actionable labels. A specific dose adjustment was classified as a strong recommendation. When the SmPC text stated an action with “must” or “mandatory,” when the label was an indication, or when the label stated an absolute contraindication, it was considered a mandatory action. Mandatory, indication, recommendation dealing with contraindication, strong recommendation, and recommendation are considered actionable labels. Negative recommendations (i.e., stating that no dose changes are required in genetic polymorphisms), were considered as pharmacogenomic information. Labels dealing with pharmacogenomic information only have that label in the tables without further content.
CPIC, DPWG, EMA, and FDA comparison
All gene–drug interactions mentioned in pharmacogenomic guidelines by the CPIC and/or DPWG were considered, except those that did not require any action. For a drug to be included in this comparison, it had to be registered by either the EMA (or if registered before 1995, FM) and/or by the FDA. Drugs were excluded (i) if they were neither registered by the EMA/FM nor by the FDA and (ii) if the guidelines required no action by both the CPIC and DPWG.
For an overview of discrepancies in label categories, a table was created where only drugs with differences in the label category by the EMA/FM and the FDA were included.
The full table was condensed by only including the strongest label for each gene–drug interaction for the guidelines and agencies with a brief description (Table S1 ). The complete tables can be provided upon request.
EMA and FDA comparison
Gene–drug interactions for which no guideline by either the CPIC or DPWG existed were collected and subdivided based on their Anatomical Therapeutic Chemical (ATC) class. ATC classes with few drugs (< 3) in the full tables were grouped together in the ATC class V (various). The compared drugs had to be approved by either the EMA/FM and/or the FDA and contain content on pharmacogenomic biomarkers (including germline and somatic gene variants) both in humans and microorganisms. Drugs were excluded if (i) they were not registered by the EMA/FM and the FDA or (ii) the SmPCs in both the EMA/FM and the FDA contained no pharmacogenomic information.
In case of multiple gene–drug interactions for a single drug, all of them were considered if each fulfilled the inclusion criteria for the EMA and FDA comparison.
The published tables were shortened from the complete tables by only including the strongest label of the actionable labels (the three different recommendations, indications, and mandatory actions) for each gene–drug interaction for the agencies with a brief description and excluding nonactionable labels (no information or pharmacogenomic information) and can be found in the Supplementary Material (Tables S2–S11 ). The complete tables can be provided by the authors upon request. Again, a table was created only including drugs with differences in the label category by the EMA/FM and the FDA.
Pie charts
To identify and compare the proportion of types of information included in the labels between the two regulatory agencies pie charts were created. Nonactionable labels were also considered; therefore, all drugs from the complete table that were approved by both the EMA/FM and the FDA were included.
Venn diagram
To compare the overlap of recommendations between the CPIC and DPWG guidelines and the regulatory agencies, two different Venn diagrams were created using an online tool of the Bioinformatics & Evolutionary genomics group of Ghent University.16 For the first Venn diagram, the drugs had to be covered by all four parties (CPIC, DPWG, EMA/FM, and FDA). If the drug approval was prior to 1995 and both the FIDMD and MEB had labels, the strongest one was chosen as the representative for EMA (Pharmacogenomic information < Recommendation < Strong recommendation < Recommendation dealing with contraindication < Indication < Mandatory). Therefore, the EMA is partly represented by FM.
The second Venn diagram was created for the comparison of the EMA and FDA labels. Again, only drugs that were registered by both the EMA/FM and FDA and had actionable labels in at least one agency were included. The EMA is again partly represented by FM.
Results
Comparison of pharmacogenomic labels among the CPIC, DPWG, EMA/FM, and FDA
We compared pharmacogenomic guidelines and drug labeling. Two comparisons were made, the first one focusing on drugs that have guidelines by the CPIC and/or DPWG, and a second screen for drugs lacking CPIC or DPWG guidelines. Distribution of different types of labels, as defined below, was compared, as well as actionable labels specifically. Flow charts for the selection of gene–drug interactions are given in Figures 1 and 2.
Figure 1.
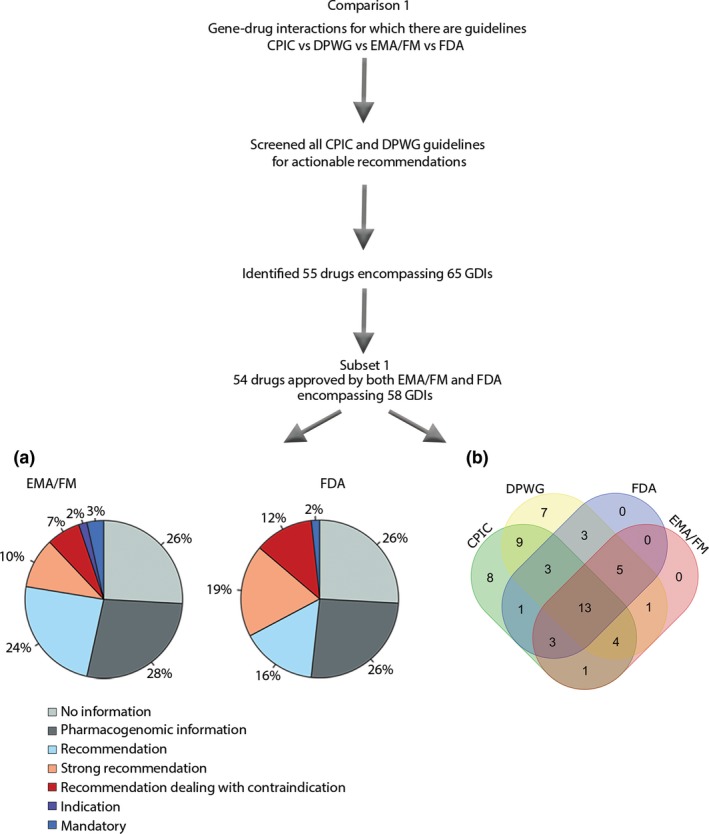
Selection of drugs for comparison 1, comparing CPIC, DPWG, EMA/FM, and FDA, was based on all CPIC and DPWG guidelines that contain recommendations (55 drugs with 65 GDIs). The distribution of label categories was compared in a subset of these GDIs (subset 1), only including those that are approved by both the EMA/FM and the FDA, leaving 58 GDIs. (a) Pie charts showing the distribution of different label categories between the EMA/FM and the FDA for 58 GDIs that have guidelines by the CPIC and/or DPWG. (b) Venn diagram of the same 58 GDIs that have guidelines by the CPIC and/or DPWG and are approved by both the EMA/FM and the FDA, with actionable labels (mandatory, indication, recommendation dealing with contraindication, or (strong) recommendation). For 13 of the GDIs (abacavir, allopurinol, atomoxetine, azathioprine (2 genes), capecitabine, carbamazepine, citalopram, codeine, mercaptopurine (2 genes), and thioguanine (2 genes)) there are common guidelines or actionable labels by all consortia and agencies. For 9 (15.5%) of the GDIs (clomipramine – CYP2D6, doxepin, imipramine (2 genes), nortriptyline, paroxetine, simvastatin, tacrolimus, and voriconazole) guidelines are provided by both the CPIC and DPWG but no actionable labels by the EMA/FM or the FDA. A further eight (13.8%) and seven (12.1%) GDIs that have guidelines by the CPIC or DPWG, respectively, are unaccounted for in both the EMA/FM and the FDA SmPCs (atazanavir, atorvastatin, clomipramine, doxepin, efavirenz, flecainide, lamotrigine, metoprolol, ondansetron, peginterferon, ribavirin, tamoxifen, and trimipramine (2 genes) and venlafaxine). CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; EMA/FM, European Medicines Agency/FIDMD+MEB; FDA, US Food and Drug Administration; GDI, gene–drug interaction.
Figure 2.
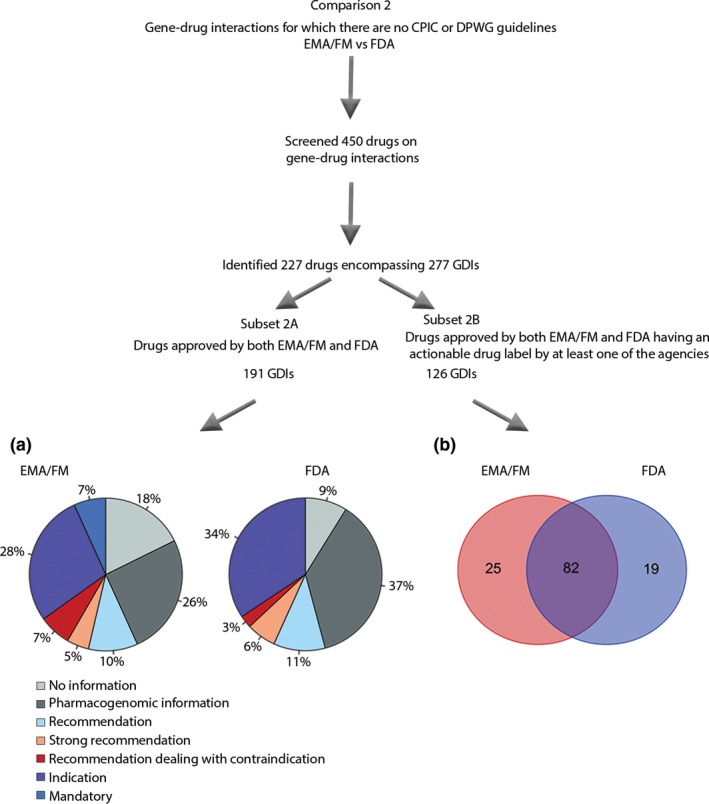
Comparison 2, comparing the EMA/FM and the FDA, consisted of drugs that do not have guidelines. Of 450 screened drugs, 227 drugs encompassing 277 GDIs were identified. Again, the distribution of label categories was compared in a subset of these GDIs, only including those that are approved by both the EMA/FM and the FDA, leaving 191 gene–drug interactions (subset 2A), shown in the pie chart. Subset 2B, including only drugs that have an actionable label by at least one of the agencies, was used to compare the overlap in actionable labels among 126 GDIs. (a) Pie charts showing the proportions of different label categories for the 191 GDIs of the screened drugs approved by both the EMA/FM and the FDA. (b) Venn diagram of 126 GDIs of drugs that are approved by both the EMA/FM and the FDA but for which no guideline is available by the CPIC and DPWG, with actionable labels (mandatory, indication, recommendation dealing with contraindication, or (strong) recommendation). CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; EMA, European Medicines Agency; FDA, US Food and Drug Administration; GDI, gene–drug interaction; SmPC, Summary of Product Characteristics.
We grouped the European agencies EMA, FIDMD, and MEB together as the EMA/FM. The analysis comparing the drugs, where actionable guidelines had previously been given by the CPIC and/or DPWG, revealed an initial subset of 55 different drugs with pharmacogenomic information, encompassing 65 gene–drug interactions (Table S1 ). The majority of these drugs are registered prior to the EMA founding in 1995, and for those we used the SmPCs of FIDMD and MEB instead. For the 54 drugs (corresponding to 58 gene–drug interactions) that are approved by both the EMA/FM and FDA, the distribution of the different label categories among the agencies being (i) no information, (ii) pharmacogenomic information, (iii) recommendation, (iv) strong recommendation, (v) recommendation dealing with contraindication, (vi) indication, and (vii) mandatory are shown in Figure 1. The categories mandatory, indication, recommendation dealing with contraindication, strong recommendation, and recommendation (category > 3) are considered actionable labels. Additionally, the drugs and their labels by each agency are shown in Table S1 . All discrepancies in label categories between the EMA/FM and FDA of the drugs having guidelines from the CPIC and/or DPWG are summarized in Table 1, containing 28 drugs.
Table 1.
Differences in label categories between EMA (or FIDMD/MEB) and FDA for gene–drug interactions that have a label by at least one agency, as well as guidance by the CPIC and/or DPWG
| Drug | Gene | Institution | Therapeutic recommendation | ATC code |
|---|---|---|---|---|
| Abacavir | HLA‐B | EMA | Mandatory | J |
| FDA | Recommendation dealing with contraindication | |||
| Amitriptyline | CYP2C19 | EMA | N.A. | N |
| FIDMD | Strong recommendation | |||
| MEB | Strong recommendation | |||
| FDA | No information | |||
| CYP2D6 | EMA | N.A. | ||
| FIDMD | Strong recommendation | |||
| MEB | Strong recommendation | |||
| FDA | Pharmacogenomic information | |||
| Aripiprazole | CYP2D6 | EMA | Pharmacogenomic information | N |
| FDA | Strong recommendation | |||
| Atomoxetine | CYP2D6 | EMA | N.A. | N |
| FIDMD | Recommendation | |||
| MEB | Recommendation | |||
| FDA | Strong recommendation | |||
| Atorvastatin | SLCO1B1 | EMA | N.A. | C |
| FIDMD | Pharmacogenomic information | |||
| MEB | Pharmacogenomic information | |||
| FDA | No information | |||
| Carbamazepine | HLA‐B | EMA | N.A. | N |
| FIDMD | Recommendation | |||
| MEB | Recommendation | |||
| FDA | Recommendation dealing with contraindication | |||
| Clomipramine | CYP2C19 | EMA | N.A. | N |
| FIDMD | Pharmacogenomic information | |||
| MEB | Pharmacogenomic information | |||
| FDA | No information | |||
| Clopidogrel | CYP2C19 | EMA | Pharmacogenomic information | B |
| FDA | Recommendation | |||
| Codeine | CYP2D6 | EMA | N.A. | R |
| FIDMD | Pharmacogenomic information | |||
| MEB | Recommendation dealing with contraindication | |||
| FDA | Recommendation dealing with contraindication | |||
| Doxepin | CYP2C19 | EMA | N.A. | N |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Pharmacogenomic information | |||
| CYP2D6 | EMA | N.A. | ||
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Pharmacogenomic information | |||
| Eliglustat | CYP2D6 | EMA | Recommendation dealing with contraindication | A |
| FDA | Strong recommendation | |||
| Escitalopram | CYP2C19 | EMA | N.A. | N |
| FIDMD | Strong recommendation | |||
| MEB | Strong recommendation | |||
| FDA | Pharmacogenomic information | |||
| Fluvoxamine | CYP2D6 | EMA | N.A. | N |
| FIDMD | Pharmacogenomic information | |||
| MEB | Pharmacogenomic information | |||
| FDA | Recommendation | |||
| Halogenated volatile anesthetics (enflurane, isoflurane) or succinyl choline | RYR1/CACNA1S | EMA | N.A. | N |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | No information | |||
| FDA | Recommendation dealing with contraindication | |||
| Haloperidol | CYP2D6 | EMA | N.A. | N |
| FIDMD | Recommendation | |||
| MEB | Recommendation | |||
| FDA | No information | |||
| Imipramine | CYP2D6 | EMA | N.A. | N |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Pharmacogenomic information | |||
| Irinotecan | UGT1A1 | EMA | N.A. | L |
| FIDMD | Strong recommendation | |||
| MEB | Recommendation | |||
| FDA | Recommendation | |||
| Ivacaftor | CFTR | EMA | Indication | R |
| FDA | Mandatory | |||
| Mercaptopurine | TPMT | EMA | Recommendation | L |
| FDA | Strong recommendation | |||
| NIUDT15 | EMA | Recommendation | ||
| FDA | Strong recommendation | |||
| Metoprolol | CYP2D6 | EMA | N.A. | C |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Pharmacogenomic information | |||
| Oxcarbazepine | HLA‐B | EMA | N.A. | N |
| FIDMD | Recommendation | |||
| MEB | Recommendation | |||
| FDA | Recommendation dealing with contraindication | |||
| Phenytoin | CYP2C9 | EMA | N.A. | N |
| FIDMD | Recommendation | |||
| MEB | Pharmacogenomic information | |||
| FDA | Pharmacogenomic information | |||
| HLA‐B | EMA | N.A. | ||
| FIDMD | Pharmacogenomic information | |||
| MEB | Recommendation | |||
| FDA | Recommendation | |||
| Propafenone | CYP2D6 | EMA | N.A. | C |
| FIDMD | Pharmacogenomic information | |||
| MEB | Pharmacogenomic information | |||
| FDA | Recommendation | |||
| Sertraline | CYP2C19 | EMA | N.A. | N |
| FIDMD | Recommendation | |||
| MEB | Pharmacogenomic information | |||
| FDA | No information | |||
| Simvastatin | SLCO1B1 | EMA | Pharmacogenomic information | C |
| FDA | No information | |||
| Thioguanine | TPMT | EMA | N.A. | L |
| FIDMD | Recommendation | |||
| MEB | Recommendation | |||
| FDA | Strong recommendation | |||
| NUDT15 | EMA | N.A. | ||
| FIDMD | Recommendation | |||
| MEB | Recommendation | |||
| FDA | Strong recommendation | |||
| Tramadol | CYP2D6 | EMA | N.A. | N |
| FIDMD | Pharmacogenomic information | |||
| MEB | Pharmacogenomic information | |||
| FDA | Recommendation dealing with contraindication | |||
| Warfarin | CYP2C9 | EMA | N.A. | B |
| FIDMD | No information | |||
| MEB | N.A. | |||
| FDA | Strong recommendation | |||
| VKORC1 | EMA | N.A. | ||
| FIDMD | No information | |||
| MEB | N.A. | |||
| FDA | Strong recommendation |
ATC, Anatomical Therapeutic Chemical; CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; EMA, European Medicines Agency; FDA, US Food and Drug Administration; FIDMD, Federal Institute for Drugs and Medical Devices; MEB, Medicines Evaluation Board; N.A., not available.
Despite having guidelines by the CPIC or DPWG, 26% of the 58 gene–drug interactions had no pharmacogenomic information in the SmPCs by the EMA/FM or FDA (Figure 1 a). In addition, for about 27% of the gene–drug interactions, these agencies only had pharmacogenomic information without specific recommendations. Dose recommendations are given by the EMA/FM for 24% of the 58 gene–drug interactions, as compared with 16% by the FDA. On the other hand, the FDA had a higher proportion of gene–drug interactions with strong recommendations, 19% as compared with 10% by the EMA/FM, respectively. We classified label information as requiring mandatory action if the SmPC contained text for (i) indication, (ii) mandatory dosing requirements, or (iii) absolute contraindications. Only 12% of these 58 gene–drug interactions required mandatory action. All contraindications in Figure 1 a are absolute contraindications, except for the FDA's label for oxcarbazepine, which contained a relative contraindication, bringing the percentage of absolute contraindications to 10%.
Particular differences among these drugs between the EMA/FM and FDA were seen for psychoactive drugs. Whereas the EMA/FM only has nonactionable labels for aripiprazole and fluvoxamine, the FDA has a strong recommendation; similarly, only the EMA/FM provides strong recommendations for amitriptyline, escitalopram, haloperidol, phenytoin, and sertraline (Table 1).
Analysis of differences in pharmacogenomic information given by all agencies
The distribution of the 58 gene–drug interactions with actionable labels that have guidelines by the CPIC and/or DPWG and are approved by both the EMA/FM and FDA were compared as visualized in the Venn diagram in Figure 1 b. As seen, only for 13 gene–drug interactions (22.4%) common guidelines or actionable labels by all consortia and agencies are present. This compares with 10 of 54 drugs (18%). For nine (15.5%) of the gene–drug interactions guidelines are provided by both the CPIC and DPWG but no actionable labels by the EMA/FM or FDA exist. A further eight (13.8%) and seven (12.1%) gene–drug interactions that have guidelines by the CPIC or DPWG, respectively, are unaccounted for in both the EMA/FM and FDA.
Comparison between the EMA/FM and FDA
The CPIC and DPWG only provide pharmacogenomic labels for a fraction of the drugs having pharmacogenomic information in their SmPCs. Consequently, we analyzed the SmPCs from a further 450 drugs, first approved between 1941 and 2018, identified from (i) the FDA table concerning pharmacogenomic drug labels,14 (ii) from the EMA, as presented by Ehmann et al.,10 (iii) from the EMA European Public Assessment Reports website,17 and (iv) from SmPCs of all new drugs registered in 2015–2018 by the EMA (n = 140) and the FDA (n = 172). Hereby, we identified 227 drugs encompassing 277 gene–drug interactions with pharmacogenomic information or actionable labels. In Tables S2–S11 , all gene–drug interactions with an actionable label by at least one of the agencies are presented, sorted by ATC category. After excluding drugs that were not approved by both the EMA/FM and FDA, 153 drugs encompassing 191 gene–drug interactions were compared on label distribution. After removing drugs that had no actionable labels by either agency, 126 gene–drug interactions were compared on overlap in actionable labels. As pointed out before, because drugs registered before 1995 have no EMA SmPC, we grouped the European agencies EMA, FIDMD, and MEB together as EMA/FM.
The distribution of the label categories among the 191 gene–drug interactions with actionable information in the SmPCs by either the FDA and/or EMA/FM are summarized in Figure 2 a. For the EMA/FM, 42% of these SmPCs contained mandatory action requirements (indication, absolute contraindication, or mandatory) as compared with 37% by the FDA. All contraindications in Figure 2 a are absolute contraindications. The relatively greater number of drugs requiring mandatory action in this comparison as compared with the one from Figure 1 a can be explained by the high contribution of novel anticancer drugs with indication labels.
No pharmacogenomic information is present in 18% of the SmPCs by the EMA/FM as opposed to 9% by the FDA. There is also discordance in the proportion of labels with pharmacogenomic information only, with 26% for the EMA/FM as compared with 37% for the FDA. Additionally, there are more contraindications for the EMA/FM than for the FDA (7% as compared with 3%). This is partly attributed to contraindications in the EMA/FM for chloroquine, hydroxychloroquine, and sulfasalazine for patients with a G6PD deficiency, whereas the FDA labels only provide recommendations for these drugs. Regarding, for example, metoclopramide, nitrofurantoin, sevoflurane, sulfadiazine, and sulfamethoxazole, the FDA SmPCs have pharmacogenomic information only, whereas the EMA/FM states contraindications instead.
We examined to what extent the information categories in the labels from the FDA and the EMA/FM changed during 2015–2018. We found a relatively constant number of labels with actionable information (about 5–10 per year) for 2015–2017, whereas for 2018 a pronounced increase in actionable labels are presented by both the EMA/FM and the FDA (10–15 per year). The proportions can be seen in Figure S1 . The additional drug labels for 2018 mainly refer to specific somatic genome‐based indications for anticancer drugs.
Specific differences in SmPC labels between the FDA and EMA/FM
We identified the explicit differences in the SmPCs regarding drug labels between the FDA and the EMA/FM among the 191 gene–drug interactions having pharmacogenomic information or labels by one or both of the agencies. In 54 cases, the FDA and the EMA/FM showed discrepancies in label categories for gene–drug interactions, as summarized in Table 2. Although the other gene–drug interactions are in the same label category, there are qualitative differences. For example, for erlotinib, the EMA/FM does not specify the types of EGFR mutation, whereas the FDA does. A second example is lapatinib, where the EMA/FM states a combination therapy with trastuzumab in HR‐negative disease, whereas the FDA suggests a combination therapy with letrozole in HR‐positive disease.
Table 2.
Differences in label categories for additional gene–drug interactions without CPIC and/or DPWG guidelines that have a label by at least one agency, comparing the EMA (or FIDMD/MEB) and the FDA
| Drug | Gene | Institution | Therapeutic recommendation | ATC code |
|---|---|---|---|---|
| Acetylsalicylic acid/clopidogrel | G6PD | EMA | Mandatory | B |
| FDA | No information | |||
| Amifampridine | NAT2 | EMA | Pharmacogenomic information | N |
| FDA | Strong recommendation | |||
| Binimetinib | BRAF V600 | EMA | Mandatory | L |
| FDA | Indication | |||
| Brivaracetam | CYP2C19 | EMA | Pharmacogenomic information | N |
| FDA | Recommendation | |||
| Caplacizumab | Hemophilia, coagulation factor deficiencies | EMA | Recommendation | B |
| FDA | Pharmacogenomic information | |||
| Carvedilol | CYP2D6 | EMA | N.A. | C |
| FIDMD | Recommendation | |||
| MEB | No information | |||
| FDA | Pharmacogenomic information | |||
| Ceftriaxone | G6PD, nonspecific (congenital methemoglobinemia) | EMA | N.A. | J |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Chloroquine | G6PD | EMA | N.A. | P |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | N.A. | |||
| FDA | Recommendation | |||
| Clozapine | CYP2D6 | EMA | N.A. | N |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Cobimetinib hemifumarate | BRAF V600 | EMA | Mandatory | L |
| FDA | Indication | |||
| Dabrafenib | BRAF V600 | EMA | Mandatory | L |
| FDA | Indication | |||
| G6PD | EMA | No information | ||
| FDA | Recommendation | |||
| RAS | EMA | Recommendation | ||
| FDA | Strong recommendation | |||
| Dapsone | G6PD | EMA | N.A. | D |
| FIDMD | Recommendation | |||
| MEB | N.A. | |||
| FDA | Pharmacogenomic information | |||
| Nonspecific (congenital methemoglobinemia) | EMA | N.A. | ||
| FIDMD | No information | |||
| MEB | N.A. | |||
| FDA | Recommendation | |||
| Durvalumab | CD274 (PD‐L1) | EMA | Indication | L |
| FDA | Pharmacogenomic information | |||
| Eluxadoline | SLCO1B1 | EMA | Recommendation | A |
| FDA | No information | |||
| Elvitegravir/Cobicistat/Emtricitabine/Tenofovir alafenamide fumarate | HIV mutations | EMA | Indication | J |
| FDA | Pharmacogenomic information | |||
| Encorafenib | BRAF V600 | EMA | Mandatory | L |
| FDA | Indication | |||
| Evolocumab | PCSK9 | EMA | Indication | C |
| FDA | No information | |||
| Flurbiprofen | CYP2C9 | EMA | N.A. | M |
| FIDMD | N.A. | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Gemtuzumab ozogamicin | CD33 | EMA | Indication | L |
| FDA | No information | |||
| Glibenclamide | β‐cell ATP‐sensitive potassium channel and chromosome 6q24‐related transient neonatal diabetes mellitus | EMA | Indication | A |
| FDA | No information | |||
| Goserelin | ESR, PGR | EMA | N.A. | L |
| FIDMD | No information | |||
| MEB | Indication | |||
| FDA | Pharmacogenomic information | |||
| Hydroxychloroquine | G6PD | EMA | N.A. | J |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation | |||
| FDA | Recommendation | |||
| Ibrutinib | Chromosome 17p | EMA | Pharmacogenomic information | L |
| FDA | Indication | |||
| Ipilimumab | Microsatellite Instability, Mismatch Repair | EMA | No information | L |
| FDA | Indication | |||
| Isoniazid, Pyrazinamide, and Rifampin | nonspecific (NAT) | EMA | N.A. | J |
| FIDMD | Recommendation | |||
| MEB | Strong recommendation | |||
| FDA | Pharmacogenomic information | |||
| Lenalidomide | Chromosome 5q | EMA | No information | L |
| FDA | Indication | |||
| Mepivacaine | Nonspecific (Congenital Methemoglobinemia) /G6PD | EMA | N.A. | N |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Metoclopramide | CYB5R | EMA | N.A. | A |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation dealing with contraindication | |||
| FDA | Pharmacogenomic information | |||
| G6PD | EMA | N.A. | ||
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Midostaurin | FLT3 mutation | EMA | Mandatory | L |
| FDA | Indication | |||
| Necitumumab | EGFR | EMA | Indication | L |
| FDA | No information | |||
| Neratinib | ERBB2 (HER2) | EMA | Mandatory | L |
| FDA | Indication | |||
| Nitrofurantoin | G6PD | EMA | N.A. | J |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation dealing with contraindication | |||
| FDA | Pharmacogenomic information | |||
| Nivolumab | BRAF | EMA | Pharmacogenomic information | L |
| FDA | Indication | |||
| Microsatellite Instability, Mismatch Repair | EMA | No information | ||
| FDA | Indication | |||
| Olaparib | BRCA/ERBB2 (HER2)/ESR, PGR | EMA | Mandatory | L |
| FDA | Indication | |||
| Pantoprazole | CYP2C19 | EMA | No information | A |
| FDA | Recommendation | |||
| Pembrolizumab | Microsatellite Instability, Mismatch Repair | EMA | No information | L |
| FDA | Indication | |||
| Peramivir | Influenza virus genotype | EMA | Recommendation | J |
| FDA | Pharmacogenomic information | |||
| Pertuzumab | ERBB2 (HER2) | EMA | Mandatory | L |
| FDA | Indication | |||
| Piroxicam | CYP2C9 | EMA | N.A. | M |
| FIDMD | N.A. | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Ranolazine | CYP2D6 | EMA | Recommendation | C |
| FDA | No information | |||
| Rucaparib | UGT1A1 | EMA | Recommendation | L |
| FDA | No information | |||
| BRCA | EMA | Mandatory | ||
| FDA | Indication | |||
| Sevoflurane | Nonspecific (Genetic Susceptibility to Malignant Hyperthermia) | EMA | N.A. | N |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation dealing with contraindication | |||
| FDA | Pharmacogenomic information | |||
| Succinylcholine | BCHE | EMA | N.A. | V |
| FIDMD | N.A. | |||
| MEB | Recommendation | |||
| FDA | Strong recommendation | |||
| Sulfadiazine | G6PD | EMA | N.A. | J |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation | |||
| FDA | Pharmacogenomic information | |||
| Sulfamethoxazole and Trimethoprim | G6PD | EMA | N.A. | J |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation | |||
| FDA | Pharmacogenomic information | |||
| Sulfasalazine | G6PD | EMA | N.A. | A |
| FIDMD | Recommendation dealing with contraindication | |||
| MEB | Recommendation dealing with contraindication | |||
| FDA | Recommendation | |||
| Trametinib | BRAF V600 | EMA | Mandatory | L |
| FDA | Indication | |||
| Trastuzumab | ERBB2 (HER2) | EMA | Mandatory | L |
| FDA | Indication | |||
| ESR, PGR | EMA | Indication | ||
| FDA | Pharmacogenomic information | |||
| Tretinoin | PML‐RARA | EMA | N.A. | D |
| FIDMD | N.A. | |||
| MEB | No information | |||
| FDA | Recommendation | |||
| Vandetanib | RET | EMA | Recommendation | L |
| FDA | No information | |||
| Vemurafenib | RAS | EMA | Recommendation | L |
| FDA | Pharmacogenomic information | |||
| BRAF V600 | EMA | Mandatory | ||
| FDA | Indication | |||
| Vincristine | BCR‐ABL1 (Ph+) | EMA | N.A. | L |
| FIDMD | No information | |||
| MEB | No information | |||
| FDA | Indication | |||
| Vortioxetine | CYP2D6 | EMA | Recommendation | N |
ATC, Anatomical Therapeutic Chemical; CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; EMA, European Medicines Agency; FDA, US Food and Drug Administration; FIDMD, Federal Institute for Drugs and Medical Devices; MEB, Medicines Evaluation Board; N.A., not available.
The overlap in actionable labels for drugs registered by both the EMA/FM and the FDA was investigated. We included the 126 gene–drug interactions where at least one of the agencies had such labels in the SmPCs in question and the data are shown in a Venn diagram (Figure 2 b). Of the 126 gene–drug interactions, 82 (65.6%) had actionable labels by both the EMA/FM and the FDA, whereas the EMA/FM and the FDA had 25 (20.0%) and 19 (15.2%) additional actionable gene–drug interactions, respectively (Table S12 ). When looking at the overlap in actionable labels, it was found that drugs in ATC group L (antineoplastic and immunomodulating agents) did overlap significantly more (75%) than drugs in groups A (alimentary tract and metabolism), J (antiinfectives for systemic use), and N (nervous system; 53–58%), most likely due to the fact that most of the new anticancer drugs have indication labels.
Comparison of the drugs with actionable labels by all agencies
We compared the consensus of drug SmPC pharmacogenomic labeling by all agencies investigated, namely the CPIC, DPWG, EMA/FM, and FDA. As shown in Table 3, such consensus was only found for 10 (18%) of the 54 having any actionable label by all agencies. Comparing the FDA with the EMA/FM reveals that among the total number of gene‐drug interactions with actionable labels (54 + 126 = 180) there was only a 54.3% concordance between the agencies. [Correction added on 10th January, 2020, after first online publication: In the preceding sentence, “the total number of drugs with actionable labels” has been changed to “the total number of gene‐drug interactions with actionable labels”]. Indeed taken together, this strongly indicates a lack of true consensus for pharmacogenomic advice and/or labels in the drug SmPCs and indicates a necessity to revisit and reconsider previous decisions made in this respect.
Table 3.
Drugs that have guidelines/actionable labels by all four agencies (CPIC, DPWG, EMA (or FIDMD/MEB))
| Drug | Gene | CPIC | DPWG | EMA | FIDMD | MEB | FDA |
|---|---|---|---|---|---|---|---|
| Abacavir | HLA‐B | Abacavir is not recommended due to the risk of hypersensitivity reactions | Contraindicated due to the risk of hypersensitivity reactions | Mandatory: HLA‐B*5701 status must always be documented prior to initiating therapy | Recommendation dealing with contraindication | ||
| Allopurinol | HLA‐B | Contraindicated due to the risk of hypersensitivity reactions | Consider alternative drug (febuxostat) or dose adjustment | Recommendation: Screening for HLA‐B*5801 should be considered before starting treatment with allopurinol in patient subgroups where the prevalence of this allele is known to be high | Recommendation: Immediately discontinue at first signs of skin rash or allergic reactions | ||
| Atomoxetine | CYP2D6 | Consider dose adjustment | In case of efficacy with adverse reactions, lower the dose and monitor if efficacy is maintained or consider alternative (clonidine) | N.A. | Recommendation: A lower initial dose and a slower titration may be considered | Recommendation: Consider a lower starting dose and slower up titration of the dose | Strong recommendation: Specific dose adjustment |
| Azathioprine | TPMT | Consider alternative drug or dose adjustment | Consider alternative drug or dose adjustment | N.A. | Recommendation: Dose adjustment | Recommendation: Dose adjustment | Recommendation: Alternative drug or dose reduction is recommended |
| NUDT15 | Consider alternative drug or dose adjustment | Consider alternative drug or dose adjustment | N.A. | Recommendation: Dose adjustment and monitoring of blood levels | Recommendation: Dose adjustment | Recommendation: Consider alternative drug | |
| Capecitabine/5‐fluorouracil | DPYD | Select alternative drug or dose adjustment | Select alternative drug or dose adjustment | Mandatory: Patients with partial G6PD deficiency must be treated with extreme caution and frequent monitoring with dose adjustment according to toxicity | Recommendation: Select alternative drug | ||
| Carbamazepine | HLA‐B | If patient is carbamazepine‐naive, do not use carbamazepine | Choose alternative drug | N.A. | Recommendation: Do not use carbamazepine unless no alternative drug is available | Recommendation: Do not use carbamazepine unless no alternative drug is available | Recommendation dealing with contraindication |
| Citalopram | CYP2C19 | Consider dose adjustment or alternative drug | Consider dose adjustment | N.A. | Strong recommendation: Specific dose adjustment | Strong recommendation: Specific dose adjustment | Strong recommendation: Specific dose adjustment |
| Codeine | CYP2D6 | Avoid codeine use due to potential for toxicity | Codeine is contraindicated in CYP2D6 ultra‐rapid metabolizers due to the risk of overdose | N.A. | Pharmacogenomic information | Recommendation dealing with contraindication | Recommendation dealing with contraindication |
| Mercaptopurine | TPMT | Consider alternative drug or dose adjustment | Select alternative drug or dose adjustment | Recommendation: Consider a substantial dose reduction | Strong recommendation: Specific dose adjustment | ||
| NUDT15 | Dose adjustment or alternative drug | Select alternative drug or dose adjustment | Recommendation: Dose adjustment | Strong recommendation: Specific dose adjustment | |||
| Thioguanine | TPMT | Consider dose adjustment | Select alternative drug or dose adjustment | N.A. | Recommendation: Consider dose adjustment | Recommendation: Dose reduction is usually necessary. Monitor blood count closely. | Strong recommendation: Specific dose adjustment |
| NUDT15 | Consider dose adjustment | Consider alternative drug or dose adjustment | N.A. | Recommendation: Genotypic analysis should be considered before initiation of thiopurine therapy to determine the NUDT15 genotype | Recommendation: Dose adjustment | Strong recommendation: Specific dose adjustment |
CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; EMA, European Medicines Agency; FIDMD, Federal Institute for Drugs and Medical Devices; MEB, Medicines Evaluation Board; N.A., not available.
Discussion
The results presented show a considerable extent of variability in the information for different pharmacogenomics biomarkers by the CPIC, DPWG, as well as by the regulatory bodies of the EMA/FM and the FDA. Such variability in judgment between the clinical and regulatory agencies can be anticipated to be inherent to the different manners by which they evaluate available information. The data in the literature are often difficult to assess and it is commonly paired with obstacles to compare different studies in order to reach a consensus. This depends to a great extent on (i) differences or absence of definition of the patient populations, (ii) the limited power of many studies due to a low sample size, and (iii) the low number of patients carrying the variant alleles in particular. Moreover, most positive association studies lack validation of findings in an independent patient population. Furthermore, different decisions made in various groups of experts are partly dependent on the issue that there are no firm facts that have to be fulfilled for each type of label and many decisions are based on more subtle data and different interpretations of the published results. For the future, it is anticipated that larger, better designed studies should contribute to a reduction in the number of discrepant assessments regarding clinically valuable pharmacogenomic labels.
The clinical implementation of pharmacogenomic labels would considerably gain from a higher extent of consensus between the different agencies and consortia and attempts should be made to harmonize where possible and the summary here presented would facilitate such work. It is evident that the CPIC and DPWG guidelines are very thorough and constructed by many different experts in the field, showing a high level of consistency.18 However, the lack of compliance in the labels provided by the regulatory agencies, as shown here, is disappointing. It is also a complicating factor that the labels defined by the FDA compared with the EMA/FM among the 184 gene–drug interactions (58 in comparison 1 and 126 in comparison 2) with actionable labels investigated only have 54.3% concordance. The lack of concordance in drug labeling between different agencies was previously noted for 10 of the most prescribed psychiatric drugs elsewhere,19 further supporting the need for increased integration between agencies.
In general, an important note to be made is that there are sometimes differences in the SmPCs of different companies or drug formulations, despite having the same active ingredient. Examples include the German labels for irinotecan (where some SmPCs do not mention UGT1A1) and amitriptyline (tablet formulations mention dosage reduction based on CYP2C19 and CYP2D6, whereas the oral solution does not state any action). In the past, Pfistermeister et al.20 have also shown that there are differences in labeling in SmPCs of generics with the same active ingredient. Such discrepancies further cause important inconsistencies to the label system. Furthermore, because we selected one SmPC to represent each active pharmaceutical ingredient, differences in labeling within the same drugs from different companies cannot be excluded or accounted for.
Comparisons between drug labels have previously been published3, 10, 18, 21 and many drug labels have recently been included in a registry by Pharmacogenomics Knowledgebase.22 However, in the present study, we focused on the discrepancies and the reasons behind them as well as on information given explicitly in the drug SmPCs. The Pharmacogenomics Knowledgebase registry is based on the EMA European Public Assessment Reports, whereas in this investigation we have focused on the information given in the SmPCs of the drugs (i.e., the true applicable information to the clinicians).
A limitation of previous publications in the field is the absence of consideration of European labels, because drugs registered before 1995 have not been registered by the EMA. For this reason, we introduced drug labels as provided by the Dutch (CBG‐MEB) and German (FIDMD) medical product agencies, which account for nearly 30% of the European drug labels investigated in our study. Furthermore, in a previous publication regarding the drug labels provided by the EMA, a great portion of the labels mostly concerned drug–drug interactions, rather than true pharmacogenomic data.10
A major challenge is to reach harmonization in pharmacogenomic recommendations between the consortia and agencies. A simple first step would be to consider updating the SmPCs with labels based on the drugs for which there is a consensus among guidelines by the CPIC and DPWG. Second, as the role of genetics in treatment decisions will only increase in the future, adequate preparation for such a future is essential, which involves continuous evaluation of progress in the field of pharmacogenomics and continuously reviewed recommendations in the SmPCs. Third, it is important that inconsistencies in pharmacogenomic advice given in SmPC sheets dealing with different formulations of the same active ingredients are identified and corrected. Overall, these results do indicate the necessity of construction of strategic rules for pharmacogenomic labeling common to all different regulatory agencies.
Another challenge in the pursuit of clinical implementation is to increase awareness and compliance to the pharmacogenomic labels among physicians. As has recently been indicated, there is still a lack of compliance among clinicians, even when mandatory preemptive genotyping is included.23 It is, therefore, of paramount importance that clinicians are engaged in this process and involved in the establishment and refinement of future guidelines and the identification of pharmacogenomics drug labels in collaboration with the pharmacogeneticists and regulatory agencies.
In conclusion, of 54 drugs with an actionable gene–drug interaction in the CPIC and DPWG guidelines, < 50% had actionable pharmacogenomic information in the labels in the SmPCs of the EMA/FM and the FDA. Only 18% of the cases were in agreement among the CPIC, DPWG, FDA, and EMA/FM. The consensus of actionable pharmacogenomic labels of 184 different gene–drug interactions between the FDA and EMA/FM was only 54%. We conclude that there is a need for critical evaluation of the current pharmacogenomic drug labels and for much work to harmonize the priorities, procedures, and validation of pharmacogenomic drug labels in order to take the field further for a more successful clinical implementation.
Funding
The research is supported by a grant from European Union's Horizon 2020 research and innovation programme under grant agreement no. 668353.
Conflict of Interest
The authors declared no competing interests for this work.
Author Contributions
R.S., L.S., J.S., and M.I.S. wrote the manuscript. R.S., L.S., J.S., and M.I.S. designed the research. R.S. and L.S. performed the research. R.S., L.S., and M.I.S. analyzed the data.
Supporting information
Figure S1. Comparison of the pharmacogenomic labeling for novel drug approvals of drugs approved by both the European Medicines Agency and US Food and Drug Administration in 2015–2018.
Supplementary Tables S1–S12. Supplementary Tables S1‐S12.
Acknowledgment
We thank Dr. Falk Ehmann, of the EMA, for valuable information.
References
- 1. Ingelman‐Sundberg, M. , Sim, S.C. , Gomez, A. & Rodriguez‐Antona, C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 116, 496–526 (2007). [DOI] [PubMed] [Google Scholar]
- 2. Eichelbaum, M. , Ingelman‐Sundberg, M. & Evans, W.E. Pharmacogenomics and individualized drug therapy. Annu. Rev. Med. 57, 119–137 (2006). [DOI] [PubMed] [Google Scholar]
- 3. Lauschke, V.M. , Zhou, Y. & Ingelman‐Sundberg, M. Novel genetic and epigenetic factors of importance for inter‐individual differences in drug disposition, response and toxicity. Pharmacol. Ther. 197, 122–152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matthaei, J. et al Heritability of metoprolol and torsemide pharmacokinetics. Clin. Pharmacol. Ther. 98, 611–621 (2015). [DOI] [PubMed] [Google Scholar]
- 5. Ingelman‐Sundberg, M. , Mkrtchian, S. , Zhou, Y. & Lauschke, V.M. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genom. 12, 26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van der Wouden, C.H. et al Development of the PGx‐passport: a panel of actionable Germline genetic variants for pre‐emptive pharmacogenetic testing. Clin. Pharmacol. Ther. 106, 866–873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Relling, M.V. & Klein, T.E. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 89, 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swen, J.J. et al Pharmacogenetics: from bench to byte an update of guidelines. Clin. Pharmacol. Therapeut. 89, 662–673 (2011). [DOI] [PubMed] [Google Scholar]
- 9. Drozda, K. , Pacanowski, M.A. , Grimstein, C. & Zineh, I. Pharmacogenetic labeling of FDA‐approved drugs: a regulatory retrospective. JACC Basic Transl. Sci. 3, 545–549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehmann, F. et al Pharmacogenomic information in drug labels: European Medicines Agency perspective. Pharmacogenomics J. 15, 201–210 (2015). [DOI] [PubMed] [Google Scholar]
- 11. Swen, J.J. et al Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med. 4, e209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CBG‐MEB . Geneesmiddeleninformatiebank <https://www.geneesmiddeleninformatiebank.nl/nl/>. Accessed January 1, 2019.
- 13. Fachinfo Service <https://www.fachinfo.de/>. Accessed January 1, 2019.
- 14. US Food and Drug Administration . Table of Pharmacogenomic Biomarkers in Drug Labeling <https://www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling>.
- 15. US Food and Drug Administration (FDA) . Drugs@FDA <https://www.accessdata.fda.gov/scripts/cder/daf/>. Accessed June 15, 2019.
- 16. Bioinformatics & Evolutionary Genomics . Venn Diagram tool. Ghent University; <http://bioinformatics.psb.ugent.be/webtools/Venn/>. [Google Scholar]
- 17. European Medicines Agency . European Public Assessment Reports (EPAR) <https://www.ema.europa.eu/en/search/search/field_ema_web_categories%25253Aname_field/Human?search_api_views_fulltext=epars>. Accessed July 18, 2018.
- 18. Bank, P. et al Comparison of the guidelines of the clinical pharmacogenetics implementation consortium and the Dutch pharmacogenetics working group. Clin. Pharmacol. Ther. 103, 599–618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfistermeister, B. et al Different indications, warnings and precautions, and contraindications for the same drug ‐ an international comparison of prescribing information for commonly used psychiatric drugs. Pharmacoepidemiol. Drug Saf. 22, 329–333 (2013). [DOI] [PubMed] [Google Scholar]
- 20. Pfistermeister, B. et al Inconsistencies and misleading information in officially approved prescribing information from three major drug markets. Clin. Pharmacol. Ther. 96, 616–624 (2014). [DOI] [PubMed] [Google Scholar]
- 21. Imatoh, T. , Sai, K. & Saito, Y. Pharmacogenomic information in the warning section of drug labels: a comparison between labels in the United States and those in five other countries/regions. J. Clin. Pharm. Ther. 43, 493–499 (2018). [DOI] [PubMed] [Google Scholar]
- 22. Pharmacogenomics Knowledgebase (PharmGKB) . Drug Label Information and Legend <https://www.pharmgkb.org/page/drugLabelLegend>.
- 23. Roberts, J.P. Pharmacogenomics: better drugs through better screening Science <https://www.sciencemag.org/features/2018/09/pharmacogenomics-better-drugs-through-better-screening>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of the pharmacogenomic labeling for novel drug approvals of drugs approved by both the European Medicines Agency and US Food and Drug Administration in 2015–2018.
Supplementary Tables S1–S12. Supplementary Tables S1‐S12.


