Abstract
The cellular complexity and functional diversification of the human immune system necessitates the use of high-dimensional single-cell tools to uncover its role in multifaceted diseases including many rheumatic diseases as well as other autoimmune and inflammatory disorders. Next-generation immune monitoring technologies, such as mass cytometry (CyTOF), and analogous imaging approaches, including Multiplexed Ion Beam Imaging (MIBI), have evolved from their low-dimensional counterparts, flow cytometry and immunohistochemistry. Here, we introduce the underlying technologies and discuss aspects necessary for their successful implementation, including study design principles and analytical tools for the discovery of stratifying features. We highlight successful studies leveraging these technologies to identify functional biomarkers and potential therapeutic targets in several rheumatic diseases and conclude with a perspective on recent developments and future directions.
Introduction
Technological advances are a major driver of scientific insight across many areas of science. Given the complexity of the human immune system, immunology in particular, has benefitted from innovations in various single-cell technologies1. Such technologies now facilitate the in-depth study of immune cell composition, activation and their relation to disease. Dysregulation and aberrant activation of regulatory processes within the immune system are believed to play a crucial part in the development of various rheumatic diseases (RDs) and autoimmunity in general. RDs comprise a heterogeneous set of disorders including diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), Sjörgen’s syndrome (SjS) and systemic sclerosis (SSc). Many of these RDs share characteristics that point towards the immune system as a central player in their etiology. Firstly, genome-wide association studies (GWAS) have uncovered the HLA antigen presenting machinery as a major genetic risk factor for the development of many RDs2–4. Antigen presentation is often followed by other immune-related genetic associations, e.g. genes regulating cytokine production5. Genetic contribution to disease risk varies across different RDs, but in general, a combination of genetic predisposition and additional environmental insults are thought to be key events in RD etiology. For example, viral infections with the Epstein-Barr virus (EBV) and associated immune activation have been found to be associated with many RDs2,3,6–8. Inflammation and other processes can in turn prompt protein modifications (e.g. citrullination [ref:5]) or epigenetic modifications4, potentially creating novel autoantigens. Given an inflammatory microenvironment, antigen presenting cells may display such modified antigens, thus initiating an adaptive immune response. For many RDs, these are characterized by the interplay of T cells, in particular follicular T helper (TFH) cells, and B cells2,3,5. Production of autoantibodies by terminally differentiated plasma cells is a shared feature of many RDs that contributes to disease pathology and is often of high diagnostic relevance3,5,6. Lastly, immune-targeted interventions such as modulation of tumor necrosis factor (TNF) signaling or B cell depletion are therapeutically successful approaches across many RDs, again underlining the importance of the immune system in these diseases.
Given this interplay of external factors with innate and adaptive immunity, the ongoing interaction and communication within the immune system as well as the pathogenic properties of certain subsets, systems-level analysis of the immune system in RD promises better understanding of these pathological processes9–11, including: the discovery of novel biomarkers to predict treatment response and to guide therapeutic decision making12,13, the identification of therapeutic cell targets to reduce risk of infection associated with broadly immunosuppressive therapy, as well as to address patients not responding to currently available therapeutics. Importantly, identified biomarkers are often not only predictive, but also intrinsically linked to the disease mechanism and can therefore serve as new therapeutic targets as well14–16.
The complexity of human immune cell compositions, their numerous functional states, and localizations requires adequate methodologies for their comprehensive assessment. In this context, ‘comprehensive immune monitoring’ refers to the ability to robustly identify all major immune cell lineage and their most important functional subsets and to assign all cells in a given sample accordingly17. Of note, robustly determining the absence of immune populations can also provide valuable insight, e.g. to reveal populations with altered trafficking properties or to monitor the success of B cell depleting antibodies. Given their high-dimensional capabilities combined with the throughput and robustness to analyze samples across large cohorts, mass cytometry (CyTOF) and closely related imaging technologies, such as multiplexed ion beam imaging (MIBI), are particularly suited for the comprehensive immune monitoring of biospecimens from patients with RD18.
In this Review, we introduce the underlying technologies of these approaches, as well as recent developments which have greatly accelerated their adoption in clinical settings. Further, we provide a practical guideline and considerations for the design of immune monitoring studies and subsequent approaches for the comprehensive and unbiased analysis of these datasets. We conclude with examples of successful applications of these technologies as drivers for novel insights in RDs and provide our view on future possibilities and challenges involved in high-dimensional immune monitoring.
Mass cytometry for next generation immune-monitoring
For many years, immune monitoring and much of immunological research has relied on flow cytometry and immunohistochemistry (IHC) or immune fluorescence (IF) approaches to capture and quantify cellular heterogeneity and its relation to disease. The low number of parameters that could be analyzed simultaneously forced restricted analyses in terms of cellular properties and composition. Consequently, most experiments could only target a single cell type with additional answers coming at the sacrifice of time and more clinical material. More recently, single-cell parametrization has increased through the development of novel fluorophores and laser systems, driving the discovery of new immune cell sub-classes as well as important functional states. However, physical limitations have impaired the addition of further parameters in fluorescence-based cytometry due to the crowding of the visible spectrum of wavelengths, resulting in spectral overlap between different analysis channels in complex cytometry experiments that still fail to comprehensively capture a snapshot of the whole immune state simultaneously. A solution to this problem was found by substituting the fluorescence-based reporters used in flow cytometry for non-biological elemental isotopes and an inductively coupled plasma mass spectrometry (ICPMS)-based readout, subsequently termed mass cytometry or cytometry by time-of flight (CyTOF)19–21.
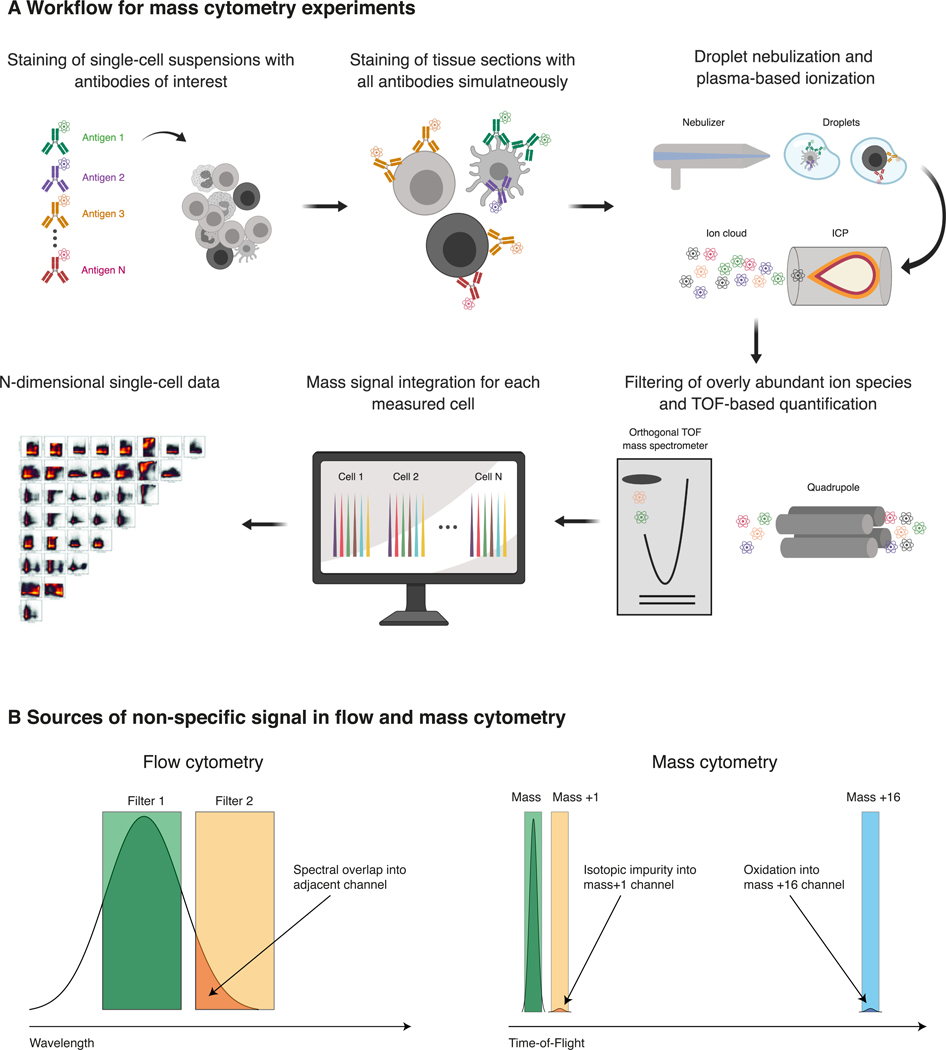
For mass cytometry, instead of fluorophores, antibodies are tagged with heavy-metal reporter ions through covalent conjugation with chelating polymers22–25. Following staining with these antibodies, single-cell suspensions are introduced into the CyTOF analyzer where they are first nebulized into droplets and subsequently vaporized, atomized, and ionized through introduction to an inductively coupled plasma (ICP) (Figure 1A). The resulting ion cloud is then mass-filtered to remove biologically abundant ion species and finally analyzed by time-of-flight (TOF) mass spectrometry to quantify the abundance of all the isotopic reporter masses in parallel, thus enabling quantification of bound antibodies and therefore expression of the target of interest (Figure 1A).
Figure 1: Principles of mass cytometry.
(A) Experimental workflow for analyzing single-cell suspensions by mass cytometry. In a first step, single-cell suspensions are incubated with heavy-metal labeled antibodies and other reporter probes of interest (e.g. MHC multimers to detect antigen-specific cells or probes discriminating live from dead cells). Antibodies against intracellular proteins can be used following common fixation and permeabilization protocols. Following a series of washing steps, cells are introduced into the CyTOF mass cytometer where the suspension is first nebulized into small droplets and subsequently introduced into an ICP that breaks down droplet-contents into a cloud of elemental ions. Ion species with low atomic masses (e.g. hydrogen and carbon) are removed through a quadrupole and the remaining higher atomic mass ions (which were conjugated to the respective probes) are subsequently quantified using an orthogonal TOF mass spectrometric detection system. Ion counts are then integrated to derive levels of bound antibodies and thus single-cell target-abundances. This high-dimensional data is then exported for further downstream analysis. (B) In flow cytometry (left), the major source of non-specific signal (in some cases >50% of the specific signal) is spectral overlap. Fluorophore emission spectra are usually broad and overlap into adjacent analysis channels, resulting in unspecific signal in this channel. Additional sources are cellular autofluorescence and fluorophore degradation (not shown). Considerable effort has to be invested in panel design to account for these effects and signals have to be corrected computationally. In mass cytometry (right), antibodies are conjugated to elemental isotopes which have non-overlapping masses that can be resolved through their time of flight. Minor sources of overlap (usually ~1%) are isotopic impurity of the metal stocks and their oxidation, thus facilitating straightforward, “plug-and-play” panel design of large panels (up to 50 antibodies).
Some of the key characteristics of flow and mass cytometry are directly compared in Table 1. While staining procedures and sample handling are comparable to flow cytometry, mass cytometry drastically reduces issues related to spectral overlap between different analysis channels (Figure 1B). The heavy-metal isotopes used to tag antibodies for mass cytometry possess non-overlapping atomic masses which can be accurately resolved and quantified through the above-mentioned time-of-flight mass spectrometry detection system.
Table 1. Key characteristics of currently available flow and mass cytometry analysis platforms.
This table has been adapted and updated from Bendall et al. (2012) Trends in Immunology 152.
| Flow Cytometry | Mass Cytometry | ||
|---|---|---|---|
| Measurement basis | Fluorescent probes | Stable mass isotope probes | |
| Sources of non-specific signal (% of specific signal) | High (10–50%) | Spectral overlap | |
| Intermediate (5–10%) | Autofluorescence, Fluorophore degradation | ||
| Low (<5%) | Isotopic impurity, Spectral overlap, Oxidation | ||
| Maximum no. of measurements | 20 (theoretically ~40) | 60 (theoretically ~120) | |
| Panel design complexity (no. of probes) | Easy | < 8 | < 40 |
| Moderate | 8–12 | 40–60 | |
| Hard | > 12 | ||
| Relative probe sensitivity (arbitrary units) | 0.1–10 | 1–3 | |
| Sampling efficiency | > 90% | ~ 50% | |
| Throughput: Measured cells/s (typical) | 500 – 40 000 (5000) | 50 – 1 000 (500) |
The availability of many different heavy metal isotopes increases the multiplexing capacity of mass cytometry to currently ≥60 separate analysis channels. This number is steadily increasing, for example through advances in conjugation chemistry approaches for the chelation of non-lanthanide metals such as yttrium, palladium, indium, bismuth and platinum26–28. It should be mentioned that the employed heavy-metal isotopes are not commonly found in unstained samples, thus eliminating biological background and issues related to autofluorescence. Rare exceptions are the treatment of cancer patients with the platinum-containing cytostatic drug cisplatin or the use of gadolinium-containing contrast agents employed in medical imaging. Interestingly, residual heavy-metals might be detectable in cells isolated from these patients shortly after administration, which has also been leveraged to study the biodistribution of these compounds in patients29.
Since its inception, there have been numerous improvements that have increased the utility of mass cytometry for widespread adaptation in immune monitoring and phenotyping21. First, adopting cellular barcoding approaches to mass cytometry enables the staining, processing and acquisition of multiple samples as a single composite sample. This drastically reduces technical variation between samples, reagent consumption, and analysis time which is especially beneficial in clinical settings where large groups of samples are to be compared. Barcoding can minimize batch effects and thus allows for large screens e.g. of phosphorylation states and their perturbations30. Furthermore, it improves the identification and exclusion of cellular doublets crucial for single-cell analysis31. Barcoding is now not only available for fixed samples but also for live cells including cells of non-hematopoietic origin32–34. Furthermore, normalization methods using bead-standards or biological control samples greatly improve mass cytometry comparability across time, batches and study sites35–37.
Mass cytometry can be employed to analyze a wide variety of cellular features of interest. Antibodies targeting surface molecules can be used to obtain lineage identity and in-depth phenotypes of heterogeneous cell populations. Furthermore, the chemically stable elemental reporters reagents are broadly compatible with common fixation and permeabilization procedures, facilitating quantification of a variety of intracellular targets in combination with the cellular identity30. For example, quantification of transcription factors can give insight into master regulators and their involvement in fate decisions and the maintenance of lineage programs38. Furthermore, intracellular cytokine staining can be used to analyze high-dimensional patterns of cytokine co-production across a wide variety of cell types39,40. Another possible intracellular readout are highly multiplexed phosphorylation levels reporting on functional cell states and their responses to external stimuli30. Additional features regularly assessed in mass cytometry are DNA content through the use of metal-containing intercalators22 and cellular viability through covalent binding of cisplatin or analogous palladium-containing compounds to membrane compromised cells34,41. Further, mass cytometry can reveal cell cycle states through the combination of a small number of antibodies and the incorporation of 5-iodo-2-deoxyuridine (IdU) which can be directly analyzed by CyTOF42. This can be further extended to simultaneously capture de novo transcriptional and translational activity in single cells using 5-bromo-2-uridine (BrU) and puromycin, respectively43.
Another exciting development is the use of reporter probes for enzymatic activities or cellular states such as cellular hypoxia which can be assessed using tellurium containing probes44. Phagocytic activity of differentially polarized myeloid cells has recently been assessed using metal-labeled targets to quantify their uptake45. Furthermore, making use of mRNA hybridization and rolling circle signal amplification, RNA transcripts have been analyzed together with proteomic targets by mass cytometry46,47. Lastly, this technology allows the multiplexed analysis of a wide array of antigen-specific T cells through the use of metal-conjugated tetramers48–52. Taken together, mass cytometry enables in-depth cellular characterization across a variety of biological functions which importantly, can be assessed simultaneously at a single-cell level, thus providing insights into their co-regulation and potential relevance for disease.
Emergence of high-dimensional imaging technologies
Like conventional flow cytometry, mass cytometry is uniquely suited to perform highly multiplexed, single-cell analysis on a wide range of parameters from cell suspensions such as blood or other liquid biopsies. Peripheral blood, for instance, is available for many RD research scenarios. Still, RD typically manifests in solid tissue itself, thus tissue-based imaging methods are required to investigate cells in their native or pathological context. Spatial analysis provides insight into the cellular microenvironment as well as cell-cell interactions, both important contributors to cellular identity and functional state in assessing biological contexts53.
IHC on formalin fixed paraffin embedded (FFPE) samples has been the predominant methodology for pathological analysis of human tissues, followed by IF. Relying on chromogenic substrates and fluorescence emission respectively, both technologies have a limited multiplexing capacity of approximately up to 4 simultaneous measurements under routine conditions. To address these limitations, several approaches have recently been developed with the aim of increasing the number of parameters represented in these images. One set of approaches relies on the serial acquisition of a small set of markers across many cycles. Usually, these technologies employ fluorescently labeled antibodies which are read in multiple cycles of staining, imaging and quenching54–56. A recent variation of this, termed CODEX, restricts cycling to several rounds of imaging, fluorophore reporter removal and subsequent acquisition of additional reporters57. While these fluorescence-based technologies do have the ability to acquire high-dimensional images, certain limitations and challenges remain. Cyclic staining, with repeated exposure to reporter stripping chemicals, can lead to changes in epitope accessibility and altered tissue morphology making optimization of staining order challenging and reassembly of cyclic image stack less robust. Tissues, particularly those that are archival FFPE, suffer from background signal due to autofluorescence, limiting assay sensitivity and often requiring additional analytical compensation58. Additionally, chromogenic and fluorophore dyes are often not chemically stable making stability for long-term storage challenging. Further, the nature of cyclic methods is incompatible with subsequent re-acquisitions under different imaging conditions (i.e. a first pass low resolution scan followed by a higher resolution acquisition). Analogous to the development of mass cytometry from flow cytometry, two related technologies, termed imaging mass cytometry (IMC) [ref: 59] and multiplexed ion beam imaging (MIBI) [ref: 60,61], have been developed to replace the fluorescence detection with elemental mass reporters for epitope quantification in an imaging context.
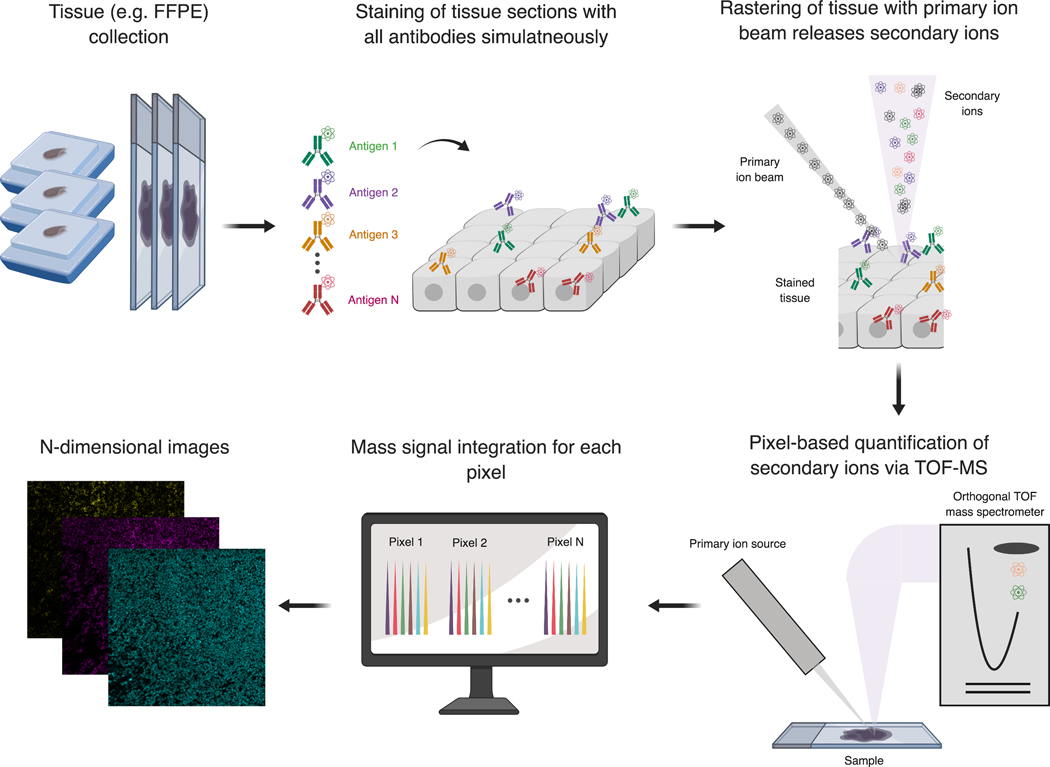
For both technologies, samples including FFPE and cryopreserved tissues are mounted onto a slide and stained with heavy-metal isotope-tagged antibodies or other reporter probes similar to IHC or IF procedures (Figure 2). In contrast to cyclic approaches, samples can be stained with all antibodies and reporter tags at once, no serial staining steps are necessary. Stained slides are then introduced into the respective analyzers. In the IMC, following staining, a laser system is used to ablate the stained tissue pixel by pixel. The resulting ablated material is then introduced into a CyTOF analyzer where it is ionized through an ICP and the elemental masses are quantified similar to whole cells by mass cytometry. The current commercial IMC implementation enables 1 μm image resolution and acquires approximately 100 such 1μm pixels per second62,63. In terms of sensitivity, approximately 50 copies of an epitope are required per pixel for minimal detection.
Figure 2: MIBI for high-dimensional imaging analysis of tissue sections.
Tissue sections (e.g. FFPE blocks) are placed on a slide with a conductive surface and subsequently stained with all antibodies and other reporter-probes in a single staining step. Stained sections are then placed into the MIBI analyzer and introduced into a vacuum chamber. The MIBI instrumentation rasters the tissue sections with a primary ion beam. Upon collision with the stained section, secondary ions (including those introduced through the binding of the heavy-metal tagged antibodies) are liberated from the sample. This cloud of secondary ions is then focused through a series of lenses and introduced into an orthogonal TOF mass spectrometer. Detected signal is then integrated and translated into ion counts per pixel. These represent multi-dimensional images that can be directly visualized or further analyzed using various image analysis pipelines. Parts of this figure were adapted from Keren et al.61.
In contrast to laser-ablation based IMC, MIBI utilizes the principle of secondary ionization mass spectrometry. For MIBI, stained slides are introduced into an analysis chamber under vacuum where elemental reporters are sputtered with submicron resolution by rastering pixel by pixel with a primary ion beam (Figure 2). As the primary ions collide with the sample, they liberate secondary ions, including the heavy-metal isotope reporters introduced through antibody staining from the tissue. While the laser-based IMC approach completely ablates tissue at each pixel, preventing reanalysis and detracting from image quality in thicker tissue sections, only few hundred nm of tissue are liberated with each MIBI scan. This allows the same cells in the same tissue section to be re-analyzed multiple times, e.g. to create low-resolution overviews of large tissue areas, identify specific regions of interest and subsequently perform high-resolution analysis of these areas, or even create 3-dimensional reconstructions.
In MIBI, the liberated secondary ions, already in the vacuum, are directly introduced into the detection system without further ionization or transfer which increases its sensitivity to as little as a single copy of an epitope per pixel. While the first MIBI implementation made use of a magnetic sector mass spectrometer, next-generation instrumentation (termed MIBI-TOF) now makes use of a full TOF detection system (Figure 2), thus allowing a large range of atomic masses to be analyzed, including naturally occurring elements such as phosphorus or iron which have important biological functions61. This current MIBI-TOF instrumentation can acquire pixel resolutions of ~250 nm at a rate as high 10’000 pixels per second61,64.
Given the similarities of the staining procedures to IHC and IF, there is a wide variety of commercially available antibodies which can be adopted for their use in IMC and MIBI. Besides antibodies, alternative reporter probes as employed in mass cytometry can be adapted to these imaging technologies. For example, simultaneous imaging of mRNA and protein epitopes has recently been demonstrated by IMC65. To identify broader tissue structures and extracellular features, a ruthenium-based dye has been proposed, resulting in counterstaining analogously to hematoxylin routinely used in IHC66. In situ detection of antigen specific T cells has been previously demonstrated in an IHC and IF context and should therefore be also feasible by IMC and MIBI67–69.
Important factors for the implementation of these technologies in clinical settings involving large groups of patients are the throughput and robustness of these platforms. Two recent publications used IMC to create an image-based map of type 1 diabetes (T1D) spanning cohorts of 12–18 patients and multiple areas within each sample62,63. Demonstrating throughput and robustness for MIBI, a recent study imaged FFPE samples from 41 patients with triple negative breast cancer61. In this example 800 μm2 images each containing 2048×2048 pixels from all 41 patients were acquired in less than two weeks and new MIBI-TOF instrumentation and ion sources promise to increase this throughput by another order of magnitude64. Together, we believe that high-dimensional imaging methods including IMC and MIBI provide interesting possibilities to study RD pathology and identify relevant cellular interactions and perturbations directly in the tissue microenvironment.
Analytical tools for the discovery of biomarkers and stratifying populations
The analysis of highly multiplexed datasets comprising large groups of samples and dozens of measurements provides a unique opportunity to gain insight into biological variation and its importance in a therapeutic context. However, they also pose additional challenges since their high-dimensional properties make comprehensive manual analysis, e.g. in two-dimensional space through manual gating approaches, virtually impossible. We here provide an introduction of some commonly employed analytic approaches that can be used as a starting point for a more unbiased and comprehensive analysis of datasets from such high-dimensional immune monitoring studies70–73.
Image segmentation and analysis of spatial relationships
As a first level of analysis, multiplexed imaging data can be visualized in its native image format or as multi-color overlays to emphasize the spatial relationships between different epitopes and cell types. Beyond this, several approaches can be employed to extract single-cell data from high-dimensional images. For example, deep-learning-based segmentation approaches74 (see Glossary) have recently been applied to MIBI-TOF images61,75. In this example, a classifier was trained to discriminate nuclear from non-nuclear pixels which can then be used to automatically identify nuclei and thus cells from a large set of images. Other studies have employed a combination of Ilastik-based classification76 and CellProfiler-guided segmentation77 and many more deep-learning approaches have been proposed for cellular segmentation of high-dimensional image sets62,63,78,79. Once imaging data has been segmented, downstream analysis as described below can be performed analogously to other types of single-cell data. It should be noted that image segmentation is not limited to the identification of single cells. Potentially, larger structures such as extracellular pathological deposits but also smaller subcellular structures like nuclei and other cell compartments could be automatically identified. Furthermore, information about cellular location and proximity to other cells can be assigned to segmented data and leveraged in the downstream analysis. Recent examples applied to high-dimensional images are the use of Delaunay triangulation80 to identify cellular niches in a mouse model of lupus57, spatial enrichment analysis to identify a hierarchical organization of tumor and immune cells in triple negative breast cancer61 and cellular neighborhood analysis of pancreatic islands from patients with type 1 diabetes62.
Visualization of high-dimensional single-cell data
As a first step, single-cell proteomic data (either directly obtained by mass cytometry or through image segmentation as described above) is usually transformed using arcsinh or related logarithmic approaches to account for differential variance in protein expression levels observed between low and high expressing populations81–83. Following this pre-processing phase, high-dimensional datasets are often projected into a human interpretable lower dimensional space with the aim of preserving as much of the inherent high-dimensional structure as possible. An effective approach optimized to group cells based on the similarity of their high-dimensional expression profiles is the t-Stochastic Neighbor Embedding (tSNE) algorithm84,85. tSNE projects high-dimensional data onto a (usually) two-dimensional map, thus providing a readily interpretable overview of populations contained in the dataset. The tSNE algorithm has been successfully applied to a wide range of single-cell measurements and it is now integrated into many major cytometry data processing platforms86. Recently, several extensions and modifications of the original tSNE algorithm have further broadened its applicability87. Real-time visualizations of embeddings provide users with an feedback which can be used to interactively optimize parameters88 and a hierarchical step-wise application of tSNE has been shown to improve the identification of rare populations89. Furthermore artificial neural networks (NN, see Glossary) can approximate the tSNE embedding function, thus enabling the projection and comparison of additional data which was not part of the initial embedding90. Alternatively, a novel embedding technique for dimension reduction termed uniform manifold approximation and projection (UMAP) has been proposed91,92. In comparison to tSNE, UMAP improves the preservation of global structure in high-dimensional datasets and provides shorter calculation times, both factors contributing to its rapid adoption.
An alternative approach, especially suited to simultaneously comparing the characteristics of multiple populations between a set of samples, is spanning-tree progression analysis of density normalized events (SPADE)93. SPADE provides an overview of all populations present in a sample as clusters on a minimum spanning tree which can be color-coded by any marker of interest as well as differential expression when one or more reference samples are included86. Further data analysis and visualization approaches make use of force-directed layouts to provide low dimensional representations of the high-dimensional dataset. The Vortex clustering environment combines a clustering algorithm with a single-cell representation of cellular heterogeneity through force-directed layouts94. Flowmap follows a similar concept, except it incorporates the use of time-course data when building the model95. Incorporation of prior biological knowledge about lineage-defining expression patters can be further achieved through the use of Scaffold maps38. Here, high-dimensional datasets are clustered and subsequently mapped around previously defined landmark populations in a force-directed layout. As such, new data from different platforms can be integrated and compared to previous analyses.
Population identification and differential abundance analysis
In order to make the analysis of high-dimensional datasets more reproducible and to reduce analysis-related variability, many algorithms have been developed for the automated population identification through clustering96. A prominent example that has high performance and computational efficiency is FlowSOM, an adaption of the principle of self-organizing maps to cytometry data97. Following population assignment, potential associations of cluster frequencies or expression characteristics can be derived. To statistically compare cluster characteristics between clinical groups or biological scenarios, Citrus uses a hierarchical clustering step and subsequent regularized regression to identify stratifying features between cohorts98 and has been integrated into complementary visualization approaches like Scaffold99.
While clustering can be a useful approach to identify groups of cells and populations, questions arise as to the right choice of clustering parameters, the definition and correct number of clusters in the datasets as well as potential loss of statistical power through over-clustering. Therefore, several approaches have been developed to identify differentially abundant cells or stratifying signatures without the need for clustering. One solution is to assign cells to so called hyperspheres and then test for differential abundances for each hypersphere100. Alternatively, an approach termed CellCNN relies on a representation learning approach to identify cell events associated with differential clinical outcome or other characteristics of interest101.
Cellular differentiation and beyond
Cellular developments and lineage decisions can be traced using trajectory algorithms which order cells according to progressive changes in their phenotype. Initial examples of such algorithms applied to mass cytometry data were Wanderlust102 and Wishbone103, however there is now a large number of such trajectory algorithms available104. Furthermore, single-cell regulatory networks have been modeled and quantified using a conditional density based approach termed DREMI/DREVI [ref: 105] and cellular diversity can be quantified by calculating an Inverse Simpson Index106.
In conclusion, the above-mentioned analytical tools provide researches with interpretable data representations and correlations with clinical features, thus enabling the identification of cellular disease signatures and potential novel therapeutic targets.
Practical considerations for high-dimensional immune monitoring studies
Conception and planning of large immune monitoring studies involves several layers of complexity including proper choice and appropriate size of experimental and control groups. Most of these are dependent on the specific disease context and biological hypotheses. Given this context-dependency, such considerations cannot be comprehensively covered in this Review. Instead, we provide a set of practical considerations which cover commonly encountered questions when planning studies involving mass cytometry and MIBI.
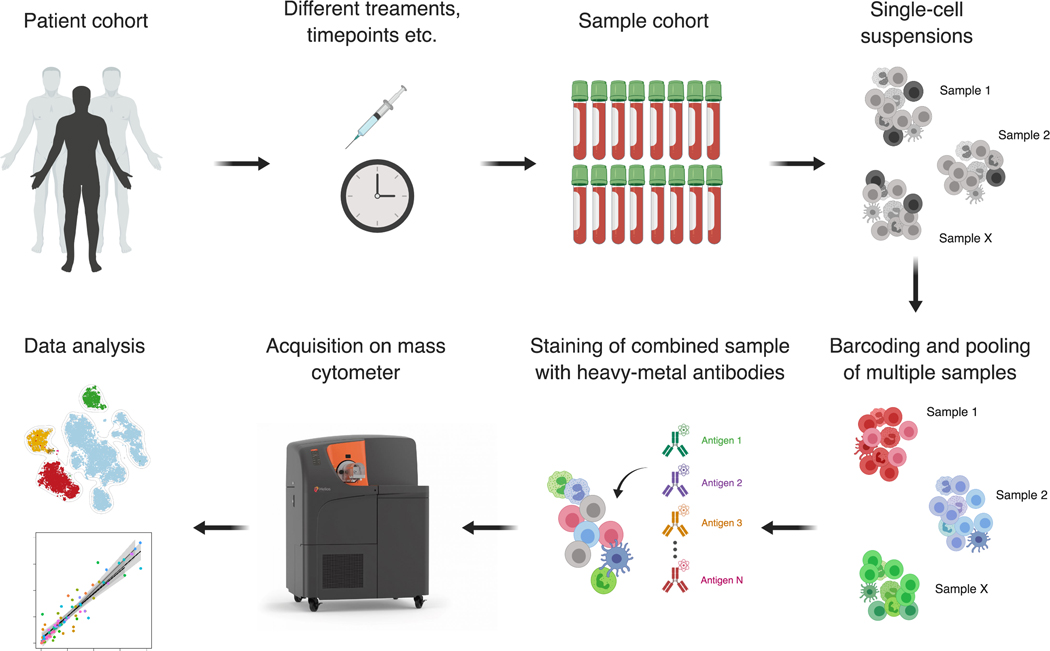
Studies involving human subjects often leverage existing collections of samples that have been acquired over several years (Figure 3). Cell suspensions that have been viably cryopreserved (i.e. with DMSO solutions) can be directly entered into the sample preparation pipeline for mass cytometry while archival FFPE tissue samples from standard pathology workflows have been shown to be compatible with MIBI. If sample collection is still ongoing, several considerations can be made. Single-cell suspensions can be stored viably, which allow functional assays such as stimulation of intracellular cytokine production or induction of protein phosphorylation events. While fixation prior to storage can help to recover more cryo-sensitive populations and prevent freeze-thaw dependent changes in their functional state, fixation-sensitive epitopes might be less accessible for antibody staining, and thus reduce overall analytical quality. It is also advisable to anticipate the inclusion of a separate validation cohort by gathering samples not included in the initial analysis which can be subsequently used to independently confirm initial findings. Mass cytometry is destructive, and cells cannot be recovered post-acquisition, however there a numerous examples where populations of interest can be identified in high-dimensional space and prospectively isolated by FACS for subsequent orthogonal analysis14,102,107,108. For MIBI, serial sections of the same tissue samples could also be used for confirmatory orthogonal assays such as traditional IHC or other imaging modalities or bulk biomolecule analysis.
Figure 3. Conducting large-scale immune monitoring studies using mass cytometry.
Patient cohorts are selected with regard to the experimental questions to be addressed. Cells that were samples at different timepoints can be cryopreserved (either live or fixed) and collected prior to their analysis. In order to reduced technical variation, single cell suspensions from multiple donors can be barcoded and pooled prior to staining and acquisition. In this process, a unique heavy-metal tag (or a combination of several tags) is attached to all cells of a sample. These tagged samples can then be pooled and processed (stained and acquired) as a single sample. Following acquisition, cells can be assigned back to their original sample and analyzed further.
Following sample curation, intra-assay comparison, questions of panel design and the choice of antibodies and other probes have to be considered109. We advocate for a backbone panel of antibodies able to capture and quantify a broad range of immune cell types beyond the hypothesized target population(s) of interest. This backbone can be supplemented with more hypothesis-specific targets. A recent publication proposed a validated and commercially available set of antibodies that can be easily implemented for such comprehensive immune monitoring in RD17.
Next, antibodies against chosen targets have to be distributed to the available mass reporter channels. Panel design considerations have been outlined previously110,111. In short, a minor source of cross-channel contamination can result from initial isotopic impurities which were not completely removed during the purification process, typically generating a signal in the mass +1 channel and restricted to around ≤ 3% of the parent isotope (see Figure 1B). Further, elements can oxidize and generate signal in the mass +16 channel with similar extend to isotopic contamination levels. Both phenomena are often insignificant relative to technical variation or compared to fluorescence-based signal overlap. Moreover, it can be practically resolved through panel design, reagent titration or through recently developed, bead-based, compensation approaches that could be applied to both mass cytometry but also imaging applications and which have been shown to further improve data accuracy112.
While the list of commercially available heavy-metal tagged antibodies is steadily growing, there is usually a need to perform a set of conjugations in-house. Protocols for heavy-metal conjugation of antibodies which can be used in mass cytometry or imaging are available and can be performed in a few hours28,113. Antibody specificity should always be confirmed using biological controls, i.e. samples known to express the respective epitope and such known to not express it. As with many other biological assays, batch processing is recommended to minimize technical variation. For mass cytometry, the use of barcoding approaches allows for the combined staining and acquisition of multiple samples as once30–34. Cells can then be assigned back to their original sample in silico. An analogous approach for imaging is the use of tissue microarrays which combine cores of multiple patient samples onto a single slide.
If data is generated across long periods of time or multiple instruments, the use of standards can improve sample comparability. Metal containing beads can normalize machine performance across time for mass cytometry applications35 while repeat aliquots of standard sample(s) analyzed with each run can be used in suspension all well as imaging technologies36.
In order to improve reproducibility and accelerate research efforts, raw data should be made available in public repositories with published studies. Mass cytometry raw data is often shared using Cytobank86, Immport114 and flowrepository115 and the MIFlowCyt standard116 can be adopted to mass cytometry experiments to provide crucial information about experimental design, further allowing meta-analysis approaches117. Analogous databases have been proposed for biomedical imaging data in general118.
Immune monitoring technologies enable discovery in rheumatic and autoimmune diseases
Employing these experimental and analytical approaches, several studies have started to explore the potential of these technologies to gain insights into disease mechanisms of various RD, some examples of which will be discussed here.
RA and spondyloarthritis
Mass cytometry was used to study the activation and cytokine production profile of T cells isolated from peripheral blood or directly from synovial fluid and inflamed tissues of patients with RA and spondyloarthritis. Employing this high-dimensional approach enabled the identification of a pathologically expanded population of PD-1hiCXCR5negCD4pos T cells in RA patients seropositive for rheumatoid factor or anti-citrullinated peptide antibody119. In-depth phenotyping of these cells revealed expression of several proteins enabling B cell help, including IL-21, CXCL13, ICOS and MAF and discriminated them from traditional follicular helper T cells while mechanistic downstream experiments demonstrated the ability of this population to promote plasma cell differentiation. In a related study, peripheral blood of patients with RA and control subjects with osteoarthritis was analyzed by mass cytometry120. The authors found that RA patients had an expanded population of effector memory CD4+ T cells displaying a CD27- HLA-DR+ phenotype, suggesting chronic activation of these cells in RA.
Leveraging mass cytometry to study immune activation and cytokine production in spondyloarthritis, the authors identified increased frequencies of a variety of immune cell types producing the cytokine GM-CSF in the blood and joints of spondyloarthritic patients121. Amongst the cell types producing GM-CSF in these patients were CD4+ and CD8+ T cells, γδ T cells as well as innate lymphoid cells (ILCs).
Together, these studies provide mechanistic hypotheses for T cell-mediated pathological processes in RA and spondyloarthritis. In RA, the identified tissue-infiltrating T cell subset offers a mechanism for the recruitment of additional immune cells through their production of CXCR5 and IL-21. Identification of such pathological processes additionally provides potential targets for therapeutic intervention. For example, GM-CSF expression by T cells as identified in spondyloarthritis has been suggested as a key mediator and therapeutic target in other autoimmune diseases122,123. Besides these soluble mediators, expression of surface epitopes such as PD-1 on the RA-associated population could be targeted to interfere with T-B-cell interactions using existing therapeutics.
SLE and SjS
In-depth functional and phenotypic analysis by mass cytometry has also been employed to study immune cell responses and cytokine networks underlying in SLE and SjS. Performing a comprehensive analysis of immune cell subset responses to TLR ligands in SLE patients enabled the identification of a disease-associated chemokine signature upon Toll-like receptor (TLR) engagement124. More specifically, CD14high monocytes from patients with SLE showed an inflammatory chemokine signature, characterized by MCP-1, MIP-1β and TNF-α. In a separate study, this monocytic SLE-signature was also found to be present in pediatric SLE patients and to express IL1RA [ref:125]. Interestingly, the presence of this immune signature correlated with clinical disease activity and could be abrogated through blockade of IFNAR or JAK inhibition, thus providing mechanistic insights into emerging therapeutics for SLE currently evaluated in clinical trials126,127. Focusing on B cell biology as a contributor to SLE, a recent study identified several cell subtype alterations during treatment with belimumab, an antibody neutralizing the soluble B-cell activating factor (BAFF) that has been approved for the treatment of SLE128. High-dimensional phenotyping of B cells enabled the longitudinal monitoring of a broad range of B cell subsets, including age-associated B cells (ABCs). ABCs are a T-bet dependent, alternative-differentiation state of B cells described to appear not only with increasing age but also upon repeated viral infections and in patients with autoimmunity, including SLE129,130. Using mass cytometry and defining ABCs as CD11cposCD21neg B cells, Ramsköld et al. found that a decrease in the frequency of this population to correlates with early clinical improvement. In addition to this population, the authors identified overall pre-treatment B cell counts as a predictive feature for response to belimumab therapy.
Mass cytometry applied to blood samples and salivary gland biopsies of patients with primary Sjörgen’s syndrome (pSjS) revealed a multi-population disease signature, dominated by activated CD8+ T cell populations and terminally differentiated plasma B cells131. Importantly, this immunological signature could predict pSjS diagnosis as well as stratify patients based on clinical features and disease activity and thus pose as potential targets for future clinical intervention.
Unifying many of these findings across multiple autoimmune and rheumatic diseases, a recent study used mass cytometry and metal-conjugated HLA-II tetramers to perform in-depth phenotyping of antigen-specific autoimmune T cells in celiac disease and other autoimmune and rheumatic diseases. The antigen-specific T cells were found to display a phenotype strikingly similar to the one described in RA by Rao et al. [ref: 119], including expression of multiple activation markers, PD1, IL-21 and absence of CXCR5. Interestingly, T cells with this particular phenotype were enriched across many disorders besides celiac disease, including systemic sclerosis and SLE132, thus positioning these cells as key disease-driving T cells across many autoimmune and rheumatic diseases.
Novel insights into RDs
These first studies illustrate some key advantages of mass cytometry for in-depth phenotyping of immune cell subsets and how it can provide valuable insights into disease mechanisms in RD. Firstly, multiple studies found evidence of chronic and/or elevated T cell activation, e.g. expansion of CD27- memory T cells in RA and multifaceted T cell activation in spondyloarthritis and SjS. While previous studies have also shown immune activation in several RDs, a unique advantage of defining high-dimensional protein co-expression patterns is to more precisely identify cellular signatures to better understand their functional context and to potentially serve as specific therapeutic targets. More specifically, several of the previously mentioned studies underline the importance of TFH – B cell interplay and plasma cell differentiation and they define surface phenotypes of cell populations involved in these processes. Plasma cells ultimately contribute to autoantibody production, an important pathogenic and diagnostic factor for many RDs and thus interfering with plasma cell development might provide a therapeutic strategy for these diseases.
Another emerging theme from these studies are altered cytokine expression profile by both innate immune cells in SLE and several populations of adaptive immune cells in RA and spondylarthrosis, pointing towards the importance of immune communication pathways which could be targeted therapeutically. Investigation of such high-dimensional cytokine production profiles and other intracellular processes in combination with complex surface phenotypes is made possible as heavy-metal reporter conjugated antibodies are compatible with commonly used fixation methods.
While its clinical applicability in the context of cancer has been previously demonstrated, we are not aware of any published studies employing MIBI in RD research. However, as we discuss the following section, we expect major insights to be gained by studying tissue-based cellular interactions, including sites of active disease.
Together, these studies provide first examples of how increased parametrization allows the identification of complex cellular features that are correlated with disease activity and clinical improvement as well as the discovery of cellular signatures that could be used diagnostically or that can serve therapeutic targets across a range of RDs.
Future research directions and challenges for rheumatic and autoimmune diseases
As laid-out above, continued analytical and methodological developments in both mass cytometry and related imaging technologies like MIBI make them core technologies for many immune monitoring and related studies. Naturally, technological improvements are continuously ongoing, and based on these we outline future research directions.
First, the ability to extract and recognize biologically informative but complex information from these high-dimensional datasets will be further enhanced through developments in the fields of artificial intelligence and machine learning (see Glossary). For example, the use of autoencoders and related tools as already demonstrated on single-cell data133 could be directly applied to multiplexed imaging information to enhance identification of relevant features there too134,135.
Another exciting area of ongoing development for high-dimensional imaging is the transition from imaging a two-dimensional plane to three-dimensional structures, providing the opportunity to leverage health and dysfunctional spatial information. For instance, given that as little as ~100nm of tissue is consumed with each MIBI scan, multiple scans of the same region could be used to reconstruct three-dimensional structures with highly resolved multiplexed infomation61,136,137.
For mass cytometry, recent studies have expanded its applicability into new research areas. For example, comprehensive immune phenotyping by mass cytometry was performed on as little as 100 μl of blood from newborns to follow early immune development138. This study additionally performed analysis of plasma protein levels from these samples, highlighting the benefits of integrating multiple types of data. Such multi-omics approaches combine transcriptomic, epigenetic, metabolomic, microbiome and clinical phenotypes that together can reveal otherwise hidden correlations. Computational analysis of such integrated datasets remains challenging but is an area of active research139–141. Using probes against chromatin modification marks, researchers have also directly obtained single-cell epigenetic information using mass cytometry, observing changes with immunological age142. Multiple autoimmune diseases and especially RDs such as SLE, RA and SSc have suspected epigenetic contributions to their etiology and it would be of high interest to study these on the single-cell level in combination with immune cell phenotype(s) and abundance4,143,144.
A major research interest in RD is the identification of biomarkers that stratify patients into clinically relevant groups. For example, early discrimination of patients likely to respond well to a given therapeutic option from patients unlikely to benefit, as has been shown in the context of cancer therapy145, could contribute to more personalized and tailored therapeutic schemes. Importantly, such in-depth monitoring of patients receiving various therapies could pinpoint the mechanism of action of currently approved drugs and thus open opportunities to directly modulate a specific cellular function instead of broadly targeting complete immune cell lineages.
Furthermore, while many autoimmune and RD are chronic diseases that are characterized by long-term progressing organ damage, they often in addition display short-term disease exacerbations termed flares. Analysis and comparison of affected tissues spanning this cycle and including samples from times of disease respite as well as samples taken right before the appearance of clinical episodes and during ongoing flares could give insight to the reservoir of pathologic cells and together enable the identification of biomarkers that signify the timing of such flares. Novel microneedle sampling devices could facilitate such longitudinal studies by potentially enabling at-home blood collections146,147. Skin or synovial biopsies or samples acquired using other recently developed tissue sampling techniques such as fine needle aspirates of various tissues148 or microneedle patches that longitudinally sample cells in a minimally invasive manner from the skin149 could be imaged directly or analyzed as a single-cell suspension by mass cytometry.
The holy grail of medical intervention in any autoimmune syndrome would be prevention of the disease before onset or alternatively, early detection of damage to avoid disease exacerbation and the onset of comorbidities150. In SLE for example, anti-nuclear antibodies have been found up to 9 years before disease onset in some patients6. Analogously, anti-citrullinated protein antibodies can precede the onset of RA151 and it would be of great interest investigate if associated cellular signatures that predict imminent disease onset in subjects at risk of disease development can be identified. Taken together, we believe that there are unique opportunities to leverage comprehensive immune phenotyping through technologies such as mass cytometry and MIBI to find stratifying and predicting immune signatures, reveal novel therapeutic targets and identify biomarkers for therapeutic success.
Conclusions
Many RDs are characterized by a heterogeneous clinical presentation and complex pathobiology. The systems immunology perspective offered through the use of mass spectrometry-powered single-cell technologies in combination with innovative analytical approaches now provides a unique opportunity to elucidate these mechanisms. Here, we discussed the premise of these technologies as well as important practical considerations for the planning of such projects. As illustrated by several recent studies, these technologies enable the discovery of disease associated cellular signatures that give insight into disease pathology and additionally offer perspectives for future treatments of RDs.
Key points.
Human immune monitoring using systems immunology approaches allows new insights into pathological processes and therapeutic opportunities for many RDs
Mass spectrometry-based single-cell approaches with elemental reporters such as mass cytometry and MIBI are amenable to study a wide array of clinical samples
These technologies enable in-depth analysis of the cellular phenotype and functional state on a single-cell level
MIBI and related approaches use analogous concepts to image cells in their histological context
Combination of these technologies with data driven analytical approaches can give predictive insights into disease mechanisms
Acknowledgements
F.J.H was supported by the EMBO organization (EMBO Long-Term Fellowship ALTF 1141–2017), the Novartis Foundation for medical-biological Research (16C148) and the Swiss National Science Foundation (SNF Early Postdoc Mobility P2ZHP3–171741). S.C.B. is supported by the Damon Runyon Cancer Research Foundation Fellowship (DRG-2017–09), the NIH 1DP2OD022550–01, 1R01AG056287–01, 1R01AG057915–01, 1-R00-GM104148–01, 1U24CA224309–01, 5U19AI116484–02, U19 AI104209, and a Translational Research Award from the Stanford Cancer Institute.
Glossary
- Machine learning
Machine learning (ML) constitutes a subset of the field of artificial intelligence. In ML, algorithms parse data to learn from it in order to perform certain task such as classification or predicting of certain outcomes. Instead of supplying a rigid set of instructions, patterns are identified (learned) from the supplied data
- Artificial neural networks
Artificial neural networks (NN) are a subset of ML frameworks that are inspired by the biological structure of the brain. NN consist of (potentially many) layers of nodes which are connected and can transmit information to each other through edges. A node that receives input through one or multiple edges then applies a given transformation on this input and returns the respective output via its edges. In order to perform classification or prediction tasks, the strength of the connections (called weights) are iteratively adjusted based on the input data
- Deep learning
Deep learning uses NN to learn from large amounts of data. It usually refers to the NN having multiple layers of nodes that can be adjusted to enable learning
Footnotes
Declaration of Interests
S.C.B. is an inventor of the MIBI technology and a scientific founder of the IONpath Inc., the company commercializing it.
References
- 1.Robinson WH & Mao R Technological advances transforming rheumatology. Nat. Rev. Rheumatol. 11, 626–628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mavragani CP & Moutsopoulos HM Sjögren’s Syndrome. Annu. Rev. Pathol. Mech. Dis. 9, 273–285 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Brito-Zerón P et al. Sjögren syndrome. Nat. Rev. Dis. Prim. 2, 16047 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Angiolilli C et al. New insights into the genetics and epigenetics of systemic sclerosis. Nat. Rev. Rheumatol. 14, 657–673 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Smolen JS et al. Rheumatoid arthritis. Nat. Rev. Dis. Prim. 4, 18001 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kaul A et al. Systemic lupus erythematosus. Nat. Rev. Dis. Prim. 2, 16039 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Tsokos GC, Lo MS, Reis PC & Sullivan KE New insights into the immunopathogenesis of systemic lupus erythematosus. Nat. Rev. Rheumatol. 12, 716–730 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Gatto M, Zen M, Iaccarino L & Doria A New therapeutic strategies in systemic lupus erythematosus management. Nat. Rev. Rheumatol. 15, 30–48 (2019). [DOI] [PubMed] [Google Scholar]
- 9.von Herrath MG & Nepom GT Lost in translation: barriers to implementing clinical immunotherapeutics for autoimmunity. J. Exp. Med. 202, 1159–1162 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis MM A prescription for human immunology. Immunity 29, 835–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis MM & Brodin P Rebooting Human Immunology. Annu. Rev. Immunol. 36, 843–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson WH & Mao R Biomarkers to guide clinical therapeutics in rheumatology? Curr. Opin. Rheumatol. 28, 168–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ermann J, Rao DA, Teslovich NC, Brenner MB & Raychaudhuri S Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat. Rev. Rheumatol. 11, 541–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudillière B et al. Clinical recovery from surgery correlates with single-cell immune signatures. Sci. Transl. Med. 6, 255ra131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good Z et al. Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat. Med. 24, 474–483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmann FJ et al. Comprehensive Immune Monitoring of Clinical Trials to Advance Human Immunotherapy. Cell Rep. 28, 819–831.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair N et al. Mass cytometry as a platform for the discovery of cellular biomarkers to guide effective rheumatic disease therapy. Arthritis Res. Ther. 17, 127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandura DR et al. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal. Chem. 81, 6813–6822 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Ornatsky O et al. Highly multiparametric analysis by mass cytometry. J. Immunol. Methods 361, 1–20 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–96 (2011).21551058 [Google Scholar]
- 22.Ornatsky OI et al. Study of Cell Antigens and Intracellular DNA by Identification of Element-Containing Labels and Metallointercalators Using Inductively Coupled Plasma Mass Spectrometry proliferation in clinical samples is important for diagnostic. Anal. Chem. 80, 2539–2547 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Ornatsky OI et al. Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J. Anal. At. Spectrom. 23, 463 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lou X et al. Polymer-based elemental tags for sensitive bioassays. Angew. Chemie - Int. Ed. 46, 6111–6114 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majonis D et al. Synthesis of a functional metal-chelating polymer and steps toward quantitative mass cytometry bioassays. Anal. Chem. 82, 8961–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mei HE, Leipold MD & Maecker HT Platinum-conjugated antibodies for application in mass cytometry. Cytom. Part A 89, 292–300 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Han G et al. Atomic mass tag of bismuth-209 for increasing the immunoassay multiplexing capacity of mass cytometry. Cytom. Part A 91, 1150–1163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han G, Spitzer MH, Bendall SC, Fantl WJ & Nolan GP Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat. Protoc. 13, 2121–2148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Q et al. Biodistribution of cisplatin revealed by imaging mass cytometry identifies extensive collagen binding in tumor and normal tissues. Sci. Rep. 6, 36641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodenmiller B et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zunder ER et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mei HE, Leipold MD, Schulz AR, Chester C & Maecker HT Barcoding of Live Human Peripheral Blood Mononuclear Cells for Multiplexed Mass Cytometry. J. Immunol. 194, 2022–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai L, Ong R, Li J & Albani S A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF). Cytometry. A 87, 369–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann FJ, Simonds EF & Bendall SC A Universal Live Cell Barcoding-Platform for Multiplexed Human Single Cell Analysis. Sci. Rep. 8, 10770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finck R et al. Normalization of mass cytometry data with bead standards. Cytometry. A 83, 483–94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinsteuber K et al. Standardization and quality control for high-dimensional mass cytometry studies of human samples. Cytometry. A 89, 903–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leipold MD et al. Comparison of CyTOF assays across sites: Results of a six-center pilot study. J. Immunol. Methods 453, 37–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spitzer MH et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartmann FJ et al. High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J. Exp. Med. 213, 2621–2633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galli E et al. GM-CSF and CXCR4 define a T helper cell signature in multiple sclerosis. Nat. Med. 1 (2019). doi: 10.1038/s41591-019-0521-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP & Bodenmiller B A platinum-based covalent viability reagent for single-cell mass cytometry. Cytom. Part A 81 A, 467–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ & Nolan GP Single-cell mass cytometry adapted to measurements of the cell cycle. Cytom. Part A 81 A, 552–566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimmey SC, Borges L, Baskar R & Bendall SC Parallel analysis of tri-molecular biosynthesis with cell identity and function in single cells. Nat. Commun. 10, 1185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar LJ et al. Identification of Hypoxic Cells Using an Organotellurium Tag Compatible with Mass Cytometry. Angew. Chemie Int. Ed. 53, 11473–11477 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Schulz D, Severin Y, Zanotelli VRT & Bodenmiller B In-Depth Characterization of Monocyte-Derived Macrophages using a Mass Cytometry-Based Phagocytosis Assay. Sci. Rep. 9, 1925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frei AP et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods 13, 269–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duckworth AD et al. Multiplexed profiling of RNA and protein expression signatures in individual cells using flow or mass cytometry. Nat. Protoc. 14, 901–920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newell EW, Sigal N, Bendall SC, Nolan GP & Davis MM Cytometry by Time-of-Flight Shows Combinatorial Cytokine Expression and Virus-Specific Cell Niches within a Continuum of CD8 + T Cell Phenotypes. Immunity 36, 142–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newell EW et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat. Biotechnol. 31, 623–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newell EW & Davis MM Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat. Biotechnol. 32, 149–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leong ML & Newell EW Multiplexed Peptide-MHC Tetramer Staining with Mass Cytometry. in Methods in molecular biology (Clifton, N.J.) 1346, 115–131 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Simoni Y et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Bodenmiller B Multiplexed Epitope-Based Tissue Imaging for Discovery and Healthcare Applications. Cell Syst. 2, 225–238 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Gerner MY, Kastenmuller W, Ifrim I, Kabat J & Germain RN Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerdes MJ et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. U. S. A. 110, 11982–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin J-R et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goltsev Y et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis AS et al. Characterizing and Diminishing Autofluorescence in Formalin-fixed Paraffin-embedded Human Respiratory Tissue. J. Histochem. Cytochem. 62, 405–423 (2014).24722432 [Google Scholar]
- 59.Giesen C et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–22 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Angelo M et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keren L et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damond N et al. A Map of Human Type 1 Diabetes Progression by Imaging Mass Cytometry. Cell Metab. (2019). doi: 10.1016/J.CMET.2018.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang YJ et al. Multiplexed In Situ Imaging Mass Cytometry Analysis of the Human Endocrine Pancreas and Immune System in Type 1 Diabetes. Cell Metab. 29, 769–783.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keren L et al. MIBI-TOF: A multi-modal multiplexed imaging platform for tissue pathology. (in Rev. [Google Scholar]
- 65.Schulz D et al. Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell Syst. 6, 25–36.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Catena R, Montuenga LM & Bodenmiller B Ruthenium counterstaining for imaging mass cytometry. J. Pathol. 244, 479–484 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Skinner PJ, Daniels MA, Schmidt CS, Jameson SC & Haase AT Cutting edge: In situ tetramer staining of antigen-specific T cells in tissues. J. Immunol. 165, 613–7 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Steinert EM et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S et al. Simian Immunodeficiency Virus-Producing Cells in Follicles Are Partially Suppressed by CD8 + Cells In Vivo. J. Virol. 90, 11168–11180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mair F et al. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur. J. Immunol. 46, 34–43 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Saeys Y, Gassen S. Van & Lambrecht BN Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 16, 449–62 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Chester C & Maecker HT Algorithmic Tools for Mining High-Dimensional Cytometry Data. J. Immunol. 195, 773–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newell EW & Cheng Y Mass cytometry: blessed with the curse of dimensionality. Nat. Immunol. 17, 890–895 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Moen E et al. Deep learning for cellular image analysis. Nat. Methods 1 (2019). doi: 10.1038/s41592-019-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Valen DA et al. Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments. PLOS Comput. Biol. 12, e1005177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sommer C, Straehle C, Kothe U & Hamprecht FA Ilastik: Interactive learning and segmentation toolkit. in 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 230–233 (IEEE, 2011). doi: 10.1109/ISBI.2011.5872394 [DOI] [Google Scholar]
- 77.Dao D et al. CellProfiler Analyst: interactive data exploration, analysis and classification of large biological image sets. Bioinformatics (2016). doi: 10.1093/bioinformatics/btw390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schapiro D et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haberl MG et al. CDeep3M—Plug-and-Play cloud-based deep learning for image segmentation. Nat. Methods 15, 677–680 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gabriel KR & Sokal RR A New Statistical Approach to Geographic Variation Analysis. Syst. Zool. 18, 259 (1969). [Google Scholar]
- 81.Finak G, Perez J-M, Weng A & Gottardo R Optimizing transformations for automated, high throughput analysis of flow cytometry data. BMC Bioinformatics 11, 546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bagwell CB Hyperlog?A flexible log-like transform for negative, zero, and positive valued data. Cytom. Part A 64A, 34–42 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Parks DR, Roederer M & Moore WA A new ‘Logicle’ display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry. A 69, 541–51 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Maaten L. Van Der & Hinton G Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 85.Amir ED et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kotecha N, Krutzik PO & Irish JM Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. Chapter 10, Unit10.17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van der Maaten L Barnes-Hut-SNE. 1–11 (2013). [Google Scholar]
- 88.Pezzotti N et al. Approximated and User Steerable tSNE for Progressive Visual Analytics. 1–15 (2015). [DOI] [PubMed] [Google Scholar]
- 89.van Unen V et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat. Commun. 8, 1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho H, Berger B & Peng J Generalizable and Scalable Visualization of Single-Cell Data Using Neural Networks. Cell Syst. 7, 185–191.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McInnes L & Healy J UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv (2018). [Google Scholar]
- 92.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. (2018). doi: 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- 93.Qiu P et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Samusik N, Good Z, Spitzer MH, Davis KL & Nolan GP Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zunder ERR, Lujan E, Goltsev Y, Wernig M & Nolan GPP A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell 16, 323–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weber LM & Robinson MD Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry. A 89, 1084–1096 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Van Gassen S et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry. A 87, 636–45 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ & Nolan GP Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl. Acad. Sci. U. S. A. 111, E2770–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Spitzer MH et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell 168, 487–502.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lun ATL, Richard AC & Marioni JC Testing for differential abundance in mass cytometry data. Nat. Methods 14, 707–709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arvaniti E & Claassen M Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun. 8, 14825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bendall SC et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157, 714–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Setty M et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 34, 637–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saelens W, Cannoodt R, Todorov H & Saeys Y A comparison of single-cell trajectory inference methods: towards more accurate and robust tools. bioRxiv 276907 (2018). doi: 10.1101/276907 [DOI] [PubMed] [Google Scholar]
- 105.Krishnaswamy S et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 346, 1250689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Strauss-Albee DM et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci. Transl. Med. 7, 297ra115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Good Z et al. Proliferation tracing with single-cell mass cytometry optimizes generation of stem cell memory-like T cells. Nat. Biotechnol. (2019). doi: 10.1038/s41587-019-0033-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aghaeepour N et al. GateFinder: projection-based gating strategy optimization for flow and mass cytometry. Bioinformatics 34, 4131–4133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spitzer MH & Nolan GP Mass Cytometry: Single Cells, Many Features. Cell 165, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leipold MD, Newell EW & Maecker HT Multiparameter Phenotyping of Human PBMCs Using Mass Cytometry. Methods Mol. Biol. 1343, 81–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takahashi C et al. Mass cytometry panel optimization through the designed distribution of signal interference. Cytom. Part A 91, 39–47 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Chevrier S et al. Compensation of Signal Spillover in Suspension and Imaging Mass Cytometry. Cell Syst. 6, 612–620.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hartmann FJ et al. Scalable Conjugation and Characterization of Immunoglobulins with Stable Mass Isotope Reporters for Single-Cell Mass Cytometry Analysis. Methods Mol. Biol. 1989, 55–81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bhattacharya S et al. ImmPort: disseminating data to the public for the future of immunology. Immunol. Res. 58, 234–239 (2014). [DOI] [PubMed] [Google Scholar]
- 115.Spidlen J, Breuer K, Rosenberg C, Kotecha N & Brinkman RR FlowRepository: A resource of annotated flow cytometry datasets associated with peer-reviewed publications. Cytom. Part A 81A, 727–731 (2012). [DOI] [PubMed] [Google Scholar]
- 116.Lee JA et al. MIFlowCyt: The minimum information about a flow cytometry experiment. Cytom. Part A 73A, 926–930 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu Z et al. MetaCyto: A Tool for Automated Meta-analysis of Mass and Flow Cytometry Data. Cell Rep. 24, 1377–1388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Williams E et al. Image Data Resource: a bioimage data integration and publication platform. Nat. Methods 14, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rao DA et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fonseka CY et al. Mixed-effects association of single cells identifies an expanded effector CD4+ T cell subset in rheumatoid arthritis. Sci. Transl. Med. 10, eaaq0305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Al-Mossawi MH et al. Unique transcriptome signatures and GM-CSF expression in lymphocytes from patients with spondyloarthritis. Nat. Commun. 8, 1510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Noster R et al. IL-17 and GM-CSF expression are antagonistically regulated by human T helper cells. Sci. Transl. Med. 6, 241ra80 (2014). [DOI] [PubMed] [Google Scholar]
- 123.Hartmann FJ et al. Multiple sclerosis-associated IL2RA polymorphism controls GM-CSF production in human TH cells. Nat. Commun. 5, 5056 (2014). [DOI] [PubMed] [Google Scholar]
- 124.O’Gorman WE et al. Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J. Allergy Clin. Immunol. 136, 1326–1336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O’Gorman WE et al. Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. J. Autoimmun. 81, 74–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.NCT02535689. Available at: https://clinicaltrials.gov/ct2/show/NCT02535689.
- 127.NCT02446899. Available at: https://clinicaltrials.gov/ct2/show/NCT02446899.
- 128.Ramsköld D et al. B cell alterations during BAFF inhibition with belimumab in SLE. EBioMedicine 0, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rubtsova K, Rubtsov AV, Cancro MP & Marrack P Age-Associated B Cells: A T-bet-Dependent Effector with Roles in Protective and Pathogenic Immunity. J. Immunol. 195, 1933–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang S et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun. 9, 1758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mingueneau M et al. Cytometry by time-of-flight immunophenotyping identifies a blood Sjögren’s signature correlating with disease activity and glandular inflammation. J. Allergy Clin. Immunol. 137, 1809–1821.e12 (2016). [DOI] [PubMed] [Google Scholar]
- 132.Christophersen A et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat. Med. 1 (2019). doi: 10.1038/s41591-019-0403-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lopez R, Regier J, Cole MB, Jordan MI & Yosef N Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Falk T et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). [DOI] [PubMed] [Google Scholar]
- 135.Gupta A et al. Deep Learning in Image Cytometry: A Review. Cytom. Part A (2018). doi: 10.1002/cyto.a.23701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rovira-Clave X et al. Subcellular localization of drug distribution by super-resolution ion beam imaging. bioRxiv 557603 (2019). doi: 10.1101/557603 [DOI] [Google Scholar]
- 137.Coskun AF et al. Ion beam subcellular tomography. bioRxiv 557728 (2019). doi: 10.1101/557728 [DOI] [Google Scholar]
- 138.Olin A et al. Stereotypic Immune System Development in Newborn Children. Cell 174, 1277–1292.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pedersen HK et al. A computational framework to integrate high-throughput ‘-omics’ datasets for the identification of potential mechanistic links. Nat. Protoc. 2018 1312 13, 2781 (2018). [DOI] [PubMed] [Google Scholar]
- 140.Ghaemi MS et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 35, 95–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang S, Chaudhary K & Garmire LX More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front. Genet. 8, 84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cheung P et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell 173, 1385–1397.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Z & Zhang R Epigenetics in autoimmune diseases: Pathogenesis and prospects for therapy. Autoimmun. Rev. 14, 854–863 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Jeffries MA & Sawalha AH Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev. Clin. Immunol. 11, 45–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Krieg C et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 24, 144–153 (2018). [DOI] [PubMed] [Google Scholar]
- 146.Blicharz TM et al. Microneedle-based device for the one-step painless collection of capillary blood samples. Nat. Biomed. Eng. 2, 151–157 (2018). [DOI] [PubMed] [Google Scholar]
- 147.Josyula VS, Lakshmikanth T, Mikes J, Chen Y & Brodin P Systems-level immunomonitoring using self-sampled capillary blood. bioRxiv 694521 (2019). doi: 10.1101/694521 [DOI] [Google Scholar]
- 148.Tatovic D et al. Fine-Needle Aspiration Biopsy of the Lymph Node: A Novel Tool for the Monitoring of Immune Responses after Skin Antigen Delivery. J. Immunol. 195, 386–392 (2015). [DOI] [PubMed] [Google Scholar]
- 149.Mandal A et al. Cell and fluid sampling microneedle patches for monitoring skin-resident immunity. Sci. Transl. Med. 10, eaar2227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Munroe ME et al. Discerning Risk of Disease Transition in Relatives of Systemic Lupus Erythematosus Patients Utilizing Soluble Mediators and Clinical Features. Arthritis Rheumatol. 69, 630–642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van der Woude D et al. Epitope spreading of the anti-citrullinated protein antibody response occurs before disease onset and is associated with the disease course of early arthritis. Ann. Rheum. Dis. 69, 1554–1561 (2010). [DOI] [PubMed] [Google Scholar]
- 152.Bendall SC, Nolan GP, Roederer M & Chattopadhyay PK A deep profiler’s guide to cytometry. Trends Immunol. 33, 323–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]