Abstract
SARS-CoV-2 is a new generation of coronavirus, which was first determined in Wuhan, China, in December 2019. So far, however, there no effective treatment has been found to stop this new generation of coronavirus but discovering of the crystal structure of SARS-CoV-2 main protease (SARS-CoV-2 Mpro) may facilitate searching for new therapies for SARS-COV-2. The aim was to assess the effectiveness of available FDA approved drugs which can construct a covalent bond with Cys145 inside binding site SARS-CoV-2 main protease by using covalent docking screening. We conducted the covdock module MMGBSA module in the Schrodinger suite 2020-1, to examine the covalent bonding utilizing. Besides, we submitted the top three drugs to molecular dynamics simulations via Gromacs 2018.1. The covalent docking showed that saquinavir, ritonavir, remdesivir, delavirdine, cefuroxime axetil, oseltamivir and prevacid have the highest binding energies MMGBSA of –72.17, −72.02, −65.19, −57.65, −54.25, −51.8, and −51.14 kcal/mol, respectively. The 50 ns molecular dynamics simulation was conducted for saquinavir, ritonavir and remdesivir to evaluate the stability of these drugs inside the binding pocket of SARS-CoV-2 main protease. The current study provides a powerful in silico results, means for rapid screening of drugs as anti-protease medications and recommend that the above-mentioned drugs can be used in the treatment of SARS-CoV-2 in combined or sole therapy.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, covalent docking, drug repurposing, MD simulation, PCA, Mpro
1. Introduction
SARS-CoV-2 also called ‘severe acute respiratory syndrome coronavirus 2’ abbreviated SARS-CoV-2 was recognized to be the causative of atypical pneumonia (Joshi, 2020; Pant et al., 2020) outbreak in Wuhan, China (Hasan, 2020; S. A. Khan et al., 2020). The virus belongs to the family known as ‘coronaviruses’ because of the crown-like appearance of spikes glycoproteins on the envelope under an electron microscope (Y Chen et al., 2020). World health organization (WHO) recently announced that the virus transforms from epidemic to pandemic, which requires urgent intervention to prevent the growing spread of the virus across the globe (Chan et al., 2020). The total confirmed cases 2,347,884 with 738,923 cases in the United State of America (USA) alone and the total death of 161,138 (as of April 19), with mortality estimated within 2% and about 3.4%, according to estimates of approved cases and death worldwide (N. Chen et al., 2020). The most familiar is a virus that arose from the Rhinolophus bat which is > 96% homologous with the modern SARS-CoV-2 virus and it is just 79% homologous with the initial SARS-CoV (Fisher & Heymann, 2020). The fast-growing number of infected cases globally urged the World Health Organization to announce a state of global health emergency to correlate scientific and medical disciplines to develop rapidly an effective treatment for patients, (Morse et al., 2020; Sarma et al., 2020) elderly patients and people with severe underlying health diseases like heart diseases, lung illness, and diabetic patients, for instance, appear to be at greater risk of revealing severe SARS-CoV-2 requires immediate intervention rather than waiting virus vaccine which may require 1 year to be available (Enayatkhani, 2020). While drug repurposing could be a short-term and fast resolution to handle SARS-CoV-2 patients (Elfiky, 2020; R. J. Khan et al., 2020; Kumar et al., 2019), repurposing existing drugs can offer a good choice to overcome the virus and offer better risk-versus trade-off as compared with discovering new drug and can help overcome time waiting for new therapy rather than use the available resources (Elmezayen, 2020; Muralidharan, 2020) One successful repurposing drug story includes duloxetine which originally developed for depression and FDA approved as the first-in-class choice for stress urinary incontinence (Sweeney & Chancellor, 2005), duloxetine initially created as antidepressant also is now passed to Phase III clinical trials as a first-in-class treatment for premature ejaculation (McMahon, 2012) and thalidomide, which had a tragic start as an over-the-counter sedative for morning sickness in pregnancy is now being applied to manage leprosy and multiple myeloma (Hideshima & Anderson, 2002).
As a result of, a newly issued X-ray crystal of SARS-CoV-2 Main protein (Mpro), we planning to use computational approaches (Cameron et al., 2013) to contribute to find an effective treatment for SARS-Cov-2.Thereby, computational analyses speed up these approaches since they allow to handle millions of data simultaneously (Gupta et al., 2020). Molecular docking includes a set of computational methods and algorithms that aimed to identify novel relationships between chemical ligands and targets through using the modelling of their direct physical interaction (Aanouz, 2020; Ekins et al., 2007). In present study, we attend to evaluate some of approved drugs to be as covalent binders, irreversible interactions, which can provide a powerful strategy to fight against epidemic viruses. And molecular dynamics simulations can give a more detail for the image which got from molecular covalent docking.
2. Materials and methods
2.1. Covalent virtual screening
The crystal structure of FDA-available covalent drugs which available in Table 1 were selected based on the review of Kumalo et al. (2015) and some of the antiviral drugs that can form a covalent bond to the target protein. And we aim also here to redirect them for other indications specially to see their possibility to fight against SARS-CoV-2. Thence, we searched about the chosen drugs in PubChem (https://pubchem.ncbi.nlm.nih.gov/) to identify the possibility of the selected drugs to be as covalent binders toward SARS-CoV-2 Mpro. Where, PubChem provides detailed information about the selected drugs rather than other repositories, especially, the property of drug to form covalent bond. Before starting covalent docking, we downloaded the selected drugs one by one and optimize them by using the Ligprep (Lim et al., 2020) based on the OPLS_2005 force field and generated possible state employing Epik in the Schrodinger 2020 (Elfiky 2020; S. A. Khan et al.,2020). In the next step, the structure of SARS-CoV-2 Mpro (6LU7) was downloaded from Protein Data Bank (http://www.rcsb.org/pdb) (Jin et al. , 2020). The protease structure was optimized by adding hydrogens, removing water molecules and optimizing charge using the Protein Preparation Wizard module (Kumalo et al., 2015) in Schrodinger suite 2020-1. The covalent docking protocol was preferred since the cysteine 145 residue which is available in the binding site of SARS-CoV-2 Mpro considered as a vital residue that can form a covalent bond with the drug if it can interact covalently. The different mechanisms for the Cys145-FDA drug were mentioned in Table 1 based on the nature of the drug so that the reactive functional group on the ligand and receptor residue are identified and the bond is formed between the correct atoms. These covalently docked complexes were created using Covdock in Schrodinger suite 2020-1. Finally, we selected the lowest the MMGBSA value for each drug as a propriate conformation of the drug inside the binding pocket.
Table 1.
Selected FDA drugs with PubChem ID, molecular weight covalent docking results showing type of based reaction for constructing covalent bond, docking score, Glide Score and RMSD.
| Entry name | PubChem ID | Molecular weight | Type of reaction | Docking score (kcal/mol) | Gscore (kcal/mol) | MMGBSA dG bind (kcal/mol) | RMSD (Å) |
|---|---|---|---|---|---|---|---|
| saquinavir | 441243 | 670.8 | Nucleophilic addition to a double bond | –9.856 | –10.449 | –72.17 | 0.039 |
| ritonavir | 392622 | 720 | Nucleophilic addition to a double bond | –8.361 | –8.834 | –72.02 | 0.047 |
| remdesivir | 121304016 | 602.6 | Nucleophilic addition to a triple bond | –7.9 | –7.925 | –65.19 | 0.036 |
| delavirdine | 6321416.1 | 456.6 | Nucleophilic addition to a double bond | –6.756 | –6.128 | –57.65 | 0.035 |
| cefuroxime axetil | 6321416 | 510.5 | Nucleophilic addition to a double bond | –6.801 | –6.794 | –54.25 | 0.047 |
| oseltamivir | 65028 | 312.4 | Nucleophilic addition to a double bond | –7.142 | –7.014 | –51.8 | 0.026 |
| prevacid | 3883 | 369.4 | Nucleophilic substitution | –6.652 | –6.222 | –51.14 | 0.02 |
| PRD_002214 (ref) | Nucleophilic addition to a double bond | –6.99 | –6.866 | –50.69 | 0.039 | ||
| protonix | 4679 | 383.4 | Nucleophilic substitution | –4.838 | –3.137 | –50.19 | 0.045 |
| lopinavir | 92727 | 628.8 | Nucleophilic addition to a double bond | –7.399 | –7.441 | –49.84 | 0.042 |
| nelfinavir– | 64143 | 567.8 | Nucleophilic addition to a double bond | –5.263 | –6.355 | −49.44 | 0.039 |
| ceftriaxone | 5479530 | 554.6 | Nucleophilic addition to a double bond | –6.41 | –7.59 | –47.06 | 0.021 |
| orlistat | 3034010 | 495.7 | Nucleophilic addition to a double bond | –6.751 | –6.779 | –45.72 | 0.03 |
| meropenem | 441130 | 383.5 | Nucleophilic addition to a double bond | –5.159 | –5.289 | –45.25 | 0.021 |
| floxuridine | 5790 | 246.19 | Nucleophilic addition to a double bond | –5.784 | –5.383 | –42.66 | 0.034 |
| exemestane | 60198 | 296.4 | Michael addition | –4.543 | –4.543 | –32.02 | 0.049 |
| dutasteride | 6918296 | 528.5 | Nucleophilic addition to a double bond | –5.517 | –5.927 | –40.19 | 0.049 |
| decitabine | 451668 | 228.21 | Nucleophilic addition to a double bond | –6.497 | –6.916 | –37.9 | 0.029 |
| bortezomib | 387447 | 384.2 | Boronic acid addition | –8.091 | –8.091 | –38.49 | 0.033 |
| omnicef | 6915944 | 395.4 | Nucleophilic addition to a double bond | –6.516 | –6.427 | –37.94 | 0.046 |
| tipranavir | 54682461 | 602.7 | Nucleophilic substitution | –5.158 | –3.253 | –37.54 | 0.04 |
| ribavirin– | 37542 | 244.2 | Nucleophilic addition to a double bond | –5.968 | –5.404 | –36.62 | 0.042 |
| baloxavir | 124081876 | 483.5 | Nucleophilic addition to a double bond | –3.837 | –3.637 | –35.2 | 0.024 |
| proscar | 57363 | 372.5 | Nucleophilic addition to a double bond | –4.607 | –6.893 | –35.18 | 0.037 |
| etravirine | 193962 | 435.3 | Nucleophilic addition to a triple bond | –5.853 | –5.707 | –35.17 | 0.029 |
| darunavir | 213039 | 547.7 | Nucleophilic addition to a double bond | –6.513 | –6.772 | –34.1 | 0.046 |
| fosamprenavir | 131536 | 585.6 | Nucleophilic addition to a double bond | –5.689 | –5.908 | –32.8 | 0.035 |
| saxagliptin | 11243969 | 315.4 | Nucleophilic addition to a triple bond | –5.96 | –4.219 | –32.67 | 0.039 |
| warfarin | 54678486 | 308.3 | Nucleophilic addition to a double bond | –5.285 | –5.319 | –32.65 | 0.036 |
| ceclor | 51039 | 367.8 | Nucleophilic addition to a double bond | –6.066 | –7.678 | –32.48 | 0.033 |
| penicillin | 6869 | 334.4 | Nucleophilic addition to a double bond | –5.401 | –5.933 | –32.39 | 0.306 |
| vildagliptin | 6918537 | 303.4 | Nucleophilic addition to a double bond | –5.241 | –4.159 | –32 | 0.033 |
| cephalexin | 27447 | 347.4 | Nucleophilic addition to a double bond | –5.751 | –6.686 | –31.02 | 0.036 |
| propylthiouracil | 657298 | 170.23 | Nucleophilic addition to a double bond | –4.769 | –3.623 | –28.29 | 0.039 |
| vigabatrin | 5665 | 129.16 | Nucleophilic addition to a double bond | –4.226 | –4.266 | –27.65 | 0.045 |
| carbidopa | 34359 | 226.23 | Nucleophilic addition to a double bond | –6.069 | –6.626 | –26.85 | 0.046 |
| isoniazid | 3767 | 137.14 | Nucleophilic addition to a double bond | –4.214 | –3.12 | –25.95 | 0.047 |
| mercaptopurine | 667490 | 152.18 | Nucleophilic addition to a double bond | –2.415 | –3.455 | –25.57 | 0.04 |
| efavirenz | 64139 | 315.67 | Nucleophilic substitution | –4.565 | –4.565 | –25.37 | 0.043 |
| gemcitabine | 60750 | 263.2 | Nucleophilic addition to a double bond | –5.327 | –5.333 | –25.26 | 0.044 |
| eflornithine | 3009 | 182.17 | Nucleophilic addition to a double bond | –4.599 | –5.52 | –22.38 | 0.032 |
| azvudine | 24769759 | 286.22 | Nucleophilic addition to a double bond | –3.889 | –3.889 | –36.46 | 0.04 |
| D-cycloserine | 6234 | 102.09 | Nucleophilic addition to a double bond | –3.315 | –3.562 | –20.6 | 0.032 |
| disulfiram | 3117 | 296.5 | Nucleophilic addition to a double bond | –3.027 | –5.252 | –20.35 | 0.022 |
| aspirin | 2244 | 180.16 | Nucleophilic addition to a double bond | –3.841 | –3.841 | –18.73 | 0.041 |
| fosfomycin | 446987 | 138.0 | Epoxide opening | –3.214 | –3.467 | –14.49 | 0.048 |
| favipiravir | 492405 | 157.1 | Nucleophilic substitution | –2.855 | –2.865 | –8.62 | 0.036 |
| indinavir | 5362440 | 613.8 | Nucleophilic addition to a double bond | –6.067 | –6.458 | –36.34 | 0.035 |
2.2. Prime MM-GBSA
The chosen drugs binding energies were calculated using Prime MM‑GBSA modules (Vijayakumar et al., 2014) in the Schrödinger (2020). The best poses of selected drugs—SARS-CoV-2 Mpro—were chosen to obtain the binding free energy calculation. Prime MMGBSA is a method that combines Optimized Potential for Liquid Simulations‑All Atoms (OPLSAA) force field, molecular mechanics energies (EMM), an SGB solvation model for polar solvation (GSGB), and a non‑polar solvation term (GNP) composed of the non‑polar solvent accessible surface area and van der Waals interactions. The total binding free energy: Δ Gbind = Gcomplex – (G-protein + Gligand).
2.3. Molecular dynamics simulation
Molecular dynamics simulations are a decision-making process for inspections of protein-drug complexes’ stabilities (Al-Khafaji & Taskin Tok, 2020b). It is used to clarify the dynamic behavior at an atomic level of biological systems, which is hard to handle in labs (Shukla et al., 2019). In the current study, we conducted molecular dynamics simulations for the top three drugs based on MMGBSA values. The got protein-drug complex structures from covalent docking were submitted to MD simulations (saquinavir, ritonavir, and remdesivir with SARS-CoV-2 Mpro). We hired Gromacs 2018.1 to run 50 ns MD simulations (Abraham et al., 2015). Charmm 27 force field for all atoms were chosen to run MD simulation (Bjelkmar et al., 2010). We used Swiss PARAM to produce the topologies of drugs (Zoete et al., 2011). All protein-drug systems were solvated with three-point transferable intermolecular potential (TIP3P) and their charges were neutralized via adding Na or Cl ions. In the following step, the protein-drugs systems were energetically minimized through the steepest descent algorithm at a tolerance value of 1000 kJ/mol.nm. Then the equilibration with position restraint on the protein molecules for 0.1 ns using NVT and NPT ensembles were done. Electrostatic interactions were evaluated by Particle Mesh Ewald summation (Darden et al., 1993). We performed the molecular dynamics simulation with no restraint on the protein molecules or ligand to determine the stability in the final step (time step of 0.015 ns). RMSD, RMSF, Rg, and number of hydrogen bonds were chosen to analyze MD trajectories by using GROMACS utilities.
2.4. Principal component analysis
The principal component analysis (PCA) approach was employed to calculate eigenvectors and eigenvalues and their projection along with the first two principal components (Al-Khafaji & Taskin Tok, 2020a). This approach is based on the protocol of GROMACS 2018.1 (Abraham et al., 2015). It is used to simplify the effect of drugs on the dynamic motion of the targeted protein where it extracts the dynamic motions in simulations that are required for their biological function (Amadei et al., 1993). We got the principal component analysis from the MD trajectories. A series of eigenvectors and eigenvalues were generated by diagonalizing the matrix. We chose trajectories of the protein backbone of the complexes to get 2 D-projection of motion of trajectory.
3. Results
To assess the possibility of selected FDA drugs to work as treatment of SARS-CoV-2, the covalent docking was utilized to screen the selected library and rank them according to their binding affinities. The calculated binding free energies of some available drugs using docking score, Glide Gscore, and ensemble-average MM/GBSA are shown in Table 1.
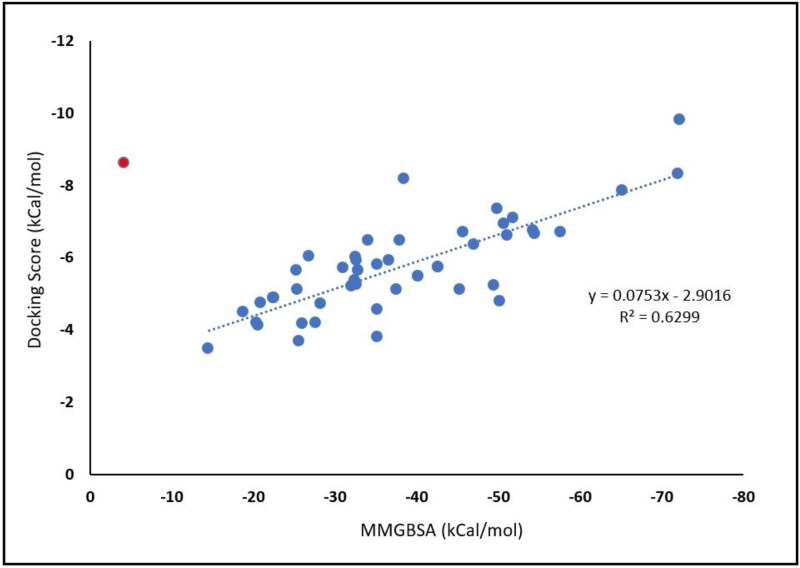
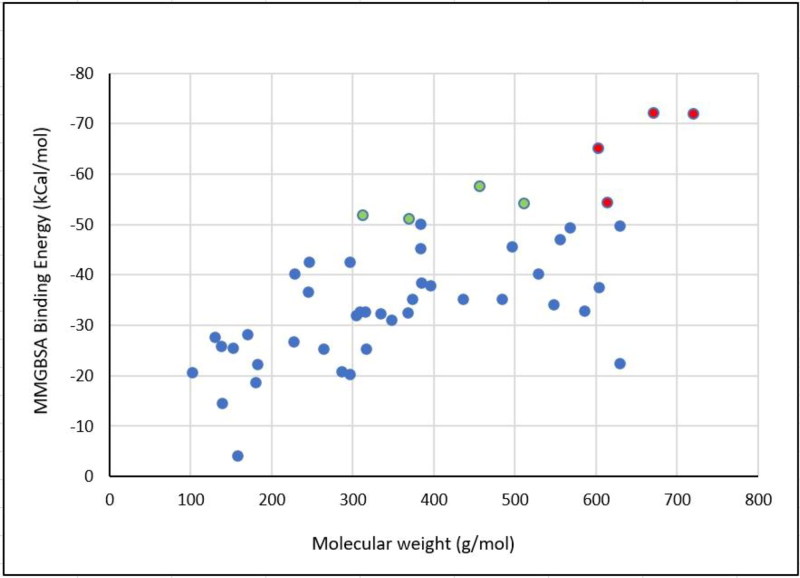
To validate our covalent docking results, the correlation between MMGBSA and docking score was constructed (Figure 1). The binding energy MMGBSA-docking score correlation shows a good correlation (R2 = 0.6299). Based on this correlation, we chose the top three ranked-MMGBSA and docking score values of FDA drugs for dissection their binding modes inside the binding site. Covalent docking showed that saquinavir, ritonavir, remdesivir, delavirdine, cefuroxime axetil, oseltamivir, and prevacid have −72.17, −72.02, −65.19, −57.65, −54.25, −51.8, and −51.14, respectively. Where all the top-eight FDA drugs show a higher affinity to form a covalent, irreversible bond with Cys145 of SARS-CoV-2 Mpro. Here we investigated the role of molecular weight upon the affinity of selected drugs to bind covalently to Cys145, whereas 57% of selected drugs which have molecular weight over than 600 g/mol (Figure 2). and have higher free binding energy MMGBSA than −50 kcal/mol. While the ratio of selected drugs decreased to be 16% of drugs which can form covalent bonding with over than −50 kcal/mol. Surprisingly, none of the selected drugs which have a molecular weight below than 300 g/mol can form a good affinity of binding energy. This indicates that higher molecular weight covalent warheads can form stable and efficient binding energy.
Figure 1.
The correlation between MMGBSA binding energies and docking score.
Figure 2.
Effect of molecular weight upon the MMGBSA binding energy.
3.1. Docked complex analyses
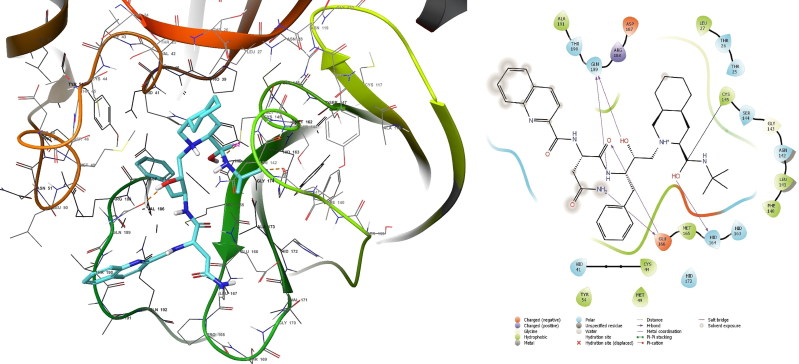
What stands out in the Table 1 is saquinavir has the highest binding affinity (lowest binding energy MMGBSA of 72.17 kcal/mol). Therefore, the deep examination of saquinavir is needed. Figure 3 shows that saquinavir not only formed covalent bond of 1.81 Å with Cys145 but also formed five hydrogen bonds inside the pocket of SARS-CoV-2 Mpro. In the second rank ritonavir as presented in Table 1 has −72.02 kcal/mol.
Figure 3.
Covalent docking analysis of saquinavir inside the SARS-CoV-2 Mpro.
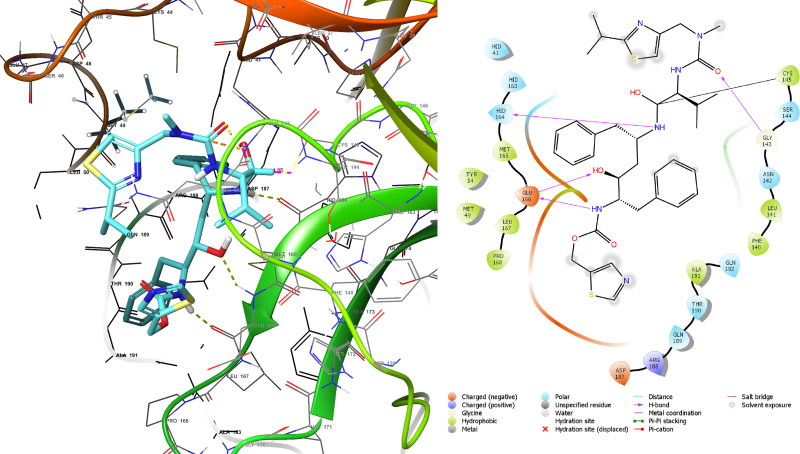
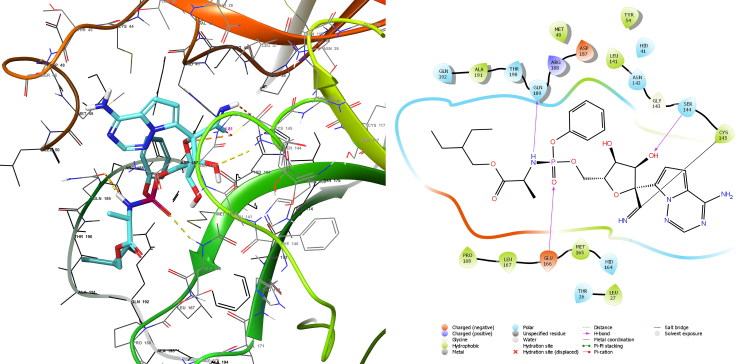
This high affinity resulted from covalent bond between ritonavir and Cys145 of 1.82 Å (Figure 4) through nucleophilic addition to double bond reaction besides it interacted within binding site by forming three hydrogen bonds. Despite remdesivir formed covalent bond with Cys145 of 1.82 Å and three hydrogen bonds (Figure 5) and this is similar to Ritonavir, but the MMGBSA value is lower than that of ritonavir this may due to nature of reaction which in remdesivir nucleophilic addition to triple bond.
Figure 4.
Covalent docking analysis of ritonavir inside the SARS-CoV-2 Mpro.
Figure 5.
Covalent docking analysis of remdesivir inside the SARS-CoV-2 Mpro.
3.2. Molecular dynamic simulation
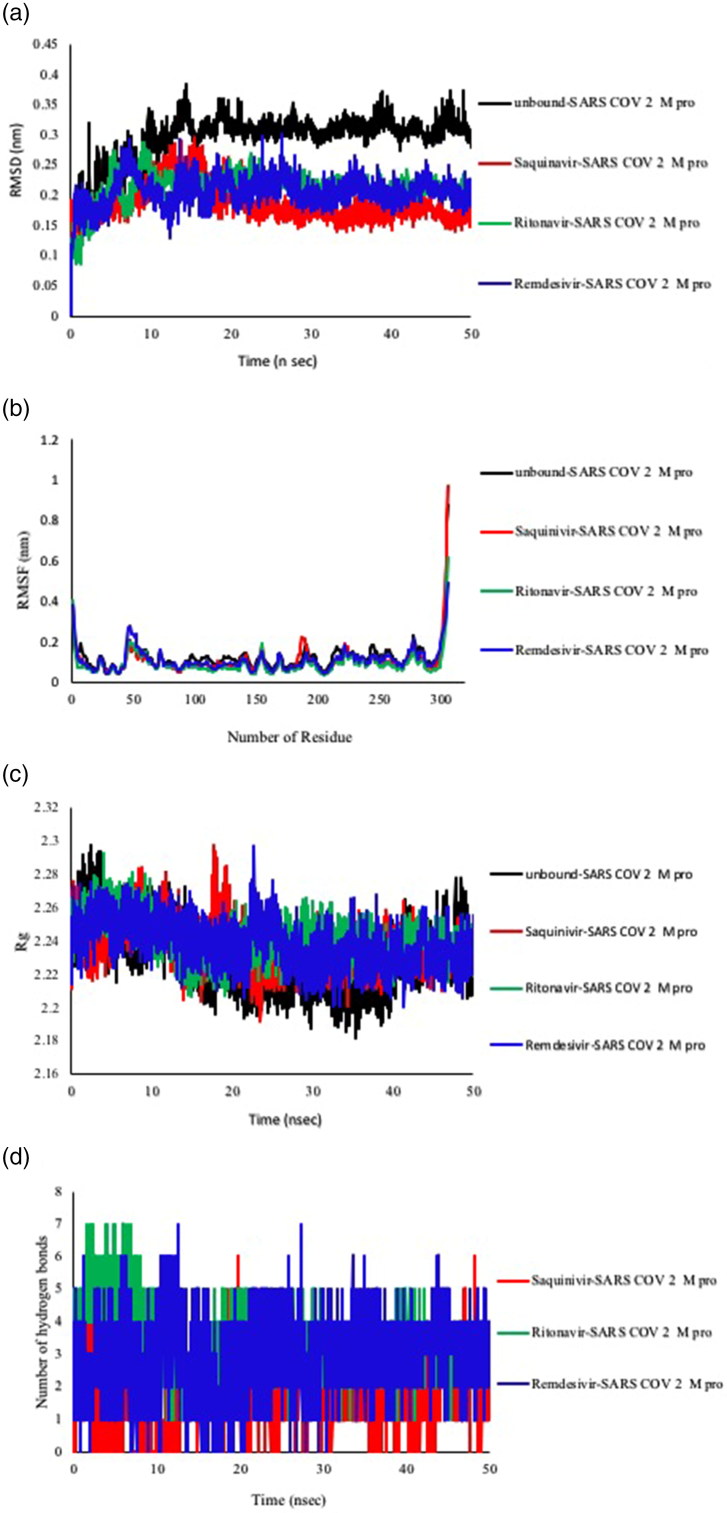
The effect of drug-protein interactions upon dynamics of biological system is a fundamental in drug discovery thereby we used RMSD to investigate the influence of saquinavir, ritonavir, and remdesivir upon the stability of SARS-CoV-2 Mpro. We utilized Gromacs to execute the MD simulations of 50 ns for three drug-protein systems besides of apo protein. RMSD fluctuations for both apo and hollo forms were measured and presented. The RMSD was calculated to assess the overall dynamics, stability, and convergence of the various systems and the results are presented in Figure 6(a).
Figure 6.
Analysis of RMSD, RMSF, Rg and Hydrogen bonding of saquinvir-SARS-CoV-2 Mpro, ritonavir- SARS-CoV-2 Mpro and remdesivir- SARS-CoV-2 Mpro. (a)The protein’s backbone RMSD values with respect to time. (b) RMSF of the protein’s backbone with respect to position of residue in the protein. (c) Rg of the protein backbone over the entire time of MD simulation. (d) The number of hydrogen bonds through all the time of MD simulation.
Figure 6(a) shows there is a significant decrease in the RMSD value when SARS-CoV-2 main protease whether it bound to saquinavir, ritonavir, or remedisivir. Further analysis revealed that the RMSD average of apo SARS-CoV-2 main protease was 0.294 nm but when it bound to saquinavir, ritonavir, and remedisivir the RMSD averages were.01865, 0.2130, and 0.2053 nm, respectively. Another significant aspect of MD simulation is the flexibility of protein’s backbone which can be assessed through measuring RMSF value. The results of the comparative analysis between these drugs and their effects upon SARS-CoV-2 main protease are illustrated in Figure 6(b). Closer scrutiny of Figure 6(b) exhibits the binding of saquinavir diminished the fluctuations of the protein’s backbone. And behavior is also can be seen from Figure 6(b) where the binding of both ritonavir and remedisivir led to reducing the flexibility of the protein. The radius of gyration (Rg) is a definition of system’s density, and substantially influences the folding rate and stability of proteins. Rg was employed to assess the compactness of all complexes. In this work, Rg values are in agreement with RMSF values where there are no significant differences between apo form and hollo forms as presented in Figure 6(c). This reveals that protein remained stable and compact all through the 50 ns time. The number of intermolecular hydrogen bonds is a vital aspect to give an impression about the stability of drug and protein. Also, the number of hydrogen bonds is relevant to the binding scores of molecular docking process. We calculated the hydrogen bonds over time to validate the stable interactions between top three drugs and their corresponding target. As seen in the Figure 6(d) the Ritonavir has highest order of hydrogen bonding (the average =3.35), while saquinavir has an average of 1.68.
3.3. Principle component analysis (PCA)
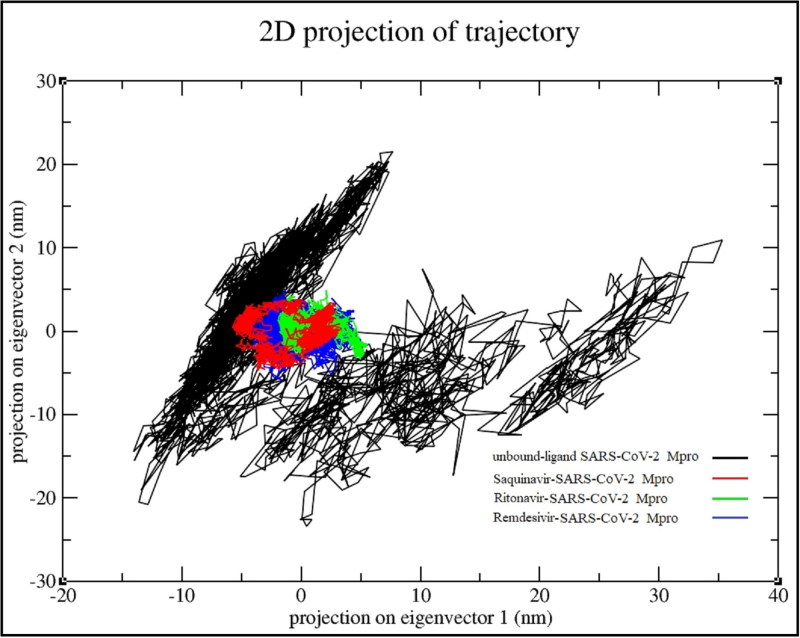
The essential dynamic method is a tool to explore the dynamical behavior in the space of SARS-CoV-2 main protease combined with saquinavir, ritonavir, and remedisivir. Basically, the comparison of the drug-bound SARS-CoV-2 Mpro and drug-unbound was made as reference. In order to further understand the configurational space, we selected the first two principal components (PC1, PC2) to analyze their projection of trajectories during the simulations of ligand free and ligand bound SARS-CoV-2 Mpro of the phase space (shown in Figure 7). During the four system simulations, the results clearly show that the unbound ligand protein covered a wider region of phase space, while all three drug-protein system occupied a smaller region of phase space. Especially, saquinavir reduced the essential dynamics to lowest degree of functional motions as compared with another drugs. Moreover, the PCA results suggest that the drug-bound SARS-CoV-2 Mpro is more stable than ligand-unbound SARS-CoV-2 Mpro form of SARS-CoV-2 Mpro. In short, the PCA results are also in agreement with the RMSD and RMSF results, which enhance the validity of the performed analysis.
Figure 7.
Two-dimensional projection of motion of trajectory of SARS-CoV-2 Mpro bound with drugs over the PC1 and PC2.
4. Discussion
SARS-CoV-2 causes major pandemic health issue since its spread across the world and can infect humans mainly respiratory system causing severe pneumonia with no vaccine and drug treatment available. Prior study that have referred to the significance of molecular docking to determine effective treatment in short time (Wu, et al., 2020). In reviewing the literature, we took the advantage of possibility of FDA available drugs that can be as a covalent warhead to inhibit the SARS-CoV-2 Mpro with Cys145. An initial objective of the project was to identify effective and applicable treatment. The present study focused on the main protease (Mpro), especially PDB ID (6LU7) as a potential target for several marketed drugs as possible therapeutic option to combat the virus to see the capability of these drugs to bind with the cysteine 145 residue which is available in the binding site of SARS-CoV-2 main protease. The Mpro in SARS-CoV-2 is necessary for the proteolytic maturation of the virus targeting this protein to limit the expansion of infection by hindering the cleavage of the viral polyprotein. The most interesting finding was that the both saquinavir and ritonavir have the same affinity (binding energy MMGBSA ≈ −72 kcal/mol) to block binding site of SARS-CoV-2 with irreversible interactions. This study supports evidence from previous observations that lopinavir/ritonavir can inhibit SARS-CoV-2 (Lim et al., 2020). Another important outcome was that the remdesivir comes in second rank with binding affinity (MMGBSA = −65 kcal/mol ). Whereas previous research has established that remdesivir can inhibit SARS-CoV-2 M proenzyme through docking results. Another significant finding, delavirdine computationally showed the ability to form irreversible covalent bond of −57.65 kcal/mol MMGBSA binding energy. Moreover, indinavir and cefuroxime axetil exhibited the possibility to halt the pocket of SARS-CoV-2 Mpro by forming stable interaction through covalent bonding and hydrogen bonding (MMGBSA binding energies ≈ −54 Kcal/mol). Furthermore, oseltamivir and prevacid exhibited same affinity to bind with SARS-CoV-2 protease through one covalent bond and one hydrogen bond (≈ −51 Kcal/mol), which is a good explanation for the activity of oseltamivir (Peeri et al., 2020). In this study, results obtained show the binding affinity of drugs to an active site depends on several factors mainly the ability of a compound to form a covalent bond with amino acid residues of the Mpro (Cys145) and length of a covalent bond, the number of H-bonds that can form with a pocket of the active site and type of nucleophilic addition of unsaturated bonds. In this concept, the structural features in saquinavir and ritonavir like free amine group (-NH2), hydroxyl groups (-OH), carbonyl groups (C=O) in addition to ether group play key structural feature to form H-bond. Results show the promising activities for antiretroviral drugs saquinavir that used for HIV/AIDS more than ritonavir and remdesivir followed by lopinavir as the best drugs to bind covalently toward SARS-CoV-2 Mpro with lowest energy of binding. As discussed above, saquinavir, ritonavir, remdesivir, delavirdine, cefuroxime axetil, oseltamivir, and prevacid showed the ability to block the binding site of SARS-CoV-2 Mpro by forming covalent bond and stabilized with hydrogen bonding.
These outcomes are contrary to that of Kandeel and Al-Nazawi (2020), they found that ribavirin, telbivudine, vitamin B12, and nicotinamide can be can form non-bonding interactions. Whilst our results showed that, saquinavir, ritonavir, and remdesivir can form irreversible interactions, which are considered an effective way for viral infections.
In the present work, comparative molecular dynamics simulations were conducted to evaluate the effects of saquinavir, ritonavir, and remdesivir on the conformational and dynamical demeanor in either apo or hollo forms to understand the inhibitory possibility at atomic level. An initial mission of the project was to identify rapid, effective, safe, and available for large proportions of people, so we run covalent docking and MMGBSA to sift the possibility of these drug to form irreversible interactions. Therefore, we had chosen these drugs because they carry covalent warheads that can bind covalently to the target. But the docking results are not adequate, so we run MD simulations examine how much these drugs are able to form stable interactions with targeted protein. One interesting finding is the RMSD of protein backbone in apo status has higher average value, whereas the binding with top three drugs diminished these RMSD average. Another important finding was that the RMSF of protein’s backbone be less flexible when it compared to apo form of SARS-CoV-2 main protease. It is somewhat surprising that the binding of three drugs have not noted in Rg values. It is not yet clear whether the top three drugs can show an evidence to inhibit the targeted protein, thereby we lean on the principal component analysis to analyze the MD trajectories to judge without dispute. The most obvious finding to emerge from 2D PCA analysis is that the binding of under investigated drugs caused stately impact on essential dynamics of protein by reducing its essential dynamics to its least possible motions. These results seem to be consistent with RMSD results. A possible explanation for this might be that binding of the drugs make the binding site much narrower due to hydrogen bindings this makes the 3D structure of targeted protein more rigid so it will lose its biological functions. It is possible, therefore, that using these available drugs in two ways: either alone or in combined way. These findings suggest the possible use of nominated drugs against SARS-CoV-2 in short time as approved by covalent docking screening. Also, the present results are significant in at least directing the clinicians to use these safe drugs to stop development of Corona virus and second giving hope that available drugs that can be efficient treatment.
5. Conclusions
Coronavirus today emphasizes as a potential threat to all people worldwide. Although extensive researches have been directed to stop SARS-CoV-2, but till now there is no medication. Meanwhile, the spreading with complicated crisis requires immediate therapy to overcome the spread and minimize mortality of SARS-CoV-2. The aim of the present study was to discover effective treatment through repurposing some of available FDA-approved drugs against SARS-CoV-2 Mpro. Where, they can provide covalent warheads in virtual screening. The most prominent finding to emerge from this study is that the affinity of covalent binder toward SARS-CoV-2 Mpro is ranked: saquinavir > ritonavir > remdesivir > delavirdine > cefuroxime axetil > oseltamivir = prevacid. One of the more noteworthy findings in this study is that MD simulation analysis that saquinavir, ritonavir, and remdesivir can form stable interaction inside the binding site of SARS-CoV-2 Mpro. Also, they restricted the essential motions of protein. Overall, the results of screening toward Mpret encourage for further clinical evaluations. They can be easy to reach and exploit as persuasive treatments for SARS-CoV-2.
Acknowledgements
Authors thankful for staff membered of Gaziantep University Institute of Health Sciences, Department of Bioinformatics and Computational Biology.
Disclosure statement
The authors declare no conflict of interest.
References
- Aanouz I. (2020). Moroccan Medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics, 38, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., & Lindahl E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Al-Khafaji K., & Taskin Tok T. (2020. a). Amygdalin as multi-target anticancer drug against targets of cell division cycle: Double docking and molecular dynamics simulation. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1742792 [DOI] [PubMed] [Google Scholar]
- Al-Khafaji K., & Taskin Tok T. (2020. b). Understanding the mechanism of Amygdalin’s multifunctional anti-cancer action using computational approach. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1736159 [DOI] [PubMed] [Google Scholar]
- Amadei A., Linssen A. B., & Berendsen H. J. (1993). Essential dynamics of proteins. Proteins, 17(4), 412–425. 10.1002/prot.340170408 [DOI] [PubMed] [Google Scholar]
- Bjelkmar P., Larsson P., Cuendet M. A., Hess B., & Lindahl E. (2010). Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. Journal of Chemical Theory and Computation, 6(2), 459–466. 10.1021/ct900549r [DOI] [PubMed] [Google Scholar]
- Cameron, D., Bodenreider, O., Yalamanchili, H., Danh, T., Vallabhaneni, S., Thirunarayan, K., Sheth, A. P., & Rindflesch, T. C. (2013). A graph-based recovery and decomposition of Swanson's hypothesis using semantic predications. Journal of biomedical informatics, 46(2), 238–251. 10.1016/j.jbi.2012.09 [DOI] [PMC free article] [PubMed]
- Chan J. F.-W., Yuan S., Kok K.-H., To K. K.-W., Chu H., Yang J., Xing F., Liu J., Yip C. C.-Y., Poon R. W.-S., Tsoi H.-W., Lo S. K.-F., Chan K.-H., Poon V. K.-M., Chan W.-M., Ip J. D., Cai J.-P., Cheng V. C.-C., Chen H., … Yuen K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., & Zhang L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., & Guo D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., & Pedersen L. (1993). Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Ekins S., Mestres J., & Testa B. (2007). In silico pharmacology for drug discovery: Methods for virtual ligand screening and profiling. British Journal of Pharmacology, 152(1), 9–20. 10.1038/sj.bjp.0707305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D., & Heymann D. (2020). Q&A: The novel coronavirus outbreak causing COVID-19. BMC Medicine, 18(1), 57 10.1186/s12916-020-01533-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. K. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T., & Anderson K. C. (2002). Molecular mechanisms of novel therapeutic approaches for multiple myeloma. Nat. Rev. Cancer, 2(12), 927–937. 10.1038/nrc952 [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., … Yang H. (2020). Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature, 1–27. [DOI] [PubMed] [Google Scholar]
- Joshi R. S. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., & Al-Nazawi M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. [DOI] [PubMed] [Google Scholar]
- Khan R. J. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumalo H., Bhakat S., & Soliman M. (2015). Theory and applications of covalent docking in drug discovery: Merits and pitfalls. Molecules, 20(2), 1984–2000. 10.3390/molecules20021984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Gupta S., Chand Yadav T., Pruthi V., Kumar Varadwaj P., & Goel N. (2019). Extrapolation of phenolic compounds as multi-target agents against cancer and inflammation . J. Biomol. Struct. Dyn., 37(9), 2355–2369. 10.1080/07391102.2018.1481457 [DOI] [PubMed] [Google Scholar]
- Lim J., Jeon S., Shin H.-Y., Kim M. J., Seong Y. M., Lee W. J., Choe K.-W., Kang Y. M., Lee B., & Park S.-J. (2020). Case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. Journal of Korean Medical Science, 35(6), 1–6. 10.3346/jkms.2020.35.e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C. G. (2012). Dapoxetine: A new option in the medical management of premature ejaculation. Ther. Adv. Urol., 4(5), 233–251. 10.1177/1756287212453866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse J. S., Lalonde T., Xu S., & Liu W. R. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan N. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. [DOI] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeri N. C., Shrestha N., Rahman M. S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., & Haque U. (2020). The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? International Journal of Epidemiology, 1–10. 10.1093/ije/dyaa033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kuamr S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B.. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, Maestro. (2020). LLC New York, NY, USA, 2019.
- Shukla R., Munjal N. S., & Singh T. R. (2019). Identification of novel small molecules against GSK3β for Alzheimer’s disease using chemoinformatics approach. Journal of Molecular Graphics & Modelling, 91, 91–104. 10.1016/j.jmgm.2019.06.008 [DOI] [PubMed] [Google Scholar]
- Sweeney, D. D., & Chancellor, M. B. (2005). Treatment of stress urinary incontinence with duloxetine hydrochloride. Reviews in urology, 7, 81–86. [PMC free article] [PubMed]
- Vijayakumar B., Parasuraman S., Raveendran R., & Velmurugan D. (2014). Identification of natural inhibitors against angiotensin I converting enzyme for cardiac safety using induced fit docking and MM-GBSA studies. Pharmacognosy Magazine, 10(Suppl 3), S639–S644. 10.4103/0973-1296.139809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., & Li H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 1–23. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Cuendet M. A., Grosdidier A., & Michielin O. (2011). SwissParam: A fast force field generation tool for small organic molecules. Journal of Computational Chemistry, 32(11), 2359–2368. 10.1002/jcc.21816 [DOI] [PubMed] [Google Scholar]