Abstract
Emergent novel SARS-CoV-2 is responsible for the current pandemic outbreak of severe acute respiratory syndrome with high mortality among the symptomatic population worldwide. Given the absence of a current vaccine or specific antiviral treatment, it is urgent to search for FDA-approved drugs that can potentially inhibit essential viral enzymes. The inhibition of 3CLpro has potential medical application, due to the fact that it is required for processing of the first translated replicase polyproteins into a series of native proteins, which are essential for viral replication in the host cell. We employed an in silico approach to test if disulfiram, as well as its metabolites, and captopril could be used as potential antiviral drugs against COVID-19. We provide data on the potential covalent interaction of disulfiram and its metabolites with the substrate binding subsite of 3CLpro and propose a possible mechanism for the irreversible protease inactivation thought the reaction of the aforementioned compounds with the Cys145. Although, captopril is shown to be a potential ligand of 3CLpro, it is not recommended anti-COVID-19 therapy, due to the fact that it can induce the expression of the viral cellular receptor such as, angiotensin-converting enzyme ACE-2, and thus, making the patient potentially more susceptible to infection. On the other hand, disulfiram, an alcoholism-averting drug, has been previously proposed as an antimicrobial and anti-SARS and MERS agent, safe to use even at higher doses with low side effects, it is recommended to be tested for control of SARS-CoV-2 infection.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, COVID-19, 3CL main protease, thiol-reacting drugs, disulfiram
1. Introduction
With more than 2,000 000 cases currently reported worldwide (Dong et al., 2020) (https://arcg.is/0fHmTX April 14, 2020), emerging novel respiratory coronavirus SARS-CoV-2 (Zhou et al., 2020) is responsible of a pandemic outbreak of atypical pneumonia, which can result into a severe acute respiratory stress syndrome (ARSS) and even death in 2-3% of all infected individuals (Walls et al., 2020; Zhou et al., 2020; Zhu et al., 2020). A member of the viral family Coronaviridae, the virus infectious particle consists of an enveloped proteinaceous capsid containing a positive-sense single-stranded RNA genome (Boopathi et al., 2020; Zhou et al., 2020; Zhu et al., 2020). Transmitted by contaminated droplets, the virus enters the cell by binding of its transmembrane trimeric spike (S) glycoprotein to the host cellular receptor, angiotensin-converting enzyme 2 (ACE2) (Hasan et al., 2020; Walls et al., 2020), making individuals of 65 years of age and older, as well as patients with hypertension and diabetes the most susceptible population (Mahase, 2020; Walls et al., 2020; Wang et al., 2020). Upon entrance to the susceptible cell by the mechanism of endocytosis, acidification of this organelle promotes conformational changes on the S protein, exposing its fusogenic domain, which leads to fusion of the viral envelope and the endosomal membrane, followed by the release of the viral ssRNA into the cytoplasm (Colson et al., 2020; Liu et al., 2020; Walls et al., 2020; Wang et al., 2020) . Next, ribosomes translate the 5′-segment of the viral genome that contains ORF1a and ORF1b into the two overlapping replicase polyproteins, termed pp1a and pp1ab (Zhou et al., 2020; Zhu et al., 2020) that are later cleaved into 16 different functional proteins (non-structural proteins, NSP1-16) by its two virally-encoded proteases, papain-like protease (PLpro) and 3 C-like protease (3CLpro) (Liu et al., 2020; Woo et al., 2005; Zhang et al., 2020). Thus, functional activity of these viral proteases is essential for viral replication inside the infected cell; and consequently, they represent potential antiviral targets (Khan et al., 2020; Zhang et al., 2020). The structure of the SARS-CoV-2 main protease, 3CLpro was recently resolved and is available at the Protein Data Bank (PDB) (code 6lu7) (Zhang et al., 2020). 3CLpro is a cysteinyl protease that contains a Cys-His catalytic dyad, which initially self-cleaves from the polyprotein replicase complex; and then, it specifically digests the peptide bond at ten conserved Gln sites of the C-terminal end of the viral polyproteins (Hegyi & ZIebuhr, 2002; Liu et al., 2020; Woo et al., 2005). The structure of 3CLpro can be used for an initial in silico high-throughput screening of potential interacting molecules, many of which have conceivable inhibitory effect that could further be validated by in vitro and in vivo viral infection experiments (Ton et al., 2020). Besides its functional need for viral transcription and replication, 3CLpro lacks homology to any human protease, and thus, inhibitory and experimentally validated molecules that specifically bind to this viral enzyme could be a safe, therapeutic target (Zhang et al., 2020). Given the urgency of developing effective anti-SARS-CoV-2 therapeutic agents that can rapidly reduce viral load, and its associated inflammatory response in infected patients, there is the need to investigate the effect of currently existing FDA-approved drugs, which are applied in other medical pathologies, and their potential interactions with essential SARS-CoV-2 enzymes, such as the main viral protease. Currently, the most promising repurposing drug is the nucleoside-analog remdesivir, originally designed for the treatment of Ebola virus infections, which is now being tested in animal trials in MERS-CoV challenge in macaques (Yuen et al., 2020). Additionally, it has been suggested by in silico studies a design of novel multi-epitope vaccine candidate against COVID-19 (Enayatkhani et al., 2020) and the usage of other potential anti-coronaviral drugs that target several viral proteins such as 2′-O-ribose methyltransferase (Boopathi et al., 2020); N-protein (Khan et al., 2020); envelope protein ion channel (Gupta et al., 2020); the spike protein (Aanouz et al., 2020); MERS-CoV polymerase (Elfiky & Azzam, 2020); and for 3CLpro: disomin, hesperidine, MK-3207, dihydroergocristina, bolazine, R228, ditercalinium, etoposide, teniposide, UK-432097, irinotecan, lumacaftor, velpastasvir, eluxadoline y ledipasvir (Chen et al., 2020); three FDA approved drugs (Remdesivir, Saquinavir and Darunavir) and two natural compounds (flavone and coumarine derivatives) (Khan et al., 2020); the effect of synergism of the drugs, lopinavir, oseltamivir and ritonavir (Muralidharan et al., 2020); two available drugs (Talampicillin and Lurasidone) and two novel drug-like compounds (ZINC000000702323 and ZINC000012481889) (Elmezayen et al., 2020); Cobicistat, ritonavir, lopinavir, and darunavir (Pant et al., 2020); and those published in a rapid screening of compounds (Ton et al., 2020), could potentially develop as therapeutics against COVID-19.
In the present paper, we describe an in silico selective screening of some thiol-reacting FDA-approved drugs that bind to the main active site cavity of 3CLpro and could be further evaluated by in vitro and in vivo experiments in specialized laboratories. From these candidates, disulfiram (DSF) appears to be the most viable as an antiviral agent, given its extensive use for the treatment of chronic-alcoholism, with only few reported side effects (Yoshimura et al., 2014). Furthermore, this is not the first time that disulfiram has been considered for a new medical use. It has been proposed as an antimicrobial agent against pathogenic bacteria and human parasites (Dalecki et al., 2015; Díaz-Sánchez et al., 2016; Galkin et al., 2014; Zaldívar-Machorro et al., 2011). Moreover, it also possesses antiviral activity against hepatitis C virus (Lee et al., 2016); and remarkably, it was recently suggested as an inhibitor of the papain-like proteases of related SARS-CoV-1 and MERS-CoV (Lin et al., 2018).
2. Materials and methods
Our inclusion criteria for the selection of drugs for potential COVID-19 therapy were: (1) The drug is a FDA-approved that can potentially be repurposed as an ready-to-use therapeutic antiviral; (2) Its use has been extensively studied, and there is sufficient literature on its pharmacology; (3) The drug has few side effects in long-term administration, with not known direct fatalities associated to it, and not additional extensive toxicological studies are needed; (4) Drug can interact with active site of SARS-CoV-2 main protease and reacts with thiol group of its catalytic cysteine, producing an irreversible covalent-inhibition; (5) Administered drug shows efficient distribution through multiple organs, as recent publications suggest the possibility that SARS-CoV-2 has tropism for multiple tissues, beyond the pneumocytes, including heart and blood vessels, liver, intestine, neural cortex and brain stem (Wadman et al., 2020); (6) Drug penetrates the cell membrane, as its antiviral target, the protease acts early during replication in the host cytoplasm; and (7) When administered, drug is metabolized and excreted slowly; and possible metabolites have also potential inhibitory activity.
The binding of DSF, its active over thiol groups metabolites (N,N-diethyldithiocarbamate (DDC); S-methyl N,N-diethyldithiocarbamoyl sulfide (MeDDTC); S-methyl-N,N-diethyldithiocarbamoyl sulfoxide (MeDDTC-SO); S-methyl-N,N-diethyldithiocarbamoyl sulfone (MeDDTC-SO2); S-methyl-N,N-diethylthiocarbamoyl sulfoxide (MeDDC-SO) and S-methyl-N,N-diethylthiocarbamoyl sulfone (MeDDC-SO2)), and captopril (Figure 1) into SARS-CoV-2 main protease enzyme was performed with USCF-Chimera using the Autodock Vina menu (https://www.cgl.ucsf.edu/chimera/). These drugs were selected by their potential capacity to covalently block the catalytic Cys of many enzymes, and conceivable, main Cys-protease of SARS-CoV-2 (Zhang et al., 2020). The crystalline structure of SARS-CoV-2 3CLpro main protease (SARS-CoV-2mp) (PDB code: 6lu7) was used as the receptor and as ligands, the available structures of compounds in Data Bases: captopril (ZINC code: 57001); DSF (Zinc code: 1529266); DDC (PubChem code: 28343); MeDDC-SO (Pubchem code: 3035711); MeDDC-SO2 (Pubchem code: 3552889). MeDDTC, MeDDTC-SO and MeDDTC-SO2 metabolites were drawn using MolView tool (http://molview.org/) and subsequently, ligand structures were energy minimized in the Avogadro software (v 1.2.0) using the chemical and MMF94 force fields. The resulting optimized structures were used to perform the ligand-SARS-CoV-2 protease molecular coupling. The docking parameters used for different ligands were the same, using the default options: Number of binding modes (1-10, default 9); Exhaustiveness of search (1-8, default 8); Maximum energy difference (kcal/mol) (1-3, default 3); binding modes with scores not within this range of the best score were discarded. First, a search volume that corresponds to the entire protein was used to find the principal potential binding cavities and given that most solutions match into the active site, subsequently, the volume was adjusted to this region. Binding sites for captopril, disulfiram and its metabolites in SARS-CoV-2mp enzyme were inferred by means of nine best-scored solutions with the corresponding ligands. The most abundant auto-validated complex model in which potential reactive groups were closer in distance is depicted. Docking protein-ligand results were plotted using LipPlot software (Laskowski & Swindells, 2011), running the PDB model with default run parameters for searching polar and hydrophobic interactions between·CLpro and ligands.
Figure 1.
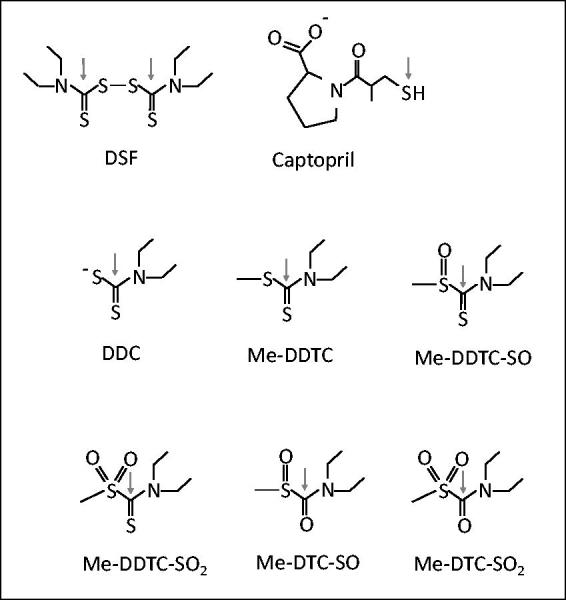
Structure of the drugs used as ligands for the SARS-CoV-2 main protease 3CLpro. Arrow indicates the thiol-reactive group. Disulfiram (DSF); N,N-diethyldithiocarbamate (DDC); S-methyl N,N-diethyldithiocarbamoyl sulfide (MeDDTC); S-methyl-N,N-diethyldithiocarbamoyl sulfoxide (MeDDTC-SO); S-methyl-N,N-diethyldithiocarbamoyl sulfone (MeDDTC-SO2); S-methyl-N,N-diethylthiocarbamoyl sulfoxide (MeDDC-SO) and S-methyl-N,N-diethylthiocarbamoyl sulfone (MeDDC-SO2).
3. Results and discussion
3.1. Disulfiram and captopril docking
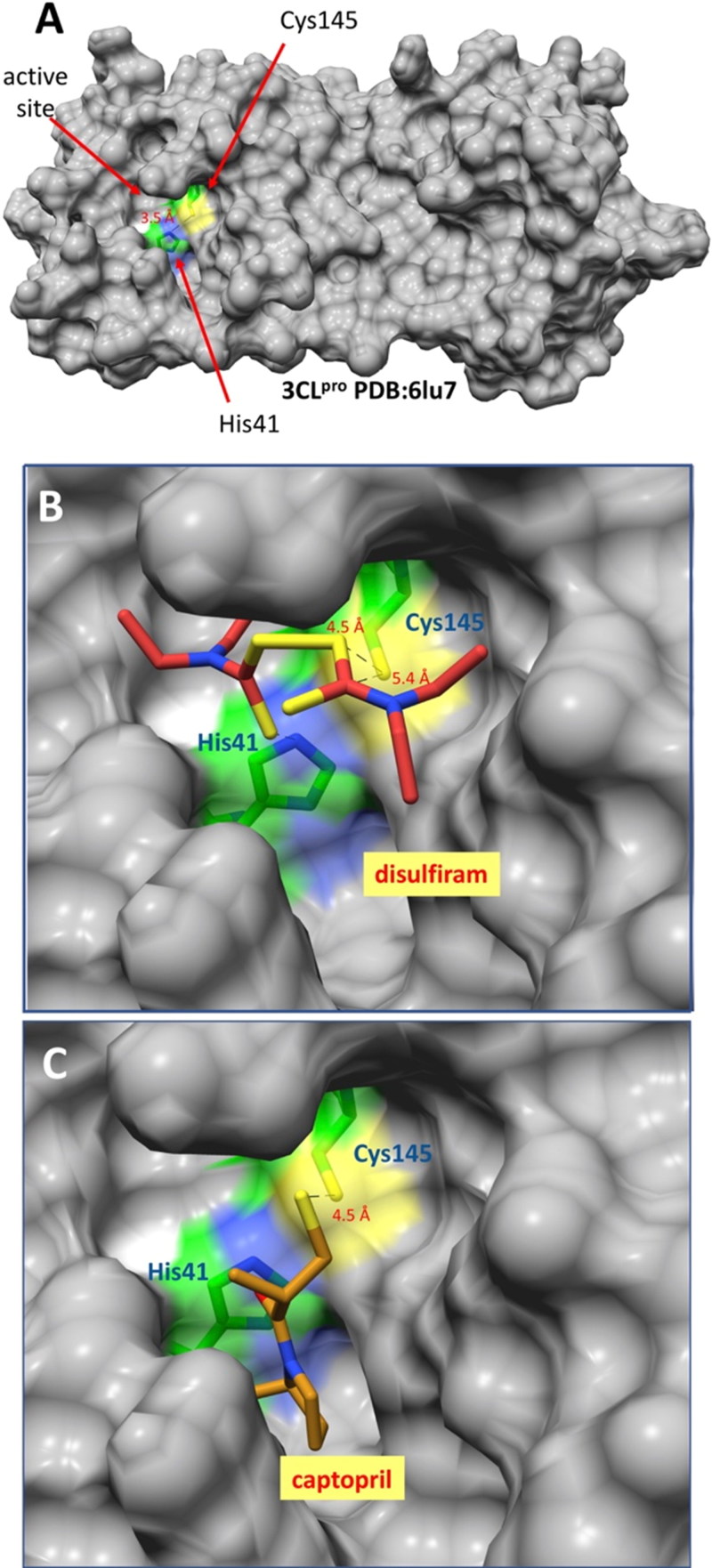
The binding potential of two FDA-approved drugs that contain groups which can react with surface exposed thiols groups of Cys residues on the major protease of SARS-CoV-2 was inferred. As reported earlier seen in the crystal structure (Zhang et al., 2020), this protease possesses a Cys residue (145) in the active site in a close proximity (3.5 Å) to the catalytic His; thus, His41 may participate in its activation, by deprotonation of sulfhydryl group, and therefore, in its reactivity (Figure 2A). From the docking experiments, it was observed that both disulfiram and captopril bind at the active site of the enzyme with relative proximity to the thiol group of Cys145 (Figure 2B and C).
Figure 2.
Binding models of DSF and captopril into SARS-CoV-2 main protease 3CLpro. (A) Depiction of the overall structure showing the Cys145 and His41 catalytic residues located in the active site cavity. (B) SARS-CoV-2 3CLpro-DSF and (C) SARS-CoV-2 3CLpro-captopril model complexes.
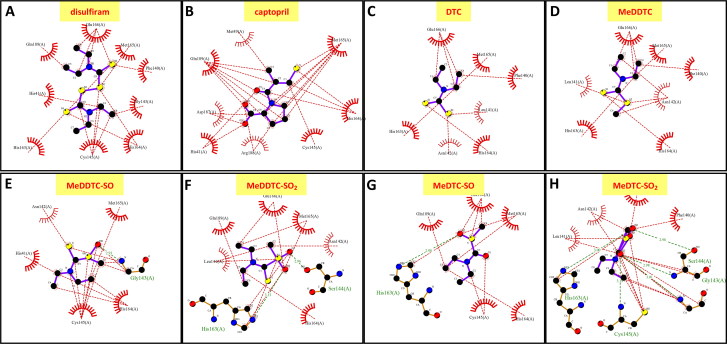
The carboxyl group of captopril even interacts with the imidazole ring of His163 (3.3 Å), and potentially with nearby His41 and Asn142 after formation of the expected mixed-disulfide bond with the drug (Figure 3A, B and Table 1). The closest thionyl group carbon of bound DSF is 3.8 Å from the sulfur of the Cys145 thiol group, in a geometry appropriate to receive an attack by the latter; also, one of the DSF disulfide sulfurs is found at 4.1 Å of the Cys145 thiol group. The rest of the DSF molecule is bound by nonpolar interactions (Figure 3A and Table 1).
Figure 3.
LigPlot analysis of the binding of ligands in SARS-CoV-2 main protease 3CLpro. (A) DSF; (B) captopril; (C) diethyldithiocarbamate (DDC); (D) S-methyl N,N-diethyldithiocarbamoyl sulfide (MeDDTC); (E) S-methyl-N,N-diethyldithiocarbamoyl sulfoxide (MeDDTC-SO); (F) S-methyl-N,N-diethyldithiocarbamoyl sulfone (MeDDTC-SO2); (G) S-methyl-N,N-diethylthiocarbamoyl sulfoxide (MeDTC-SO) and (H) S-methyl-N,N-diethylthiocarbamoyl sulfone (MeDTC-SO2).
Table 1.
Docking analysis of ligand binding in SARS-CoV-2 main protease 3CLpro.
| Ligand | Score (kcal/mol) | Number of hydrophobic interactions with active site cavity | Aminoacids involved and number of hydrophobic interactions | Number of H-bonds with active site cavity | Aminoacids involved in H-bonding |
|---|---|---|---|---|---|
| DSF | −4.0 | 16 | Glu166 (4) Met165 (1) Phe140 (1) Gly143 (1) His164 (2) Cys145 (4) His163 (1) His41 (1) Gln189 (1) |
0 | none |
| Captopril | −4.7 | 22 | Met165 (6) His164 (3) Cys145 (1) Arg188 (2) Asp187 (2) His41 (2) Met49 (1) Gln189 (5) |
2 | His163 |
| DDC | −2.7 | 9 | Glu166 (3) Met165 (1) Phe140 (1) Leu141 (1) His164 (1) Asn142 (1) His165 (1) |
0 | none |
| MeDDTC | −2.9 | 11 | Glu166 (3) Met165 (1) Phe140 (1) Arg142 (3) His163 (1) Leu141 (1) His163 (1) |
0 | none |
| MeDDTC-SO | −3.5 | 16 | Met165 (1) Gly143 (4) His164 (3) Cys145 (6) His41 (1) Asn142 (1) |
1 | Gly143 (1) |
| MeDDTC-SO2 | −3.8 | 12 | Glu166 (2) Met165 (2) Asn142 (1) His164 (1) His163 (3) Leu141 (2) Gln189 (1) |
2 | Ser144 (1) His163 (1) |
| MeDTC-SO | −3.2 | 10 | Cys145 (2) His163 (1) His164 (1) Met165 (2) Glu166 (3) Gln189 (1) |
1 | His163 (1) |
| MeDTC-SO2 | −4,3 | 17 | Phe140 (1) Leu141 (2) Asn142 (2) Gly143 (4) Ser144 (2) Cys145 (4) His163 (1) Met165 (1) |
5 | Gly143(1) Ser144 (2) Cys145 (1) His163 (1) |
Both DSF and captopril bind at the active site of 3LCpro (6lu7), at the position where the N3 inhibitor was observed bound in the crystal structure (Zhang et al., 2020). The N3 inhibitor is an α-ketoamide, which reacts irreversibly with Cys145 of 3LCpro after binding into the shallow substrate subsite cavity (Zhang et al., 2020). Although the mechanism of inhibition of 3LCpro by the N3 compound is not known, it is inferred to be competitive; hence, it directly interferes with the fitness of the substrate. The models infer that both DSF and captopril occupy the same subsite as N3, and obstruct the function of the catalytic dyad Cys145 and His41; and consequently, we predict that binding of these two molecules, as well as the DSF metabolites, will result in competitive inhibition of the viral protease. The fact that DSF acts as a competitive inhibitor on the papain-like protease of SARS-CoV-1 (Lin et al., 2018), together with the high similarity of this protease with the 3LCpro of SAR-CoV-2, support this hypothesis.
The use of DSF as antiviral against COVID-19 is promising for many reasons: Upon ingestion, one fifth of the administered dose is active even after one week and hence classified as a long-lasting drug (Clayton & Clayton, 1981). This is of particular interest to avoid long administration periods. The pharmacokinetics studies of DSF indicate that a relatively small fraction of the compound is excreted in urine after 6 h of administration (5-20%) and maintaining a half-life of 6 h in the plasma (Ellenhorn, 1987). While the rest is transformed to other metabolites: MeDDTC-SO; MeDDTC-SO2; MeDTC-SO; and MeDTC-SO2 (Zaldívar-Machorro et al., 2011). Therefore, the administration of DSF, produces six other thiol-reactive species that could additionally reacts with Cys thiol groups. We docked these metabolic products of DSF in SARS-CoV-2 main protease 3CLpro (Figure 4).
Figure 4.
Binding models of disulfiram metabolites into SARS-CoV-2 main protease 3CLpro. (A) DDC; (B) MeDDTC; (C) MeDDTC-SO; (D) MeDDTC-SO2; (E) MeDTC-SO and (F) MeDTC-SO2.
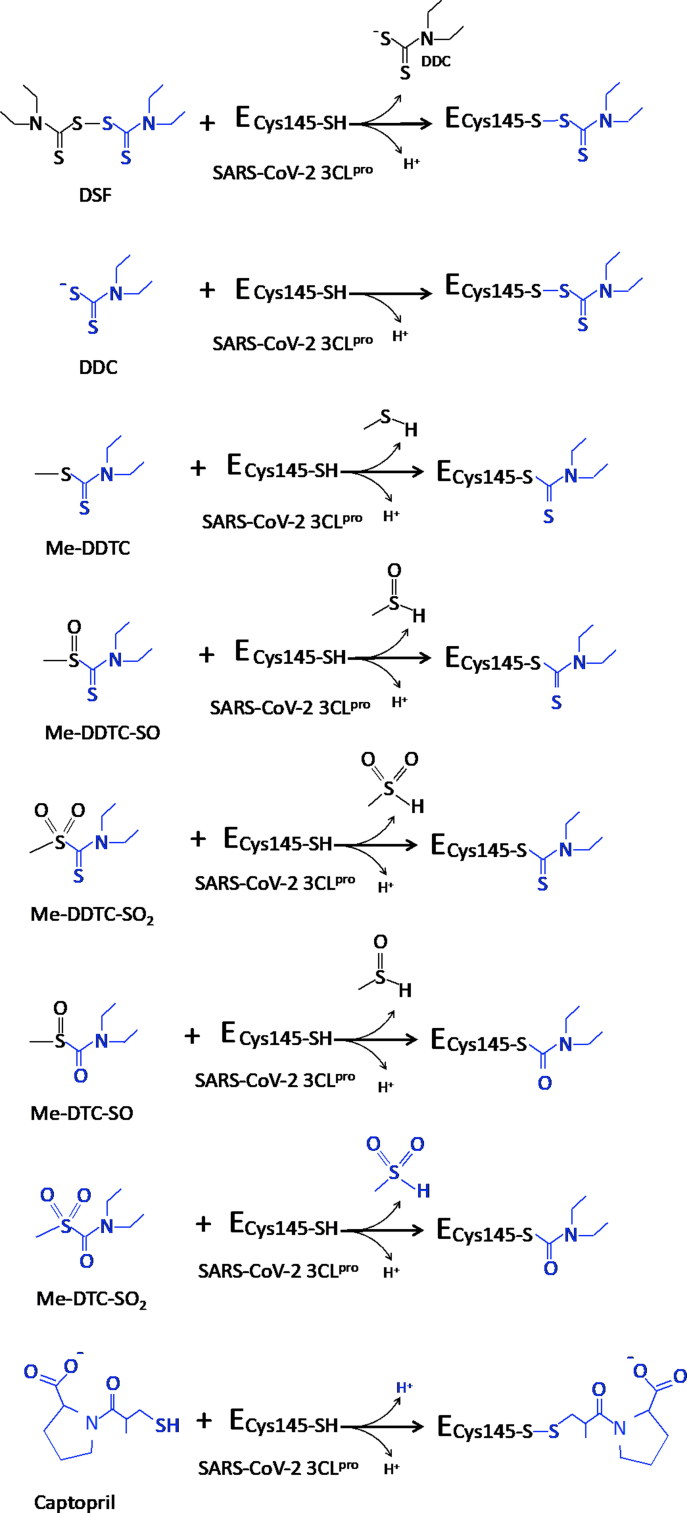
It is well known that disulfiram and its metabolites; and probably captopril as well, do not exert their inhibition through a single binding step. These drugs first bind non-covalently to the target enzyme sites and then, a reaction between critical groups results into a more stable covalently bound enzyme-inhibitor complex. In this regard, antimicrobial and antiviral compounds that covalently inhibit their target proteins are more effective than reversible inhibitors. Therefore, the use of drugs that possess groups capable of reacting covalently with the SARS-CoV-2 essential proteins are desired. DSF and its metabolites react with critical cysteine residues that participate in the stability and activity of enzymes recognized as pharmacological targets for the treatment of microbial and viral diseases and acute infections (McCance-Katz et al., 2014). Presumably, the inhibitory potential of these compounds on 3CLpro is not through a one-step mechanism, but rather by initial binding to the protein followed by a nucleophilic attack by thiol group of Cys145 and, thus, they cause irreversible covalent inactivation of the enzyme (Figure 5). This predicted attack would enhance the estimated energy score obtained in the docking studies, as in our interaction models only the reversible binding step was considered. Overall, the final covalent stabilization step would surely contribute to acquire a more satisfactory dissociation constant value.
Figure 5.
Possible mechanism of reaction of compounds upon binding to SARS-CoV-2 main protease 3CLpro.
Regarding the potential for DSF and its metabolites to form covalent derivatives with the SARS-CoV-2 protease, it was recently found that DSF is an inhibitor of the MERS-CoV and SARS-CoV-1 papain-like proteases, through a non-competitive and competitive mechanisms, respectively (Lin et al., 2018). In this same publication, it was observed a time-dependent inactivation of the recombinant enzyme, which was not reversed after removal of the excess of DSF, The residual inhibition observed is through a covalent bonding of DSF, which must involve the thiol group of the catalytic Cys residue, probably forming a covalent adduct with Cys112 (number of SARS-CoV-1 PLpro), the latter mechanism deduced from crystallography and docking. Finally, it was argued that the combined use of DSF and 6-thioguanine or mycophenolic acid offers a potential therapeutic use for clinical treatment of these emerging coronavirus infections. The SARS-CoV-1 protease conserves the catalytic residues, and probably the mechanism, of the SARS-CoV-2 enzyme; for this reason, we predict the formation of stable and irreversible 3CLpro-inhibitor (DSF and its metabolites) complexes.
The other thiol-reacting compound modeled in this study is, captopril, a drug that acts as an inhibitor of the enzyme ACE-I, a Zn-metallo-dipeptidase that converts angiotensin-I to angiotensin-II, an enzymatic function which is essential for the regulation of blood pressure. The free thiol group of captopril blocks the catalytic Zn+2-center of ACE-I. Captopril has also been suggested as a potential antibiotic, capable of effectively inhibiting a Zn-metal succinylase/dipeptidase, by means of the blocking of its catalytic Zn-center (Gillner et al., 2009; Starus et al., 2015). The thiol group present in captopril has the potential to form mixed disulfides bond with functionally relevant Cys residues. We hypothesize that after an initial binding step, captopril, could bind covalently with Cys145 of 3CLpro (Figure 5). The administration of a single dose of captopril provides pharmacological activity in a short time (10-30 min) at the cellular level (Mousavi et al., 2018). This is due to the fact that it is transported mainly by plasma proteins (albumin) and is absorbed between 60-75% of the total in the form of captopril-cysteine disulfide and the captopril disulfide dimer. These metabolites can undergo a reversible interconversion with a half-life of approximately two hours. Although, the potential binding capacity of captopril over 3CLpro is shown here, with the current data we cannot suggest its clinical therapeutic use against COVID-19 (Table 2), since prolonged administration of this drug induces an overexpression of the angiotensin-II converting enzyme (ACE-2), the main receptor used by SARS-CoV-2 to enter human cells (Walls et al., 2020); thus, it could potentially increase the susceptibility to infection and exacerbate symptoms and medical complications. However, it could help in combination with other drugs, such angiotensis II receptor blockers (Messerli et al., 2020), administrated for a short period during the acute or critical phase of the infection, preferably, in patients not pharmacologically treated for hypertension.
Table 2.
Possible side effects and recommended doses of the thiol-reacting drugs identified for SARS-CoV-2 main protease 3CLpro.
| Drug | Disulfiram | Captopril |
|---|---|---|
| Possible side effects | Contraindicated in hypersensitivity, liver and kidney failure | Contraindicated in events of hypotension, liver, kidney and heart failure, angioedema and hypersensitivity |
| Treatment for | Alcoholism | Hypertension |
| Doses per day | 800 mg/200 mg | 25 mg |
| References | (Yoshimura et al., 2014) | (Mousavi et al., 2018) |
| Recommended to be tested for SARS-CoV-2 | Yes | No |
In contrast, DSF can potentially be tested for clinical therapeutic use against COVID-19 in a safer manner. It is in the list of FDA-approved drugs and is used to facilitate the treatment of chronic alcoholism, as it targets the hepatocyte mitochondrial acetaldehyde dehydrogenase (ALDH-2), an enzyme to which the drug and its metabolites acts by blocking its catalytic cysteine (Lipsky et al., 2001). Hence, it is clear that DSF and its metabolites can penetrate human cells; and consequently, have a high potential to block the proteolytic function of the 3CLpro, essential for SARS-CoV-2 replication. It is known that DSF and its metabolites not only block aldehyde dehydrogenase-2, as it has also been recognized as an inhibitor of other cysteine-dependent enzymes, such as: (1) the betaine aldehyde dehydrogenase from P. aeruginosa (PaBADH, an ALDH9 member) (Velasco-García et al., 2003; Zaldívar-Machorro et al., 2011) and from Amaranthus hypochondriacus leaves (ALDH10) (Velasco-García et al., 2003; Velasco-García et al., 2006); (2) the carboxikinase from Giardia lamblia (Galkin et al., 2014); (3) urease from Citrullus vulgaris (Díaz-Sánchez et al., 2016), an enzyme that possesses a Cys residue critical on the closure of the active site flap, a conformational transition that is essential for its function (Macomber et al., 2015); and perhaps, more relevant to this study (4) the papain-like protease of the related coronaviruses SARS-CoV-1 and MERS-CoV (Lin et al., 2018). Given this inhibitory activity, it was also proposed as an antimicrobial agent against P. aeruginosa (Zaldívar-Machorro et al., 2011); M. tuberculosis, in a co-administration with copper ions (Dalecki et al., 2015); and Giardia lamblia trophozoites (Galkin et al., 2014). Additionally, it was proposed as an antiviral agent for the treatment of hepatitis C virus (Lee et al., 2016); and relevantly for repurposing for human administration, it has been used in clinical trials for treatment of HIV infection (Trial ID: NCT00002065 and NCT03198559); Glioblastoma (Trial ID: NCT03151772); Breast Neoplasms (Trial ID: NCT03323346) and Prostatic Neoplasms (Trial ID: NCT02963051).
4. Conclusions
The current in silico study provide evidence that FDA-approved, DSF could potentially be develop into new antiviral agents for the treatment of COVID-19. Here we provide in silico data to show that the catalytic Cys145 of SARS-CoV-2mp could be blocked and inactivated by DSF and its thiol-reactive derivatives metabolites.
Glossary
Abbreviations
- 3CLpro
3C-like protease
- ACE-2
Angiotensin-converting enzyme 2
- ACE-I
Angiotensin-converting enzyme 1
- ALDH
Aldehyde dehydrogenase
- ALDH-2
Acetaldehyde dehydrogenase 2
- ARSS
Severe Acute Respiratory Stress Syndrome
- CLpro
C-like protease
- COVID-19
Coronavirus Disease 2019
- DDC
N,N-diethyldithiocarbamate
- DSF
Disulfiram
- FDA
Food and Drug Administration
- MeDDC-SO
S-methyl-N,N-diethylthiocarbamoyl sulfoxide
- MeDDC-SO2
S-methyl-N,N-diethylthiocarbamoyl sulfone
- MeDDTC
S-methyl N,N-diethyldithiocarbamoyl sulfide
- MeDDTC-SO
S-methyl-N,N-diethyldithiocarbamoyl sulfoxide
- MeDDTC-SO2
S-methyl-N,N-diethyldithiocarbamoyl sulfone
- MERS
Middle East Respiratory Syndrome
- MERS-CoV
Middle East Respiratory Syndrome Coronavirus
- NSP
Non-structural proteins
- ORF1a
Open Reading Frame 1a
- pp1a and pp1ab
two overlapping replicase polyproteins
- PaBADH
Betaine aldehyde dehydrogenase from Pseudomonas aeruginosa
- PDB
Protein Data Bank
- PLpro
Papain-like protease
- RNA
Ribonucleic acid
- SARS-CoV-1
Severe Acute Respiratory Stress Syndrome Coronavirus 1
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SARS-CoV-2mp
SARS-CoV-2-main protease
- ssRNA
Single-stranded ribonucleic acid
- WHO
World Health Organization
Acknowledgements
This work was not funded by any grant, but we would like to acknowledge Consejo Nacional de Ciencia y Tecnología and to Programa para el Desarrollo Profesional Docente from the Mexican Government.
Disclosure statement
No potential conflict of interest is reported by the authors.
References
- Aanouz I., Belhassan A., & El Khatabi K. (2020). Moroccan medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics. doi: 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A., & Kolandaivel P. (2020). Novel 2019 Coronavirus Structure, Mechanism of Action, Antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. doi: 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yiu C.-P., & Wong K.-Y. (2020). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9, 129. doi: 10.12688/f1000research.22457.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton G. D., & Clayton F. E. (1981). Patty’s industrial hygiene and toxicology (3rd ed.). John Wiley & Sons. [Google Scholar]
- Colson P., Rolain J.-M., Lagier J.-C., Brouqui P., & Raoult D. (2020). Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. International Journal of Antimicrobial Agents, 55(4), 105932. doi: 10.1016/j.ijantimicag.2020.105932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalecki A. G., Haeili M., Shah S., Speer A., Niederweis M., Kutsch O., & Wolschendorf F. (2015). Disulfiram and copper ions kill Mycobacterium tuberculosis in a synergistic manner . Antimicrobial Agents and Chemotherapy, 59(8), 4835–4844. doi: 10.1128/AAC.00692-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Sánchez Á., Alvarez-Parrilla E., Martínez-Martínez A., Aguirre-Reyes L., Orozpe-Olvera J., Ramos-Soto M., Núñez-Gastélum J., Alvarado-Tenorio B., & de la Rosa L. (2016). Inhibition of urease by disulfiram, an FDA-approved thiol reagent used in humans. Molecules, 21(12), 1628. doi: 10.3390/molecules21121628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., & Gardner L. (2020). An interactive web-based dashboard to track COVID-19 in real time. The Lancet Infectious Diseases, 20(5), 533–534. 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 20, 1–12. doi: 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenhorn M. J., & Barceloux D. G. (1987). Medical toxicology - diagnosis and treatment of human poisoning (p. 1512). Elsevier Science Publishing [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., & Şahin A. T. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 20, 1–12. doi: 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., & Faezi S. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 15, 1–19. doi: 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin A., Kulakova L., Lim K., Chen C. Z., Zheng W., Turko I. V., & Herzberg O. (2014). Structural basis for inactivation of Giardia lamblia carbamate kinase by disulfiram. The Journal of Biological Chemistry, 289(15), 10502–10509. 11;doi: 10.1074/jbc.M114.553123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillner D., Armoush N., Holz R. C., & Becker D. P. (2009). Inhibitors of bacterial N-succinyl-L,L-diaminopimelic acid desuccinylase (DapE) and demonstration of in vitro antimicrobial activity. Bioorganic & Medicinal Chemistry Letters, 19(22), 6350–6352. 15doi: 10.1016/j.bmcl.2009.09.077 [DOI] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., & Donde R. (2020). In-silico to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 15, 1–11. doi: 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., & Hussain A. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 22, 1–9. doi: 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., & ZIebuhr J. (2002). Conservation of substrate specificities among coronavirus main proteases. The Journal of General Virology, 83(Pt 3), 595–599. doi: 10.1099/0022-1317-83-3-595 [DOI] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., & Amera G. M. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 20, 1–14. doi: 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., & Ashraf S. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 13, 1–10. doi: 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., & Swindells M. B. (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. doi: 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Lee Y.-M., Duh Y., Wang S.-T., Lai M. M. C., Yuan H. S., & Lim C. (2016). Using an old drug to target a new drug site: Application of disulfiram to target the Zn-Site in HCV NS5A protein. Journal of the American Chemical Society, 138(11), 3856–3862. doi: 10.1021/jacs.6b00299 [DOI] [PubMed] [Google Scholar]
- Lin M.-H., Moses D. C., Hsieh C.-H., Cheng S.-C., Chen Y.-H., Sun C.-Y., & Chou C.-Y. (2018). Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes. Antiviral Research, 150, 155–163. doi: 10.1016/j.antiviral.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky J. J., Shen M. L., & Naylor S. (2001). In vivo inhibition of aldehyde dehydrogenase by disulfiram. Chemico-Biological Interactions, 130-132(1–3), 93–102. doi: 10.1016/S0009-2797(00)00225-8 [DOI] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., & Wang M. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery, 6, 16. doi: 10.1038/s41421-020-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L., Minkara M. S., Hausinger R. P., & Merz K. M. (2015). Reduction of urease activity by interaction with the flap covering the active site. Journal of Chemical Information and Modeling, 55(2), 354–361. 23;doi: 10.1021/ci500562t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. (2020). Coronavirus covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate. BMJ (Clinical Research ed.).), 368, m641. doi: 10.1136/bmj.m641 [DOI] [PubMed] [Google Scholar]
- McCance-Katz E. F., Gruber V. A., Beatty G., Lum P., Ma Q., DiFrancesco R., Hochreiter J., Wallace P. K., Faiman M. D., & Morse G. D. (2014). Interaction of disulfiram with antiretroviral medications: Efavirenz increases while atazanavir decreases disulfiram effect on enzymes of alcohol metabolism. The American Journal on Addictions, 23(2), 137–144. doi: 10.1111/j.1521-0391.2013.12081.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli F. H., Siontis G. C., & Rexhaj E. (2020). COVID-19 and renin angiotensin blockers: Current evidence and recommendations. Circulation, 13. doi: 10.1161/CIRCULATIONAHA.120.047022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi M., Razavianzadeh N., Armin M., & Fadaei Dashti M. (2018). Sublingual versus oral captopril for decreasing blood pressure in hypertension urgency: A randomized clinical trial. Iranian Red Crescent Medical Journal, 20(6), e61606. doi: 10.5812/ircmj.61606 [DOI] [Google Scholar]
- Muralidharan N., Sakthivel R., & Velmurugan D. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 16, 1–6. doi: 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Pant S., Singh M., & Ravichandiran V. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 20, 1–15. doi: 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starus A., Nocek B., Bennett B., Larrabee J. A., Shaw D. L., Sae-Lee W., Russo M. T., Gillner D. M., Makowska-Grzyska M., Joachimiak A., & Holz R. C. (2015). Inhibition of the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase from Neisseria meningitidis by L-Captopril. Biochemistry, 54(31), 4834–4844. 11;doi: 10.1021/acs.biochem.5b00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A. T., Gentile F., & Hsing M. (2020). Rapid identification of potential inhibitors of SARS‐CoV‐2 main protease by deep docking of 1.3 billion compounds. Molecular Informatic, 39, 200002. doi: 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-García R., Zaldívar-Machorro V. J., Mújica-Jiménez C., González-Segura L., & Muñoz-Clares R. A. (2006). Disulfiram irreversibly aggregates betaine aldehyde dehydrogenase-a potential target for antimicrobial agents against Pseudomonas aeruginosa. Biochemical and Biophysical Research Communications, Biochem. Biophys. Res. Commun., 341(2), 408–415. https://doi.org/ 10.1016/j.bbrc.2006.01.003 16426571 [DOI] [PubMed] [Google Scholar]
- Velasco-García R., Chacón-Aguilar V. M., Hervert-Hernández D., & Muñoz-Clares R. A. (2003). Inactivation of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa and Amaranthus hypochondriacus L. leaves by disulfiram. Chemico-Biological Interactions, 143–144, 149–158. 1; doi: 10.1016/S0009-2797(02)00199-0 [DOI] [PubMed] [Google Scholar]
- Wadman M., Couzin-Frankel J., & Kaiser J. (2020). How does coronavirus kill? Clinicians trace a ferocious rampage through the body, from brain to toes. Science, 17. doi: 10.1126/science.abc3208 [DOI] [PubMed] [Google Scholar]
- Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., & Veesler D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, 181(2), 281–292.e6. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., & Xiao G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. doi: 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. Y., Huang Y., Lau S. K. P., Tsoi H-w., & Yuen K-y. (2005). In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiology and Immunology, 49(10), 899–908. doi: 10.1111/j.1348-0421.2005.tb03681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A., Kimura M., Nakayama H., Matsui T., Okudaira F., Akazawa S., Ohkawara M., Cho T., Kono Y., Hashimoto K., Kumagai M., Sahashi Y., Roh S., & Higuchi S. (2014). Efficacy of disulfiram for the treatment of alcohol dependence assessed with a multicenter randomized controlled trial. Alcoholism, Clinical and Experimental Research, 38(2), 572–578. doi: 10.1111/acer.12278 [DOI] [PubMed] [Google Scholar]
- Yuen K.-S., Ye Z.-W., Fung S.-Y., Chan C.-P., & Jin D.-Y. (2020). SARS-CoV-2 and COVID-19: The most important research questions. Cell & Bioscience, 10, 40. doi: 10.1186/s13578-020-00404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldívar-Machorro V. J., López-Ortiz M., Demare P., Regla I., & Muñoz-Clares R. A. (2011). The disulfiram metabolites S-methyl-N,N-diethyldithiocarbamoyl sulfoxide and S-methyl-N,N-diethylthiocarbamoyl sulfone irreversibly inactivate betaine aldehyde dehydrogenase from Pseudomonas aeruginosa, both in vitro and in situ, and arrest bacterial growth. Biochimie, 93(2), 286–295. doi: 10.1016/j.biochi.2010.09.022 [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., & Hilgenfeld R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.).), 368(6489), 409–412. 24doi: 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., … Shi Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., & Tan W. (2020). A novel coronavirus from patients with pneumonia in China, 2019 . The New England Journal of Medicine, 382(8), 727–733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]