Abstract
Plant elicitor peptide 1 (Pep1) is a versatile immune modulator involved in plant defense against herbivores and pathogens. A recent study uncovers that arabidopsis (Arabidopsis thaliana) releases Pep1 from the carboxyl-terminus of the tonoplast-resident precursor protein, PROPEP1, cleaved by Ca2+-activated metacaspases upon cell membrane rupture in the damaged tissues.
Keywords: Plant immunity, wounding, plant elicitor peptide 1 (Pep1), metacaspase
Plant extracellular peptides are critical regulators of various physiological processes, including growth, reproduction, and responses to environmental cues. Most peptides are initially synthesized as precursor proteins and are released to apoplasts as biologically active peptides after the maturation process. Once exposed on cell surfaces, the peptide could be recognized by plasma membrane (PM)-localized receptors to initiate downstream signaling events [1].
The secreted peptides usually contain a secretion signal at the amino-terminus of the precursor proteins to guide secretion. In contrast, there are a small number of peptides, whose precursors are absent of secretion sequence. They are presumed to be enclosed in cell interiors under normal physiological conditions and are released out of cells upon cell damages. Interestingly, this type of peptides identified so far are involved in plant immunity, and categorized as the damage-associated molecular patterns (DAMPs) [2], which are referred to host-derived molecules that are released when cells suffer from attacks of microbes or herbivores, or mechanical damages. Among these peptides, plant elicitor peptide 1 (Pep1) has been widely studied due to its conserved functions in resistance to herbivores and pathogens across different plant species. Pep1 is derived from the carboxyl-terminus of the propeptide, PROPEP1 [3]. Subcellular localization analysis indicated that PROPEP1 localizes on the tonoplast or in the cytosol as predicted for the lack of amino-terminal canonical signal sequences for secretion [4]. However, Pep1 is recognized by the PM-localized receptors, PEP RECEPTOR 1 (PEPR1) and PEPR2, via their extracellular leucine-rich repeat (LRR) domains [5]. How plants switch on the Pep1 signaling upon herbivore or pathogen attacks remains largely unknown. Recently, Hander et al. [6] revealed an elegant mechanism underlying Pep1 maturation in arabidopsis (Arabidopsis thaliana) upon wounding.
Hander et al. detected a specific PROPEP1 cleavage after 30 seconds with a peak at 5 minutes upon plant tissue physical damages, followed by a general protein degradation [6]. This finding suggests that PROPEP1 is rapidly cleaved by an unknown protease(s), whose activity might be induced during cell damages. Using a pharmacological approach coupled with gene co-expression patterns, Hander et al. narrowed down the candidate protease to METACASPASE4 (MC4), one of the type II MCs, which are mainly activated through autocatalytic cleavage of the p20 inhibitory domain upon Ca2+ binding. The in vitro biochemical analysis and genetic studies demonstrated that MC4 preferentially cleaved PROPEP1 at arginine (R) 69 to produce Pep1 [6]. In addition, Hander et al. found all eight arabidopsis PROPEPs and a tomato (Solanum lycopersicum) PROPEP ortholog are cleaved by the active MC4 in vitro, suggesting that PROPEP processing by a metacaspase is conserved across plant species. Of note, PROPEP3 does not contain an arginine ahead of the Pep3 sequence, but it is still cleaved by MC4 with a lower activity in an in vitro assay. Therefore, other factors, besides the conserved arginine residue, might also mediate the cleavage of PROPEPs. Although at a reduced level, Pep1 is still processed in the roots of mc4 loss-of-function mutants, implying that other undefined proteases may have a redundant function with MC4 in mediating PROPEP1 cleavage [6].
Hander et al. went on to examine the spatiotemporal dynamics of how cytosolic Ca2+ ([Ca2+]cyt) fluxes, MC4 activation, and Pep1 maturation are connected upon cell damage with an in vivo laser confocal imaging assay assisted by immunoblots [6]. Under the real-time confocal scanning, arabidopsis root cells were subtly damaged with the multiphoton laser ablation and footnoted with propidium iodide (PI) entry into the cells, and the Ca2+ fluxes and Pep1 subcellular localization were traced with a Cameleon Ca2+ probe and yellow fluorescent protein fused with Pep1, respectively. It appears that PROPEP1 is sequestered on the vacuolar membrane in intact cells but processed and released upon a substantial increase of [Ca2+]cyt only in the damaged cells (Figure 1). Intriguingly, Hander and colleagues also observed that portions of PROPEP1 remain uncut even when cells are disrupted. They reasoned that a threshold of [Ca2+]cyt may be required for potentiating MC4 beyond the basal activity, and such a threshold may prevent adventitious MC4 activation triggered by intracellular Ca2+ fluctuation. Hander et al. also indicated that the increased [Ca2+]cyt spreads from the damaged cells outward surrounding intact cells. Consistent with this observation, a recent report demonstrated that mechanical damage of arabidopsis leaves causes a release of cytosolic glutamate, which triggers a [Ca2+]cyt “wave” that rapidly propagates to distant tissues [7]. However, no PROPEP1 cleavage was detected in intact cells surrounding the subtle damaged cells [6]. This is consistent with the previous observation that Pep-PEPR signaling is involved in plant systemic immunity, but the Pep peptides might not be mobile [8].
Figure 1. Maturation and Release of Peps in Response to Wounding and Pathogen Infection in Plants.
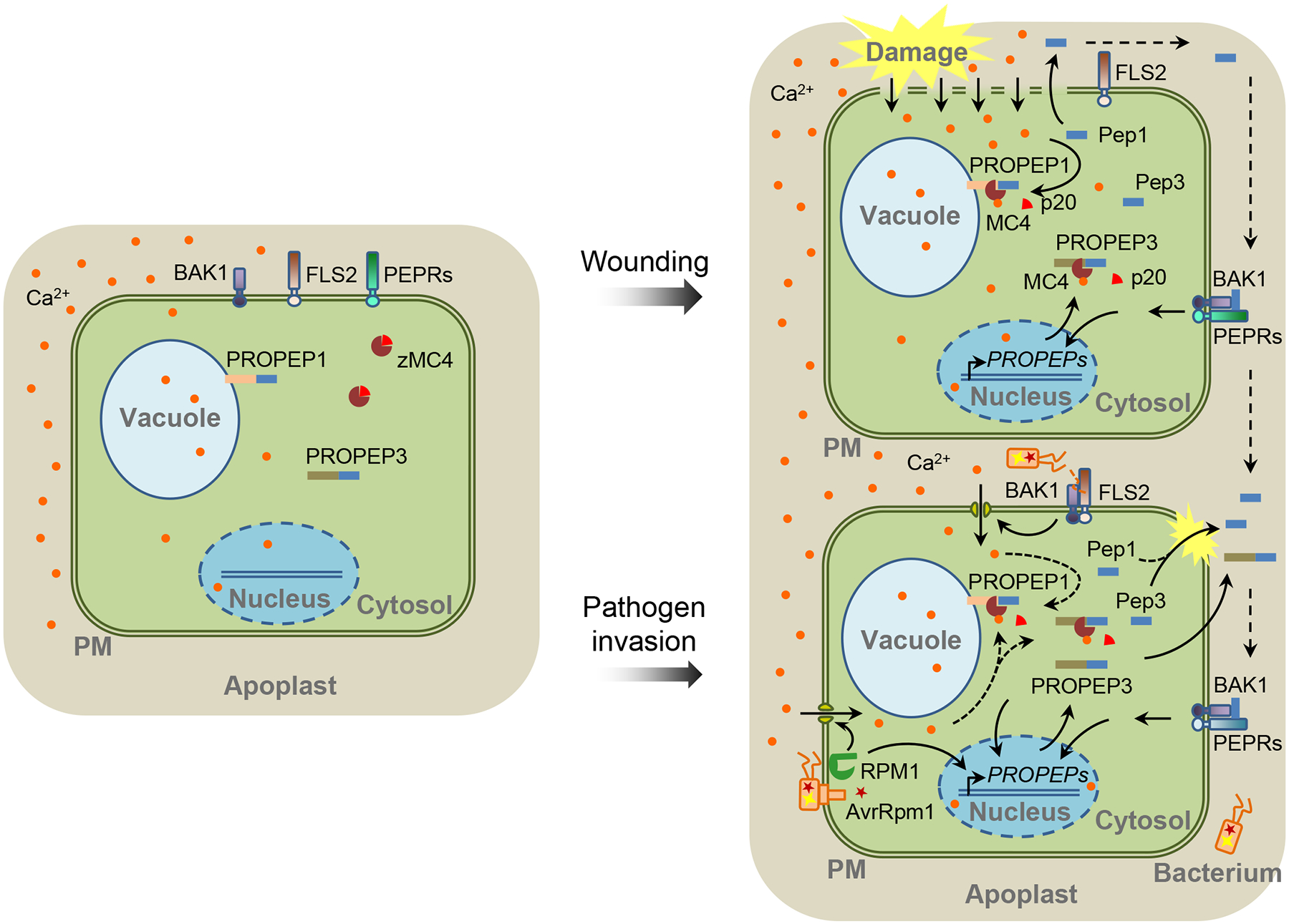
In undisturbed cells, zymogen METACASPASE 4 (zMC4), and likely other MCs, keep inactive in the cytosol. The PROPEP1 and PROPEP3 were reported to reside on the tonoplast and in the cytosol, respectively [4] (left). Cell damage leads to substantial increase of cytosolic calcium (Ca2+) ([Ca2+]cyt), which activates MC4 through autocatalytic cleavage of the p20 inhibitory domain, leading to the cleavage of PROPEPs and generation of Pep peptides. Peps as well as unprocessed PROPEPs might be released out of cells and reach the surfaces of surrounding cells. On the cell surface, they are perceived by the PEP RECEPTOR 1 (PEPR1)/PEPR2 and coreceptor BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) and induce downstream signaling events, including the transcription of PROPEP genes (right top). Upon the bacterial pathogen infections, plant cells induce an increase of [Ca2+]cyt and PROPEP transcription through the perception of pathogen-derived PATHOGEN-ASSOCIATED MOLECULAR PATTERNs (PAMPs) (such as flagellin) or effectors (such as AvrRpm1) by receptor proteins (such as the flagellin receptor FLAGELLIN SENSING 2 [FLS2] and the AvrRpm1 receptor RESISTANCE TO P. SYRINGAE PV MACULICOLA 1 [RPM1]). The [Ca2+]cyt increase may potentially activate MC4-mediated cleavage of PROPEPs (right bottom). PM: plasma membrane.
In addition to wounding, pathogen infection also stimulates Pep signaling (Figure 1). In arabidopsis, infection with either virulent or avirulent bacterial pathogens induced the cleavage of PROPEP3, especially in the immunocompromised bak1 mutants [9]. It remains to be determined whether the pathogen infection-induced PROPEP processing relies on Ca2+-dependent MC4 activation. An observation supporting this hypothesis is that self-processing of MC4 was detected in arabidopsis leaves in response to an avirulent bacterial pathogen [10]. DAMPs, including PROPEP genes, are induced by multiple plant pathogen-associated molecular patterns (PAMPs). Thus, DAMP signaling is considered to function as an amplifier of PAMP-triggered immunity (PTI) [2]. It will be interesting to address how Peps are processed to amplify PTI since cell damage is usually not observed during PTI. However, most PAMPs do trigger a rapid increase of [Ca2+]cyt concentration, which may reach a threshold to activate MCs.
What is the biological importance of the MC-mediated PROPEP cleavage? For most peptides, protease-mediated cleavage is an essential post-translational process to generate biologically active peptides [1]. However, Hander et al. showed that Pep1 activity in root growth inhibition does not depend on its cleavage by MC4. So, the authors suggest that MC4-dependent PROPEP1 cleavage might mainly unleash Pep1 from the tonoplast membrane of damaged cells and facilitate its extracellular release and motility into the surrounding tissue [6]. This apparently is not the case for the cytoplasm-localized PROPEP3. Of particular note, unprocessed PROPEP3 may also be released when cells are not totally disrupted in arabidopsis seedlings, especially in the bak1 mutants following Pep treatment, or wild-type seedlings suffering from cell wall damage [9, 11]. PROPEP3 was suggested to be released into the extracellular space [9, 11], where it might also be cleaved by an unknown extracellular protease(s). Thus, MC4-mediated PROPEP cleavage may not be a prerequisite for the extracellular release of Pep3. Further studies are required to elucidate the complex mechanisms underlying the generation of DAMPs from PROPEPs.
As crucial regulators of plant immunity, protease activities appear to be tightly and specifically controlled under every conceivable condition. As indicated by Hander et al., plants set a checkpoint to keep metacaspase inactivation, which switches off the Pep1 signaling to avoid any detrimental effects on plant growth [6]. Besides MCs, other proteases are also involved in the maturation process of immunomodulatory peptides. For example, two maize papain-like cysteine proteases (PLCPs), CP1 and CP2, mediate the cleavage of a propeptide PROZIP1 to release Zip1, leading to the activation of plant resistance to the biotrophic fungus Ustilago maydis [12]. Similar to MCs, PLCPs also contain an auto-inhibitory domain and are produced as inactive zymogens. However, different from MC4, CP1 and CP2 are secreted into apoplasts. The following work will be to determine whether and how the pathogen invasion leads to the PLCP secretion, activation and subsequent PROZIP1 cleavage. Furthermore, some plant proteases and glycosylated proteins are associated with cell walls. Immunomodulation through the protease-mediated release of peptide elicitors upon plant cell wall perturbations may represent another mechanism to activate plant immunity [11].
Ultimately, understanding of extracellular peptide maturation via various proteases in fine-tuning plant immunity will provide insights into the precise regulation of plant immune signaling, and also offer strategies for the genetic engineering of immunomodulatory peptides to improve plant disease resistance.
Acknowledgement
We thank Libo Shan for critical reading of this manuscript. P.H. was supported by National Science Foundation (NSF) (IOS-1252539). S.H. was supported by the National Natural Science Foundation of China (No. 31500971).
Reference
- 1.Olsson V et al. (2019) Look closely, the beautiful may be small: precursor-derived peptides in plants. Annu. Rev. Plant Biol 70, 153–186 [DOI] [PubMed] [Google Scholar]
- 2.Gust AA et al. (2017) Sensing danger: key to activating plant immunity. Trends Plant Sci. 22, 779–791 [DOI] [PubMed] [Google Scholar]
- 3.Huffaker A et al. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U. S. A 103, 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartels S et al. (2013) The family of Peps and their precursors in Arabidopsis: differential expression and localization but similar induction of pattern-triggered immune responses. J. Exp. Bot 64, 5309–5321 [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi Y et al. (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hander T et al. (2019) Damage on plants activates Ca(2+)-dependent metacaspases for release of immunomodulatory peptides. Science 363, eaar7486. [DOI] [PubMed] [Google Scholar]
- 7.Toyota M et al. (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–11158 [DOI] [PubMed] [Google Scholar]
- 8.Ross A et al. (2014) The Arabidopsis PEPR pathway couples local and systemic plant immunity. EMBO J. 33, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada K et al. (2016) Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 35, 46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe N and Lam E (2011) Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 66, 969–982. [DOI] [PubMed] [Google Scholar]
- 11.Engelsdorf T et al. (2018) The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal 11, pii: eaao3070. [DOI] [PubMed] [Google Scholar]
- 12.Ziemann S et al. (2018) An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 4, 172–180 [DOI] [PubMed] [Google Scholar]


