Abstract
The default network (DN) is a brain network with correlated activities spanning frontal, parietal, and temporal cortical lobes. The DN activates for high-level cognition tasks and deactivates when subjects are actively engaged in perceptual tasks. Despite numerous observations, the role of DN deactivation remains unclear. Using computational neuroimaging applied to a large dataset of the Human Connectome Project (HCP) and to two individual subjects scanned over many repeated runs, we demonstrate that the DN selectively deactivates as a function of the position of a visual stimulus. That is, we show that spatial vision is encoded within the DN by means of deactivation relative to baseline. Our results suggest that the DN functions as a set of high-level visual regions, opening up the possibility of using vision-science tools to understand its putative function in cognition and perception.
Keywords: deactivation, default network, Human Connectome Project, population receptive field, ultra-high-field fMRI
Introduction
Functional magnetic resonance imaging (fMRI) allows the measurement of activation and deactivation in large-scale brain networks relative to a baseline (Gusnard and Raichle 2001). While the roles of sensory, attention, and limbic networks have been identified (Yeo et al. 2011), much less is known about the role of the default network (DN). Nevertheless, this network encompasses wide, evolutionarily recent swaths of frontal, parietal, and temporal lobes. DN activation reflects high-level cognition such as mind-wandering (Mason et al. 2007; Christoff et al. 2009), leading different authors to conclude that the DN is a domain general associative network (Gusnard and Raichle 2001; Raichle 2015). But our limited knowledge about its role partly resides in the fact that most studies to date show that the DN activates when humans or monkeys passively rest in a scanner (Raichle et al. 2001; Mantini et al. 2011), making experimental inferences based on its activation hard or impossible.
Conversely, deactivation of the DN can be robustly elicited by different visual tasks, including visual attention (Mayer et al. 2010; Ossandón et al. 2011) as well as visual working memory and episodic memory (Mayer et al. 2010; Foster et al. 2012; Lee et al. 2017; Sormaz et al. 2018) or visual perception tasks (González-García et al. 2018). Based on these results, we hypothesized that DN deactivation may be driven by the presentation of localized visual stimuli.
To test our hypothesis, we took advantage of a computational neuroimaging method previously used to model blood-oxygenation-level-dependent (BOLD) signal increases in the visual system (Dumoulin and Wandell 2008; Dumoulin and Knapen 2018). Using repeated sequences of spatially modulated visual stimuli (Fig. 1a), we estimate the spatial tuning properties of neural populations, their population receptive field (pRF). Following a traditional approach in the field, we first estimated pRFs from BOLD “activation” and will refer to these as “positive” population receptive fields (+pRFs).
Figure 1.
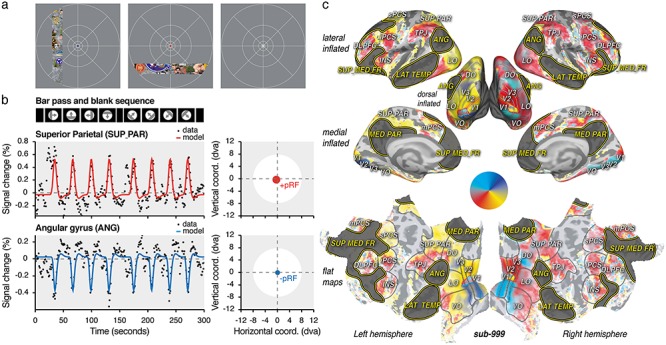
Task, models and polar angle maps. (a) HCP dataset task. About 181 subjects fixated a fixation point at the screen center and viewed a bar composed of different visual objects and noise patterns. They reported color changes of the fixation dot (see blue, red and white fixation dots, in the left, center, and right panel, respectively). (b) HCP dataset visual design and time samples. The bar traversed the screen following the same sequence across runs and subjects. The averaged time samples across the HCP subjects (“sub-999”) were modeled with isotropic Gaussian pRFs (represented as red or blue circles put in the screen coordinates, see right panels) predicting either positive (+pRF, top panel) or a negative (−pRF, bottom panel) modulations of the signal as a function of the bar position overt time. (c) Inflated and flat views of the cortex. The inflated lateral, dorsal, and medial views as well as the flat view of the cortex depict the best explained polar angle of the +pRFs obtained with sub-999. +pRF polar angle progression on the cortex (see color wheel) and anatomical references were used to determine low-level (V1/V2/V3: visual areas 1, 2, and 3; VO/LO/DO: visual occipital, lateral, and dorsal occipital areas) and high-level visual areas (SUP_PAR: superior parietal area; TPJ: temporal parietal junction; mPCS/sPCS/iPCS: medial, superior, and inferior pre-central sulcus areas; INS: insula; DLPFC: dorso-lateral prefrontal cortex). DN areas were defined using an atlas (highlighted in dark gray, ANG: angular gyrus; LAT_TEMP: lateral temporal area; MED_PAR: medial parietal area; SUP_MED_FR: superior medial frontal area). The explained variance (R2) of the model is depicted with color transparency. See Supplementary Figure S1 for individual subjects dataset polar angle maps (sub-001 and sub-002).
Combining +pRF properties on the surface of the cortex allows researchers to noninvasively reveal human retinotopic maps in both low- and high-level visual areas (Kay et al. 2015; Benson et al. 2018). In particular, this method has been used to reliably determine the linear relationship between +pRF eccentricity and size parameters (Winawer et al. 2010; Harvey and Dumoulin 2011; Sprague and Serences 2013; DeSimone et al. 2015; van Es et al. 2019). Such a relationship is inherited throughout the visual system from the organization of the retinal photoreceptors themselves (Curcio et al. 1990) and predicts that receptive field size depends on the distance between its center and the gaze position: the larger this distance, the bigger the receptive field size.
In contrast to previous work, our analysis allowed the pRF model to capture both BOLD signal increases and decreases (Fig. 1b). We observed that the DN signals are best predicted by deactivation as a function of the stimulus position, with what we call “negative” population receptive fields (−pRFs). We first analyzed the averaged time courses across 181 subjects (a total of 108 600 time samples, see “HCP dataset” in Methods), made available by the Human Connectome Project (Van Essen et al. 2013; Benson et al. 2018). All subjects viewed a bar traversing the screen in different directions (Fig. 1a) while being scanned using state-of-the-art ultra-high field (7T) methods (Benson and Winawer 2018). We present these average-subject results as our first dataset: “sub-999.” We next confirmed these results at the individual level, replicating our results with a different stimulus and 7T scanner. As the first dataset involved several hours of recording, we aimed at maximizing the data collection with two volunteers extensively scanned over 25 (sub-001, 3000 time samples) and 33 averaged runs (sub-002, 3960 time samples) of a similar retinotopy experiment. Further, to provide population-level assessments of our findings, we finally analyzed the 181 subjects individually (600 time samples per subject) and present their average results as “sub-000.” While datasets differ in several parameters (e.g., repetition time, time points per run) they were both analyzed with the same model and compared at the level its outputs (see Material and Methods).
We found, consistently across datasets, that DN deactivation signals can be explained based on the position of visual inputs, with the DN acting as a negatively modulated, high-level visual network.
Materials and Methods
Experimental Model and Subject Details
HCP Dataset
The publicly available HCP young adult 7T retinotopy dataset was used for analyses. Please see the accompanying publication for details on subjects (Benson et al. 2018). A total of 181 subjects (109 females, 72 males, age 22–35) took part in a retinotopy experiment. All subjects had normal or corrected-to-normal visual acuity. Each subject has been assigned a number (sub-003 to sub-183), the averaged time series is referred as “sub-999,” and the average of parameter estimates across subjects after fitting as “sub-000.”
Individual Subject Dataset
Two staff members (one author) from the Spinoza Centre for Neuroimaging participated in the experiment (sub-001 and sub-002, 2 males, 29 and 40 years old). Both subjects had normal vision. The experiment was undertaken with the understanding and written consent of both subjects and was carried out in accordance with the Declaration of Helsinki. Experiments were designed according to the ethical requirement specified by the Amsterdam University Medical Centre institutional review board ethics approval.
MRI Data Acquisition
HCP Dataset
Please see the accompanying publication for details on MRI acquisition (Benson et al. 2018). Briefly, structural scans were acquired at the Washington University on a customized Siemens 3T Connectom scanner (Siemens Healthcare, 0.7-mm isotropic resolution) and used as the anatomical substrate for the functional data. Functional data were acquired at the Center for Magnetic Resonance Research at the University of Minnesota on a Siemens 7T Magnetom actively shielded scanner (Siemens Healthcare) and a 32-channel receiver coil array with a single-channel transmit coil (Nova Medical). Whole-brain functional data were collected at a resolution of 1.6 mm isotropic and 1 s time repetition (TR), a multiband acceleration factor of 5, an in-plane acceleration factor of 2 and 85 slices covering the whole brain.
Individual Subject Dataset
T1-weighted structural scans were acquired at the Spinoza Centre for Neuroimaging on a Philips Achieva 3T scanner (Philips Medical Systems, 1.0 mm isotropic resolution) and a 32-channel receiver coil array with a single-channel transmit coil. Functional data were collected at the same center on a Philips Achieva 7T scanner (Philips Medical Systems) with a 32-channel receiver coil array with eight-channel transmit coil (Nova Medical). Functional data were collected at a resolution of 2.0 mm isotropic, 1.5 s TR, flip angle of 62°, in-plane anterior-posterior SENSE and SMS acceleration factors of 2 and 60 slices covering the occipital, the parietal, and partly the frontal and temporal cortical lobes. To estimate and later correct induced susceptibility distortion, we separately acquired opposite phases encoding direction images. The phases of the transmit channels were set to provide good signal homogeneity over the entire brain.
Retinotopy Tasks
HCP Dataset
Please see the accompanying publication for more details on the visual stimuli (Benson et al. 2018). Briefly, the experimental software controlling the task was implemented in Matlab (MathWorks), using the Psychophysics toolboxes (Brainard 1997; Pelli 1997). Stimuli were presented at a viewing distance of 101 cm, on a back projection screen displaying images from a NEC NP4000 projector (Tokyo, Japan) viewed through a mirror mounted on the radiofrequency coil. The screen had a spatial resolution of 1024 by 768 pixels and a vertical refresh rate of 60 Hz. Button responses were collected using a MRI compatible button box (Current Designs). The experiment analyzed here consisted of two identical runs of 300 s (300 TRs). During each run, a slowly moving bar aperture was presented on a uniform gray background (Fig. 1a). The bar aperture contained a 15 Hz dynamic texture composed of visual objects of different sizes placed on pink noise background. Visual objects of different size were placed on the pink noise background and include human and nonhuman animate objects (body parts or faces) and natural or artificial imamate objects from Kriegeskorte et al. (2008). The bar aperture was constrained within a virtual circle of about 8° of visual angle (dva), had a width of ~ 2 dva, and a duty cycle of ~3 s. The runs started with a blank period of 16 s, followed by four visual stimulation periods of 32 s each, a blank period of 12 s, four visual stimulation periods of 32 s each, and a final blank period of 16 s. In the visual stimulation periods, the bar aperture moved in the same direction during the first 28 s before being blanked for 4 s. The bar aperture movement direction was sequentially 0° (right), 90° (up), 180° (left), 270° (down), 45° (right-up), 315° (left-up), 225° (left-down), and 135° (right-down). A semitransparent central fixation dot (0.15 dva radius) and fixation grid were superimposed on the display throughout the run. The fixation dot color changed every 1–5 s randomly between three colors (black, white, and red). Subjects were instructed to maintain fixation on the dot and to report the color change via a button press.
Individual Subject Dataset
The experimental software controlling the task was implemented in Python using Psychopy toolbox (Peirce 2008). Stimuli were presented at a viewing distance of 225 cm, on a 32-inch LCD screen (BOLDscreen, Cambridge Research Systems) situated at the end of the bore and viewed through a mirror. The screen had a spatial resolution of 1920 by 1080 pixels and a refresh rate of 120 Hz. Button responses were collected using a MRI compatible button box (Current Designs). The experiment consisted of 25 (sub-001) and 34 (sub-002) identical runs of 180 s (120 TRs) collected within two to three experimental sessions. During each run, a slowly moving bar aperture was presented on a uniform gray background. The bar aperture contained small drifting Gabor elements (100% contrast; standard deviation of the Gaussian window: 0.2 dva). Gabor elements were updated every 0.5 s at a random location uniformly distributed over the bar aperture, with a random orientation, a random spatial (between 0.5 and 5.0 cycles per dva) and temporal frequency (between 7 and 12 Hz). Gabor elements were in grayscale, except during the middle 0.5 s of each TR, when they were colored in cyan/magenta or blue/yellow. The bar aperture could either be horizontally or vertically oriented and their centers moved on every TR by discrete steps to traverse the entire screen perpendicular to the bar direction (17.8 by 10 dva). The vertically oriented bar aperture had a width of ~2.25 dva and traverse the screen in 20 steps. The horizontally oriented bar aperture had a width of ~1.25 dva and traverse the screen in 13 steps. The runs started with a blank period of 24 s, followed by horizontally and vertically oriented stimulation periods of 22.5 and 33 s, respectively, a blank period of 22.5 s, vertically and horizontally oriented stimulation periods of 33 and 22.5 s, respectively, and a final blank period of 22.5 s. In the stimulation periods, the bar aperture moved in the same direction before being blanked during the last 3 s. The bar aperture movement direction was sequentially 270° (down), 180° (left), 0° (right), and 90° (up). A central fixation bull’s eye dot (0.1 dva radius) was superimposed on the display throughout the runs. Subjects were instructed to keep their eyes on the fixation dot and to report after each TR the ratio between the cyan/magenta and blue/yellow colored Gabor elements via button presses. The difficulty of the task was titrated at a performance of 79% (three-up and one-down staircases) across the visual field by controlling the ratio between the colored elements independently at three equally divided distances of the bar aperture center from the fixation dot.
fMRI Preprocessing
HCP Dataset
Brain surfaces were reconstructed from the structural scans using the HCP Pipelines (Glasser et al. 2013) and aligned on the surface using “Multimodal Surface Matching All” registration (Robinson et al. 2014, 2018; Glasser et al. 2016). The data were processed using the HCP pipelines (Glasser et al. 2013) to correct head motion and EPI spatial distortion. The data were later registered with the HCP standard surface space. The functional data were denoised for spatially specific structured noise using multirun “sICA-þFIX” (Glasser et al. 2018). The HCP pipeline produces for each subject Connectivity Informatics Technology Initiative files (CIFTI) containing 91 282 grayordinates covering both cortical and subcortical brain regions with 2.0 mm spatial resolution. As we were interested to cortical regions, we converted the CIFTI files to Geometry format under the Neuroimaing Informatics Technology Initiative (GIfTI) hemisphere surface files containing 32 492 vertices per hemisphere. Low frequency drifts of the time series were removed using a third order Savitzky–Golay filter (window length of 210 s). Arbitrary functional signal units were converted to BOLD percent signal change and averaged across runs and across subjects (sub-999).
Individual Subject Dataset
We used fMRIPrep (Esteban et al. 2019) to preprocess the functional scans. Briefly, based on the estimated susceptibility distortion, an unwarped BOLD reference was calculated for a more accurate coregistration with the anatomical reference. The BOLD reference was then coregistered to the T1-weighted images. Coregistration head-motion parameters with respect to the BOLD reference were next estimated. The functional time series were next resampled to “fsaverage6” space and saved as GIfTI hemisphere surface files composed of 81 924 vertices. Low frequency drifts of the functional time series were removed using a third order Savitzky–Golay filter (window length of 210 s and polynomial order of 3). Arbitrary functional signal units were converted to BOLD percent signal change and averaged across runs. Registrations to T1-weighted images and “fsaverage6” were inspected; one run was excluded due to incorrect registration.
PRF Model
We analyzed time series of both datasets using an isotropic Gaussian pRF model (Dumoulin and Wandell 2008). The model was implemented using “Popeye” (DeSimone et al. 2015) and included the position (x, y), the size (standard deviation of the Gaussian), as well a signal amplitude (beta) and signal baseline as parameters. The time series were fitted in two parts. The first part was a coarse spatial grid search of six linear steps constraining the position and size parameters of the model. A linear regression between the predicted and measure time series signal was used to determine the baseline and amplitude parameters. The best-fitting parameters of the first step were next used as the starting point of an optimization phase to produce finely tuned estimates. The visual stimulus was downsampled by a factor 4 for the first of these fitting stages. The grid search position parameters were distributed linearly within 150% of the stimulus size, giving 12 (“HCP dataset”) and 13.4 dva (“individual subject dataset”) to the left and right of the screen center for the vertical position parameter and 12 (HCP dataset) and 7.5 dva (“individual subject dataset”) for the vertical position parameter. The grid search size parameters were bounded between 0 and 187.5% of the smallest stimulus side, giving bounds between 0 and 15 dva (“HCP dataset”) and between 0 and ~9.4 dva. In the fine search stage, we used a downhill simplex algorithm starting from the obtained grid search parameters, leaving the parameter ranges essentially unconstrained. Estimated parameter values for grayordinates in the HCP dataset were converted to “fsaverage” using connectome workbench after fitting (Marcus et al. 2011). Estimated values for each vertex of the “fsaverage6” cortical surface were used for visualization converted to “fsaverage” using Freesurfer mri_surf2surf (Dale et al. 1999).
Data Analyses
PRF polar angle maps (Fig. 1c and Supplementary Fig. S1) were derived with the best explained pRF position parameters. The polar angle of each vertex was drawn using “Pycortex” (Gao et al. 2015) on an inflated and a flattened cortical visualization of “fsaverage.” The coefficient of determination of the model (R2) was used to determine color transparency.
PRF coefficient of determination (R2) of each vertex was computed by dividing the sum of squared differences between the best predicted model and the observed time samples by the sum of the squared demeaned observed time samples. pRF R2 values of each vertex were drawn on an inflated and a flat cortical visualization of “fsaverage” as a function of the sign of the best explained amplitude parameters (Fig. 2a and Supplementary Fig. S2). −pRFs (pRF with a negative amplitude parameter) were represented with a color scale going between black (R2 = 0) and blue (R2 = 1). +pRFs (pRF with a positive amplitude parameter) were represented with a color scale going between black and red (R2 = 1). The averaged and standard deviation of pRF R2 were computed across the vertices observed within each region of interest for +pRF and −pRF separately (Fig. 2b, left panels).
Figure 2.
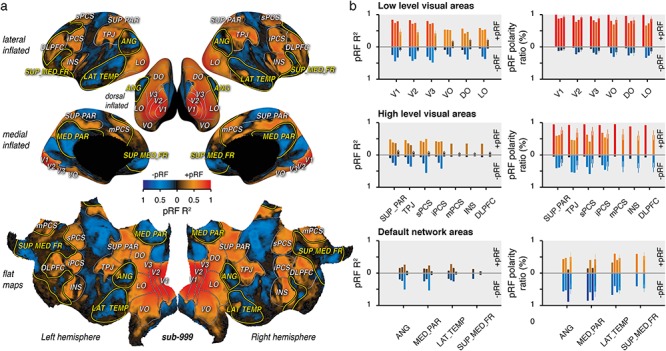
Positive and negative pRF. (a) Inflated and flat views of the cortex. The inflated lateral, dorsal, and medial views as well as the flat view of the cortex depict the coefficient of determination of the pRF model prediction (pRF R2) multiplied by the sign of the estimated amplitude parameter (color scale: +pRFs: red colors; −pRFs:blue colors), for the averaged time samples across the subjects of the HCP dataset (sub-999). (b) Averaged pRF R2 (left panels) and pRF polarity ratio (right panels) observed with +pRF (top parts) and −pRF (bottom parts), respectively, within low-level visual areas (top panels), high-level visual areas (middle panels), and default network areas (bottom panels). The results of “sub-999,” “sub-001,” “sub-002,” and “sub-000” are shown per region of interest in order from the leftmost to the rightmost bar. Note that results from “sub-001” and “sub-002” most frontal areas (mPCS, INS, DLPFC and SUP_MED_FR) are not reported. These areas were not covered with our partial field of view (see MRI Data Acquisition). Colors are determined with the scale used in panel A, error bars show the STD across the HCP subjects.
pRF polarity ratio was derived from the proportion within a region of interest of vertices with a positive or a negative amplitude parameter (Fig. 2b, right panels).
The coefficient of correlation (r) between the pRF eccentricity and pRF size (Fig. 3b, left panels) was obtained by computing for each region of interest, the covariance between these pRF parameters weighed by the explained variance (R2). The coefficient describes the degree to which the eccentricity and size parameters of a region are related or associated with each other. It varies between −1 and 1, with positive values indicating that pRF size increase as a function of the increase of the pRF eccentricity.
Figure 3.
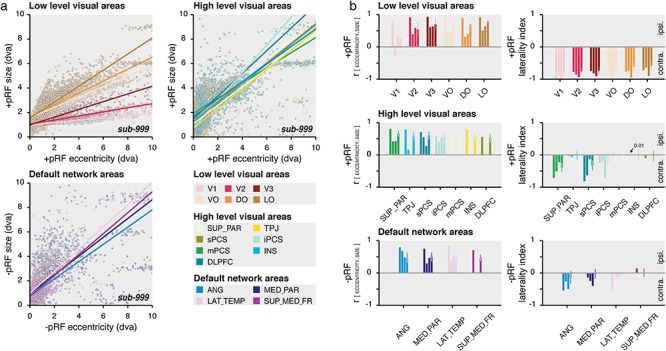
DN visual characteristics. (a) pRF eccentricity and size relationship. Size as a function of the eccentricity of +pRFs within low- (top left panel) and high-level visual areas (top right panel) and of −pRFs of the DN areas (bottom left panel), for the averaged time samples across the HCP subjects (sub-999). Dots represent individual cortical vertex (see color legend) and lines show the best explained linear function weighted by the pRF explained variance (R2). (b) Correlation and laterality index. Left panels show the correlation coefficient (r) between the pRF eccentricity and size parameters weighted by the pRF explained variance (R2) and right panels show the laterality index of +pRFs within low- (top left panel) and high-level visual areas (middle left panel) and of −pRFs of the DN areas (bottom left panel). The results of sub-999, sub-001, sub-002 and sub-000 are shown per region of interest in order from the leftmost to the rightmost bar. The most frontal areas of sub-001 and sub-002 (mPCS, INS, DLPFC and SUP_MED_FR) are not reported. Error bars show the STD across the HCP subjects.
The pRF laterality index (Fig. 3b, right panels) was derived from the ratio of pRF vertices representing the ipsilateral or contralateral visual hemifield (using the best explained pRF horizontal position parameter) relatively to the brain hemisphere each vertex belongs. The index varies between −1 (100% contralateral) and +1 (100% ipsilateral).
To avoid outlier effects, the coefficients of correlation and laterality indices were computed from vertices where the pRF was not located too close to the fixation target (<0.1 dva) or well outside the visual stimulus (>12 dva). We also excluded vertices from which the pRF size was small considering that a recorded voxel included thousands of visual neurons (<0.1 dva), and from which the pRF size was too big to properly distinguish the visual bar position over time (>12 dva).
Results from the most frontal areas (mPCS, INS, DLPFC and SUP_MED_FR) of the individual subject dataset are not reported. These areas were not well covered with the partial field of view used for the functional scans (see MRI Data Acquisition).
For statistical comparisons, we drew 10 000 bootstrap samples (with replacement) from the original pair of compared values. We then calculated the difference of these bootstrapped samples and derived two-tailed P values from the distribution of these differences.
Code Availability
HCP Retinotopy Dataset
The HCP functional dataset was published online (https://balsa.wustl.edu/study/show/9Zkk). We make available online our pRF analysis codes (https://github.com/mszinte/HCP_dataset_analysis) and our pRF parameters estimates (https://osf.io/y4hfn/).
Individual Subject Dataset
We published online the individual subject dataset imaging data (https://osf.io/5bmdn/), our pRF analysis codes (https://github.com/mszinte/Indiv_dataset_analysis/), and pRF parameters estimates (https://osf.io/5bmdn/).
Results
We first analyzed the resulting time series of the average across HCP subjects by fitting a model predicting signal modulations as a function of the bar position over time multiplied by isotropic Gaussian pRFs, of which position and size parameters can be varied (Fig. 1b). Converting these best-fitting parameters to polar angle and eccentricity (i.e., the angle and distance relative to the fixation point, see Fig. 1c) and combining these with anatomical references (Wandell et al. 2007), we determined the boundaries of low- and high-level visual areas (Fig. 1c). DN cortical areas (Fig. 1c) were, on the other hand, defined using an atlas obtained by intrinsic functional connectivity analysis of 1000 human subjects performing a resting state task (Yeo et al. 2011). We took a conservative approach to our analysis, applying the same region-of-interest definitions to the HCP (sub-999 and sub-000), as well as our individual subject datasets (sub-001 and sub-002). We further did not tailor our analyses to either our datasets or subjects specifically. Instead, we used an automated workflow to make our results robust across different individuals, datasets, scanners, and protocols.
We first quantified the quality of the model fit for both positive and negative pRFs. Figure 2a shows different cortical views of the pRF coefficient of determination (R2) derived from the average across HCP subjects (see Supplementary Fig. S2 for individual subjects dataset).
First, we found that overall V1 signal variance was extremely well explained by our model in the average HCP subject (Fig. 2b, sub-999: +pRF R2 = 0.84) and in the individual subjects dataset (+pRF R2: sub-001: 0.74; sub-002: 0.79). As expected from the limited scanning duration per participant, V1 explained variance was reduced when the analysis relied on signals from individual subjects of the HCP dataset (+pRF R2: sub-000: 0.47 ± 0.09—mean ± STD across subjects—). Positive amplitude pRF parameters best explained V1 signals across all datasets (Fig. 2b, +pRF polarity ratio: sub-999: 99.7%; sub-001: 87.7%; sub-002: 89.0%; sub-000: 93.8 ± 5.2%). Moreover, similar results were found across nodes that we defined as low-level visual areas (+pRF R2: sub-999: 0.69 ± 0.14; sub-001: 0.61 ± 0.13; sub-002: 0.63 ± 0.14; sub-000: 0.33 ± 0.14; +pRF polarity ratio: sub-999: 97.6 ± 2.1%; sub-002: 78.7 ± 7.2%; sub-000: 87.6 ± 4.4%; sub-001: 78.0 ± 7.4% —mean ± STD across areas—).
Next, we found that the average HCP subject variance within what we defined as high-level visual areas were also best explained with +pRFs (sub-999: +pRF R2 = 0.41 ± 0.06, +pRF polarity ratio: 93.1 ± 2.4%). A similar range of explained variance across high level visual areas was obtained for the individual subjects dataset (+pRF R2: sub-001: 0.39 ± 0.02; sub-002: 0.40 ± 0.08), with, however, a less pronounced proportion of +pRF than that observed for low level visual areas (+pRF polarity ratio: sub-001: 59.2 ± 9.2%; sub-002: 50.4 ± 9.0%). Explained variance dropped significantly in high-level as compared to low-level visual areas also in individual subjects of the HCP dataset (sub-000: +pRF R2 = 0.08 ± 0.02, +pRF R2 of low vs. high visual areas: P < 0.0001), with nevertheless a large predominance of +pRFs over −pRFs remaining (sub-000: +pRF polarity ratio: 66.7 ± 6.2%, +pRF polarity ratio of low vs. high visual areas: P < 0.0001). Thus, we found that both low- and high-level visual areas were best explained with pRF models predicting a positive amplitude modulation (+pRF).
We then turned our attention to the DN. There, we found that the majority of vertices (brain units) included in atlas-defined DN areas were best explained with pRF models predicting negative amplitude modulations (−pRF). This effect was observed both with the average HCP subject (Fig. 2b, sub-999: −pRF ratio: 62.5 ± 18.9%) as well as with the analysis of the individual subject datasets (sub-001: 50.8 ± 9.8%; sub-002: 77.6 ± 15.8%) and individual subject of the HCP dataset (−pRF ratio: sub-000: 51.9 ± 4.5%). DN explained variance of −pRF for individual subjects of the HCP dataset was, however, diminished (sub-000: DN −pRF R2 = 0.05 ± 0.01) as compared to that observed for +pRF of the low- (P < 0.0001) and high-level visual areas (P < 0.0001). This indicates that visual pRF signal-to-noise in the DN is lower than in the canonical visual system. This notion is confirmed by the fact that, when using the average HCP subject (−pRF R2: sub-999: 0.20 ± 0.04) or our two highly sampled individual subjects (−pRF R2: sub-001: 0.21 ± 0.03; sub-002: 0.43 ± 0.15), the explained variance of −pRFs within the DN is comparable to what observed with +pRF in high-level visual areas. Nevertheless, we found that our model of visually selective BOLD responses explained high levels of time series variance in the angular gyrus (−pRF R2: sub-999: 0.19; sub-001: 0.24; sub-002: 0.51), the medial parietal area (−pRF R2: sub-999: 0.25; sub-001: 0.18; sub-002: 0.54), the lateral temporal area (−pRF R2: sub-999: 0.18; sub-001: 0.22; sub-002: 0.26), and the superior medial frontal area (−pRF R2: sub-999: 0.18), all well-known nodes of the DN.
Together, these results suggest that the DN encoded the visual stimulus position over time by means of systematic reductions in BOLD signals relative to the baseline. We reasoned that the large datasets of high quality at our disposal, combined with our modeling methods, could allow us to determine some potential visual–spatial characteristics of this selectivity.
Specifically, we aimed at comparing some known visual characteristics of +pRFs of the visual network to what observed with −pRFs of the DN. We first studied the relationship between +pRF eccentricity and size parameters across visual areas (see Methods). As expected, we found that the size of +pRFs within the low-level visual areas depended on their eccentricity, with bigger +pRFs in the periphery than near the fovea. Across low-level visual areas, we observed a strong positive relationship between these parameters for the average HCP subject (Fig. 3, sub-999: +pRF r = 0.62 ± 0.19) and the individual subject datasets (Supplementary Fig. S3, +pRF r: sub-001: 0.44 ± 0.12, excluding V1: −0.27; sub-002: 0.51 ± 0.13). We found a similar relationship when analyzing individual subjects of the HCP dataset (+pRF r: sub-000: 0.62 ± 0.19) with positive correlations across the low-level visual areas (Ps < 0.0001). Similarly, we found positive correlations between the +pRF eccentricity and size parameters across all high-level visual areas (+pRF r: sub-999: 0.70 ± 0.10; sub-001: 0.38 ± 0.16; sub-002: 0.31 ± 0.24; sub-000: 0.57 ± 0.05, P < 0.0001).
Interestingly, the best-fitting eccentricity and size parameters of the −pRFs predicting DN deactivation was also positively correlated, even when analyzing individual subjects of the HCP dataset (−pRF r: sub-000: 0.48 ± 0.01, P < 0.0001), in both the angular gyrus (−pRF r: sub-000: 0.48 ± 0.16, P < 0.0001), the medial parietal area (−pRF r: sub-000: 0.50 ± 0.14, P < 0.0001), the lateral temporal area (−pRF r: sub-000: 0.48 ± 0.11, P < 0.0001), and the superior medial frontal area (−pRF r: sub-000: 0.47 ± 0.10, P < 0.0001). These signatures of visual–spatial structure in the DN were even more prominent in the across-participant average HCP data (Fig. 3, −pRF r: sub-999: 0.78 ± 0.06) and in our own highly sampled individual participants dataset (Supplementary Fig. S3, −pRF r: sub-001: 0.45 ± 0.21; sub-001: 0.49 ± 0.01). The strong correlations between pRF eccentricity and size we find in the DN indicate that the way the DN represents visual space is similar to that of the canonical visual system.
Next, it is well established that visual neurons in the cerebral cortex principally sample contralateral visual locations. We investigated whether −pRFs represent the visual hemifield opposite to their cerebral hemisphere by computing a laterality index (see Methods). +pRFs in low-level visual areas predominantly represent the opposite visual hemifield (+pRF laterality index: sub-999: −0.78 ± 0.10; sub-000: −0.70 ± 0.11; sub-001: −0.80 ± 0.11; sub-002: −0.93 ± 0.04). Replicating electrophysiological findings, this contralaterality was present but reduced in some high-level visual areas (+pRF laterality index: sub-999: −0.30 ± 0.33; sub-001: −0.35 ± 0.26; sub-002: −0.27 ± 0.30; sub-000: −0.13 ± 0.16), such as superior parietal and sPCS/iPCS areas (Schall 2015), with other regions such as TPJ and DLPFC representing both visual hemifields in each cerebral hemisphere (Funahashi et al. 1991).
Akin to these latter regions, we found that −pRFs lateral temporal area (−pRF laterality index: sub-999: −0.55; sub-001: −0.05; sub-002: −0.13; sub-000: −0.08 ± 0.13) and the superior medial frontal area (−pRF laterality index: sub-999: 0.14; sub-000: 0.01 ± 0.13) bilaterally represent both visual hemifields. But −pRFs in the more posterior nodes of the DN, the angular gyrus (−pRF laterality index: sub-999: −0.54; sub-001: −0.26; sub-002: −0.50; sub-000: −0.14 ± 0.20) and the medial parietal area (−pRF laterality index: sub-999: −0.14; sub-001: −0.25; sub-002: −0.40; sub-000: −0.02 ± 0.15) specifically represent the contralateral visual hemifield, making their spatial sampling similar to that of +pRFs in neighboring visual regions. Again, we observed consistent contralaterality within these regions mainly when analyzing the averaged time samples across the HCP subjects or our individual subject datasets, suggesting that such level of detail necessitates a high signal-to-noise ratio.
Discussion
We aimed at determining a putative visual role of the cortical DN. To do so, we analyzed the cortical activity assessed with ultra-high field functional imaging methods in 183 subjects viewing different sequences of a localized visual stimulus. We assumed that each cortical unit had the potential to treat the spatial content of the stimulus through a 2D-Gaussian window (pRF) of varying sizes, polar angles, and visual eccentricities. Crucially, we allowed our model to predict both BOLD activation or deactivation as a function of the spatio-temporal sequence of the stimulus. We observed that the majority of the cortical units within the DN were best explained by a model predicting negative BOLD signals, that is, by spatially specific “negative” population receptive fields.
We found that within four cortical areas of the DN, −pRFs are spatially selective and that their sizes depend on their eccentricities. These negative pRFs also predominantly represent the contralateral visual hemifield. These features resemble those observed across high-level visual areas and suggest that the DN acts as a negatively modulated high-level visual network.
Negative BOLD signals have previously been studied using combined electrophysiological and BOLD signals in V1 (Shmuel et al. 2002, 2006). It was shown that negative BOLD is associated with decreases in neural firing rate relative to a resting baseline and was evoked as a function of the retinal position of a stimulus. Such findings ruled out the possibility that negative BOLD could be due to correlated noise, head motion, imaging, or vascular steal artifacts (Shmuel et al. 2006). Other than for the early visual cortex, negative BOLD signals have often been reported in the context of DN fMRI studies. Indeed, DN areas were defined by highly correlated BOLD activation during rest (Raichle et al. 2001) and BOLD deactivation while subjects participated in demanding tasks (Mayer et al. 2010; Sestieri et al. 2011; Sormaz et al. 2018).
We propose that DN deactivation reflects the response of populations of neurons with what can be considered negative visual receptive fields. The observed DN deactivation on which our model fits rely is unlikely to reflect artifactual effects. First, DN deactivation is readily observed in direct electrical measures in humans (Miller et al. 2009; Foster et al. 2015). Second, our results demonstrate that negative BOLD signals are selective: they depend on the position of the visual stimulus over time. Third, the ordered patterns of visual–spatial representations in the DN, and their similarity with the high-level visual system, is unlikely to be the result of across-subject averaging or imaging artifact, as we replicate these patterns in individual subjects and independent experiments.
The DN is thought to constitute one extreme of a gradient leading from primary sensory and motor regions to transmodal association cortex (Buckner and Krienen 2013; Margulies et al. 2016). Our results suggest that the DN encodes the spatial content of a visual scene by negative modulations. Simultaneously, the visual and attention networks also encode similar information but with positive modulations. What could be the role of DN deactivation?
The authors have suggested that the DN has a role in high-level cognition such as mind-wandering (Weissman et al. 2006; Mason et al. 2007; Christoff et al. 2009), when attention is directed toward internally generated cognitions, and away from incoming sensory information. On the other hand, DN deactivation is observed in various visual tasks (Mayer et al. 2010; Sestieri et al. 2011; Sormaz et al. 2018). The visual selectivity of the DN that we observe here could thus potentially serve to store sensory signals before reusing such information to form visual memory-guided thought such as during mind-wandering. Importantly, visual space in this framework could serve as a shared reference frame for interactions between networks.
Second, there is abundant evidence from sensory processing that the interplay between activation and deactivation can serve to decorrelate neural responses to input patterns and increase processing efficiency by means of predictive coding (Srinivasan et al. 1982). That is, there are computational benefits to representing the same signal with both activation and deactivation. For example, deactivation can “explain away” errant computational outcomes by explicitly representing “what is not” (Goncalves and Welchman 2017). By implementing these computational strategies, the DN would be ideally suited and located to perform integration across modalities (Margulies et al. 2016) and time (Sestieri et al. 2017).
Importantly, while we tested a putative visual role of the DN, we cannot exclude the possibility that it also encodes sensory information in other modalities. Indeed, the DN has been shown to deactivate in auditory tasks (Humphreys et al. 2015; Simony et al. 2016), with even a modular reconfiguration to the auditory network as defined with connectivity methods during a challenging listening task (Alavash et al. 2019). Future work should determine the sensory reference frames of the DN, for example by adapting previously developed population receptive field models for example of audition (Thomas et al. 2015) or proprioception (Schellekens et al. 2018).
Another important aspect is that our data derived from fMRI BOLD negative signals. While previous studies have shown that these signals reliably relate to neural activity (Shmuel et al. 2002, 2006), future work could make use of electrophysiological methods to confirm or disconfirm our results with direct electrical measures or stimulation in the human (Foster and Parvizi 2017) or animal model (Mantini et al. 2011). Moreover, while 2D-Gaussian models used here explained well the variance of low level visual areas activity, more elaborated models might better apply to higher visual areas and to the DN (Kay et al. 2008, 2013; Nishimoto et al. 2011; Zuiderbaan et al. 2012). In particular, a model including a difference-of-Gaussians pRF with in addition to a negative center a positive surround would account for the observed positive overshoot of the BOLD signals in the DN (Fig. 1b), similarly to how this model accounted for the negative undershot in low-level visual areas (Zuiderbaan et al. 2012).
A dominant research direction in the study of the DN focuses on connectivity measures by correlating activation and deactivation BOLD signals during the resting state (Raichle 2015). We here used a functional connectivity atlas to determine DN cortical boundaries (Yeo et al. 2011), but stress that our findings are based on forward modeling of visual responses but not connectivity. That is, our model established the visual selectivity of many cortical locations in the DN by analyzing time-varying signals in relation to a visual stimulus alone. Yet, our results suggest that the correlation patterns between the DN and the higher-level visual system during the resting state partly reflect the use of a common sensory reference frame. This indicates that sensory organizational principles may play a large role in the structure of higher-level cognition.
We found that fMRI BOLD deactivation within the DN is well explained by negative population receptive fields tuned to circumscribed locations in visual space. Our results open up new “visual” possibilities to study the putative roles of the DN in both low- and high-level cognitive functions.
Funding
Marie Sklodowska-Curie Action Individual Fellowship (704537 to M.S.); Nederlandse Organisatie voor Wetenschappelijk Onderzoek-Chinese Academy of Sciences (NWO-CAS) (012.200.012 to T.K. ); Amsterdam Brain and Mind Project (ABMP) (20157 to T.K.).
Notes
We are grateful to the members of the Knapen and Dumoulin laboratories and especially Daan M. van Es and to Elodie Parison, Alice and Clémence Szinte for their support. Conflict of Interest: The authors declare no competing interests.
Author Contributions
M.S. and T.K. helped in conceptualization, methodology, software development, validation, formal analysis, providing resources, writing–original draft, writing–review and editing, visualization, and funding acquisition. M.S. investigation and supervised the study. T.K. contributed in data curation and project administration.
Supplementary Material
References
- Alavash M, Tune S, Obleser J. 2019. Modular reconfiguration of an auditory control brain network supports adaptive listening behavior. Proc Natl Acad Sci USA. 116:660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson NC, Jamison KW, Arcaro MJ, Vu AT, Glasser MF, Coalson TS, Van Essen DC, Yacoub E, Ugurbil K, Winawer J et al. 2018. The Human Connectome Project 7 tesla retinotopy dataset: description and population receptive field analysis. J Vis. 18:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson NC, Winawer J. 2018. Bayesian analysis of retinotopic maps. elife. 7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainard DH. 1997. The psychophysics toolbox. Spat Vis. 10:433–436. [PubMed] [Google Scholar]
- Buckner RL, Krienen FM. 2013. The evolution of distributed association networks in the human brain. Trends Cogn Sci. 17:648–665. [DOI] [PubMed] [Google Scholar]
- Christoff K, Gordon AM, Smallwood J, Smith R, Schooler JW. 2009. Experience sampling during fMRI reveals default network and executive system contributions to mind wandering. Proc Natl Acad Sci USA. 106:8719–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. 1990. Human photoreceptor topography. J Comp Neurol. 292:497–523. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis: I. Segmentation and Surface Reconstruction. NeuroImage. 9:179–194. [DOI] [PubMed] [Google Scholar]
- DeSimone K, Viviano JD, Schneider KA. 2015. Population receptive field estimation reveals new retinotopic maps in human subcortex. J Neurosci. 35:9836–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoulin SO, Knapen T. 2018. How visual cortical organization is altered by ophthalmologic and neurologic disorders. Annu Rev Vis Sci. 4:357–379. [DOI] [PubMed] [Google Scholar]
- Dumoulin SO, Wandell BA. 2008. Population receptive field estimates in human visual cortex. NeuroImage. 39:647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, Kent JD, Goncalves M, DuPre E, Snyder M et al. 2019. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods. 16:111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BL, Dastjerdi M, Parvizi J. 2012. Neural populations in human posteromedial cortex display opposing responses during memory and numerical processing. Proc Natl Acad Sci USA. 109:15514–15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BL, Parvizi J. 2017. Direct cortical stimulation of human posteromedial cortex. Neurology. 88:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster BL, Rangarajan V, Shirer WR, Parvizi J. 2015. Intrinsic and task-dependent coupling of neuronal population activity in human parietal cortex. Neuron. 86:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi S, Bruce CJ, Goldman-Rakic PS. 1991. Neuronal activity related to saccadic eye movements in the monkey's dorsolateral prefrontal cortex. J Neurophysiol. 65:1464–1483. [DOI] [PubMed] [Google Scholar]
- Gao JS, Huth AG, Lescroart MD, Gallant JL. 2015. Pycortex: an interactive surface visualizer for fMRI. Front Neuroinform. 9:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Bijsterbosch JD, Harrison SJ, Harms MP, Anticevic A, Van Essen DC, Smith SM. 2018. Using temporal ICA to selectively remove global noise while preserving global signal in functional MRI data. NeuroImage. 181:692–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M et al. 2016. A multi-modal parcellation of human cerebral cortex. Nature. 536:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR et al. 2013. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves NR, Welchman AE. 2017. “What not” detectors help the brain see in depth. Curr Biol. 27:1403–1412.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García C, Flounders MW, Chang R, Baria AT, He BJ. 2018. Content-specific activity in frontoparietal and default-mode networks during prior-guided visual perception. elife. 7:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnard DA, Raichle ME. 2001. Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci. 2:685–694. [DOI] [PubMed] [Google Scholar]
- Harvey BM, Dumoulin SO. 2011. The relationship between cortical magnification factor and population receptive field size in human visual cortex: constancies in cortical architecture. J Neurosci. 31:13604–13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys GF, Hoffman P, Visser M, Binney RJ, Lambon Ralph MA. 2015. Establishing task- and modality-dependent dissociations between the semantic and default mode networks. Proc Natl Acad Sci USA. 112:7857–7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay KN, Naselaris T, Prenger RJ, Gallant JL. 2008. Identifying natural images from human brain activity. Nature. 452:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay KN, Weiner KS, Grill-Spector K. 2015. Attention reduces spatial uncertainty in human ventral temporal cortex. Curr Biol. 25:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay KN, Winawer J, Mezer A, Wandell BA. 2013. Compressive spatial summation in human visual cortex. J Neurophysiol. 110:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Ruff DA, Kiani R, Bodurka J, Esteky H, Tanaka K, Bandettini PA. 2008. Matching categorical object representations in inferior temporal cortex of man and monkey. Neuron. 60:1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chun MM, Kuhl BA. 2017. Lower parietal encoding activation is associated with sharper information and better memory. Cereb Cortex. 27:2486–2499. [DOI] [PubMed] [Google Scholar]
- Mantini D, Gerits A, Nelissen K, Durand J-B, Joly O, Simone L, Sawamura H, Wardak C, Orban GA, Buckner RL et al. 2011. Default mode of brain function in monkeys. J Neurosci. 31:12954–12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Harwell J, Olsen T, Hodge M, Glasser MF, Prior F, Jenkinson M, Laumann T, Curtiss SW, Van Essen DC. 2011. Informatics and data mining tools and strategies for the Human Connectome Project. Front Neuroinform. 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, Bezgin G, Eickhoff SB, Castellanos FX, Petrides M et al. 2016. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 113:12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MF, Norton MI, Van Horn JD, Wegner DM, Grafton ST, Macrae CN. 2007. Wandering minds: the default network and stimulus-independent thought. Science. 315:393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer JS, Roebroeck A, Maurer K, Linden DEJ. 2010. Specialization in the default mode: task-induced brain deactivations dissociate between visual working memory and attention. Hum Brain Mapp. 31:126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KJ, Weaver KE, Ojemann JG. 2009. Direct electrophysiological measurement of human default network areas. Proc Natl Acad Sci USA. 106:12174–12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Vu AT, Naselaris T, Benjamini Y, Yu B, Gallant JL. 2011. Reconstructing visual experiences from brain activity evoked by natural movies. Curr Biol. 21:1641–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossandón T, Jerbi K, Vidal JR, Bayle DJ, Henaff M-A, Jung J, Minotti L, Bertrand O, Kahane P, Lachaux J-P. 2011. Transient suppression of broadband gamma power in the default-mode network is correlated with task complexity and subject performance. J Neurosci. 31:14521–14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JW. 2008. Generating stimuli for neuroscience using PsychoPy. Front Neuroinform. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelli DG. 1997. The VideoToolbox software for visual psychophysics: transforming numbers into movies. Spat Vis. 10:437–442. [PubMed] [Google Scholar]
- Raichle ME. 2015. The brain's default mode network. Annu Rev Neurosci. 38:433–447. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. 2001. A default mode of brain function. Proc Natl Acad Sci USA. 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EC, Garcia K, Glasser MF, Chen Z, Coalson TS, Makropoulos A, Bozek J, Wright R, Schuh A, Webster M et al. 2018. Multimodal surface matching with higher-order smoothness constraints. NeuroImage. 167:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M. 2014. MSM: a new flexible framework for multimodal surface matching. NeuroImage. 100:414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall JD. 2015. Visuomotor functions in the frontal lobe. Annu Rev Vis Sci. 1:469–498. [DOI] [PubMed] [Google Scholar]
- Schellekens W, Petridou N, Ramsey NF. 2018. Detailed somatotopy in primary motor and somatosensory cortex revealed by Gaussian population receptive fields. NeuroImage. 179:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestieri C, Corbetta M, Romani GL, Shulman GL. 2011. Episodic memory retrieval, parietal cortex, and the default mode network: functional and topographic analyses. J Neurosci. 31:4407–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestieri C, Shulman GL, Corbetta M. 2017. The contribution of the human posterior parietal cortex to episodic memory. Nat Rev Neurosci. 18:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A, Augath M, Oeltermann A, Logothetis NK. 2006. Negative functional MRI response correlates with decreases in neuronal activity in monkey visual area V1. Nat Neurosci. 9:569–577. [DOI] [PubMed] [Google Scholar]
- Shmuel A, Yacoub E, Pfeuffer J, Van de Moortele PF, Adriany G, Hu X, Ugurbil K. 2002. Sustained negative BOLD, blood flow and oxygen consumption response and its coupling to the positive response in the human brain. Neuron. 36:1195–1210. [DOI] [PubMed] [Google Scholar]
- Simony E, Honey CJ, Chen J, Lositsky O, Yeshurun Y, Wiesel A, Hasson U. 2016. Dynamic reconfiguration of the default mode network during narrative comprehension. Nat Commun. 7:12141–12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormaz M, Murphy C, Wang H-T, Hymers M, Karapanagiotidis T, Poerio G, Margulies DS, Jefferies E, Smallwood J. 2018. Default mode network can support the level of detail in experience during active task states. Proc Natl Acad Sci USA. 115:9318–9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague TC, Serences JT. 2013. Attention modulates spatial priority maps in the human occipital, parietal and frontal cortices. Nat Neurosci. 16:1879–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan MV, Laughlin SB, Dubs A. 1982. Predictive coding: a fresh view of inhibition in the retina. Proc R Soc Lond B Biol Sci. 216:427–459. [DOI] [PubMed] [Google Scholar]
- Thomas JM, Huber E, Stecker GC, Boynton GM, Saenz M, Fine I. 2015. Population receptive field estimates of human auditory cortex. NeuroImage. 105:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es DM, van der Zwaag W, Knapen T. 2019. Topographic maps of visual space in the human cerebellum. Curr Biol. 29:1689–1694.e3. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, TEJ B, Yacoub E, Ugurbil K, WU-Minn HCP Consortium . 2013. The WU-Minn Human Connectome Project: an overview. NeuroImage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandell BA, Dumoulin SO, Brewer AA. 2007. Visual field maps in human cortex. Neuron. 56:366–383. [DOI] [PubMed] [Google Scholar]
- Weissman DH, Roberts KC, Visscher KM, Woldorff MG. 2006. The neural bases of momentary lapses in attention. Nat Neurosci. 9:971–978. [DOI] [PubMed] [Google Scholar]
- Winawer J, Horiguchi H, Sayres RA, Amano K, Wandell BA. 2010. Mapping hV4 and ventral occipital cortex: the venous eclipse. J Vis. 10:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuiderbaan W, Harvey BM, Dumoulin SO. 2012. Modeling center-surround configurations in population receptive fields using fMRI. J Vis. 12:1–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


