Abstract
Biobanks have facilitated the conduct of large-scale genomics studies, but they are challenged by the difficulty of validating some phenotypes, particularly for complex traits that represent heterogeneous groups ofpatients. The guideline definition of COPD, based on objective spirometry measures, has been preferred in genome-wide association studies (GWAS) conducted with epidemiological cohorts, but spirometry measures are seldom available for biobank participants. Defining COPD based on International Classification of Disease (ICD) codes or self-reported measures is highly feasible in biobanks, but it remains unclear whether the misclassification inherent in these definitions prevent the discovery of genetic variants that contribute to COPD. We found that while there was poor agreement in classification of UK Biobank participants as having COPD based on ICD diagnosis codes, self-reported doctor diagnosis or spirometry measures, contrasting GWAS results for these definitions provided insights into what patient characteristics each trait may capture.
Introduction
COPD is a major cause of morbidity and mortality that remains a public health challenge worldwide1. Smoking is its most common risk factor, accounting for approximately 8 out of 10 COPD-related deaths in the U.S., but COPD risk is also influenced by environmental exposures such as secondhand smoke and air pollution1,2. Persistent airflow limitation is a hallmark feature of COPD, and the Global Initiative for Chronic Obstructive Lung Disease guidelines (GOLD) recommend use of spirometry, in conjunction with patient symptoms, to establish a diagnosis of COPD. Although this guideline definition is the cornerstone of diagnostic criteria for COPD in most clinical trials and research studies, in day-to-day practice, physicians and healthcare providers often rely on patient history and clinical exam features to diagnose and treat COPD3,4. Genome-wide association studies (GWAS) of COPD and related traits, including lung function and smoking, have identified many disease-associated loci and have contributed to the growing recognition that multiple pathobiological mechanisms underlie the lung function changes that receive the diagnostic label of COPD. Uncovering these so-called endotypes is a goal of precision medicine efforts that seek to improve patient outcomes with targeted preventive and treatment strategies.
The creation of biobanks that link electronic health record (EHR) data to DNA and other biospecimens have facilitated the conduct of large-scale genomics studies for over a decade5–8. Biobanks have been used to successfully identify loci associated with various diseases, and they have been leveraged to develop novel approaches, such as phenome-wide association studies (PheWAS)9. A major limitation of biobanks is that much of their phenotype data is biased, as EHR data was not collected for research purposes. This limitation is more pronounced for complex disease such as COPD that consist of highly heterogeneous subjects. International Classification of Diseases (ICD) codes are commonly used to assign affection status to biobank subjects due to the simplicity and convenience of their use, often under the rational that the large sample sizes available in biobanks will counterbalance problems arising from misclassification. In the case of COPD, ICD codes and self-reported data can be obtained with relatively low cost and effort, but they are subject to misclassification. Spirometry data is objective but is not usually available for general population studies, and when it is present in EHRs, it is biased by indication, as spirometry tests are only ordered for specific patients. The UK Biobank, which aims to investigate genetic and nongenetic determinants of a wide range of diseases of middle and old ages10, is an exception: it is the largest spirometric study ever conducted in the UK, with measures available for a large proportion of lifelong non-smokers11.
Although previous studies have noted poor agreement among different COPD definitions3,4,12–15, few have evaluated how the different definitions affect GWAS results. Borlée et al. 16 found that most associations between COPD and demographic risk factors were similar across different COPD definitions, despite variation in prevalence estimates. Such consistency of associations, however, may not be expected in GWAS because the precision and accuracy of phenotype have a considerable impact on the ability to detect genetic signals that explain only a small fraction of disease susceptibility. The UK Biobank provides an unprecedented opportunity to assess the impact of different case definitions on the identification of COPD-related genetic loci due to its wide range of diagnostic measures and the availability of spirometry data for most genotyped subjects. Here, we used UK Biobank data to perform GWAS of COPD defined in three ways: 1) based on ICD diagnosis codes, 2) based on self-reported doctor diagnosis, and 3) according to GOLD criteria. Comparison of GWAS results revealed insights into what each of these traits may represent, demonstrating that genetic studies can shed light on inconsistencies observed among definitions of a complex trait.
Methods
Data and COPD definitions
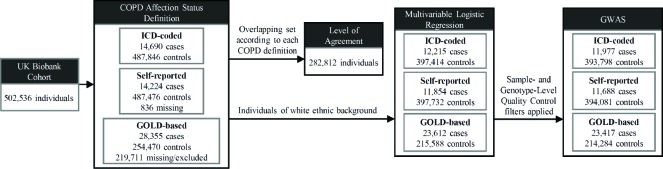
UK Biobank data from a total sample comprising 502,536 individuals aged 37-73 at recruitment was obtained (Figure 1). COPD was defined based on ICD diagnosis codes (ICD-coded COPD), self-reported doctor diagnosis (self-reported COPD), and pre-bronchodilator spirometry measures (GOLD-based COPD) as follows:
Figure 1:
COPD assignment and statistical analysis workflow.
- ICD-coded COPD: participants with any of the following primary and secondary ICD codes were regarded as a COPD case:
- ICD-9: 491 (Chronic bronchitis), 492 (Emphysema), and 496 (Chronic airways obstruction, not elsewhere classified) from UK Biobank data-fields 41203 and 41205.
- ICD-10: J41 (Simple and mucopurulent chronic bronchitis), J42 (Unspecified chronic bronchitis), J43 (Emphysema), and J44 (Other chronic obstructive pulmonary disease) from UK Biobank data-fields 41202 and 41204.
Controls were subjects without any of these codes. This definition resulted in 14,690 cases and 487,846 controls.
Self-reported COPD: affection status was defined according to information obtained from a verbal interview conducted during the initial assessment and an online follow-up questionnaire, which were available for 501,700 individuals. Participants with any non-cancer illness codes corresponding to ‘COPD’, ‘emphysema/chronic bronchitis’, and their child categories were considered as COPD cases (UK Biobank data-field 20002). Participants were also classified as having COPD if they provided a positive answer to any question in UK Biobank data-fields 22128 (Doctor diagnosed emphysema, Online follow-up), 22129 (Doctor diagnosed chronic bronchitis, Online follow-up), and 22120 (Doctor diagnosed COPD, Online follow-up). Controls were remaining subjects with completed verbal interview and online questionnaire data. This definition resulted in 14,224 cases and 487,476 controls.
GOLD-based COPD: following quality control filters of spirometry data undertaken as described in Shrine et al.17, the ‘best measure’ per individual of forced expiratory volume in 1 second (FEV1) and forced vital capacity (FVC) were selected. This yielded measures for 353,469 individuals, from which cases were defined on the basis of pre-bronchodilator evidence of moderate-to-severe airflow limitation by the modified GOLD criteria as described in Hobbs et al.18 Controls were defined as persons with normal spirometry measures. This definition resulted in 28,355 cases and 254,470 controls.
Additional phenotypes were extracted from participants’ responses to a computer-assisted interview, self-completed questionnaires, and physical measures at the visit based on UK Biobank data-fields 31 (sex), 50 (height), 20003 (age), 20160 (ever smoked), 21000 (ethnic background), 21001 (body mass index), and 22006 (genetic ethnic grouping).
Statistical analysis
The demographic characteristics of participants were summarized by proportion or mean. Fleiss’ Kappa19 (κ) was calculated to represent a level of agreement using the common set of individuals among the COPD definitions (n = 282,812). Multivariable logistic regressions were performed to evaluate the associations between participant characteristics and the three definitions of COPD. For each COPD definition, a GWAS was performed using the subset of case-control samples who self-identified as White British and had very similar genetic ancestry based on a principal component analysis of genotypes, while excluding subjects who had a mismatch between self-reported and genetic sex as determined by chromosomal make-up, sex chromosome configurations that were not XX or XY, or had non-normal heterozygosity and missing rates according to measures provided by the UK Biobank team20. These procedures resulted in 11,977 cases and 393,798 controls for ICD-coded COPD GWAS, 11,688 cases and 394,081 controls for self-reported COPD GWAS, and 23,417 cases and 214,284 controls for GOLD-based COPD GWAS (Figure 1). At the genotype level, variants with minor allele frequency (MAF) < 0.01 or imputation INFO score measure < 0.3 were excluded. Association testing was performed in a generalized mixed model framework using SAIGE21 to account for relatedness and fine-scale population structure, while including as covariates age, age-squared (age2), sex, height, smoking status (ever versus never), and 4 principal components. We used FUMA22 to obtain functional characteristics of genetic loci based on GWAS summary statistics. GWAS results were contrasted by comparing genome-wide significant loci directly or by measuring pairwise genetic correlations. Genetic correlations between the different case definitions were estimated using linkage disequilibrium (LD) score regression23 using the effects of all SNPs with INFO score > 0.9 and pre-calculated LD scores based on 1000 Genomes Project data for European populations24.
Results
Demographic characteristics
Characteristics of participants summarized using all available records for each COPD definition are provided in Table 1. More than 94% of participants self-identified as White. Among all participants, two thirds were overweight or obese, and nearly 60% had a positive smoking history. The proportion of cases was similar between the ICD-coded (2.92%) and self-reported (2.84%) COPD groups, and much higher in the GOLD-based COPD group (10.03%). Par-ticipants classified as having COPD were older, more often male, and more often a former or current smoker across all definitions. COPD cases showed a greater proportion of asthma diagnosis than controls in all definitions. Lung function was decreased in cases versus controls for each COPD definition, with the GOLD-based group having the greatest difference between case and control medians of percent predicted FEV1 and FEV1/FVC ratio.
Table 1:
Demographic characteristics of the participants according to different COPD definitions
| Total | ICD-coded COPD | Self-reported COPD | GOLD-based COPD | ||||
|---|---|---|---|---|---|---|---|
| Control | Case | Control | Case | Control | Case | ||
| N | 502,536 | 487,846 | 14,690 | 487,476 | 14,224 | 254,470 | 28,355 |
| Age, mean (SD) | 56.5 (8.1) | 56.4 (8.1) | 61.4 (6.3) | 56.4 (8.1) | 59.4 (7.2) | 55.8 (8.1) | 59.2 (7.4) |
| Male % | 45.6 | 45.3 | 55.0 | 45.5 | 49.3 | 43.2 | 53.2 |
| Ethnic background∗ % | |||||||
| White | 94.1 | 94.0 | 96.3 | 94.1 | 96.7 | 96.7 | 94.6 |
| Mixed | 0.6 | 0.6 | 0.5 | 0.6 | 0.5 | 0.6 | 0.5 |
| Asian or Asian British | 2.0 | 2.0 | 1.1 | 2.0 | 0.9 | 0.8 | 1.9 |
| Black or Black British | 1.6 | 1.6 | 0.6 | 1.6 | 0.6 | 0.7 | 1.4 |
| Chinese | 0.3 | 0.3 | 0.1 | 0.3 | 0.1 | 0.3 | 0.2 |
| Others | 0.9 | 0.9 | 0.5 | 0.9 | 0.5 | 0.6 | 0.8 |
| Not Available | 0.6 | 0.5 | 0.8 | 0.5 | 0.6 | 0.3 | 0.5 |
| Body mass index (kg/m2) % | |||||||
| Underweight (< 18.5) | 0.5 | 0.5 | 1.6 | 0.5 | 1.3 | 0.4 | 1.0 |
| Normal (18.5-25) | 32.3 | 32.5 | 26.1 | 32.5 | 28.3 | 34.0 | 33.7 |
| Overweight (25-30) | 42.2 | 42.4 | 36.2 | 42.4 | 39.0 | 43.4 | 41.3 |
| Obese (≥ 30) | 24.3 | 24.0 | 34.7 | 24.2 | 30.7 | 22.2 | 24.0 |
| Not Available | 0.6 | 0.6 | 1.3 | 0.5 | 0.6 | 0.1 | 0.1 |
| Ever smoker % | 59.8 | 59.0 | 87.1 | 59.3 | 77.2 | 58.8 | 72.7 |
| Asthma† % | 7.4 | 6.6 | 35.8 | 6.9 | 25.9 | 4.9 | 20.2 |
| Lung function | |||||||
| FEV1 predicted percentage, median | 92.2 | 92.5 | 70.7 | 92.4 | 80.3 | 96.8 | 67.4 |
| FEV1/FVC ratio, median | 0.77 | 0.77 | 0.68 | 0.77 | 0.72 | 0.78 | 0.64 |
Ethnicity categories follow the tree structure of ethnic background in the UK Biobank touchscreen questionnaire (UK Biobank data-field 21000)
Affection status was assigned based on having ICD-9 493 and/or ICD-10 J45 codes
Subject overlap according to COPD definitions
In total, 33,666 cases were identified by at least one COPD definition among the 282,812 participants whose case-control status could be ascertained according to all definitions (Figure 2). Agreement of affection status according to different COPD definitions was low: only 4.9% of participants classified as cases by one of the three definitions met the criteria of all three. Of 28,354 GOLD-based COPD cases, more than 80% were not classified as cases according to ICD codes or self-reported doctor diagnosis. Approximately half of the self-reported COPD cases and a quarter of the ICD-coded COPD cases were not classified as having COPD by the other definitions. The k statistic (SE) among the three definitions was 0.185 (0.001), indicating a poor agreement. The pairwise k statistic was as 0.293 (0.007) for the ICD-coded versus self-reported COPD, 0.198 (0.005) for the ICD-coded versus GOLD-based COPD, and 0.115 (0.005) for the self-reported COPD versus GOLD-based COPD.
Figure 2:
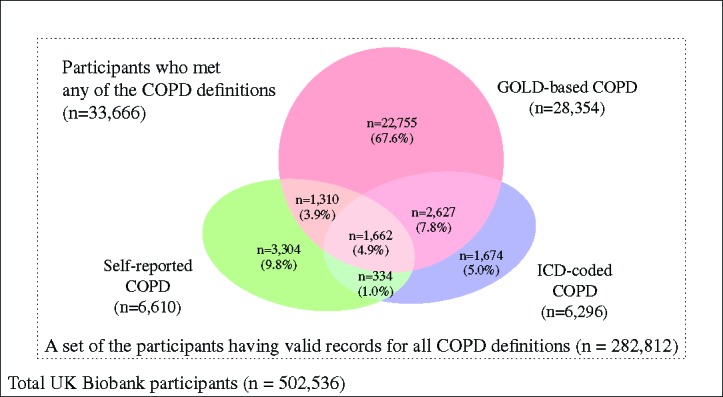
Overlap of COPD cases as classified by different COPD definitions. Of 33,666 participants identified as having COPD by at least one definition, only 1,662 participants (4.9%) met case criteria of the three definitions (k = 0:185).
Associations between participant characteristics and COPD
Logistic regression results were consistent with observed trends in demographic profiles of participants (Table 2). Participants who were male, older, and had a positive smoking history (i.e., ever smokers) had higher odds of COPD diagnosis, regardless of which definition was used. While being underweight versus having normal weight consistently raised the odds of COPD classification, the magnitude and significance of associations between COPD and other categories of body mass index varied among the definitions. Specifically, obese participants had increased odds of GOLD-based COPD classification but reduced odds of ICD-coded or self-reported COPD; overweight participants had reduced odds for ICD-coded and GOLD-based COPD but no significant difference versus normal weight for self-reported COPD.
Table 2:
Associations between participant characteristics and different COPD definitions
| ICD-coded COPD | Odds ratio (95% CI) | ||
|---|---|---|---|
| Self-reported COPD | GOLD-based COPD | ||
| Age | 1.094 (1.091-1.098)∗ | 1.048 (1.046-1.051)∗ | 1.061 (1.059-1.063)∗ |
| Gender (vs. female) | |||
| Male | 1.261 (1.214-1.309)∗ | 1.078 (1.038-1.119)∗ | 1.430 (1.391-1.471)∗ |
| Body mass index (vs. normal) | |||
| Underweight (<18.5) | 4.467 (3.788-5.237)∗ | 3.218 (2.704-3.803)∗ | 2.808 (2.397-3.275)∗ |
| Overweight (25-30) | 0.893 (0.852-0.936)∗ | 0.974 (0.930-1.019) | 0.797 (0.771-0.823)∗ |
| Obese (_30) Smoking status (vs. never) | 1.537 (1.466-1.613)∗ | 1.324 (1.261-1.390)∗ | 0.938 (0.904-0.973)∗ |
| Ever smoker | 4.359 (4.128-4.607)∗ | 2.170 (2.077-2.268)∗ | 1.842 (1.786-1.899)∗ |
Multivariable logistic regression model using age, gender, body mass index, and smoking status as independent variables. Samples were restricted to White ethnic background (UK Biobank data-field 22006), as these were the subjects used in GWAS. _P < 0:001.
GWAS results
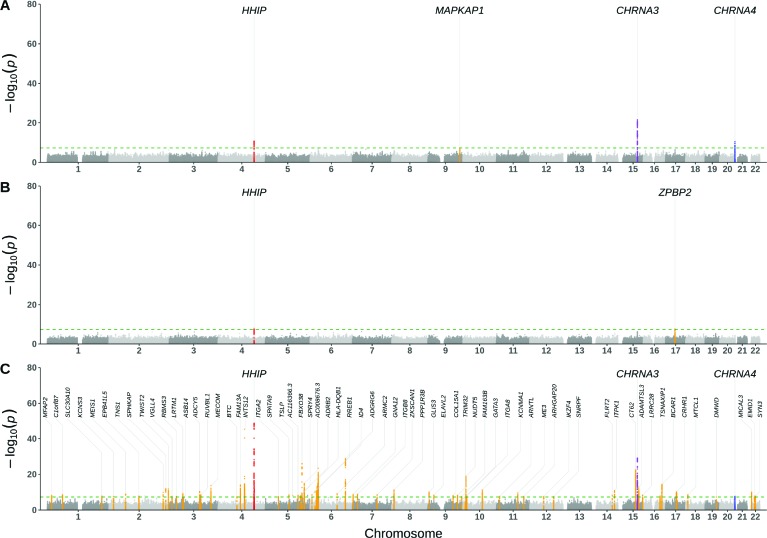
Genetic loci associated with COPD differed substantially according to case definitions (Figure 3). There were 66 loci with genome-wide significant associations for GOLD-based COPD, but only 2 and 4 for the self-reported and ICD-coded COPD, respectively. A risk locus near HHIP was the only one shared across the three GWAS. Two additional loci near CHRNA3 and CHRNA4 overlapped between the GOLD-based and ICD-coded COPD GWAS. In contrast to the low overlap of genome-wide significant loci, the estimated genetic correlation (SE) that utilized genome-wide data was higher, with measures of 0.884 (0.055) for ICD-coded versus self-reported COPD, 0.703 (0.036) for ICD-coded versus GOLD-based COPD, and 0.652 (0.050) for self-reported versus GOLD-based COPD. Consistent with these observation, the direction of associations at the lead significant loci were similar across the three COPD definitions despite their differing effect sizes (Table 3).
Figure 3:
Manhattan plots of GWAS results from three different COPD definitions: A) ICD-coded COPD (11,977 cases, 393,798 controls), B) Self-reported COPD (11,688 cases, 394,081 controls), C) GOLD-based COPD (23,417 cases, 214,284 controls). P values were two sided based on Wald statistics without multiple-testing adjustment. Each risk locus was annotated with its nearest coding gene. The green horizontal dashed line represents a genome-wide significance level (P < 5×10−8).
Table 3:
Summary statistics for the lead variants at genome-wide significant loci
| rsID | Position* | Allelet | ICD-coded COPD | Self-reported COPD | GOLD-based COPD | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF# | ORl | P | RAF | OR | P | RAF | OR | P | ||||
| ICD-coded COPD | ||||||||||||
| rs1291438511 | 15:78898723 | C/T | 0.38 | 1.14 | 2.59 x 10-22 | 0.38 | 1.06 | 8.84 x 10-6 | 0.38 | 1.11 | 1.02 x 10-24 | |
| rs1828591 | 4:145480780 | A/G | 0.39 | 0.91 | 1.62 x 10-11 | 0.39 | 0.93 | 4.53 x 10-8 | 0.40 | 0.86 | 4.37 x 10-49 | |
| rs151176846 | 20:61997500 | T/C | 0.08 | 1.18 | 2.90 x 10-11 | 0.08 | 1.07 | 6.60 x 10-3 | 0.08 | 1.11 | 2.04 x 10-8 | |
| rs4838290 | 9:128475056 | C/T | 0.64 | 1.08 | 3.99 x 10-8 | 0.64 | 1.04 | 1.65 x 10-3 | 0.64 | 1.03 | 5.20 x 10-3 | |
| Self-reported COPD | ||||||||||||
| rs6537293 | 4:145479761 | A/G | 0.39 | 0.91 | 1.91 x 10-11 | 0.39 | 0.93 | 2.44 x 10-8 | 0.40 | 0.86 | 1.34 x 10-49 | |
| rs12939457 | 17:38032188 | T/C | 0.48 | 0.96 | 1.03 x 10-3 | 0.48 | 0.93 | 2.57 x 10-8 | 0.48 | 0.98 | 1.09 x 10-2 | |
| Gold-based COPD 1 | ||||||||||||
| rs13113591 | 4:145489097 | C/T | 0.39 | 0.91 | 2.47 x 10-11 | 0.39 | 0.93 | 7.90 x 10-8 | 0.39 | 0.86 | 1.15 x 10-49 | |
| rs34712979 | 4:106819053 | G/A | 0.26 | 1.05 | 7.29 x 10-4 | 0.26 | 1.06 | 6.10 x 10-5 | 0.26 | 1.18 | 5.47 x 10-46 | |
| rs111704647 | 15:78900650 | C/T | 0.33 | 1.14 | 4.19 x 10-21 | 0.33 | 1.07 | 1.49 x 10-6 | 0.33 | 1.13 | 6.40 x 10-30 | |
| rs262126 | 6:142835364 | A/C | 0.31 | 0.97 | 6.36 x 10-2 | 0.31 | 0.97 | 2.98 x 10-2 | 0.31 | 0.89 | 1.30 x 10-29 | |
| rs7733410 | 5:147856522 | G/A | 0.44 | 0.96 | 4.12 x 10-3 | 0.44 | 0.97 | 1.55 x 10-2 | 0.44 | 0.90 | 8.35 x 10-27 | |
| rs10851839 | 15:71628370 | T/A | 0.67 | 0.98 | 1.20 x 10-1 | 0.67 | 0.97 | 1.71 x 10-2 | 0.67 | 0.90 | 2.99 x 10-23 | |
| rs57062879 | 10:12278525 | A/G | 0.51 | 0.98 | 2.14 x 10-1 | 0.51 | 0.99 | 2.39 x 10-1 | 0.52 | 0.91 | 1.08 x 10-19 | |
| rs4713572 | 6:32626952 | T/C | 0.40 | 1.05 | 8.16 x 10-5 | 0.40 | 1.04 | 5.21 x 10-3 | 0.40 | 1.09 | 2.73 x 10-18 | |
| rs10476063 | 5:156928823 | G/A | 0.33 | 1.04 | 8.42 x 10-3 | 0.33 | 1.02 | 1.13 x 10-1 | 0.33 | 1.09 | 1.33 x 10-15 | |
| rs11645016 | 16:75311828 | C/T | 0.64 | 1.03 | 4.43 x 10-2 | 0.64 | 1.04 | 6.67 x 10-3 | 0.64 | 1.09 | 4.26 x 10-15 | |
| rs1964516 | 4:89875909 | C/T | 0.51 | 1.04 | 2.14 x 10-3 | 0.51 | 1.03 | 2.00 x 10-2 | 0.51 | 1.08 | 7.98 x 10-15 | |
| rs1420472 | 3:168776326 | G/T | 0.44 | 1.02 | 1.26 x 10-1 | 0.44 | 1.02 | 1.59 x 10-1 | 0.44 | 1.08 | 2.26 x 10-14 | |
| rs1896797 | 15:84274591 | G/A | 0.49 | 0.98 | 8.15 x 10-2 | 0.49 | 0.98 | 1.72 x 10-1 | 0.49 | 0.93 | 1.41 x 10-13 | |
| rs12614274 | 2:229583850 | A/G | 0.08 | 0.95 | 2.64 x 10-2 | 0.08 | 0.97 | 2.00 x 10-1 | 0.08 | 0.88 | 1.11 x 10-12 | |
| rs35945722 | 2:239893783 | G/A | 0.19 | 0.97 | 5.76 x 10-2 | 0.19 | 1.00 | 8.12 x 10-1 | 0.19 | 0.91 | 1.24 x 10-12 | |
position based on GRCh37; tnon-risk/risk allele; $risk allele frequency; §odds ratio; Ilead variant according to GOLD-based COPD at the same locus; ¶the top 15 lead variants at genome-wide significant loci sorted according to P values
Discussion
Well-defined and reproducible case definitions are critical for the identification and replication of genetic associations, and definitions of COPD based on measures of spirometry as recommended by GOLD guidelines have thus been preferred in genetic epidemiological studies. Because spirometry data is often lacking or obtained in a highly biased fashion in EHRs and large administrative datasets, ICD codes and/or self-reported measures have been used in COPD biobank studies to assign affection status, despite the high likelihood of misclassification that doing so entails. Using a population sample of adults from the UK Biobank who have spirometry data, as well as ICD codes from medical encounters and self-reported measures of health, we assessed the impact of using three different COPD definitions on GWAS results.
The proportion of COPD cases classified according to the three definitions we obtained was consistent with previous findings in which the spirometry-based definition yielded a higher prevalence of COPD16,25,26. Although the current guidelines recommend that persistent respiratory symptoms in combination with spirometric evidence of obstructive airflow limitation should be used to establish a diagnosis of COPD, epidemiological studies generally have defined COPD with spirometry alone1. The latter approach has a high sensitivity of diagnosis, as most of true COPD patients would be identified but also an increased false positive rate, as some people with spirometric evidence of airflow obstruction are asymptomatic and lack other clinical evidence of disease. Underdiagnosis of COPD, on the other hand, is likely to occur with the definitions based solely on ICD codes and/or self-reported doctor diagnosis because people typically seek medical attention when symptoms interfere with daily activities. Additionally, published reports have failed to find evidence of airflow obstruction on spirometry for a substantial portion of COPD patients identified by ICD codes or self-reported doctor diagnosis13,27,28.
The poor agreement we observed among who was classified as a case by the three COPD definitions (κ = 0.185) was also consistent with previously published reports3,4,12–15. Less than 5% of COPD cases met all three case definitions, and there was greater agreement between the ICD-coded and self-reported CODP (κ = 0.298) compared to the other pairs of COPD definitions. Despite the disagreement among definitions, the direction of associations between participant characteristics and case assignment were similar and consistent with previous studies: a person with COPD was more likely to be older, male, and have a positive smoking history3,25. Although the association between being obese and having COPD varied according to COPD definition, the consistent positive association between being underweight and COPD diagnosis is supported by recent findings from both patient- and population-based studies3,16. Thus, each case definition may select for distinct sub-phenotypes of COPD that still capture its core clinical features.
Based on the number of genome-wide significant loci, we observed little genetic overlap across COPD definitions. The large number of loci associated with GOLD-based COPD were not identified with the other COPD definitions. As expected, these loci mapped to genes previously reported in GWAS of lung function measures and COPD such as C1orf87, VGLL4, ADCY5, BTC, RREB1, ID4, ITGB8, ARNTL, and ADAMTSL317, 29, 30. Only one locus near HHIP was identified in the three GWAS. The HHIP gene, which encodes a member of the hedgehog-interacting protein family, is a well-known COPD susceptibility locus that has been identified in GWAS and gene expression studies17,31,32 and has also been associated with COPD exacerbations2. Two additional loci near CHRNA3 and CHRNA4 were associated with both ICD-coded and GOLD-based COPD. Variants in/near the CHRNA3 and CHRNA4 genes, which encode subunits of the nicotine acetylcholine receptor superfamily, have been associated with smoking behavior and a range of complex lung diseases30,33,34. A locus that was unique to ICD-coded COPD was close to MAPKAP1, a gene associated with various smoking behavior phenotypes in recent GWAS29,33. Taken together, the loci associated with ICD-coded COPD suggest that this trait represents smoking-related COPD with symptoms that are sufficiently severe to lead people to seek emergency care. Alternatively, the results could indicate that smokers are more likely to receive ICD codes for COPD when presenting with relevant symptoms. Because smoking is such a prominent risk factor for COPD, we performed an additional GWAS of ICD-coded COPD using the subgroup of cases with a positive smoking history. This sub-analysis yielded the same genome-wide significant loci as the non-stratified ICD-coded COPD without additional signals a result that is not unexpected given that 87% of ICD-based cases had a positive smoking history. Further biobank studies with a greater number of COPD non-smokers or more detailed smoking history may shed light on which genetic associations are due to smoking status versus COPD.
Besides HHIP, there was one other genome-wide significant association in the self-reported COPD GWAS adjacent to ZPBP2. The presence of an association with this gene, which is part of the well-known asthma-associated 17q21 locus35,36, suggests that the self-reported COPD definition also captured patients who have or have had child-onset asthma. Interestingly, according to ICD codes of asthma, ICD-coded COPD had the greatest proportion of people with asthma (36% versus 26% in self-reported COPD), and yet asthma-associated loci were not present in ICD-coded COPD. Although asthma and COPD are distinct conditions, it can be difficult to differentiate them in practice due to the similarity of their signs and symptoms37,38. Recent studies have focused on asthma-COPD overlap given that patients with both diseases have traditionally been excluded from clinical trials despite having greater morbidity than those with asthma or COPD alone39. Our GWAS findings underscore the particular difficulty in classifying patients with both asthma and COPD, and they suggest that classification of asthma in adults by ICD codes yields a phenotype that is different from the childhood-onset asthma that is characterized by a strong 17q21 association signal.
In contrast to the results of overlapping genome-wide significant loci, there was substantial genetic correlation among COPD definitions when the effects of all well-imputed SNPs that did not reach genome-wide significance were considered. Observed correlations suggest that a large fraction of genetic liability is shared across the different definitions, even if loci did not reach genome-wide significance due to ‘noisy’ classification schemes. For example, a case according to one definition could be a control in another, a situation that affected the ICD-coded and self-reported definitions most, as a far greater number of GOLD-based COPD cases could be assigned as controls by the other definitions. This greater potential of misclassification in the ICD-coded and/or self-reported COPD may have attenuated genetic signals more severely, leading to only a few loci reaching genome-wide significance. Further, differences in statistical power due to decreased sample sizes of ICD-based and self-reported COPD cases compared to GOLD-based cases could have yielded fewer genome-wide significant findings among loci that still show the same direction of effect and consistent odd ratios.
Given the high genetic correlations, one could argue that using phenotype information from EHRs and questionnaires is justifiable because sample sizes much larger than those restricted to having spirometry measures can be obtained. Our results, however, found that a non-negligible proportion of genetic effects were still unique to each definition. As the κ statistics indicated, COPD defined by spirometry was less similar to the other definitions. Therefore, even a highly-powered GWAS of COPD based on ICD codes or self-reported measures may not identify the genetic associations found with spirometry-based definitions.
Limitations of our study include the fact that the UK Biobank is not representative of the general population with respect to a number of health-related characteristics where the participants are, on average, more health-conscious40. This could introduce a bias toward underreporting COPD, which would especially impact the definition based on ICD codes and self-reported doctor diagnosis, as these would not be obtained without patient-perceived symptoms that led to seeking of medical care. Secondly, a variety of comorbid conditions were not accounted for that could have influenced genetic signals, via over- or under-estimating differences among case definitions. Further study of the associations that were unique to GOLD-based COPD in the context of comorbidities beyond asthma may clarify how they contribute to COPD-like symptoms, or why despite having evidence of airflow obstruction, some individuals remain asymptomatic38. Finally, we restricted the genetic analyses to participants of European ancestry, and thus, our results may not be generalizable across other racial/ethnic groups.
In summary, we found poor agreement between ICD-coded, self-reported and GOLD-based COPD definitions, and considerable differences in genomic risk loci identified via GWAS of UK Biobank participants classified according to each definition. Although use of ICD codes and self-reports are convenient and efficient for phenotype classification in COPD, even large sample sizes achieved by their use may not yield association signals as strong as those of more objective criteria such as lung function measures. Nonetheless, comparison of GWAS results obtained for different COPD definitions revealed insights into what each of these traits may represent and how genetics studies can shed light on complex phenotypes.
Acknowledgments
This work was supported by National Institutes of Health (NIH) K08 HL136928 (BDH), R01 HL133433 (BEH), and R01 HL141992 (BEH). This research has been conducted using the UK Biobank Resource under Application Number 40375.
Figures & Table
References
- 1.Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease 2017 Report. GOLD Executive Summary. American Journal of Respiratory and Critical Care Medicine. 2017 Jan;195(5):557–582. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 2.United States Surgeon General. The Health Consequences of Smoking – 50 Years of Progress: A Report of the Surgeon General: (510072014-001) American Psychological Association. 2014 [Google Scholar]
- 3.Prieto-Centurion V, Rolle AJ, Au DH, Carson SS, Henderson AG, Lee TA, et al. Multicenter Study Comparing Case Definitions Used to Identify Patients with Chronic Obstructive Pulmonary Disease. American Journal of Respiratory and Critical Care Medicine. 2014 Nov;190(9):989–995. doi: 10.1164/rccm.201406-1166OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooke CR, Joo MJ, Anderson SM, Lee TA, Udris EM, Johnson E, et al. The Validity of Using ICD-9 Codes and Pharmacy Records to Identify Patients with Chronic Obstructive Pulmonary Disease. BMC Health Services Research. 2011;11:37. doi: 10.1186/1472-6963-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott SA, Owusu Obeng A, Botton MR, Yang Y, Scott ER, Ellis SB, et al. Institutional Profile: Translational Pharmacogenomics at the Icahn School of Medicine at Mount Sinai. Pharmacogenomics. 2017 Oct;18(15):1381–1386. doi: 10.2217/pgs-2017-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGregor TL, Van Driest SL, Brothers KB, Bowton EA, Muglia LJ, Roden DM. Pediatric Sample Inclusion in an Opt-out Biorepository Linking DNA to de-Identified Medical Records: Pediatric BioVU. Clinical pharmacology and therapeutics. 2013 Feb;93(2):204–211. doi: 10.1038/clpt.2012.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox N. UK Biobank Shares the Promise of Big Data. Nature. 2018 Oct;562(7726):194–195. doi: 10.1038/d41586-018-06948-3. [DOI] [PubMed] [Google Scholar]
- 8.Canela-Xandri O, Rawlik K, Tenesa A. An Atlas of Genetic Associations in UK Biobank. Nature Genetics. 2018 Nov;50(11):1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny JC, Bastarache L, Roden DM. Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annual review of genomics and human genetics. 2016 Aug;17:353–373. doi: 10.1146/annurev-genom-090314-024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Medicine. 2015 Mar;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RP, Strachan DP. Ventilatory Function as a Predictor of Mortality in Lifelong Non-Smokers: Evidence from Large British Cohort Studies. BMJ Open. 2017 Jul;7(7):e015381. doi: 10.1136/bmjopen-2016-015381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celli BR, Halbert RJ, Isonaka S, Schau B. Population Impact of Different Definitions of Airway Obstruction. European Respiratory Journal. 2003 Aug;22(2):268–273. doi: 10.1183/09031936.03.00075102. [DOI] [PubMed] [Google Scholar]
- 13.Murgia N, Brisman J, Claesson A, Muzi G, Olin AC, Torén K. Validity of a Questionnaire-Based Diagnosis of Chronic Obstructive Pulmonary Disease in a General Population-Based Study. BMC Pulmonary Medicine. 2014 Mar;14:49. doi: 10.1186/1471-2466-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohangoo AD, van der Linden MW, Schellevis FG, Raat H. Prevalence Estimates of Asthma or COPD from a Health Interview Survey and from General Practitioner Registration: What’s the Difference? European Journal of Public Health. 2006 Feb;16(1):101–105. doi: 10.1093/eurpub/cki043. [DOI] [PubMed] [Google Scholar]
- 15.Romanelli AM, Raciti M, Protti MA, Prediletto R, Fornai E, Faustini A. How Reliable Are Current Data for Assessing the Actual Prevalence of Chronic Obstructive Pulmonary Disease? PLoS ONE. 2016 Feb;11(2) doi: 10.1371/journal.pone.0149302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borlée F, Yzermans CJ, Krop E, Aalders B, Rooijackers J, Zock JP, et al. Spirometry, Questionnaire and Electronic Medical Record Based COPD in a Population Survey: Comparing Prevalence, Level of Agreement and Associations with Potential Risk Factors. PLOS ONE. 2017 Mar;12(3):e0171494. doi: 10.1371/journal.pone.0171494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, et al. New Genetic Signals for Lung Function Highlight Pathways and Chronic Obstructive Pulmonary Disease Associations across Multiple Ancestries. Nature Genetics. 2019 Mar;51(3):481. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, et al. Genetic Loci Associated with Chronic Obstructive Pulmonary Disease Overlap with Loci for Lung Function and Pulmonary Fibrosis. Nature genetics. 2017 Mar;49(3):426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleiss JL. Measuring Nominal Scale Agreement among Many Raters. Psychological Bulletin. 19720101;76(5):378. [Google Scholar]
- 20.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature. 2018 Oct;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, et al. Efficiently Controlling for Case-Control Imbalance and Sample Relatedness in Large-Scale Genetic Association Studies. Nature Genetics. 2018 Sep;50(9):1335. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional Mapping and Annotation of Genetic Associations with FUMA. Nature Communications. 2017 Nov;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An Atlas of Genetic Correlations across Human Diseases and Traits. Nature genetics. 2015 Nov;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature. 2015 Oct;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halbert RJ, Natoli JL, Gano A, Badamgarav E, Buist AS, Mannino DM. Global Burden of COPD: Systematic Review and Meta-Analysis. European Respiratory Journal. 2006 Sep;28(3):523–532. doi: 10.1183/09031936.06.00124605. [DOI] [PubMed] [Google Scholar]
- 26.Ford ES, Croft JB, Mannino DM, Wheaton AG, Zhang X, Giles WH. COPD Surveillance—United States, 19992011. CHEST. 2013 Jul;144(1):284–305. doi: 10.1378/chest.13-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein BD, Bautista A, Schumock GT, Lee TA, Charbeneau JT, Lauderdale DS, et al. The Validity of International Classification of Diseases, Ninth Revision, Clinical Modification Diagnosis Codes for Identifying Patients Hospitalized for COPD Exacerbations. Chest. 2012 Jan;141(1):87–93. doi: 10.1378/chest.11-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tálamo C, de Oca MM, Halbert R, Perez-Padilla R, Jardim JRB, Muiño A, et al. Diagnostic Labeling of COPD in Five Latin American Cities. Chest. 2007 Jan;131(1):60–67. doi: 10.1378/chest.06-1149. [DOI] [PubMed] [Google Scholar]
- 29.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. American Journal of Human Genetics. 2019 Jan;104(1):65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve NF, Guyatt AL, Jackson VE, et al. Genetic Landscape of Chronic Obstructive Pulmonary Disease Identifies Heterogeneous Cell-Type and Phenotype Associations. Nature Genetics. 2019 Mar;51(3):494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Qiu W, Sathirapongsasuti JF, Cho MH, Mancini JD, Lao T, et al. Gene Expression Analysis Uncovers Novel Hedgehog Interacting Protein (HHIP) Effects in Human Bronchial Epithelial Cells. Genomics. 2013 May;101(5):263–272. doi: 10.1016/j.ygeno.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young RP, Whittington CF, Hopkins RJ, Hay BA, Epton MJ, Black PN, et al. Chromosome 4q31 Locus in COPD Is Also Associated with Lung Cancer. European Respiratory Journal. 2010 Dec;36(6):1375–1382. doi: 10.1183/09031936.00033310. [DOI] [PubMed] [Google Scholar]
- 33.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association Studies of up to 1.2 Million Individuals Yield New Insights into the Genetic Etiology of Tobacco and Alcohol Use. Nature Genetics. 2019 Feb;51(2):237– 244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-Scale Association Analysis Identifies New Lung Cancer Susceptibility Loci and Heterogeneity in Genetic Susceptibility across Histological Subtypes. Nature Genetics. 2017 Jul;49(7):1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WO, Altmüller J, Ang W, et al. Multiancestry Association Study Identifies New Asthma Risk Loci That Colocalize with Immune Cell Enhancer Marks. Nature genetics. 2018 Jan;50(1):42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, et al. A Genome-Wide Cross-Trait Analysis from UK Biobank Highlights the Shared Genetic Architecture of Asthma and Allergic Diseases. Nature Genetics. 2018 Jun;50(6):857–864. doi: 10.1038/s41588-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heffler E, Crimi C, Mancuso S, Campisi R, Puggioni F, Brussino L, et al. Misdiagnosis of Asthma and COPD and Underuse of Spirometry in Primary Care Unselected Patients. Respiratory Medicine. 2018 Sep;142:48–52. doi: 10.1016/j.rmed.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Ho T, Cusack RP, Chaudhary N, Satia I, Kurmi OP. Under- and over-Diagnosis of COPD: A Global Perspective. Breathe. 2019 Mar;15(1):24–35. doi: 10.1183/20734735.0346-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekerljung L, Mincheva R, Hagstad S, Bjerg A, Telg G, Stratelis G, et al. Prevalence, Clinical Characteristics and Morbidity of the Asthma-COPD Overlap in a General Population Sample. Journal of Asthma. 2018 May;55(5):461–469. doi: 10.1080/02770903.2017.1339799. [DOI] [PubMed] [Google Scholar]
- 40.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. American Journal of Epidemiology. 2017 Nov;186(9):1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]