Abstract
Precision medicine focuses on developing new treatments based on an individual’s genetic, environmental, and lifestyle profile. While this data-driven approach has led to significant advances, retrieving information specific to a patient’s condition has proved challenging for oncologists due to the large volume of data. In this paper, we propose the PRecIsion Medicine Robust Oncology Search Engine (PRIMROSE) for cancer patients that retrieves scientific articles and clinical trials based on a patient’s condition, genetic profile, age, and gender. Our search engine utilizes Elasticsearch indexes for information storage and retrieval, and we developed a knowledge graph for query expansion in order to improve recall. Additionally, we experimented with machine learning and learning-to-rank components to the search engine and compared the results of the two approaches. Finally, we developed a front-facing ReactJS website and a REST API for connecting with our search engine. The development of this front-facing website allows for easy access to our system by healthcare providers.
Introduction
Precision medicine, in recent years, has become the focus of much of the recent cancer research literature. While the underlying concepts of precision medicine are readily visible in longstanding medical procedures such as blood transfusions, the field has recently experienced rapid growth due to the announcement of the Precision Medicine Initiative in 2015†. Precision medicine, as defined by the Precision Medicine Initiative, is “an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment, and lifestyle for each person” [1]. The first target of the initiative has been cancer due to the new understanding of the physiological and genetic drivers of the disease and, more importantly, its high mortality rate [2].
Since the initiative’s inception in 2015, vast amounts of literature have been published linking precision medicine treatments with various genetic, environmental, and lifestyle conditions. However, translating these treatments into clinical settings has proven challenging. One such challenge has been developing precision medicine decision support tools to aid oncologists in identifying the most up-to-date evidence-based treatments for their patients. In this case, the vast amounts of information proves to be a double-edged sword: on one hand research describing patient specific targeted treatments is potentially available to help the patient, while on the other hand, identifying these treatments given the large amount of scientific evidence can be a daunting, time-consuming task. This can drastically limit the effectiveness of these treatments. Oncologists need to keep up-to-date with the latest precision medicine information in order to determine the best possible treatment for a particular patient, but without an advanced decision support tool this may prove difficult.
The problem of inaccessibility of information can further be divided into two sub-problems based on the two primary information needs: inaccessibility of scientific articles (for identifying treatments) and the inaccessibility of clinical trials (for identifying potential experimental trials). For oncologists, both are fundamental to the treatment of their patients. The first sub-problem of inaccessibility of scientific abstracts occurs due to the amount of information in the MEDLINE‡ database and its corresponding search engine, PubMed, which provides journal citations and abstracts from biomedical literature from around the world. However, not all documents indexed in MEDLINE are relevant to an oncologist searching for precision medicine information primarily because the document doesn’t relate to the treatment, prevention, or prognosis of a patient’s condition [3;4]. The second sub-problem occurs with clinical trials. Currently, ClinicalTrials.gov§ maintains a database of privately and publicly funded clinical studies conducted around the world. However, this database suffers from similar constraints as the MEDLINE database in finding relevant clinical trials amongst all clinical trials is difficult.
In order to combat this problem, this paper proposes a search engine with three main components: an information retrieval system, a machine learning-based ranking model, and a user accessible website. The search engine ranks the relevance of a scientific abstract or clinical trial in relation to cancer precision medicine and returns a collection of documents for oncologist use.
This paper will focus on our methods of building the search engine system and several approaches we took to improve the performance of the machine learning based ranking model.
Related Work
Developing a search engine for precision medicine was a problem introduced in 2017 as part of the TREC Precision Medicine track [3]. The track is currently on its third iteration with the 2019 challenge issued earlier this year. Multiple attempts have been made in order to create a search engine that is able to retrieve relevant documents using a variety of approaches including learning-to-rank models [5], Bayesian probability graphs [6], and query expansion techniques [7;8].
Few of these current systems provide their code, and none, to our knowledge, have made a public-facing web interface. This limits the accessibility of the search engines which in turn decreases their effectiveness. In the development of PRIMROSE, we prioritized the availability of a web-based user interface in order to meet the needs of a practicing clinicians and to allow us to engage in future studies of the accessibility and usability of the interface (which is often more important than actual retrieval performance).
Data
For the storage of our data, we used an Elasticsearch version 7.1.1 cluster. Within our cluster, we created separate indexes for scientific abstracts and clinical trials. The scientific abstracts index contained a January 2017 snapshot of articles available on MEDLINE as well as American Society of Clinical Oncology (ASCO) and American Association for Cancer Research (AACR) proceedings. This data was retrieved from the 2017 TREC Precision Medicine provided files [3]. The clinical trials index contained an April 2017 snapshot of ClinicalTrials.gov and was also retrieved from the 2017 TREC Precision Medicine provided files.
We used the provided XML versions of the scientific abstracts in order to retrieve the MeSH terms, title, and abstract from the documents, and mapped this information to the mesh_terms and text fields respectively. These data subdivisions were predetermined by the TREC Precision Medicine Track and were marked as such in the provided files. For both fields, the default Elasticsearch tokenizer was used and the index was created using the default settings.
For the clinical trials, we preprocessed the provided XML files in order to only retrieve the brief_description, brief_summary, brief_title, condition, detailed_description, eligibility, gender, gender_based, keyword, maximum_age, minimum_age, and overall_status fields. These data subdivisions were predetermined by the TREC Precision Medicine Track and were marked as such in the provided files. The document was then inserted into the Elasticsearch index using the default Elasticsearch tokenizer and created the index using the default settings.
After inserting the data, we had 23,739,462 abstracts indexed in the abstracts index and 241,006 clinical trials indexed in the trials index from the 2017 dataset. From the provided TREC Precision Medicine files, we were able to get a record of documents that had been previously graded to determine whether they were precision medicine or not. There were 22,643 abstracts that had been previously judged. Of these 22,642 abstracts, 3,875 were judged as precision medicine while 18,767 abstracts were judged to be not precision medicine. The precision medicine documents were then further subdivided into relevant and partially relevant. Relevant documents contained a descriptions of the disease and gene information while partially relevant abstracts contained either the disease or the gene information or generalities of both. There were 2,022 totally relevant documents and 1,853 partially relevant documents. Clinical trials contained similar parameters with 13,019 graded documents, 1,171 precision medicine documents, and 11,848 not precision medicine documents. The precision medicine clinical trials were then further subdivided into relevant and partially relevant. There were 436 fully relevant clinical trials and 735 partially relevant clinical trials.
In order to test the recall of the search engine, we used topics provided by the 2017 TREC Precision Medicine Track. Each topic contained a disease related to cancer, one or more genes, age and gender stored as demographic information, and an other category. The topics were synthetically created by experienced precision oncologists and were often inspired by real patients [3]. The first example topic contained “Liposarcoma” as the disease, “CDK4 Amplification” as the genetic mutation, “38-year-old female” as the demographic, and “GERD” as the other category (a co-morbidity). The second sample topic contained “Colon cancer” as the disease, “KRAS (G13D), BRAF (V600E)” as the genetic mutation, “52-year-old male” as the demographic, and “Type II Diabetes, Hypertension” as the other category. The topics were stored in XML format.
Methods
A. Information Retrieval
To retrieve information from Elasticsearch, PRIMROSE queries ElasticSearch for documents matching the disease, gene, and demographic information of the topic. In this section we describe the queries for both the abstracts and trials.
A.1. Scientific Abstracts
Our baseline method for scientific abstracts was adapted from the imi_mug system [9]. The baseline query mandated that both the disease and the gene be present somewhere in the document, whether it be in the MeSH terms or in the title/abstract. The query placed a heavier weight on a matching disease than it did on a matching gene. Finally, the query searched for the gender and age demographic corresponding to the topic, but didn’t require that the gender or demographic information be in the document. The JSON for the baseline abstracts query can be seen on our github repository.¶
A.2. Clinical Trials
Our clinical trial methodology differed in the construction of the baseline query, primarily due to the availability of more information. In construction of the clinical trials baseline query, we treated the disease and gene fields the same as in the abstracts query. Since our index contained a minimum age and a maximum age range for each clinical trial and a gender field, we matched the demographic information to these fields. The JSON for the baseline trials query can be seen on our github repository.‖
A.3. Knowledge Graph Construction
After the construction of our baseline queries, we ran each topic through our knowledge graph in order to expand the disease, gene, and demographic information retrieved. In the construction of our knowledge graph, we followed the life cycle of a knowledge graph defined in Zhou et al. [7]. This life cycle follows three stages: domain knowledge modeling, knowledge acquisition, and finally knowledge integration and identifier normalization. The workflow for our knowledge graph is show in Figure 1.
Figure 1.
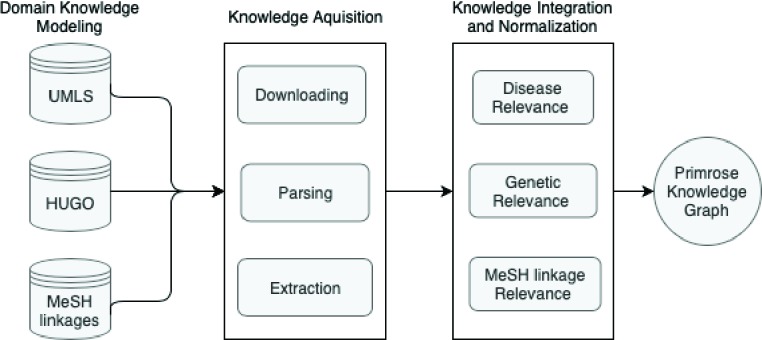
Knowledge graph for topic expansion
For the knowledge modeling part of the life cycle, we used existing databases of medical knowledge and previously defined architectures as data sources for the graph. We made use of the Unified Medical Language System (UMLS) and the HUGO Gene Nomenclature Committee (HGNC) to develop the bulk of the graph [10;11]. The UMLS exists as a repository of biomedical vocabularies and contains linkages between the concepts stored [10]. Querying the UMLS for a concept, like a cancer condition, returns related terms to the concept. The HGNC contains the approved standardized version of gene symbols, and stores the results in the Human Gene Nomenclature database. The HGNC provides two services: first it retrieves the standardized symbol for a gene and then it retrieves more information about the gene based on its symbol. Querying for a standardized symbol returns a XML document containing hgnc_id, symbol, and score as fields. Querying for more information based on the symbol or hgnc_id returns more information about the gene including its alias_symbols, location, and gene_group. We then created our own architecture for age related MeSH terms. Since Medical Subject Heading (MeSH) terms are the National Library of Medicine’s (NLM) controlled vocabulary or subject headings and can be used for age related queries, we retrieved all of the relevant age related MeSH terms from the list of existing MeSH terms as part of knowledge modeling [12]. These terms included “child”, “adolescent”, “infant”, “middle aged”, “aged”, “adult”, and “aged, 80 and over”.
Knowledge acquisition extracts knowledge relevant to the condition, genetic profile, and demographic information from the knowledge bases. The first part of this stage requires downloading the data from the appropriate part of the knowledge graph. After the data has been downloaded, it is parsed to retrieve the information requested. Once this information has been found, it is extracted from the graph. For example, querying UMLS for the disease “Liposar-coma” from the first example topic described in the Data section returns 342 results with the top 10 results being “li-posarcoma”, “Adult Liposarcoma”, “Childhood Liposarcoma”, “Liposarcoma, Dedifferentiated”, and “Liposarcoma, Pleomorphic”. Using the genetic mutation “CDK4 Amplification” from the example query, HGNC returns an XML document of 6 documents each containing the fields hgnc_id, symbol, and score. The first document contains an hgnc_id of HGNC:1773, a symbol of CDK4, and a score of 2.75. Using the symbol, CDK4, retrieved, the next part of the service returns more information about the particular gene based on its symbol such as PSK-J3 as an alias symbol, 12q14.1 as the gene location, and Cyclin dependent kinases and MicroRNA protein coding host genes as the gene_groups. We especially make use of the alias_symbols during the knowledge acquisition part of the life cycle to find related genes within the query gene’s family or gene locus.
r
Finally, the last stage of the knowledge graph is knowledge integration and normalization. The knowledge acquisition phase results in an excess of repetitive data, so the graph filters out information that is deemed repetitive. Due to the uniqueness of the format of results returned by each dataset, the graph handles each source differently. During integration, the graph assesses relevance based on proximity to the three prongs of the query - disease, genes, and demographic - and then normalizes the results. Relevance to disease is judged as , where u indicates the uniqueness of the result, while r indicates its relation to a cancer condition. Uniqueness of the result (u) occurs when the result returned contains no overlap with the entered disease concept. For example, the disease “Breast Cancer” might return “Breast cancer stage II”. This term would be deemed to be not unique since the term “Breast Cancer” appears in the returned term. The relation to a cancer condition (r) determines to degree to which something corresponds to cancer. For instance, the query “Breast cancer” might return “Breast carcinoma” and “BRCA1 Protein”. The term “Breast carcinoma” would score high on this metric since breast carcinoma is related to the name of a cancer. Conversely, “BRCA1 Protein” would score low because it describes a protein rather than the cancer. Using this methodology, we filter the returned disease results. Genetic relevance is determined by the proximity of the gene symbol to the entered query, and is calculated by genetic linkages. The first link is from gene to the gene family and the second links gene family to alias symbols. We retain all concepts that fall into these two linkages and discard the rest. Finally, we retain MeSH term linkages based on two strategies described in prior works: 1-1 age MeSH and expanded age MeSH [9;13]. These MeSH term linkage strategies provide MeSH terms that most closely represent age group being searched for. In 1-1 (one-to-one) age MeSH, the graph retains only the single best MeSH term for the age of the patient. For example, entering a 50-year-old patient would return only the “aged” MeSH term using this strategy. This MeSH term is then checked for only in the MeSH terms of the document. In expanded age MeSH, the graph uses established linkages to retain multiple MeSH terms and keywords to look for in the document. Using the expanded age MeSH strategy, the same 50-year-old patient would return the “middle aged” and “adult” MeSH terms. These MeSH terms are then added to the query in the manner specified in Kastner et al. [13]. An example of the entire integration process completed for the first example topic would return only “Liposarcoma” for the disease since the results returned from the UMLS – “liposarcoma”, “Adult Liposarcoma”, “Childhood Liposarcoma”, “Liposarcoma, Dedifferentiated”, and “Liposarcoma, Pleomorphic” – score low on the disease relevance metric due to a low uniqueness of the result. It would return cyclin dependent kinase 4, CDK4 Amplification, CDK, and PSK-J3 after running gene integration. Finally, the graph would return the “middle_aged” and “adult” MeSH terms during expanded age mesh, and just the “adult” MeSH term for 1-1 age MeSH.
The knowledge graph then normalizes to a format that can be inserted into an Elasticsearch query. By going through the normalization process, the graph reduces false positives by matching data only to relevant fields. For example, normalizing produces a query that searches for the age 53 in clinical trials only in the age related categories and not, for instance, in the brief title. For scientific abstracts, the data is linked to either the MeSH terms, the title and abstract fields, or both. For clinical trials the data is linked to the brief_title, brief_summary, mesh_terms, keyword, condition, eligibility fields or some combination of those fields. After choosing the correct field or fields for searching, the graph than determines how to prioritize matches. The graph searches for diseases both as a phrase within the specified fields and for the best match to the disease within all of the specified fields, Genes are searched for across fields and as a phrase within the document. Gene families are also searched for as phrase-prefixes within the document. After doing this, the data is then assigned a boosting factor which ElasticSearch uses when calculating the score of each document. If a document matches a particular part of the query, the boosting factor is factored into the score making matches to some parts of the query more or less valuable than others. The normalization boosting schematic prioritizes matches based on the original disease and genes found in the topic and as such assigns a higher boosting factor to these clauses of the query. Using the same first example topic, the query would generate a “Liposarcoma” best fields 1.5 boosted clause, a “Liposarcoma” phrase .3 boosted clause, a “cyclin dependent kinase 4” phrase .3 boosted clause, “CDK4” cross fields 1 boosted clause, “CDK” phrase prefix .2 boosted clause, a “PSK-J3” cross fields .2 boosted clause, and a “PSK-J3” phrase prefix .02 boosted clause.
The final part of the normalization process is the creation of partial catchers (PCs). PCs are used to include documents in the query result that contain only partial matches to the query. For example, a document might not contain the disease of interest but contains treatments for the genes of interest or vice versa. The query without a PC would discard this document despite it’s partial relevance to precision medicine. Thus, we hypothesized that the creation of PCs would increase information retrieval recall. We created two PCs: a disease PC and a gene PC. Both PCs corresponded exactly to the respective part of the original query – disease or gene – and were only modified to reduce the boosting factor of the clause.
After running a topic through the knowledge graph, we used the normalized results of the graph to modify our baseline query. For disease-expanded queries, our query is only modified with disease expansion results. For gene-expanded queries, our query is only modified with gene expanded results. For PC queries, we modified the baseline query to include PCs. We then pass the modified query to ElasticSearch and retrieve the documents that match our query.
B. Machine Learning
B.1. Data Processing
Given the availability of existing labeled data, we experimented with supervised machine learning to improve retrieval results. We used the 2017 TREC Precision Medicine track evaluation results to train our data. When we process text data, we extract the important features from the raw text and prepare an encoded input for the machine learning. First, we remove numeric terms and empty texts from the data source so that we only keep the essential text information. We then convert the precision medicine or not precision medicine ratings into two classes (relevant/not relevant to PM).
We also perform gene and cancer masking. We first extracted a list of all relevant gene and cancer names from the topics provided by the 2017, 2018 and 2019 TREC Precision Medicine Track. We then matched the exact gene and cancer names (converted to lowercase) appearing in the raw texts with our elements in our predefined list and replaced those matched names with the masked terms [gene] and [cancer]. For example, the title of one of the papers in the dataset is:
Anaplastic large cell lymphoma: redefining its morphologic spectrum and importance of recognition of the ALK-positive subset.
After gene and cancer masking, the text becomes:
anaplastic large cell [cancer]: redefining its morphologic spectrum and importance of recognition of the [gene]- positive subset.
This helps optimize the learning ability of the deep learning model since the model can focus more on learning more distinct patterns and context surrounding the words instead of the complex gene and cancer names.
After that, we encode the text using neural embeddings, for which we use two approaches: static and dynamic. For the static embeddings, a fixed document-level embedding is learned using doc2vec.∗∗doc2vec uses a neural network to learn the context of the document and phrases together. We use the default doc2vec model, which is pre-trained on a large open-domain dataset. [14].
For the dynamic embeddings, we make the use of pre-trained GloVe ** word embeddings and then fine-tune these using downstream deep learning models.
B.2. Binary Classification Approach
In this approach, we classify the documents (abstracts and trials) into two classes–relevant to precision medicine (PM) or not relevant (Not PM)–using deep learning approaches. We chose to take this approach over the feature-based model approaches such as SVM, logistic regression, etc. because deep neural networks like CNNs, though they are highly-parameterized, often outperform more traditional models even when using basic feature representations (typically word or character embeddings for NLP tasks). They thus avoid the time-consuming effort surrounding feature engineering. While it is certainly possible to encode problem-specific features into the model, this can be fairly difficult for retrieval tasks where relevance is not easily defined a priori, thus a minimal-feature approach is highly desirable. The time-consuming effort of parameter tuning as well as the computational cost of training deep learning models are further downsides, so an investigation of both types of machine learning models is justified. To optimize the computational power, the input sequences to the models are limited to 500 words because only 8% of the dataset has length beyond that limit. Short texts (<500 words) are padded with zeros and the long texts (>500 words) are truncated to 500 words. We evaluated two deep learning models:
LSTM: a bi-directional long short-term memory (LSTM) model. The LSTM memory uses 100 hidden units. Adam optimization and a learning rate of 0.1 are used for training the model.
CNN-LSTM: a convolutional neural network (CNN) combined with a bi-LSTM. The CNN has 64 kernels with a kernel size of 5. Adam optimization and a learning rate of 0.1 are used for training the model.
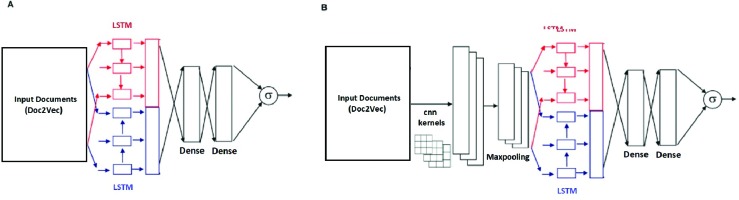
With the two basic models as shown in Figure 2, we added additional functionality to improve performance: (1) Masked deep learning models have the previously-described gene and cancer masking prior to the embedding layer. (2) An attention layer was added on top of the LSTM layer in both the LSTM and CNN-LSTM models.
Figure 2.
Deep learning models used in this paper. (A) A Bi-LSTM model, which has an input layer, a bi-directional LSTM layer, a dense layer, and an output layer. (B) A 1-layer CNN model, which has an input layer, a convolution layer, a pooling layer, a bi-directional LSTM layer, a flatten layer, a dense layer, and output layer.
B.3. Learning-to-rank Approach
In this approach, we try to directly improve ranking performance, instead of focusing on PM or Not PM classification (which plays only a part of ranking). Learning-to-rank (L2R) is a method that uses the application of supervised machine learning (ML) to perform ranking. The primary difference between L2R and traditional ML classification is that L2R calculates a score to rank a list of items so as to derive an optimal ordering of those items. L2R solves a ranking problem on a list of items such that it can optimize the ordering of those items. As such, L2R is not concerned about the exact score that each item gets, but instead focuses more on the relative ordering among all the items [15]. Therefore, L2R is very useful in information retrieval because it can improve the efficiency and precision of retrieving the top documents relevant to a topic.
With L2R, we first need to establish a scoring function and then train the model via gradient descent via a loss function defined over these scores. Hence, the model learns to predict the probability of the ranking of one document over another. In the pairwise approach, ranking is transformed into pairwise classication, comparing pairs of items one at a time. RankNet and LambdaRank are popular L2R algorithms developed by Burges and colleagues. In this research, we specifically use the LambdaRank algorithm since it has yielded better results than RankNet. LambdaRank uses the normalized discounted cumulative gain (NDCG) metric for optimization. The LambdaRank training procedure scales the gradients based on the changes in NDCG found by swapping each pair of documents.
B.4. Evaluation
For binary classification evaluation, we used the 2018 TREC PM track data for training and the 2017 track data for testing. For evaluation metrics, we use AUC, accuracy, precision, recall, and F1 for classifier performance evaluation. The performance of the individual models are reported in the Results section. The best-performing model (the LSTM with attention) was incorporated into our search engine using the LambdaRank L2R framework.
B.5. Search Engine Integration
We ran both models on the retrieved data after querying Elasticsearch with the knowledge graph in order to filter out the non-precision medicine documents. We then reranked documents based on the results of the machine learning approaches. We tested the machine learning approaches effectiveness at 4 intervals of retrieved documents: 1000, 1250, 1500, and 2000. When using the L2R rank approach, we reranked documents solely based on the results of the machine learning algorithm. For the classification approach, we settled on adding 1/2 of the range between the highest score and the lowest score in the set of retrieved documents to the score of documents classified as precision medicine by the ML classifier.
C. Web Page
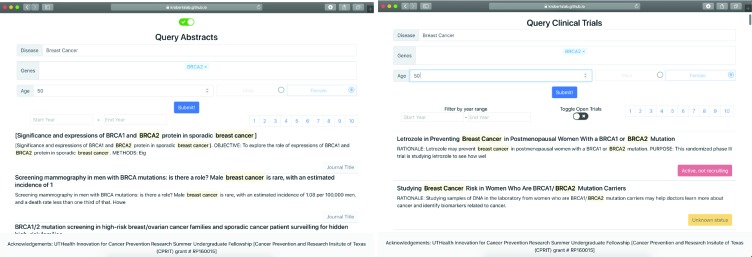
We created the web page using python’s Flask framework for the back-end part of our application and a ReactJS framework for the front-end part of the web page. Our front-end framework is available for view on Github pages, but due to security concerns we were unable to connect it with the REST portion of our application. We have provided sample data within the application for viewing purposes. Our Flask app leverages the information retrieval techniques outline above. However, it does not incorporate any machine learning in order to increase speed. The front-end framework displays both clinical trials and scientific abstracts. For scientific abstracts, the web page allows users to filter by date range and displays the journal title of the abstract. For clinical trials the web page allows users to filter by date range and displays the status of the trial. It also lets users filter results by whether the trial is open or not based on the recruitment status.
Results
A. Information Retrieval
We evaluated the precision of our search engine using the TREC 2017 PM track evaluation metrics for clinical trials and scientific abstracts. The primary metric used in evaluation of the scientific abstracts was the inferred Normalized Discounted Cumulative Gain (infNDCG) while the main metrics for clinical trials were Precision at 5 (P5), Precision at 10(P10), and Precision at 15(P15) [3]. We observed significant improvement quality of documents returned after using the Knowledge Graph. We were unable to see significant improvement using machine learning methods, however.
For the information retrieval component of our search engine, we evaluated the queries before and after the use of the Knowledge Graph and using the three different types of age MeSH strategies.
For scientific abstracts, the best query performed expansions on both the genes and the disease as well as introduced partial match catchers for both the disease and the genes. Gene expansion preformed better than disease expansion for the scientific abstracts. However, partial match catchers were more effective with diseases rather than genes. Overall, we noticed that 1-1 age MeSH strategy worked better with queries with both disease and gene expanded. We also noticed that a no MeSH strategy seemed to improve precision in cases where gene and disease were not both expanded. The results from our scientific abstract queries are show in Table 1.
Table 1.
Comparison of results for scientific abstracts based on infNDCG.
| 1-1 age MeSH | Expanded Age MeSH | No MeSH | |
| Baseline | 0.4117 | 0.4119 | 0.4130 |
| Gene Expanded | 0.4371 | 0.4351 | 0.4383 |
| Disease Expanded | 0.4178 | 0.4186 | 0.4181 |
| Disease + Gene Expanded | 0.4587 | 0.4546 | 0.4547 |
| Partial Match Catcher Expanded Disease | 0.4596 | 0.4550 | 0.4560 |
| Partial Match Catcher Expanded Gene | 0.4584 | 0.4542 | 0.4562 |
| Both Expanded Partial Match Catcher | 0.4603 | 0.4563 | 0.4562 |
For clinical trials, the best queries had either disease or gene expansion, but not both. Disease expansion performed better in terms of P5 metric than any other query. However, gene expansion performed better on the P10 and P15 metrics. Partial catchers were somewhat effective, but no clear trend emerged from the data. The results from the queries are displayed in Table 2. There was no difference between using the 1-1 age MeSH strategy and the no age MeSH strategy, which made sense due to the presence of the age range fields in our data.
Table 2.
Comparison of results for clinical trials.
| P@5 | P@10 | P@15 | |
| Baseline | 0.4276 | 0.369 | 0.3218 |
| Gene Expanded | 0.4414 | 0.4103 | 0.3609 |
| Disease Expanded | 0.4552 | 0.3828 | 0.3448 |
| Disease expanded w/ PC | 0.4414 | 0.4030 | 0.3586 |
| Disease expanded w/ PC and gene expanded | 0.2345 | 0.2103 | 0.1816 |
| Gene & Disease Expanded both w/ PC | 0.4414 | 0.4000 | 0.3494 |
B. Binary Classification
The detailed experiments for our classifier used in the learning-to-rank framework are shown in Table 3. For L2R, we also trained on the TREC 2018 dataset [3] and tested on the TREC 2017 dataset [4]. The primary evaluation metric was NDCG, which measures the closeness of the predicted ranking list with the actual labels. The NDCG of our L2R model on the testing set is 0.65 (on abstract data) and 0.69 (on clinical trial data).
Table 3.
Learning-to-rank performance on scientific abstracts and clinical trials using six different model variants.
| Scientific Abstracts | ||||||
|---|---|---|---|---|---|---|
| Model | Embedding | AUC | Acc. | Prec. | Rec. | F1 |
| LSTM | GloVe | 0.539 | 0.569 | 0.525 | 0.521 | 0.516 |
| CNN-LSTM | GloVe | 0.541 | 0.531 | 0.528 | 0.530 | 0.524 |
| CNN-LSTM | doc2vec | 0.541 | 0.575 | 0.526 | 0.520 | 0.511 |
| Masked LSTM | GloVe | 0.545 | 0.611 | 0.305 | 0.500 | 0.379 |
| Masked CNN-LSTM | GloVe | 0.549 | 0.561 | 0.536 | 0.535 | 0.535 |
| LSTM + Attention | GloVe | 0.591 | 0.552 | 0.561 | 0.564 | 0.550 |
| Clinical Trials | ||||||
| Model | Embedding | AUC | Acc. | Prec. | Rec. | F1 |
| LSTM | GloVe | 0.485 | 0.600 | 0.468 | 0.475 | 0.467 |
| CNN-LSTM | GloVe | 0.503 | 0.704 | 0.406 | 0.499 | 0.414 |
| CNN-LSTM | doc2vec | 0.497 | 0.646 | 0.471 | 0.486 | 0.457 |
| Masked CNN-LSTM | GloVe | 0.511 | 0.641 | 0.488 | 0.493 | 0.475 |
| Masked LSTM | GloVe | 0.530 | 0.392 | 0.546 | 0.532 | 0.383 |
| LSTM + Attention | GloVe | 0.500 | 0.705 | 0.353 | 0.500 | 0.414 |
C. Search Engine Integration
When machine learning was combined with the Elasticsearch queries, we noticed a general decrease in precision across all uses of the knowledge graph. The L2R queries performed generally equivalent when 1000 or less documents were returned, but decreased significantly in precision when more documents were returned. Conversely, the deep learning model maintained relatively the same precision across multiple query sizes despite an initial drop in precision for the baseline query size of 1000. Overall, the the addition of machine learning was unable to increase the score from the retrieved documents over all query sizes. A comparison of the best query when used with the machine learning approaches is shown in Table 4. Additionally, adding machine learning to the REST API decreased the end-to-end performance of the search engine.
Table 4.
Comparison of machine learning approaches on the best retrieval technique (for scientific abstracts, the Both Gene and Disease expanded w/ partial catchers (PC); for clinical trials the gene expanded query).
| Baseline | L2R | Deep Learning | |
|---|---|---|---|
| Abstracts (infNDCG) | 0.4603 | 0.4603 | 0.4544 |
| Trials (P@10) | 0.4103 | 0.4103 | 0.3897 |
D. Website
We were able to create the web interface for the search engine using ReactJS for the front-end and Flask for the creation of the REST API. We deployed the React app on Github pages††. We were unable to connect the REST API
to the deployed website due to security reasons (owing to university policies), lacking a dedicated web server. Instead, we have provided sample data for viewing. We have included screenshots of the website in Figure 3.
Figure 3.
Website Screenshots
Discussion
We were able to see a marked improvement in the retrieval of precision medicine documents using our search engine. We focused on the development of our search engine to cater to oncologists and emphasized its usability. By the development of a front-facing website with an intuitive user interface, we were able to translate preexisting knowledge into tangible solutions. The one main possible source of error was insuring that we had an accurate snapshot of MEDLINE and ClinicalTrials.gov. If we were missing documents, the evaluated accuracy of our search engine could vary, but the same general trends should still apply. Although we were able to test it internally, the inability to open our REST API for public connections is a major technical barrier that we still need to overcome, likely requiring a dedicated webserver to host the website and Elasticsearch index. We were able to get around this by testing the REST API separately from the front-facing website, but concern exists regarding scalability and accessibility. In future work, we hope to get around both the sources of error and the limitations of our study by regularly updating the information in our Elasticsearch index with the most current articles in MEDLINE and by adding additional security measures so that the REST API can be public-facing.
Conclusion
Because of the inaccessibility of precision medicine information, oncologists have been unable to use the information to actually treat their patients. We developed the PRecIsion Medicine Robust Oncology Search Engine (PRIMROSE) in order to retrieve relevant information from published scientific abstracts and clinical trials. As part of PRIMROSE, we developed a knowledge graph for query expansion and used machine learning techniques in order to improve recall. We then created a front facing website and REST API in order for an oncologist or other user to use the information. Going forward, we hope to increase recall as well as increase accessibility to the search engine.
Acknowledgements
This work was partially funded by the UTHealth Innovation for Cancer Prevention Research Summer Undergraduate Fellowship by Cancer Prevention and Research Institute of Texas (CPRIT) grant # RP160015 as well as the Patient-Centered Outcomes Research Institute (PCORI) project ME-2018C1-10963.
Footnotes
This work was completed as part of an undergraduate internship at the UTHealth School of Biomedical Informatics.
https://obamawhitehouse.archives.gov/the-press-office/2015/01/30/fact-sheet-president-obama-s-precision-medicine-initiative
https://www.nlm.nih.gov/bsd/pmresources.html/
https://clinicaltrials.gov/
https://github.com/krobertslab/primrose/blob/master/src/assets/ABSJSON.json
https://github.com/krobertslab/primrose/blob/master/src/assets/TRIALSJSON.json
http://www.davidsbatista.net/blog/2018/12/06/Word_Embeddings/
https://krobertslab.github.io/primrose/
References
- 1.What is precision medicine? - Genetics Home Reference - NIH, Aug. 2019 [Google Scholar]
- 2.Collins FS, Varmus H. A new initiative on precision medicine. New England journal of medicine. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts K, Demner-Fushman D, Voorhees EM, Hersh WR, Bedrick S, Lazar A, Pant S. Overview of the TREC 2017 Precision Medicine Track. In Proceedings of the Twenty-Sixth Text Retrieval Conference. 2017 [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts K, Demner-Fushman D, Voorhees EM, Hersh WR, Lazar A, Pant S. Overview of the TREC 2018 Precision Medicine Track. In Proceedings of the Twenty-Seventh Text Retrieval Conference. 2018 [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor SJ, Goodwin TR, Harabagiu SM. UTD HLTRI at TREC 2018: Precision Medicine Track. In Proceedings of the Twenty-Seventh Text Retrieval Conference. 2018 [Google Scholar]
- 6.Balaneshinkordan S, Kotov A. Bayesian Approach to Incorporating Different Types of Biomedical Knowledge Bases into Information Retrieval Systems for Clinical Decision Support in Precision Medicine. Journal of Biomedical Informatics. 2019:103238. doi: 10.1016/j.jbi.2019.103238. [DOI] [PubMed] [Google Scholar]
- 7.Zhou X, Chen X, Song J, Zhao G, Wu J. Team Cat-Garfield at TREC 2018 Precision Medicine Track. In Proceedings of the Twenty-Seventh Text Retrieval Conference. 2018 [Google Scholar]
- 8.Oleynik M, Faessler E, Sasso AM, Kappattanavar A, Bergner B, Da Cruz HF, Sachs J, Datta S, Böttinger E. HPI-DHC at TREC 2018 Precision Medicine Track. In Proceedings of the Twenty-Seventh Text Retrieval Conference. 2018 [Google Scholar]
- 9.López-García P. TREC 2018 Precision Medicine-Medical University of Graz. In Proceedings of the Twenty-Seventh Text Retrieval Conference. 2018 [Google Scholar]
- 10.Bodenreider O. The Unified Medical Language System (UMLS) Integrating Biomedical Terminology. 2004 doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Povey S, Lovering R, Bruford E, Wright M, Lush M, Wain H . The HUGO gene nomenclature committee (HGNC) Human Genetics. 2001;109(6):678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 12.Chapman D. Advanced search features of PubMed. Journal of the Canadian Academy of Child and Adolescent Psychiatry. 2009;18(1):58. [PMC free article] [PubMed] [Google Scholar]
- 13.Kastner M, Wilczynski NL, Walker-Dilks C, McKibbon KAnn, Haynes B. Age-specific search strategies for Medline. Journal of medical Internet research. 2006;8(4):e25. doi: 10.2196/jmir.8.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rong X. word2vec parameter learning explained. arXiv preprint arXiv:1411.2738. 2014 [Google Scholar]
- 15.Li H. A short introduction to learning to rank. IEICE TRANSACTIONS on Information and Systems. 2011;94(10):1854–1862. [Google Scholar]