Abstract
Knowledge graphs have been shown to significantly improve search results. Usually populated by subject matter experts, relations therein need to keep up to date with medical literature in order for search to remain relevant. Dynamically identifying text snippets in literature that confirm or deny knowledge graph triples is increasingly becoming the differentiator between trusted and untrusted medical decision support systems. This work describes our approach to mapping triples to medical text. A medical knowledge graph is used as a source of triples that are used to find matching sentences in reference text. Our unsupervised approach uses phrase embeddings and cosine similarity measures, and boosts candidate text snippets when certain key concepts exist. Using this approach, we can accurately map semantic relations within the medical knowledge graph to text snippets with a precision of 61.4% and recall of 86.3%. This method will be used to develop a novel application in the future to retrieve medical relations and corroborating snippets from medical text given a user query.
1. Introduction
Medical knowledge changes, evolves and shifts as new discoveries are made. In order to offer their patients appropriate care, clinicians have a strong need to stay up to date with the latest FDA approved drugs and practice guidelines from renowned medical associations. Today, information systems such as UpToDat1, Clinical Ke2 and Dyname3 are used to search for medical literature that may help pressing point of care questions. However, their content is still manually curated and maintained by teams of domain experts - they diligently scan the medical archives in search for relevant information to include in synoptic documents usually aimed at clinicians.
Given that medical information is projected to double every 73 days in 2020, whereas in 2010 it was doubling only every 3.5 year4, these curator-driven solutions are unlikely to scale. Instead, technological solutions that automate the delivery of the right snippets of information to the right person in the right context at the right time have never been so critical. To understand the types of questions that clinicians need answered, we looked at the Ely taxonom5 - a seminal paper that highlighted the types of questions that clinicians need answered when using computational systems. The 3 most common questions categories were: i) “What is the drug of choice for condition x?”, ii) “What is the cause of symptom x?”, and iii) “What test is indicated in situation x?”. However, as Del Fiol et al. observed in 2014, roughly half of all point of care questions go unanswere6. Insufficient time is often cited as the main barrier to point of care learning. Other barriers to successful point of care search often include the need to search for patient comorbidities and contexts, the ever-growing volume of medical evidence, lack of awareness of which resource to search for specialty questions, skepticism and lack of trust regarding the quality of search results, etc.7.
With that requirement in mind, the Elsevier Healthcare Knowledge Graph (HG) was created as a resource containing 400,000 medical concepts and 200,000 manually created medical relations (e.g., RHEUMATOID ARTHRITIS has drug METHOTREXATE). The primary intent of this knowledge graph is to support indexing and retrieval of relevant medical content in order to facilitate point of care search. An informal evaluation of H-Graph revealed that it could be used to answer 96% point of care questions (based on a set of 6 cardiovascular diseases and questions from the Ely taxonom5) whereas the standard reference solutions could only answer 27% of the specific question8.
Knowledge Graphs have been shown to be effective tools to address multiple search and knowledge inference problems in healthcare and biomedicin9–12. However, unless there is a team dedicated to keeping that knowledge constantly updated, alternative technological solutions are needed that ensure that important relationships such as newly approved drugs, biomarkers and results from real-world evidence studies are maintained. As such, the need to connect knowledge graph triples to text snippets supporting or denying those relations is very much a critical need. That capability is also a dependency for the more complex problem of automating the extraction of medical relations from medical text.
In this research, we present an unsupervised approach to align unstructured text snippets in medical textbooks with medical relations. This approach leverages a Medical Knowledge Graph composed of different biomedical concepts (e.g., drugs, diseases, symptoms, etc.) represented in a concept hierarchy and different biomedical relations of a fixed set of relation types (e.g., has drug, has clinical finding), as well as uses word embedding vectors (i.e., words represented in a high-dimensional vector space based on co-occurrence in literature). The key contributions are:
We identify and evaluate text snippets in two medical textbooks Ferri’s Clinical Advisor 2019 and Conn’s Current Therapy 2019 that corroborate medical relations, curated by experts in the Elsevier Healthcare Knowledge Graph (HG), for three diseases DIABETES MELLITUS, ASTHMA, and RHEUMATOID ARTHRITIS.
We investigate the coverage of our knowledge pertaining to these three diseases in HG with respect to the set of text snippets that can be mapped to corroborate a medical assertion in HG.
This research will enable the identification of corroborating text snippets from multiple literature sources for an approved medical relation in the knowledge graph. In turn, the knowledge graph can be used to drive search applications to retrieve the right snippet in a literature source given a user query (e.g., Drugs to treat Asthma in a pregnant adult). We will conclude with a brief discussion on the development of such a novel HG point of care search application.
2. Related Work
The health care community has been one of the earliest adopters of knowledge graphs. Driven by the need to create interoperable representations of knowledge that could be used at point of care, several innovations in healthcare have made use of and published data and knowledge graphs using these technologie12–15. In fact, one of the formats in which FHIR can be downloaded and created — the current HL7 proposal for healthcare interoperability — is Resource Description Framework (RDF), the W3C standard for the generation and maintenance of knowledge graphs.
Over the years, there have been several applications that have made use of these knowledge graphs to produce knowledge or empower search. PhLeGr16 used these technologies to enable federated graphical queries across pharmacological evidence from the FDA Adverse Event Reporting System and phenotypes from multiple sources. ReVeaL17 enabled cancer chemoprevention researchers to formulate complex queries against more than 80 RDF graphs to drive in silico drug discovery for breast cancer research.
Phrase (or sentence) embedding are numerical vectors that encode the semantics of a phrase. They can be generated using a weighted average of the embedding vectors for the words in the sentence. Phrase embeddings have been shown to achieve significantly better performance over sophisticated, supervised methods (e.g., Recurrent Neural Networks models) on text similarity task18. Increasingly, knowledge graphs are being combined with word and phrase embeddings to analyse and answer questions from medical text, to recommend safe therapies, to conduct pharmacovigilance using social media, or to discover novel adverse-drug reaction10, 19–22.
The main benefit of our unsupervised boosted approach is that we don’t require training data or memory-intensive models. A similar weighted average approach, where the weights were the inverse document frequencies (IDFs) for words, was used to align class labels in biomedical ontologies with metadata fields in the BioSamples repositor21.
3. Materials
3.1. HG: Elsevier Healthcare Knowledge Graph
Elsevier Healthcare Knowledge Grap23 (termed HG henceforth) is a platform built to enable advanced clinical decision support, precision medicine, and point of care content discovery. It comprises data from heterogeneous health-care information sources and relations extracted from literature. HG includes knowledge and data about diseases, drugs, order sets, guidelines, cohorts, journals and books.
HG’s core is based on UMLS terminologies and refined by subject matter experts, incorporating also medical jargon and acronyms. As of August 2019, HG consisted of 400,000 biomedical concepts classified under specific semantic groups (e.g., DRUG, DISEASE) and arranged in a parent–child poly-hierarchy. These include more than 75,000 diseases, 46,000 drugs, 63,000 procedures, and 90,000 symptoms. HG contains more than 200,000 manually generated relations between different concepts bound under a fixed set of 15 relation types (e.g., has drug, has clinical finding).
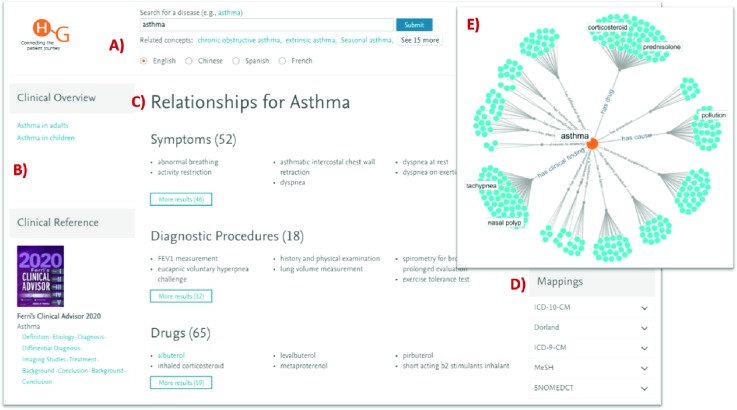
Figure 1 shows a screenshot of HGViz, an in-house visualizer, which provides a synoptic view of HG content.
Figure 1.
Elsevier Healthcare Knowledge Graph (HG): A screenshot of HGViz, an in-house visualizer, provides a synoptic view of HG content. A) For the concept ASTHMA, HG has related content in different languages, as well as content that is classified for more specific-forms of ASTHMA (e.g., CHRONIC OBSTRUCTIVE ASTHMA). B) HG links content related to ASTHMA from existing journals and books (e.g., Ferri’s Cinical Advisor 2019). C) ASTHMA is linked to other concepts in HG (e.g., drugs, symptoms, diagnostic procedures) through expert-curated semantic relations. D) The HG concept ASTHMA is mapped to related concepts in other terminologies (e.g., ICD-10, SNOMED CT). E) A hairball visualization of HG allows the user to browse all semantic relations for ASTHMA grouped according to different relation types (e.g., has clinical finding, has drug).
3.2. Medical Textbooks
Ferri’s Clinical Advisor 2019 and Conn’s Current Therapy 2019 are widely used medical textbooks published by Elsevier and accessible through the Clinical Key search engin2. The intended audience of these books are clinicians, who used it for information relevant for point of care. Ferri has 1037 chapters, of which 799 chapters cover a single disease (e.g., RHEUMATOID ARTHRITIS). The remaining chapters cover disease classes or combinations, differential diagnosis, lab tests, medical algorithms, etc. Conn has 331 chapters, of which 235 chapters cover a single disease. Over a period of 2016–2019, more than 200K unique search queries made by users against the Clinical Key search engine have led users to access Ferri and more than 100K search queries have led users to access Conn.
The chapter structure for disease chapters in both the textbooks is fairly uniform — each chapter is generally divided into multiple sections related to basic information, epidemiology and demographics, etiology, diagnosis, and treatment. However, the section titles might be different and text snippets corroborating medical relations may be under different sections with varying titles (e.g., text snippet corroborating medical relations of relation type has drug may be present in sections with titles Chronic Rx, Pharmacologic Therapy, Treatment, etc.). Additionally, disease-related information can be present in chapters related to more specific forms of the disease. For example, the medical assertion (RHEUMATOID ARTHRITIS has complication JOINT DESTRUCTION) can be corroborated by the sentence “Progressive joint destruction, particularly in affected hips, can occur even with aggressive management.” present in the chapter on JUVENILE IDIOPATHIC ARTHRITIS, a more specific form of RHEUMATOID ARTHRITIS. Finally, multiple text snippets from different, but related, sections in different, but related, chapters can support a medical assertion in HG. We will leverage the HG hierarchy and word embedding vectors to address this.
3.3. Word Embedding Vectors
In this research, we represent text phrases and sentences found in the two medical textbooks, as well as the semantic relations in HG as high-dimensional numerical vectors. We then use these embedding vectors to compute similarities between text snippets and medical relations. Toward this end, we use word embedding vectors generated previousl24 from a corpus of approximately 30 million biomedical abstracts, stored in the MEDLINE database, using the GloVe (Global Vectors for Word Representation) softwar25. More than 2.5 million words appearing in biomedical abstracts are represented as numerical vectors in 100 dimensions. These word embedding vectors were downloaded from FigShar26 under a CC BY 4.0 license and have been used previously to align biomedical metadata terms with ontology terms21, as well as to align schema elements across the vocabularies of biomedical RDF graph24.
4. Methods
4.1. Preprocessing of Unstructured Text in the Medical Journals
Each chapter in the Ferri and Conn medical textbooks was provided as a separate, self-contained XML file that was passed as input through a preprocessing pipeline with multiple operations.
The first operation converted the XML file to a JSON document, after the extraction of chapter titles and textual content, organized into sections and sub-sections, while omitting tables, figure captions, etc. As part of this operation, the nesting structure of the sections and subsections was used to extract a “breadcrumb trail” comprising chapter title, section title, sub-section title, and so on. For example, based on the nesting structure, the sentence “Most commonly used agents are methotrexate (MTX), hydroxychloroquine (HCQ), sulfasalazine (SSZ), and leflunomide (LEF).” has
the breadcrumb trail Treatment → Chronic Rx in the chapter on RHEUMATOID ARTHRITIS.
As part of the next operation in the preprocessing pipeline, we used the spaCy27 software to split section text contained within different sections and subsections into individual sentences. We use an in-house Concept Mapper API to annotate chapter titles and individual sentences for each chapter with HG concepts. For example, as shown in Figure 2A, the underlined concepts (i.e., RHEUMATOID ARTHRITIS, METHOTREXATE, etc.) in the chapter title and sentence in Ferri will be annotated by the Concept Mapper API.
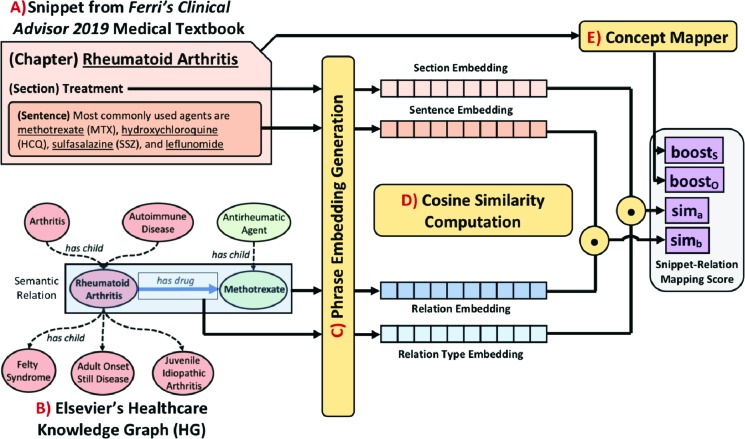
Figure 2.
Diagrammatic Representation of the Snippet–Relation Alignment Method: A) Snippet from the Ferri’s Clinical Advisor 2019 Medical Textbook showcasing chapter title, section title, and sentence Text. B) Subgraph from the Elsevier Healthcare Knowledge Graph (HG) showcasing the (RHEUMATOID ARTHRITIS has drug METHOTREXATE) semantic relation, as well as parent/child concepts for RHEUMATOID ARTHRITIS and METHOTREXATE in the HG hierarchy. C) Embedding vectors are generated using an unsupervised approach for section title, sentence text, HG relation type, and the semantic Relation. D) Two cosine similarity values are computed: section and relation type similarity, and sentence and semantic relation similarity. E) Concept Mapper annotates HG concepts in the chapter title and the sentence of the snippet (underlined here), and generates boost values. A Snippet–Relation Mapping Score is computed using the cosine similarity values and boost values.
4.2. Generating Candidate Snippets for a Semantic Relation
In Figure 2, we present a diagrammatic representation of our method to generate candidate text snippets from medical textbooks Ferri and Conn given an expert-curated medical assertion present in the Elsevier Healthcare Knowledge Graph (HG). In the subsequent subsections, we will briefly describe each component of this method in more detail.
4.2.1. Extracted Text Snippets from the Medical Textbooks
For this research, we define a text snippet as a parsed and extracted object of type mj = (chapterj , sectionj , sentencej ) from a specific medical textbook (i.e., either from Ferri or Conn), where sentencej is the parsed sentence (Section 4.1), sectionj indicates the section title where the sentence is present, and chapterj indicates the chapter title where the section is present. Most chapter titles are representative of a specific disease. An actual text snippet from a chapter on RHEUMATOID ARTHRITIS in the Ferri’s Clinical Advisor 2019 medical textbook is shown in Figure 2A.
4.2.2. Semantic Relations in HG
We define a semantic relation ei = (Si, Ri, Oi) in HG, where Si indicates the subject of the semantic relation (e.g., Si = RHEUMATOID ARTHRITIS), Ri indicates the relation type (e.g., Ri = has drug), and Oi indicates the object of the semantic relation (e.g., Oi = METHOTREXATE). In the above semantic relation, both the subject and object are distinct medical concepts (or nodes) that are present in a concept hierarchy with designated semantic types (e.g., RHEUMATOID ARTHRITIS is a child of AUTOIMMUNE DISEASE, and has JUVENILE IDIOPATHIC ARTHRITIS and
FELTY SYNDROME as its children concepts.). Figure 2B showcases a subgraph from HG, with one semantic relation and few hierarchical relations.
For this research, we only use 160,000 validated semantic relations of 15 relation types in HG.
With respect to the Elsevier Healthcare Knowledge Graph (HG), we define the following functions:
parents(Ct) retrieves the set of all parent concepts for the concept Ct in the HG hierarchy
descendants(Ct) retrieves the set of all descendant concepts for the concept Ct in the HG hierarchy.
path(Ct, Cu) retrieves all the semantic relations and hierarchical relations on the shortest undirected path between the concepts Ct and Cu in HG.
relations(Ct, Rt) retrieves the set of all semantic relations with subject Ct and relation type Rt.
4.2.3. Phrase Embedding Generation Method
In this research, we follow the approach outlined by Arora et al.18 to compute the weighted average using unigram probabilities of words in the MEDLINE corpu28. For each phrase (i.e., section title, relation type label, sentence text, semantic relation), we remove a set of 129 English stopwords (e.g., ‘and’, ‘the’) and compute a phrase embedding vector using a weighted average of words in that phrase.
In this equation, xwi indicates the word embedding vector for word wi in the phrase (Section 3.3), whereas p(wi) indicates the unigram probability of the word wi in the MEDLINE corpus of biomedical publication abstracts. α is a scalar quantity. For more frequent words, the weight α/(α + p(wi)) is smaller, so this down weights the frequent words. |s| indicates the total number of words in the phrase.
Using this method, we generate the following sets of 100-dimensional phrase embedding vectors (Figure 2C):
xsectionj : Embedding vector for a section title (e.g. Pharmacologic Therapy) in Ferri or Conn.
xsentencej : Embedding vector for a phrase or a sentence (e.g., Most commonly used agents are methotrexate (MTX), hy- droxychloroquine (HCQ), sulfasalazine (SSZ), and leflunomide (LEF).) in Ferri or Conn.
xRi : Embedding vector for a relation type (e.g., has clinical finding, has drug) in HG.
xei : Embedding vector for a semantic relation (e.g., RHEUMATOID ARTHRITIS has drug METHOTREXATE).
4.2.4. Cosine Similarity Score Computation
Using the 100-dimensional phrase embedding vectors, we compute two similarity matrices (Figure 2D):
Sima Similarity matrix between all section titles in a specific medical textbook and all relation types in HG. Each cell in this matrix sima(i, j) denotes the cosine similarity score between HG relation type Ri and section title sectionj in the medical textbook.
Simb Similarity matrix between all sentences in a specific medical textbook and all validated semantic relations in HG. Each cell simb(i, j) denotes the cosine similarity score between a semantic relation ei in HG and a sentence sentencej in the medical textbook.
4.2.5. Using the Concept Mapper to Boost the Snippet–Relation Mapping Score
As described in Section 4.1, we use the Concept Mapper API to identify HG concepts that may be present in the chapter title (concept set denoted as henceforth) and in the sentence text (concept set denoted as ). We use these concept sets to generate boost scores for certain relation–snippet mappings.
For each relation–snippet mapping, we assign a boost score boostSi if the subject Si of the HG semantic relation ei is present in the chapter title chapterj of the snippet, or if any of the descendants or direct parents for subject Si in the HG hierarchy is present in the chapter title. Similarly, we assign a boost score boostOi if the object Oi of the HG semantic relation ei is present in the sentence text sentencej of the snippet, or if any of the descendants or direct parents for object Oi is present in the sentence text. Due to the chapter structure of the medical textbooks in this research (i.e., chapters are organized according to diseases), we assign a higher boost score for the presence of subject concept in the chapter title compared to the presence of object concept in the sentence.
Finally, we generate a combined mapping score for a relation–snippet mapping as shown in the equation below.
Relation–snippet mappings are sorted according to this mapping score, and the top 5 snippets with the highest relation– snippet mapping score are selected as “candidate snippets” in a particular medical textbook (i.e., Ferri) given the particular HG relation.
4.3. Evaluation Methodology
Using the approach indicated in the previous section, we generate candidate relation–sentence mappings for three diseases: DIABETES MELLITUS, ASTHMA, and RHEUMATOID ARTHRITIS, using content from both Ferri and Conn. We further selected only those mappings with a score ≥ 2.5. The score threshold of 2.5 for the candidate mapping
selection was decided heuristically.
To evaluate the algorithm, 3 domain experts, with knowledge pertaining to biomedicine, were asked to independently review 2 random samples of these mappings — 250 relation–snippet mappings from Ferri and 250 relation–snippet mappings from Conn. Experts were asked to review each relation–snippet mapping on a 4-point scale:
0: No association between snippet and relation in Elsevier Healthcare Knowledge Graph (HG)
1: Negative association between snippet and HG relation
2: Positive association between snippet and HG relation
3: Uncertain association between snippet and HG relation
We compute inter-reviewer agreement using the Fleiss’ kappa metric (computed using the R package ‘irr’), a statistical measure for assessing the reliability of agreement between a fixed number of raters when assigning categorical ratings to a number of item29. Under the assumption of a 2.5 score threshold and assuming all HG relations are covered in one or both books, we compute precision and recall measures for each relation type.
5. Results
245,809 sentences grouped under 22,246 sections were parsed from 1,037 chapters of the Ferri’s Clinical Advisor 2019. Similarly, 88,711 sentences grouped under 5,278 sections were parsed from 331 chapters of the Conn’s Current Therapy 2019 using the preprocessing pipeline. Only 1,175 unique words, occurring 3,334 times, in Ferri (out of 34,523 unique words and 2,603,653 total words) and 914 unique words, occurring 2,495 times, in Conn (out of 28,335 unique words and 1,618,966 total words) could not be found in the vocabulary of biomedical words and did not have associated word embeddings. 5 candidate snippets (i.e., (chapter title, section title, sentence)) are generated, from both Ferri and Conn textbooks, for three diseases in Elsevier Healthcare Knowledge Graph (HG): DIABETES MELLITUS (161 semantic relations), ASTHMA (274 semantic relations), and RHEUMATOID ARTHRITIS (223 semantic relations).
3 domain experts evaluated 2 samples of 250 relation–snippet mappings generated from the two medical textbooks Ferri and Conn. Considering the 4-point evaluation scale (presented in Section 4.3), the inter-reviewer agreement, as determined by the Fleiss’ kappa metric, was 0.404 for Ferri and 0.378 for Conn. However, assuming a binary scale (i.e., only a positive association between a HG relation and a text snippet is valid for a relation–snippet mapping), Fleiss’ kappa metric for inter-reviewer agreement increased to 0.533 for Ferri and 0.492 for Conn. This metric indicates moderate agreement among the reviewers on the scoring of the relation–snippet pairs.
A consensus was reached (i.e., 2 out of 3 reviewers agreed) that 307 entries demonstrated a positive association between an HG relation and a text snippet in either Ferri or Conn (61.4%). 213 out of these 307 entries (69.8%) received a unanimous verdict from all the three reviewers. A majority of the reviewers were undecided on 57 entries (11.4%), decided that a negative association exists between a relation and a text snippet for 7 entries (1.4%), and decided that no association exists between a relation and a text snippet for 90 entries (18%). Only 39 entries out of a total of 500 relation–snippet mappings evaluated by domain experts had diverging scores among all three reviewers.
A few of these examples have been shown in Table 1. The first two rows are example mappings for the relation type has drug, where the mapped snippets are reflective of negative treatment outcomes for more specific forms of the disease (i.e., ACUTE SEVERE ASTHMA or JUVENILE IDIOPATHIC ARTHRITIS). However, the reviewers diverged on whether it can be inferred on whether ASTHMA or RHEUMATOID ARTHRITIS can generally be treated by those specific drugs. Similarly, the second set of examples for the relation type has clinical finding include snippets from the treatment-related sections. Hence, the reviewers diverged on whether to interpret NAUSEA and VOMITING as clinical findings for DIABETES MELLITUS, or as side effects for specific drugs used to treat the disease.
Table 1:
Few relation–snippet mappings where all 3 reviewer scores diverged.
| Subject | Chapter Name | Section | Sentence |
|---|---|---|---|
| asthma has drug epinephrine | Asthma | Basic Information | “Status asthmaticus, or acute severe asthma, is a refractory state that does not respond to standard therapy such as inhaled beta-agonists or subcutaneous epinephrine.” |
| rheumatoid arthritis has drug anakinra | Juvenile Idio-pathic Arthritis | Treatment | “IL-1 and IL-6 antagonists, anakinra, and tocilizumab for those with systemic JIA Meta-analysis of randomized controlled trials did not show statistically significant differences in the efficacy or safety profile of these agents.” |
| diabetes mellitus has clinical finding nausea | Diabetes Mellitus | General Rx | “Nausea is its major side effect.” |
| diabetes mellitus has clinical finding vomiting | Diabetes Mellitus | Treatment | “Major manifestations are postprandial fullness, nausea, vomiting, and bloating.” |
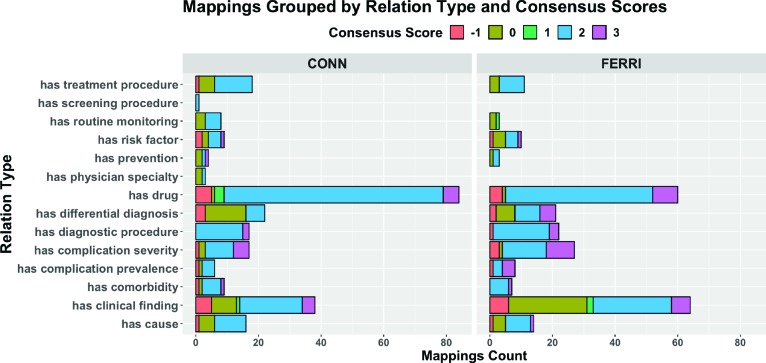
Consensus scores for relation–snippet mappings stratified according to different relation types are shown in Figure 3. It can be easily seen that HG possesses a greater number of validated relations of type has clinical finding and has drug, for the three diseases studied in this research. Moreover, while we generally obtain a consensus score of 2 (positive association) for mappings pertaining to most relation types, a large proportion of mappings where the relation is of type has differential diagnosis or has clinical finding has a consensus score of 0 (no association).
Figure 3.
Breakdown of Reviewer Scores according to Relation Types: Elsevier Healthcare Knowledge Graph (HG) contains several relation types as shown on the Y-axis. While we generally obtain a consensus of score 2 (positive association) for mappings pertaining to most relation types, a large proportion of has differential diagnosis and has clinical finding mappings achieved a consensus of 0 (no association) for both Ferri and Conn
1,015 candidate relation–snippet pairs (out of a total 3,290 candidates) from Ferri and 873 candidate relation–snippet pairs from Conn satisfy the threshold of mapping score ≥ 2.5. At a precision of 61.4% (as indicated above) for the three diseases DIABETES MELLITUS, ASTHMA, and RHEUMATOID ARTHRITIS, recall can be computed to be 86.3%. That is, 568 out of 658 semantic relations can be corroborated by at least one text snippet from either Ferri or Conn for these three diseases at a mapping score >= 2.5.
384 semantic relations can be corroborated by multiple text snippets across both the medical textbooks. For example, the Ferri statements “Most commonly used agents are methotrexate (MTX), hydroxychloroquine (HCQ), sulfasalazine (SSZ), and leflunomide (LEF).“ and “Triple therapy – MTX, HCQ, and SSZ – has been shown to be superior to MTX alone.” can both corroborate the medical assertion RHEUMATOID ARTHRITIS has drug METHOTREXATE.
6. Conclusion and Future Work
In this report we present our method for pairing/matching relations, manually curated and stored in a knowledge graph, with sentences from two point of care books: Ferri’s Clinical Advisor and Conn’s Current Therapy. We used phrase embeddings and boosting of certain keywords (e.g. ”Treatment” in section title vs ”treatment” as predicate of an relation) to improve matches. The similarity between relations and sentences was computed using cosine similarity and evaluated using a goldset, generated by 3 biomedical experts, made up of 250 relation/sentence pairs from each book. It is worth pointing out that consensus was reached between the 3 annotators only in 307 cases and inter- annotator agreement was only moderate. These numbers illustrates the difficulty in automating such mappings - if it is challenging for annotators to agree, it is even more challenging to build a computational method that tries to automate that work. Based on the consensus results, the method described here was precise in assessing the association between a relation and a snippet 61.4% of the time for the 3 tested diseases and a recall of 86.3% assuming that all relations on HG should have a matching snippet in one of the two books.
Another consideration worth making is that the 3 diseases selected for goldset generation were all immune diseases (with the exception of DIABETES MELLITUS TYPE 2, which made up some of the text from each of the books). This may mean that the method described here may not generalize to other disease types or other books, for that matter. A larger goldset would be needed to allow generalization.
There are a few other limitations of the research presented here. Elsevier Healthcare Knowledge Graph (HG) is a proprietary knowledge graph, so far, and the medical textbooks Ferri and Conn are closed domain and have a fixed disease-focused structure. In the future, we would like to expand this research to extract and align snippets from unstructured, open domain, literature, with publicly available knowledge graphs (e.g., UMLS)12. Moreover, we would like to incorporate contextualized word embeddings and graph embedding vectors to improve the performance of our methods. We also plan to explore supervised learning methods to learn the boosting and the similarity functions.
As part of future work, we are in the process of developing a novel application that is powered by HG to retrieve medical relations and corroborating snippets from medical textbooks given a user query (e.g., “Drugs to treat Asthma in a pregnant adult” or Diagnostic procedures for Rheumatoid Arthritis). Given a user query, the concept mapper will be augmented to use some of the defined H-Graph functions (e.g., path(Ct, Cu) or relations(Ct, Rt) as listed in Section 4.2.2) to identify concepts, relation types, as well as shortest paths (composed of semantic relations) between different concepts, in the user query. Given the set of semantic relations and the method described in this paper, we can also present snippets from medical textbooks that corroborate these semantic relations.
Acknowledgments
The authors would like to acknowledge Doug Anderson, David Childs, David Conrad, Cailey Fitzgerald, Dru Henke, Patti Novak, Steve Ross, Connor Skiro, Chris Stoces, Danielle Walsh, for discussions and support on this research.
Figures & Tables
References
- 1.Wolters Kluwer Health. UpToDate. https://www.uptodate.com/home. Accessed: 2019-08-09.
- 2.Elsevier Clinical Key Search Engine. https://www.elsevier.com/solutions/clinicalkey. Accessed: 2019-08-09.
- 3.EBSCO Health. Dynamed. https://www.dynamed.com. Accessed: 2019-06-09.
- 4.Densen Peter. Challenges and opportunities facing medical education. Transactions of the American Clinical and Climatological Association. 122:48–58. [PMC free article] [PubMed] [Google Scholar]
- 5.Ely John W, et al. A taxonomy of generic clinical questions: classification study. BMJ. 2000;321(7258):429–432. doi: 10.1136/bmj.321.7258.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiol Guilherme Del, Workman T Elizabeth, Gorman Paul N. Clinical questions raised by clinicians at the point of care: a systematic review. JAMA internal medicine. 2014;174(5):710–718. doi: 10.1001/jamainternmed.2014.368. [DOI] [PubMed] [Google Scholar]
- 7.Cook David A, et al. Barriers and decisions when answering clinical questions at the point of care: a grounded theory study. JAMA internal medicine. 2013;173(21):1962–1969. doi: 10.1001/jamainternmed.2013.10103. [DOI] [PubMed] [Google Scholar]
- 8.Deus Helena F., Scranton Katie, et al. From benchside to bedside and back again: evidence-based content drives the future of precision medicine. The Journal of Precision Medicine. 2019;3 [Google Scholar]
- 9.Singhal Amit. Introducing the knowledge graph: things, not strings. http://bit.ly/31Ygaie , May 2012 Accessed on 10/15/2019.
- 10.Zitnik Marinka, Agrawal Monica, Leskovec Jure. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018;34(13):i457–i466. doi: 10.1093/bioinformatics/bty294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deus Helena. Big semantic data processing in the life sciences domain. 2018 [Google Scholar]
- 12.Kamdar Maulik R, Ferna´ndez Javier D, Polleres Axel, Tudorache Tania, Musen Mark A. Enabling web-scale data integration in biomedicine through linked open data. NPJ digital medicine. 2019;2(1):1–14. doi: 10.1038/s41746-019-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SAPPHIRE (health care) - Wikipedia. https://en.wikipedia.org/wiki/SAPPHIRE_(Health_care) . (Accessed on 08/15/2019)
- 14.Tenenbaum Jessica D., et al. The biomedical resource ontology (BRO) to enable resource discovery in clinical and translational research. Journal of biomedical informatics. 2011;44(1):137–145. doi: 10.1016/j.jbi.2010.10.003. Feb. 20955817[pmid] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luciano Joanne S., et al. The translational medicine ontology and knowledge base: driving personalized medicine by bridging the gap between bench and bedside. Journal of Biomedical Semantics. 2011;2(2):S1. doi: 10.1186/2041-1480-2-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamdar Maulik R, Musen Mark A. PhLeGrA: Graph analytics in pharmacology over the web of life sciences linked open data; Proceedings of the 26th International Conference on World Wide Web; 2017. pp. 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamdar Maulik R, Zeginis Dimitris, et al. ReVeaLD: A user-driven domain-specific interactive search platform for biomedical research. Journal of biomedical informatics. 2014;47:112–130. doi: 10.1016/j.jbi.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Arora Sanjeev, Liang Yingyu, Ma Tengyu. A simple but tough-to-beat baseline for sentence embeddings; International Conference on Learning Representations; 2017. [Google Scholar]
- 19.Niu Yun, Hirst Graeme. Analysis of semantic classes in medical text for question answering; Proceedings of the Conference on Question Answering in Restricted Domains; 2004. pp. 54–61. [Google Scholar]
- 20.Nikfarjam Azadeh, Sarker Abeed, et al. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. Journal of the American Medical Informatics Association. 2015;22(3):671–681. doi: 10.1093/jamia/ocu041. 03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonc¸alves Rafael S, Kamdar Maulik R, Musen Mark A. European Semantic Web Conference. Springer; 2019. Aligning biomedical metadata with ontologies using clustering and embeddings; pp. 146–161. [Google Scholar]
- 22.Percha Bethany, Altman Russ B. A global network of biomedical relationships derived from text. Bioinformatics. 2018;34(15):2614–2624. doi: 10.1093/bioinformatics/bty114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeJong Alex, et al. Elsevier’s healthcare knowledge graph and the case for enterprise level linked data standards; Proceedings of the ISWC 2018 Posters & Demonstrations, Industry and Blue Sky Ideas Tracks co-located with 17th International Semantic Web Conference (ISWC 2018); Monterey, USA; 2018. October 8th - to - 12th, 2018. [Google Scholar]
- 24.Kamdar Maulik R. A web-based integration framework over heterogeneous biomedical data and knowledge sources. PhD thesis, Stanford University; 2019. https://purl.stanford.edu/jr863br2478 . [Google Scholar]
- 25.Pennington Jeffrey, et al. Glove: Global vectors for word representation; Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP); 2014. pp. 1532–1543. [Google Scholar]
- 26.Kamdar Maulik. Biomedical Word Vectors. 8 2019. https://figshare.com/articles/Biomedical_Word_Vectors/9598760 .
- 27.Honnibal Matthew, Montani Ines. spacy 2: Natural language understanding with bloom embeddings, convolutional neural networks and incremental parsing. To appear. 7 2017. https://spacy.io/
- 28.US National Library of Medicine. MEDLINE. https://www.nlm.nih.gov/bsd/medline.html. Accessed: 2019-06-09. [DOI] [PubMed]
- 29.Fleiss Joseph L. Measuring nominal scale agreement among many raters. Psychological bulletin. 1971;76(5):378. [Google Scholar]