Abstract
We present sig2db as an open-source solution for clinical data warehouses desiring to process natural language from prescription instructions, often referred to as “sigs”. In electronic prescribing, the sig is typically an unstructured text field intended to capture all requirements for medication administration. The sig captures certain fields that the structured data may lack such as days supply, time of day, or meal-time considerations. Our open-source software package facilitates the workflow needed to process sigs into a structured format usable by clinical data warehouses. Our solution focuses on extracting concepts from prescriptions in order to understand the intended semantics by leveraging known natural language processing tools. We demonstrate the utility of concept extraction from sigs and present our findings in processing 1023 unique sigs from 5.7 million unique prescriptions.
Introduction
Although the adoption rate for electronic prescribing has continued to rise in response to several federal legislative actions and implementations of incentive programs1, safety issues persist2, 3. One such issue is the possible disagreement between structured and unstructured prescription data3. Each electric health record (EHR) implementation of electronic prescribing varies in terms of what is available as structured data; sigs are typically implemented as unstructured free-text data. “Sig” is short for the Latin, signa (“write”) or signetur (“let it be written”); these are usually imperative sentences such as “Take one tablet daily.”
Medications have long been the subject of natural language processing research. The Third i2b2 Workshop on Natural Language Processing Challenges for Clinical Records4 focused on extracting medication information from patient discharge summaries and yielded many solutions capable of extracting drug information such as name, dose, and administrative route5–9. Medications were also the subject of the 2018 n2c2 Shared-Task and Workshop which yielded several results related to adverse drug events and medication information within EHRs10–12. Several solutions exist for using standardized vocabularies to represent medication information, such as drug name and drug classes13–15
Medication sigs and natural language processing (NLP) research have also intersected; a 2019 study analyzed drug indications and found that only 7.41% of their study’s 4.3 million prescriptions contained drug indications and of that minority, 30.35% were indicated for pain16. Sigs have also been used to calculate the morphine milligram equivalent daily dose (MEDD) for opioid prescriptions using both structured sigs and unstructured sigs through regular expressions and string matching17. The challenges of working with medication, prescription, and free-text sig data include the unpredictability of free-text17, 18; this can be challenging for solutions based on parsing and regular expressions and motivate the development of a robust solution capable of extracting relevant semantic information.
The outpatient EHR at our campus medical center, University of Kentucky Healthcare (UKHC), does not have a structured sig field; it is entirely free text and is required in order to submit an electronic prescription. Table 1 shows the top ten most common sigs in our UKHC data (2012 to 2019) and immediately demonstrates that there are several lexicographically unique ways to represent semantically equivalent sigs. Punctuation is the only difference between pairs ranked (1,4) and pairs ranked (3,6). Pairs ranked (8,9) differ by the usage of the phrases “every day” versus “once daily”. The phrase “by mouth” is sometimes specified; the others are implied to be an oral medication by use of the word tablet. The use of the integer “1” and the word “one” also varies.
Table 1:
The most popular free text sig fields in UKHC data (outpatient prescriptions); redundancies due to punctuation differences are included.
| Rank | Sig | Frequency |
|---|---|---|
| 1 | take 1 tablet daily. | 857,803 |
| 2 | take 1 tablet daily as directed | 220,765 |
| 3 | take 1 tablet twice daily. | 209,078 |
| 4 | take 1 tablet daily | 208,101 |
| 5 | take 1 tablet at bedtime. | 141,869 |
| 6 | take 1 tablet twice daily | 121,211 |
| 7 | take 1 capsule daily. | 103,654 |
| 8 | take 1 tablet by mouth once daily | 87,324 |
| 9 | take 1 tablet by mouth every day | 80,010 |
| 10 | take one tablet by mouth daily | 64,115 |
We will detail our workflow for extracting semantics from sig fields and demonstrate that semantically equivalent sigs can be grouped together for analysis. We also explore whether concept extraction causes information loss by testing if extracted concepts can successfully represent semantically equivalent sigs.
Methods
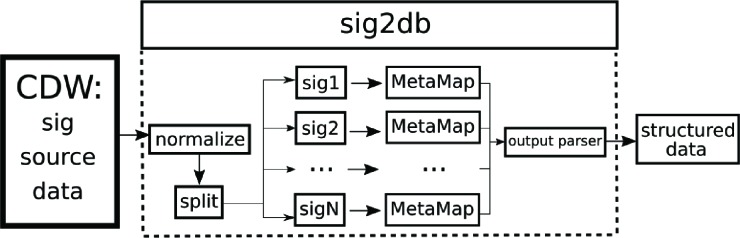
Our workflow for processing unstructured sig data is described by Figure 1. We stream source data from our clinical data warehouse and immediately normalize the raw texts and split the results into separate files in preparation for processing. For natural language processing, we use Metamap19 and in particular its ability to leverage the UMLS20 and map text to the known vocabularies 21, 22. The parameters to Metamap are configurable; we chose to default to fielded MetaMap indexing (MMI) output to assist with output parsing and we also enabled word sense disambiguation. Word sense disambiguation will resolve ambiguities within the text and select the best mapping available from candidate matches; this yields one concept per mention within the text22.
Figure 1:
A high-level overview of sig2db’s workflow
We then parse the output of MetaMap into a structured format designed for easy integration into the clinical data warehouse. An identifier uniquely identifying the sig is propagated through the process in order to link the structured results back to the source data. Although we have chosen MetaMap for our local implementation of sig2db, a call to any binary process from a different NLP tool would be able to be swapped in if it is accompanied by the needed output parser. The software is intended to help coordinate the workflow needed to convert sigs from a clinical data warehouse into structured data suitable for supplementing source data. Our code is open-source and available online23. We include database definitions for tables to which output from sig2db can be directed. These tables store a list of unique sigs with their equivalent normalized version and a list of concepts extracted from the sigs.
In Table 2, we present a selection of results from the twenty most popular sigs found within our data. Our process creates a one-to-many relationship: one sig may contain many concepts. From the table, the trigger is the text that triggered MetaMap in identifying this concept. The P.O.S. is the part of speech identified for this text. The Pref. Concept Name is the UMLS Preferred Concept Name for the concept mapped to this text. The UMLS Semantic Type is the semantic type associated with the concept. Most sigs contain at least a temporal concept and a quantitative concept. The most popular semantic types are listed in Table 3 with their corresponding frequency.
Table 2:
Example sigs and their corresponding extracted structured data
| Orignal Sig | Trigger | P.O.S. | Pref. Concept Name | UMLS Semantic Type |
|---|---|---|---|---|
| take 1 tablet daily. | take | verb | Take | Health Care Activity |
| take 1 tablet daily. | one | noun | One | Quantitative Concept |
| take 1 tablet daily. | tablet | noun | Tablet Dosage Form | Biomedical Material |
| take 1 tablet daily. | daily | adverb | Daily | Temporal Concept |
| take 1 tablet twice daily. | take | verb | Take | Health Care Activity |
| take 1 tablet twice daily. | one | noun | One | Quantitative Concept |
| take 1 tablet twice daily. | tablet | noun | Tablet Dosage Form | Biomedical Material |
| take 1 tablet twice daily. | twice daily | adverb | Twice a day | Temporal Concept |
| take 1 tablet at bedtime. | take | verb | Take | Health Care Activity |
| take 1 tablet at bedtime. | one | noun | One | Quantitative Concept |
| take 1 tablet at bedtime. | tablet | noun | Tablet Dosage Form | Biomedical Material |
| take 1 tablet at bedtime. | bedtime | noun | Bedtime (qualifier) | Temporal Concept |
| take 1 tablet by mouth once daily | take | verb | Take | Health Care Activity |
| take 1 tablet by mouth once daily | one | noun | One | Quantitative Concept |
| take 1 tablet by mouth once daily | tablet mouth | noun | Oral Tablet | Biomedical Material |
| take 1 tablet by mouth once daily | daily | adj. | Once daily | Temporal Concept |
| use two sprays in each nostril once daily | use | verb | Utilization Qualifier | Functional Concept |
| use two sprays in each nostril once daily | two | adj. | Two | Quantitative Concept |
| use two sprays in each nostril once daily | sprays nostril | noun | Sprays per Nostril | Quantitative Concept |
| use two sprays in each nostril once daily | once daily | adj. | Once daily | Temporal Concept |
| insert 1 suppository rectally at bedtime. | insert | verb | Insert | Health Care Activity |
| insert 1 suppository rectally at bedtime. | one | noun | One | Quantitative Concept |
| insert 1 suppository rectally at bedtime. | suppository | noun | Suppository | Biomedical Material |
| insert 1 suppository rectally at bedtime. | bedtime | noun | Bedtime (qualifier) | Temporal Concept |
Table 3:
Frequency of the most popular semantic types
| Rank | Semantic Type | Count | Occurs in Distinct Sigs | Percent of Sigs |
|---|---|---|---|---|
| 1 | Quantitative Concept | 1543 | 928 | 90.7 |
| 2 | Temporal Concept | 1124 | 923 | 90.2 |
| 3 | Biomedical or Dental Material | 675 | 669 | 65.7 |
| 4 | Health Care Activity | 673 | 672 | 65.4 |
| 5 | Functional Concept | 276 | 239 | 23.3 |
There are additional fields not listed in Table 2 due to space constraints: the concept unique identifier (UMLS CUI), a MetaMap relevance score, starting position of the text, length of the text, and any linked MeSH (Medical Subject Headings) tree codes24. The CUI field helps uniquely identify the concepts identified within the sigs; positional information helps disambiguate order of concepts identified.
Preprocessing is known to impact natural language processing25. Our normalization process sanitizes the text by ensuring consistent capitalization, removes punctuation, and converts integers to their word equivalent. For example, the sentence, “take 1 tablet daily”, is converted to “take one tablet daily”. For this sentence, the 1 gets assigned the semantic type of classification without normalization, but one gets assigned the semantic type of quantitative concept after normalization and better represents the intended semantics of the sig. Our initial normalization process stripped out all punctuation and initial probes of the data revealed that decimal points were erroneously being stripped from numbers and hyphens were erroneously being stripped from numerical ranges. For this reason, we skip normalization of decimal numbers and ranges; sigs containing phrases such as “2-3 times” get mapped appropriately to a concept representing “2-3 times”.
Results
We tested our workflow by processing any sig from our outpatient EHR that had more than 1000 prescriptions associated with it. In total, we processed 1023 sigs corresponding to 5.7 million prescriptions; this accounted for 30.5% of all prescriptions in our EHR. These 5.7 million prescriptions originated from 1.5 million outpatient visits. We extracted 5170 concepts from the 1023 sigs using sig2db; only 399 of these concepts were distinct. Table 4 shows the top ten most frequent concepts by reporting the raw count of concepts yielded and the count of distinct sigs from which these concepts were generated.
Table 4:
Frequency of the most popular concepts
| Rank | Semantic Type | Concept Preferred Name | Count | Percent of Sigs |
|---|---|---|---|---|
| 1 | Health Care Activity | Take | 670 | 65.5 |
| 2 | Quantitative Concept | One | 567 | 55.4 |
| 3 | Biomedical or Dental Material | Tablet Dosage Form | 313 | 30.5 |
| 4 | Temporal Concept | Daily | 253 | 24.7 |
| 5 | Biomedical or Dental Material | Oral Tablet | 177 | 17.3 |
| 6 | Temporal Concept | Twice a day | 154 | 15.1 |
| 7 | Temporal Concept | Hour | 146 | 14.3 |
| 8 | Quantitative Concept | Two | 137 | 13.4 |
| 9 | Quantitative Concept | Four | 118 | 11.5 |
| 10 | Temporal Concept | Day | 112 | 10.9 |
Since sigs are imperative sentences, the most common concept extracted was Take, occurring in 65.5% of our sigs. The most common quantitative concept was One. These trends suggest a large percentage of the sigs are simply instructing patients to take one of something. The most common biomedical or dental material concept was Tablet Dosage Form, followed by Oral Tablet, and together account for almost half of our sigs. Daily and Twice a day were the most common temporal concepts, occuring in 38.1% of our sigs.
Table 3 shows the top five most frequent semantic types by reporting the raw count of types yielded. One sig can contain multiple concepts of the same type; consequently, we also report the count of distinct sigs from which these types were generated and percentage of sigs this represents. 9.3% of the sigs did not contain a quantitative concept. Valid examples of this include, “apply sparingly to affected areas twice daily”, “inject as directed for severe hypoglycemia”, and “take as directed”. For similar reasons, 9.8% of the sigs did not contain a temporal concept, in part due to variations of “take as directed”. Some sigs appear to have omitted the temporal constraint by mistake: “take one tablet twice”, “inject one ml intramuscular”, and so on.
“As directed” is a special case which does not convey specific semantic information about the intended temporal and quantitative constraints. Variations of this sig include “as directed”, “take as directed”, “take as directed per package instructions.”, “take as directed on patient instruction card.”, and “use as directed on package”. For these medications, the structured data accompanying the sig is overwhelmingly blank, having 66.4% of the fields, such as day supply, blank. Given this, if a “directed” concept represents the sig, it implies we should not expect much value of the structured data.
We immediately noticed that the mapping for sigs containing “as directed” were consistently incorrect. “As directed” gets mapped to a concept with a preferred name of “Reproductive Human Cells, Tissues, and Cellular and Tissue-Based Products from Known Donor to Directed Recipient” and a semantic type of “Pharmacologic Substance” which are both obviously incorrect. This is a failure of the word-sense disambiguation component, which attempts to identify the best matched concept; a better mapping with the preferred name of “Direct” and a semantic type of “Qualitative Concept” is scored slightly less than the erroneous mapping.
MetaMap can block unwanted mappings as a configuration option and because our unwanted mappings are consistent, we are free to remove the erroneous concept from consideration. The existence of an obviously unwanted mapping motivated a manual review of the 50 most popular sigs. These 50 are only 4.89% of processed sigs but they correspond to 3.4 million prescriptions (59.6%).
Information Loss
There is a risk of losing information when converting a free-text sig into structured data; specifically, if text is matched to an incorrect concept, the intended semantics of the sig may not be accurately represented. We asked a healthcare data analyst familiar with the electronic prescription system to write a sig given a set of extracted concepts produced by sig2db. The analyst-generated sig was compared to the original sig and assessed if it captured the same semantic information. The exact phrasing of the analyst-generated sig and the original sig did not have to match; only the semantics mattered in judging equivalence. The comparison is straight-forward due to the short nature of sigs. Consequently, another bad mapping was discovered in four of the top 50 (8%) and is detailed in Table 5. Out of the 1023 sigs, 73 (7.1%) contained this erroneous concept.
Table 5:
Examples of consistently incorrect mapping yielding “One time” concept
| Orignal Sig | Trigger | Pref. Concept Name | UMLS Semantic Type |
|---|---|---|---|
| take 1 tablet 3 times daily. | take | Take | Health Care Activity |
| take 1 tablet 3 times daily. | one times | One time | Intellectual Product |
| take 1 tablet 3 times daily. | tablet | Tablet Dosage Form | Biomedical Material |
| take 1 tablet 3 times daily. | three | Three | Quantitative Concept |
| take 1 tablet 3 times daily. | daily | Daily | Temporal Concept |
| take 1 tablet 3 times daily as needed. | take | Take | Health Care Activity |
| take 1 tablet 3 times daily as needed. | one times | One time | Intellectual Product |
| take 1 tablet 3 times daily as needed. | tablet | Tablet Dosage Form | Biomedical Material |
| take 1 tablet 3 times daily as needed. | three | Three | Quantitative Concept |
| take 1 tablet 3 times daily as needed. | daily as needed | Daily as Required | Temporal Concept |
| take 1 capsule 3 times daily | take | Take | Health Care Activity |
| take 1 capsule 3 times daily | one times | One time | Intellectual Product |
| take 1 capsule 3 times daily | capsule | Capsule (Pharmacologic) | Biomedical Material |
| take 1 capsule 3 times daily | three | Three | Quantitative Concept |
| take 1 capsule 3 times daily | daily | Daily | Temporal Concept |
| take one tablet four times daily | take | Take | Health Care Activity |
| take one tablet four times daily | one times | One time | Intellectual Product |
| take one tablet four times daily | tablet | Tablet Dosage Form | Biomedical Material |
| take one tablet four times daily | four | Four | Quantitative Concept |
| take one tablet four times daily | daily | Daily | Temporal Concept |
| take one tablet by mouth four times a day | take | Take | Health Care Activity |
| take one tablet by mouth four times a day | one times | One time | Intellectual Product |
| take one tablet by mouth four times a day | tablet mouth | Oral Tablet | Biomedical Material |
| take one tablet by mouth four times a day | four | Four | Quantitative Concept |
| take one tablet by mouth four times a day | day | Day | Temporal Concept |
All sigs from Table 5 exhibit the same issue: the trigger is “one times” and is mapped to a concept with a semantic type of “Intellectual Product”. MetaMap selected this over the concept “One”, which was the second most popular concept in our results, because the intellectual product concept scored better. MetaMap allows filtering by semantic type and because “intellectual product” is not relevant for sigs, we can safely exclude it from consideration.
Discussion
Our workflow transforms free-text sigs into structured data which can readily be used by clinical data warehouses. By focusing on extraction of concepts, we enable semantic analysis and unlock data that would otherwise be unavailable in our EHR. Our results indicate that the over-whelming majority of sigs contain temporal and quantitative concepts which assist in understanding the intended prescription instructions.
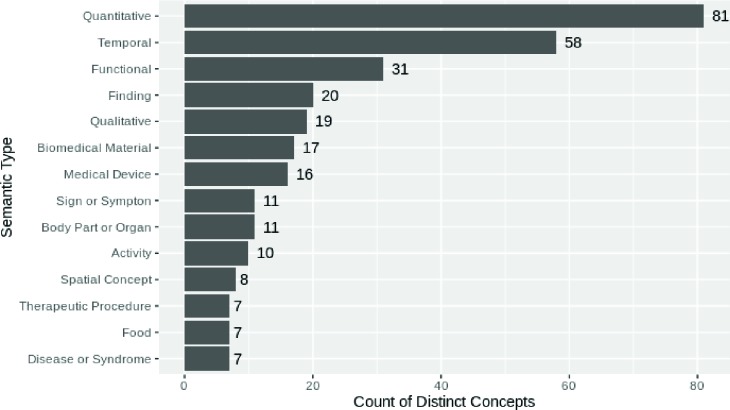
Converting free-text into concepts reduces the complexity of interpretation of text by condensing a high-volume, high-variety data set into one with manageable cell sizes should one need to manually map concepts to a known truth. For example, quantitative and temporal concepts could be mapped to canonical values where concepts such as “daily” has a value of 1, “twice daily” a value of 2, “weekly” has a value of 1/7, and so on. In conjunction with mapping quantitative concepts such as “One” to 1, “Two” to 2, and so on, this supports logic to determine quantity per day of medications. The relatively small frequencies shown in Figure 2 per each type of semantic type show that creating such mappings is a reasonable task.
Figure 2:
Semantic types and how many distinct concepts each type
One of our motivations for initiating this project was to generate data not found in our clinical data warehouse. Time of day for medication administration is not available in our outpatient EHR’s structured data. Table 6 shows the frequency of “time of day” concepts within the 1023 sigs processed and frequency within actual prescriptions from our outpatient EHR. Of those 1023, 76 matched the “Bedtime (qualifier value)” concept; the 433,409 prescriptions for bedtime medications correspond to 2,456 distinct drugs which could now be studied or reported upon according to administration time.
Table 6:
Frequency for concepts with desired time of day for medication administrationt
| Rank | Concept Preferred Name | Sig Total | Prescription Total |
|---|---|---|---|
| 1 | Bedtime (qualifier value) | 76 | 433,409 |
| 2 | Morning | 21 | 97,711 |
| 3 | Night time | 8 | 24,189 |
| 4 | Evening | 9 | 14,930 |
| 5 | Evening meal | 3 | 11,437 |
| 6 | Daily before breakfast | 3 | 6,125 |
| 7 | Once a day, at bedtime | 2 | 2,747 |
| 8 | Daily with breakfast | 1 | 2,540 |
| 9 | Late | 2 | 2,037 |
| 10 | Afternoon | 1 | 894 |
| 11 | Every morning | 1 | 892 |
Food considerations are also possible to extract with the semantic type of “food”. Table 7 shows the frequency of food-based concepts and their frequency within the sig and prescription data.
Table 7:
Frequency for concepts with food considerations
| Rank | Concept Preferred Name | Sig Total | Prescription Total |
|---|---|---|---|
| 1 | Food | 24 | 67,965 |
| 2 | Drink(dietarysubstance) | 9 | 24,332 |
| 3 | Tea | 1 | 9,233 |
| 4 | Juice | 1 | 9,233 |
| 5 | Soft food | 1 | 1,672 |
| 6 | Beverages | 1 | 591 |
| 7 | Snacks | 1 | 457 |
The existence of sigs with flexibility and ranges in the instructions complicates calculating quantity per day. For example, “apply 2-3 times per day” allows for either two doses or three doses. A maximum quantitative value would represent the dosage ceiling; a minimum would represent the smallest amount taken.
A previous study found that only 7.41% of prescriptions had drug indications and of that minority, 30.35% were indicated for pain 16. The semantic nature of our solution allows us to explore semantic types as a surrogate pointer to medication indications; reasonable semantic types for concepts that might be indications include signs/symptoms, findings, and diseases/syndromes. For concepts with signs/symptoms as their semantic type, pain was only represented in 13% of the sigs. For concepts with finding as their semantic type, pain accounted for 41.5% of records. Table 8 shows an example sig with pain as an indication and the resulting concepts extracted with sig2db. In this example, pain is simply a stand-alone concept and the only context for a finding of pain within a prescription sig would be for an indication.
Table 8:
Example of a sig having pain as an indication
| Orignal Sig | Trigger | Pref. Concept Name | UMLS Semantic Type |
|---|---|---|---|
| take one tablet every six hours as needed for pain. | take | Take | Health Care Activity |
| take one tablet every six hours as needed for pain. | one | One | Quantitative Concept |
| take one tablet every six hours as needed for pain. | tablet | Tablet Dosage Form | Biomedical Material |
| take one tablet every six hours as needed for pain. | six | Six | Quantitative Concept |
| take one tablet every six hours as needed for pain. | hours | Hour | Temporal Concept |
| take one tablet every six hours as needed for pain. | as needed | As Needed | Qualitative Concept |
| take one tablet every six hours as needed for pain. | pain | Pain, NOS | Finding |
We are not limited to studying indications for pain. Table 9 lists concepts extracted with a semantic type of finding, sign or symptom, and disease or syndrome; most of the findings are indications with the notable exceptions of “Severe (severity modifier)” and “Does chew”. These are findings about other mentions in the sig, such as a chewable pill or a severe headache.
Table 9:
Frequency for concepts with a semantic type of finding
| Rank | Semantic Type | Concept Preferred Name | Sig Total | Prescription Total |
| 1 | Finding | Pain NOS | 33 | 130,616 |
| 2 | Finding | Nausea | 10 | 22,911 |
| 3 | Finding | Does chew (finding) | 5 | 21,493 |
| 4 | Finding | Blood pressure finding | 8 | 18,720 |
| 5 | Finding | Cough | 6 | 11,026 |
| 6 | Finding | Severe (severity modifier) | 2 | 8,443 |
| 7 | Finding | Insomnia | 2 | 5,532 |
| 8 | Finding | Constipation | 3 | 4,122 |
| 9 | Finding | Vomiting | 2 | 2,546 |
| 10 | Finding | Soft stool (finding) | 1 | 1,236 |
| 1 | Sign or Symptom | Wheezing | 4 | 9,740 |
| 2 | Sign or Symptom | Spasm | 3 | 6,084 |
| 3 | Sign or Symptom | Chest Pain | 2 | 6,031 |
| 4 | Sign or Symptom | Headache | 3 | 5,052 |
| 5 | Sign or Symptom | Signs and Symptoms, Respiratory | 2 | 3,776 |
| 6 | Sign or Symptom | Breakthrough Pain | 1 | 3,031 |
| 7 | Sign or Symptom | Respiratory distress | 1 | 1,011 |
| 8 | Sign or Symptom | Severe diarrhea | 1 | 829 |
| 9 | Sign or Symptom | Labored breathing | 1 | 575 |
| 1 | Disease or Syndrome | Migraine Disorders | 5 | 11,449 |
| 2 | Disease or Syndrome | Hypoglycemia | 2 | 8,443 |
| 3 | Disease or Syndrome | Hyperglycemia | 2 | 7,445 |
| 4 | Disease or Syndrome | Asthma | 1 | 1,086 |
| 5 | Disease or Syndrome | Diaper Rash | 1 | 648 |
| 6 | Disease or Syndrome | Malnutrition | 1 | 90 |
Concepts for “as needed” or “PRN” medications are also extracted as temporal concepts. All of these examples demonstrate that additional data can be extracted from prescription sigs. These concepts all belong to standardized vocabularies and could act as a supplemental data source to the original prescription data. Additionally, the concepts allow for grouping individuals with prescriptions with semantically similar sigs for comparative analysis; this would not be possible with the unprocessed free-text sig values.
Conclusion
We presented our open-source sig2db software for processing natural language prescription sig data. The end result is a structured data set intended to complement existing data within clinical data warehouses; this extracted semantic data can be used for reporting, research, or quality assurance. The concepts are associated with semantic types. Semantic types, such as temporal concepts or quantitative concepts, can be used to figure out quantity per unit of time. Time of day, such as morning or bedtime, can be determined by the extracted concepts. Other semantic types, such as findings, signs or symptoms, and disease or syndromes, can be used to understand the context of the medication administration such as the intended treatment indication. Food considerations, such as “take with food,” can be determined by analyzing concepts with a semantic type of food.
We described in detail the challenges of processing sigs due to their entirely free-text nature. Bad mappings can occur; if known in advance or discovered through output analyses, these bad mappings are easily avoidable by banning the consideration of troublesome concepts and matches. Our solution leveraged Metamap to extract concepts, but other concept extraction tools exist and can be plugged into our solution. An output parser for the chosen tool would need to be added before integrating results into the clinical data warehouse.
We plan to review a larger selection of sigs in order to measure information loss and to gain knowledge of any additional sub-optimal mappings. We also plan to develop algorithms capable of leveraging extracted concepts to estimate intended medication quantity per day. Extracted concepts belong to standardized vocabularies included in the UMLS; this gives us structural information, such as cross-concept relationships, which we plan to explore as future work with the goal of supplementing prescription data. Sigs are free-text fields and manifest in a variety of ways; our extraction from sigs will support merging semantically equivalent sigs together and consequently grouping patients together according to similarity of prescriptions. The semantic output of sig2db and the easy integration into clinical data warehouses will enable clinical research with sig data.
Acknowledgment
The project described was supported by the NIH National Center for Advancing Translational Sciences through grant number UL1TR001998. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Figures & Tables
References
- 1.Joseph SB, Sow MJ, Furukawa MF, Posnack S, Daniel JG. E-prescribing adoption and use increased substantially following the start of a federal incentive program. Health Affairs. 2013;32(7):1221–1227. doi: 10.1377/hlthaff.2012.1197. [DOI] [PubMed] [Google Scholar]
- 2.Schiff GD, Hickman TTT, Volk LA, Bates DW, Wright A. Computerised prescribing for safer medication ordering: still a work in progress. BMJ Qual Saf. 2016;25(5):315–319. doi: 10.1136/bmjqs-2015-004677. [DOI] [PubMed] [Google Scholar]
- 3.Lanham AE, Cochran GL, Klepser DG. Electronic prescriptions: opportunities and challenges for the patient and pharmacist. Advanced Health Care Technologies. 2016;2:1. [Google Scholar]
- 4.Uzuner O¨, Solti I, Cadag E. Extracting medication information from clinical text. Journal of the American Medical Informatics Association. 2010;17(5):514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. Journal of the American Medical Informatics Association. 2010;17(5):524–527. doi: 10.1136/jamia.2010.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dele´ger L, Grouin C, Zweigenbaum P. Extracting medical information from narrative patient records: the case of medication-related information. Journal of the American Medical Informatics Association. 2010;17(5):555–558. doi: 10.1136/jamia.2010.003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spasic´ I, Sarafraz F, Keane JA, Nenadic´ G. Medication information extraction with linguistic pattern matching and semantic rules. Journal of the American Medical Informatics Association. 2010;17(5):532–535. doi: 10.1136/jamia.2010.003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doan S, Bastarache L, Klimkowski S, Denny JC, Xu H. Integrating existing natural language processing tools for medication extraction from discharge summaries. Journal of the American Medical Informatics Association. 2010;17(5):528–531. doi: 10.1136/jamia.2010.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meystre SM, Thibault J, Shen S, Hurdle JF, South BR. Textractor: a hybrid system for medications and reason for their prescription extraction from clinical text documents. Journal of the American Medical Informatics Association. 2010;17(5):559–562. doi: 10.1136/jamia.2010.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christopoulou F, Tran TT, Sahu SK, Miwa M, Ananiadou S. Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. Journal of the American Medical Informatics Association. 2019 doi: 10.1093/jamia/ocz101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y, Meystre SM. Ensemble method–based extraction of medication and related information from clinical texts. Journal of the American Medical Informatics Association. 2019 doi: 10.1093/jamia/ocz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju M, Nguyen NT, Miwa M, Ananiadou S. An ensemble of neural models for nested adverse drug events and medication extraction with subwords. Journal of the American Medical Informatics Association. 2019 doi: 10.1093/jamia/ocz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak J, Murphy SP, Willaert BN, Kremers HM, Yawn BP, Rocca WA, et al. Using RxNorm and NDF-RT to classify medication data extracted from electronic health records: experiences from the Rochester Epidemiology Project; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2011. p. 1089. [PMC free article] [PubMed] [Google Scholar]
- 14.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. Journal of the American Medical Informatics Association. 2014;21(5):858–865. doi: 10.1136/amiajnl-2013-002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salazar A, Karmiy SJ, Forsythe KJ, Amato MG, Wright A, Lai KH, et al. How often do prescribers include indications in drug orders? Analysis of 4 million outpatient prescriptions. American Journal of Health-System Pharmacy. 2019;76(13):970–979. doi: 10.1093/ajhp/zxz082. [DOI] [PubMed] [Google Scholar]
- 17.Goud A, Kiefer E, Keller MS, Truong L, SooHoo S, Riggs RV. Calculating maximum morphine equivalent daily dose from prescription directions for use in the electronic health record: a case report. JAMIA Open. 2019 doi: 10.1093/jamiaopen/ooz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajeevan N, Niehoff KM, Charpentier P, Levin FL, Justice A, Brandt CA, et al. Utilizing patient data from the veterans administration electronic health record to support web-based clinical decision support: informatics challenges and issues from three clinical domains. BMC medical informatics and decision making. 2017;17(1):111. doi: 10.1186/s12911-017-0501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proceedings of the AMIA Symposium; American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aronson AR. Bethesda, MD: NLM, NIH, DHHS; 2006. Metamap: Mapping text to the umls metathesaurus; pp. 1–26. [Google Scholar]
- 22.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.sig2db. Bitbucket.org. 2019. Available from: https://bitbucket.org/_harris/sig2db/
- 24.Nelson SJ, Johnston WD, Humphreys BL. Relationships in the Organization of Knowledge. Springer; 2001. Relationships in medical subject headings (MeSH) pp. 171–184. [Google Scholar]
- 25.Uysal AK, Gunal S. The impact of preprocessing on text classification. Information Processing & Management. 2014;50(1):104–112. [Google Scholar]