Abstract
Endometriosis is a complex and high impact disease affecting 176 million women worldwide with diagnostic latency between 4 to 11 years due to lack of a definitive clinical symptom or a minimally invasive diagnostic method. In this study, we developed a new ensemble machine learning classifier based on chromosomal partitioning, named GenomeForest and applied it in classifying the endometriosis vs. the control patients using 38 RNA-seq and 80 enrichment-based DNA-methylation (MBD-seq) datasets, and computed performance assessment with six different experiments. The ensemble machine learning models provided an avenue for identifying several candidate biomarker genes with a very high F1 score; a near perfect F1 score (0.968) for the transcriptomics dataset and a very high F1 score (0.918) for the methylomics dataset. We hope in the future a less invasive biopsy can be used to diagnose endometriosis using the findings from such ensemble machine learning classifiers, as demonstrated in this study
Introduction
Endometriosis is a complex yet common gynecological disorder of reproductive-aged women. It is characterized by the presence of endometrial tissue outside of the uterine cavity. Endometriosis is a high impact disease, commonly associated with chronic pelvic pain and infertility. Therefore, it significantly impairs mental and physical quality of patient’s life and their work performance is seriously compromised. About 176 million women worldwide are suffering from endometriosis and about 8.5 million women solely in the North Americ1. Endometriosis affects reproductive aged women (5-10%), women with subfertility (20-30%), and women with chronic pelvic pain and infertility (40-60%)2. About 70% of teens who are suffering from pelvic pain are later diagnosed with endometriosi3. In the U.S., endometriosis is a leading cause of hysterectomies (approximately 600,000 cases) performed every yea4. The total cost (direct and indirect) of endometriosis has been estimated at €30 billion in Europe and $22 billion in the U.S. each year, and direct costs have increased graduall5.
Though laparoscopy is currently the gold standard diagnostic approach for endometriosi6, it is an invasive procedure and may not be appropriate for all women with a history and physical examination indicative of endometriosis. There are many studies that assessed the diagnostic value of biomarkers for endometriosis in endometrial tissue, menstrual or uterine fluids and immunologic markers in blood or urine for clinical use as a diagnostic test for endometriosis; however, no reliable biomarkers were recommende7. Due to the lack of reliable recommended biomarkers, the current diagnostic latency is on average 4 to 11 year8. To reduce the sufferings and expenses related to the disease, an early intervention is essential. Studies have shown that endometriosis patients have an altered methylome and transcriptome, which could lead to the identification of biomarkers for developing a minimally invasive diagnostic technique for endometriosi9.
Applications of machine learning methods on microarray expression data or next generation sequencing data have been advanced over the last several decades for discovery of biological pattern10. For microarray expression data, use of both supervised and unsupervised machine learning methods have shown great succes11, including the application of: (a) clustering techniques such as hierarchical clustering and K-means clustering for identifying the groups of genes that share similar functions or expression12, and (b) disease vs. healthy classification tasks using various methods such as Decision Trees, Random Forests, Artificial Neural Networks (ANN), Support Vector Machines (SVM) and Bayesian Network13. Although the application of machine learning classifiers on transcriptomics or methylomics data had limited succes14,15, the classification difference of gene expressions in transcriptomics data or the difference of DNA-methylation in methylomics data between disease vs. healthy can provide avenues for the development of endometriosis diagnostic metho9,16.
In our previous works, we have successfully demonstrated the application of various machine learning techniques for classifying the endometriosis patients vs. the control patients using both transcriptomics and methylomics dat17,18. In this work, we describe our new ensemble technique called GenomeForest based on chromosomal partitioning. We systematically examined how well the newly developed ensemble technique perform in classifying endometriosis and control samples using both transcriptomics and methylomics data. The assessment was done from three different perspectives: (a) evaluation of classification performances of the GenomeForest ensemble classifier, (b) implication of three different normalization techniques, and (c) implication of differential analysis. The results were compared with the results from our prior work on the application of various machine learning techniques on the same dataset published elsewher17,18.
Methods
Subjects and Tissue Collection
The women participated in this study were aged between 18-49 years and all undergoing a laparoscopy procedure, either diagnostic laparoscopy for pain or infertility or seeking laparoscopic sterilization. Prior to surgery, the physician obtained informed consent following the IRB protocol. Endometrial biopsies were collected under general anesthesia prior to surgery. During laparoscopy, the physician thoroughly examined the peritoneal cavity and visually confirmed the presence or absence of endometriosis. Endometriosis patients had visually and histologically confirmed endometriosis. The control population were visually confirmed to be free of endometriosis. The tissue samples were processed for generating high-throughput mRNA transcriptomics data (RNA-Seq) and DNA methylomics data (MBD- seq). Our transcriptomics dataset includes 38 single-end RNA-seq samples (22 controls and 16 endometriosis) and the methylomics dataset includes 80 MBD-seq DNA methylation samples (36 controls and 44 endometriosis). More details can be found in our earlier publicatio18 on the same datasets.
Transcriptomics and Methylomics Data Preprocessing
We preprocessed the transcriptomics and methylomics data using several widely accepted bioinformatics tools. For transcriptomics data, we used FastQC, Cutadapt, Bowti2, TopHat and HTSeq in different steps of the preprocessing. We used h38 as reference genome. After getting the read count data from HTSeq, the rest of the analysis was performed using R packages. Low count genes were removed using the filtering criterion: keep the genes that have at least 1 count per million (cpm) reads mapped in at least n samples, where n is the smallest group size. For DNA- methylation data, we used FastQC, Cutadapt, Bowti2, Samtools, Picard and R packages for preprocessing the data. We segmented the genome sequence into tiling windows of size 1,000 bases, which is widely used and recorded the number of reads that are mapped to each tiling windows/regions. Read count are the number of aligned reads that uniquely map to the h38 reference genome. Very low count regions were filtered out using the filtering criterion: keep the regions that have non-zero counts per million (cpm) reads mapped in at least n samples, where n is the smallest group size. More details about the data preprocessing are available in our earlier publicatio18.
The read count data was normalized using three different techniques: (a) logarithm of counts per million (logCPM) of trimmed mean of M values (TMM)19, (b) Quantile normalization (qNorm)20, and (c) Voom normalization (vNorm)19. For differential analysis, a generalized linear model (GLM) followed by likelihood ratio test was applied using the edgeR package to identify differentially expressed genes (DEGs) in the transcriptomics data and differentially methylated regions (DMRs) in the methylomics data. The significance of the genes/methylated regions were defined by using an adjusted p-value cutoff set at 5% using the false discovery rate (FDR) method for multiple testin21.
In the methylomics data analysis, our goal is to identify the methylated regions of interest (MROI) and find the nearby genes. Mapping of an MROI to the reference annotation file helped us to extract the nearest genes from that MROI. Our goal is to identify the genomic features such as the protein coding genes, long intervening noncoding RNA (lincRNA) genes, microRNA (miRNA) genes, Ribosomal ribonucleic acid (rRNA) genes, small nucleolar RNA (snoRNA) genes, and small nuclear RNA (snRNA) genes. The distance threshold for the MROI position to the genomic region was set to 10,000 bps. An MROI can be in the upstream/downstream region, or it can fall into a gene.
GenomeForest
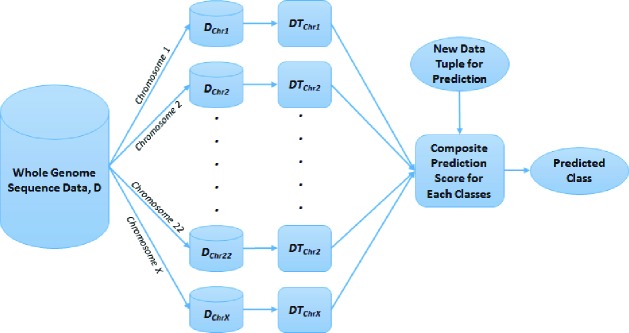
In machine learning, an ensemble is a set of k base classifier models (M1, M2, M3,…., Mk) for the purpose of creating an improved composite classification model (M). A set of k training datasets (D1, D2, D3,…., Dk) are created from the master dataset (D), where Mi is created by training a classifier model on Di (1 ≤ i ≤ k). For classifying a new data tuple, the ensemble model M generates a class prediction based on the votes of the base classifiers. We developed an ensemble method called GenomeForest (Figure 1), in which each of the classifier is a decision tree classifier representing a classification model for each pair of chromosomes (up to 23) so that the collection is a forest representing the whole genome. C4.522 is a popular algorithm for decision tree construction that uses entropy minimization or information gain for attribute selection criteria. We used an improved version of C4.5 (called C5.0/see523) for constructing the decision tree in this study. Confidence factor is used as a parameter for tree pruning in C5.0. The default value for confidence factor is 25% or 0.25. If the value of confidence factor is smaller than 0.25, it causes more pruning and vice versa.
Figure 1.
GenomeForest – an Ensemble of Decision Trees Representing Classifier for Each Chromosome for a Whole Genome Sequencing Dataset
In GenomeForest, given a whole genome sequencing dataset (D), up to 23 training datasets (DChr1, DChr2,…….., DChr22, DChrX) are created by partitioning the dataset, D, in which a training dataset DChrN (where N in ChrN is the chromosome number 1, 2, 3,….., 22, and X) contains the attributes corresponding to chromosome N. A decision tree model DTChrN is trained by using a training dataset DChrN. A composite prediction score (PS) for each class is calculated by using voting (score for each DTChrN model is 1) or weighted by various performance measures such as accuracy, sensitivity, specificity, precision, F1 score, area under the curve (AUC) of receiver operating characteristics graph, and Matthews correlation coefficient (MCC). The final predicted class (PC) for a new data tuple is the class with the highest total prediction score. There are two class labels (endometriosis vs. control) in our transcriptomics and methylomics datasets. As such, the formula for calculating the composite or total prediction scores for the endometriosis and the control classes are presented in Formulas 1 and 2. Formula 3 identifies the final predicted class.
| (1) |
| (2) |
| (3) |
Cross Validation and Model Performance
For model validation and comparing results between the experiments (described below), we applied the leave-one-out cross validation for computing the performance measures. This ensures two things: (1) the record used for model validation is not used for model construction, and (2) all records are used for model validation. This technique is useful for dataset with smaller number of samples such as in our study. The final model is constructed using all records. We computed several model performance measures: accuracy, sensitivity, specificity, precision, F1 score, Matthews correlation coefficient (MCC), and area under the receiver operating characteristics curve (AUC); the leave-one-out cross validation approach was used for calculating these measures.
Machine Learning Experimental Approach
We performed six different experiments using the GenomeForest ensemble classifier as shown in Table 1. Performance measures of each model were computed using the cross-validation approach described above. We used the default value of confidence factor (0.25) so that the decision tree is optimally pruned. For each of the GenomeForest experiment, we applied seven different criteria (such as accuracy, sensitivity, specificity, precision, F1 score, MCC, and AUC) for ranking the decision tree models (DTChrN), used the highest ranked models (topN = 1, 2, 3,……, 23) in the ensemble process, and eight different criteria for scoring (including accuracy, sensitivity, specificity, precision, F1 score, MCC, AUC and voting). This experimental approach produced up to 1,288 (7 x 23 x 8) GenomeForest ensemble models for each category of GenomeForest experimental approach as listed in Table 1.
Table 1.
Machine Learning Experimental Approach using GenomeForest
| Experiment Name | Experiment Name | Experiment Name |
|---|---|---|
| (1) TMM + GenomeForest | (3) vNorm + GenomeForest | (5) qNorm + GLM + GenomeForest |
| (2) qNorm + GenomeForest | (4) TMM + GLM + GenomeForest | (6) vNorm + GLM + GenomeForest |
In experiment (1-3) (Table 1), we applied different normalization techniques on the raw read counts of genes/methylated regions and then applied GenomeForest. In experiment (4-6), differential analysis using GLM was performed first on each partitions of the dataset (DChrN) to reduce features, such as genes in the transcriptomics data and genomic regions in the methylated data. After that, we applied different normalization techniques on the raw read counts of differential genes/methylated regions and then applied GenomeForest.
The datasets were filtered for low read count genes for the transcriptomics datasets and for low read count methylated regions for the methylomics dataset. For the transcriptomics dataset, GenomeForest experiments were conducted in two scenarios: (a) all genes including protein coding, lincRNA gene, miRNA gene, rRNA gene, etc. are present in the dataset, and (b) only protein coding genes are present in the dataset. For the methylomics dataset, all methylated regions except lower read counts were present.
The results of the six experiments using the GenomeForest models were compared with the same set of experiments using the regular decision tree models as well as three machine learning models using an enhanced algorithm for detecting biomarkers named Biosigne24; the models in Biosigner include Partial Least Squares Discriminant Analysis (PLSDA), Random Forest (RF), and Support Vector Machine (SVM). Biosigner is an enhanced algorithm for detecting biomarkers. The details of the work on same dataset using decision tree and Biosigner experiments were published in Akter et al. (2019)18.
Results
Transcriptomics GenomeForest Results
After pre-processing of the 38 RNA-seq data, we created a dataset containing the read counts of 58,050 genes in which 18,852 genes were protein-coding. After filtering for low count genes, 14,154 genes were included in the dataset in which 11,687 of them were protein coding genes.
We applied the six experimental approaches using the ensemble-based GenomeForest algorithm on both protein coding and non-protein coding genes (denoted as “all genes” in this article) that includes 14,154 genes and on protein coding genes only that includes 11,687 genes. For each experimental approach, we created up to 1,288 GenomeForest models for various combinations of ranking matrices, value of topN and scoring matrices as described in the Method section. Out of the up to 1,288 GenomeForest models in each experimental approach, the performance measures of one of the best GenomeForest model are presented in Table 2. Within the six experimental approaches using all genes, the best performance was obtained for the “qNorm + GenomeForest” experiment by using the top 18 decision tree models (DTChrN) ranked by the sensitivity measures and using precision as the scoring criteria. This experiment achieved the accuracy of 94.7%, sensitivity of 100%, specificity of 90.9%, precision of 88.9%, F1 score of 0.941 and the MCC of 0.899. Within the six experimental approaches using protein coding genes, the best performance was obtained for the “vNorm + GenomeForest” experiment by using all 23 decision tree models (DTChrN) and using F1 score as the scoring criteria. This experiment achieved the accuracy of 97.4%, sensitivity of 93.8%, specificity of 100%, precision of 100%, F1 score of 0.968 and MCC of 0.947. A total of 73 genes were identified by the individual decision tree models across all 23 pairs of chromosomes from the “vNorm + GenomeForest” experiment using the protein coding genes and three of these were differentially expressed and downregulated. We compared these 73 genes with the gene list found from the decision tree and Biosigner models in our previous stud18; NOTCH3, B4GALNT1 and GTF3C5 were found common between GenomeForest and decision tree genes. All three genes were found downregulated in the differential analysis. Only NOTCH3 was found common between GenomeForest and Biosigner.
Table 2.
GenomeForest Performance Measures Using Transcriptomics Data
| Gene Feature Set | Experiment Name | topN | Ranking Metric Name | Scoring Metric Name | Accuracy | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|---|---|---|---|
| All | TMM + GenomeForest | 23 | NA | F1 Score | 0.895 | 0.938 | 0.864 | 0.833 | 0.882 | 0.792 |
| All | qNorm + GenomeForest | 18 | Sensitivity | Precision | 0.947 | 1.000 | 0.909 | 0.889 | 0.941 | 0.899 |
| All | vNorm + GenomeForest | 21 | AUC | AUC | 0.947 | 0.938 | 0.955 | 0.938 | 0.938 | 0.892 |
| All | TMM + GLM + GenomeForest | 1 | F1 Score | F1 Score | 0.737 | 0.813 | 0.682 | 0.650 | 0.722 | 0.489 |
| All | qNorm + GLM + GenomeForest | 1 | F1 Score | F1 Score | 0.816 | 0.813 | 0.818 | 0.765 | 0.788 | 0.626 |
| All | vNorm + GLM + GenomeForest | 1 | F1 Score | F1 Score | 0.816 | 0.813 | 0.818 | 0.765 | 0.788 | 0.626 |
| Protein Coding | TMM + GenomeForest | 18 | F1 Score | F1 Score | 0.947 | 1.000 | 0.909 | 0.889 | 0.941 | 0.899 |
| Protein Coding | qNorm + GenomeForest | 23 | NA | MCC | 0.947 | 1.000 | 0.909 | 0.889 | 0.941 | 0.899 |
| Protein Coding | vNorm + GenomeForest | 23 | NA | F1 Score | 0.974 | 0.938 | 1.000 | 1.000 | 0.968 | 0.947 |
| Protein Coding | TMM + GLM + GenomeForest | 2 | F1 Score | Voting | 0.895 | 1.000 | 0.818 | 0.800 | 0.889 | 0.809 |
| Protein Coding | qNorm + GLM + GenomeForest | 2 | MCC | Voting | 0.842 | 0.938 | 0.773 | 0.750 | 0.833 | 0.702 |
| Protein Coding | vNorm + GLM + GenomeForest | 5 | F1 Score | F1 Score | 0.868 | 0.813 | 0.909 | 0.867 | 0.839 | 0.729 |
Performance Comparisons of Models Using Transcriptomics Data
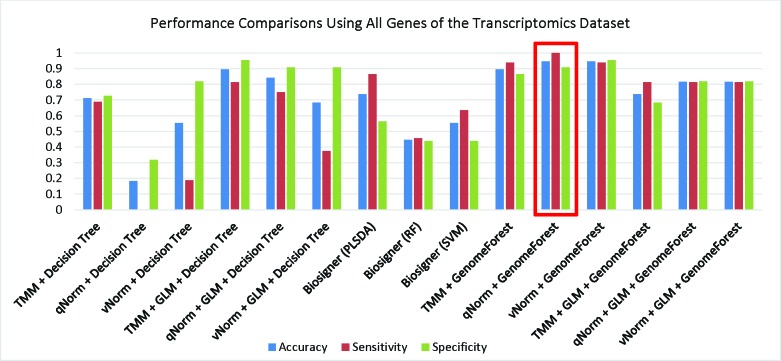
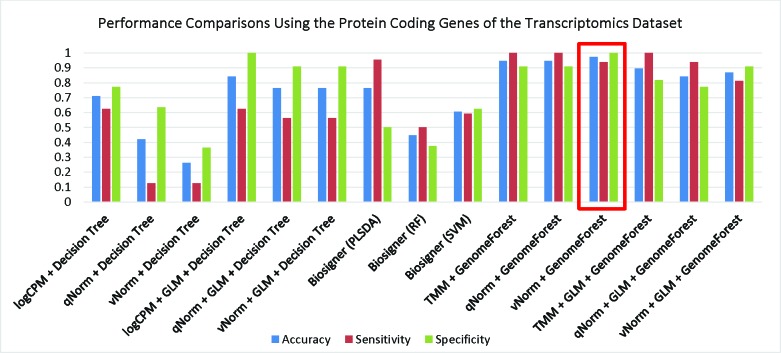
We have compared the performance of GenomeForest with our earlier machine learning classifie18 applied on the same dataset. A bar chart comparison of accuracy, sensitivity and specificity for experiments using all genes are presented in Figure 2. In this scenario, the “qNorm + GLM + GenomeForest” and “vNorm + GLM + GenomeForest” experiments have a balanced accuracy, sensitivity and specificity but does not outperform all of the experiments. The “qNorm + GenomeForest” experiment produced the highest accuracy and specificity among all the experiments and outperformed all of the experiments by F1 score and MCC. A bar chart comparison of accuracy, sensitivity and specificity for experiments using the protein coding genes are presented in Figure 3. In this scenario, the “Biosigner (SVM)” method has a balanced accuracy, sensitivity and specificity but does not outperform all of the experiments. The “vNorm + GenomeForest” experiment produced the highest accuracy and sensitivity among all of the experiments and outperformed all experiments based on F1 score and MCC. In both scenarios, GLM was useful for improving the overall performance in case of the decision tree application but GenomeForest was able to produce the best performance without using GLM.
Figure 2.
Performance Comparisons Using All Genes of the Transcriptomics Dataset
Figure 3.
Performance Comparisons Using the Protein Coding Genes of the Transcriptomics Dataset
Methylomics GenomeForest Results
We had 80 enrichment-based DNA-methylation (MBD-seq) samples where 77 samples met the quality control criteria (35 controls and 42 endometriosis). After pre-processing the data, we created a dataset containing the read counts of 3,088,281 methylated regions. After applying filtering criteria for lower read counts, 2,577,382 methylated regions were included in the dataset for further analysis.
We applied the six experimental approaches using the ensemble-based GenomeForest algorithm on the methylomics dataset that includes 2,577,382 methylated regions. For each experimental approach, we created up to 1,288 GenomeForest models for various combination of ranking matrices, values of topN and scoring matrices as described in the Method section. Out of the up to 1,288 GenomeForest models in each experimental approach, the performance measures of one the best GenomeForest model is presented in Table 3. Within the six experimental approaches, the best performance was obtained for both “qNorm + GenomeForest” and “vNorm + GenomeForest” experiments by using the top 22 decision tree models (DTChrN) ranked by the F1 score and also using F1 score as the scoring criteria. Both experiments achieved the accuracy of 90.9%, sensitivity of 92.9%, specificity of 88.6%, precision of 90.7%, F1 score of 0.918 and the MCC of 0.817. A total of 109 MROIs were identified by the individual decision tree models across all 23 pairs of chromosomes from the “qNorm + GenomeForest” experiment. We found 24 genes within the distance of 10,000 bps from those 109 MROIs, in which the biotypes of the genes were as follows: protein coding (n=7), lincRNA (n=3), antisense (n=3), sense intronic (n=1), snoRNA (n=1), snRNA (n=2), and pseudogene (n=7). The location of the regions from the genes were upstream (n=15), downstream (n=4) or overlapping (n=5). We compared these 24 genes with the gene list found from the decision tree model18 and found MFSD14B to be common.
Table 3.
GenomeForest Performance Measures Using Methylomics Data by Leave-One-Out Cross Validation
| Experiment Name | topN | Ranking Metric Name | Scoring Metric Name | Accuracy | Sensitivity | Specificity | Precision | F1 Score | MCC |
|---|---|---|---|---|---|---|---|---|---|
| TMM + GenomeForest | 23 | Accuracy | Accuracy | 0.870 | 0.929 | 0.800 | 0.848 | 0.886 | 0.740 |
| qNorm + GenomeForest | 22 | F1 Score | F1 Score | 0.909 | 0.929 | 0.886 | 0.907 | 0.918 | 0.817 |
| vNorm + GenomeForest | 22 | F1 Score | F1 Score | 0.909 | 0.929 | 0.886 | 0.907 | 0.918 | 0.817 |
| TMM + GLM + GenomeForest | 2 | Specificity | Voting | 0.831 | 0.881 | 0.771 | 0.822 | 0.851 | 0.659 |
| qNorm + GLM + GenomeForest | 7 | AUC | AUC | 0.818 | 0.810 | 0.829 | 0.850 | 0.829 | 0.636 |
| vNorm + GLM + GenomeForest | 1 | F1 Score | F1 Score | 0.792 | 0.810 | 0.771 | 0.810 | 0.810 | 0.581 |
Performance Comparisons of Models Using Methylomics Data
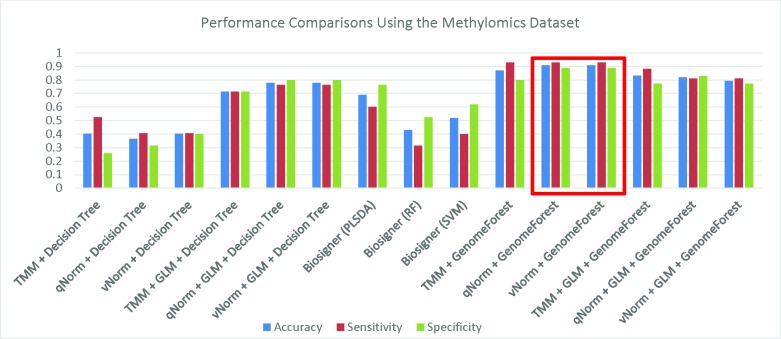
We have compared the performance of GenomeForest with our earlier machine learning classifie18 applied on the same dataset. A bar chart comparison of accuracy, sensitivity and specificity for experiments using the methylomics dataset are presented in Figure 4. The “qNorm + GLM + GenomeForest”, and “vNorm + GLM + GenomeForest” experiments have a balanced accuracy, sensitivity and specificity but does not outperform all of the experiments. Both the “qNorm + GenomeForest” and “vNorm + GenomeForest” experiments produced the highest accuracy, sensitivity and specificity among all the experiments and outperformed all of the experiments by F1 score and MCC. In our earlier study, we have shown that GLM was useful to improve the overall performance in case of decision tree applicatio18 but GenomeForest was able to produce the best performance without the help of GLM.
Figure 4.
Performance Comparisons Using the Methylomics Dataset
Discussion
This work achieves our aim of broadly evaluating the newly developed ensemble approach, named GenomeForest, under different experimental scenarios. Also, we have successfully shown that it can improve model performances in classifying endometriosis and control samples using whole genome transcriptomics and methylomics data. Our newly proposed classifier outperforms various machine learning classifier algorithms (decision tree and Biosigner) on the same datasets that have been published earlie18.
First, the ensemble technique can achieve a high classification accuracy. For whole genome sequencing data, we applied a logical partitioning approach based on each chromosome. We trained decision tree models on each chromosomal partition of the dataset and developed an ensemble approach for creating a composite collection of a forest model consisting of a set of decision trees representing the whole genome. We named this ensemble classification algorithm as GenomeForest. We experimented with various ranking and scoring techniques using GenomeForest and found that F1 score is best for both ranking and scoring. This model was able to outperform its counterpart.
Second, differential analysis using the GLM is widely used to identify the DEGs from the transcriptomics datasets and DMRs from the methylomics datasets. In our previous wor18, we have shown that differential analysis was useful for improving the performance of decision tree application by reducing the features (genes/genomic regions). However, in this work, we have shown that GenomeForest was able to produce the best performance without the help of differential analysis. We also evaluated different normalization techniques as a classifier’s performance may vary depending on the normalization techniques. We found that qNorm performed the best when all genes were considered and vNorm performed the best when only the protein-coding genes were considered in the transcriptomics dataset. For the methylomics dataset, both qNorm and vNorm normalizations performed the best in the GenomeForest application.
Third, the ensemble machine learning classifiers can be trained for creating highly accurate models for classifying endometriosis with high sensitivity and specificity thus creating the opportunity for precision medicine application for endometriosis. Mainly because of the complexity in diagnosis techniques, there is a delay from symptom onset to diagnosis ranging from 4 to 11 years which is very high. The ensemble machine learning models in this study achieved a very high F1 score; a near perfect F1 score (0.968) for the transcriptomics dataset and a very high F1 score (0.918) for the methylomics dataset. Although the predicted markers require further experimental and clinical validations, we hope in the future a less invasive biopsy can be used to diagnose endometriosis using ensemble machine learning classifiers-based findings as demonstrated in this study.
Fourth, we experimented if ensemble of a few top ranked chromosomes (ranked by different performance measures) could classify the disease samples from the controls. We observed that the best ensemble model used various numbers of decision tree models in different scenarios: (a) for the transcriptomic dataset using all genes, the best ensemble model was created using top 18 chromosomes ranked by sensitivity and scored by precision, (b) for the transcriptomic dataset using protein coding genes only, the best ensemble model was created using all chromosomes, (c) for the methylation data, the best model was created using the top 22 chromosomes ranked and scored by F1. Therefore, we concluded that instead of using few chromosomes, ensemble of most of the chromosomes in the GenomeForest could give us the best classification model.
Fifth, GenomeForest can assist in the identification of candidate biomarkers of endometriosis using transcriptomics and methylomics data. In our previous study, we discussed the candidate biomarker genes of endometriosis extracted from decision tree and Biosigner model18. All the machine learning models (GenomeForest, decision tree and Biosigner) have identified NOTCH3 as a candidate biomarker. It is also differentially expressed and downregulated in our study. The NOTCH3 signaling may play a major role in oncogenesis, tumor maintenance, and resistance to chemotherap25. NOTCH3 is associated with breast cancer developmen26 and pancreatic ductal adenocarcinoma (PDAC)27, lung carcinogenesi28 and endometrial carcinom29. Dysregulation and decrease in NOTCH signaling pathway is also associated with endometriosi30,31. B4GALNT1 and GTF3C5 were identified by the decision tre18 and GenomeForest experiments. Trimarchi et al. (2017) identified B4GALNT1 to be related to endometrial cance32. B4GALNT1 is associated with two pathways: Glycosphingolipid biosynthesis - ganglio series and Sphingolipid metabolism, and diseases named Spastic Paraplegia 26 and autosomal recessive. GTF3C5 was reported as differentially expressed between endometrioid endometrial cancer and non-endometrioid endometrial cance33. Some other candidate biomarker genes are also related to different types of cancer. For example, ZBTB8A may be involved in gastric carcinoma, gastric adenocarcinoma cell differentiation, cancer invasion and metastasi34. Aghajanova et al. (2011) identified H1FX to be differentially expressed in the comparison of severe versus mild endometriosis samples in the mid-secretory phase endometriu35. Other known functions of H1FX are Cancer, Cell-To-Cell Signaling and Interaction, and Skeletal and Muscular Disorders. AIMP1 is a cytokine that is specifically induced by apoptosis, and involved in the control of angiogenesis, inflammation, and wound healing. It is also involved in the stimulation of inflammatory responses after proteolytic cleavage in tumor cells. Baek et al. (2018) identified KLC4 to be associated with human lung cancer cell line36. XRCC2 promotes colorectal cancer cell growth, regulates cell cycle progression, and apoptosi37 and mutations in XRCC2 can increase the risk of breast cance38. The aberrant expression of MED19 is involved in tumorigenesis and it promotes the proliferation of breast cance39. Tamaresis et al. (2014) identified NRXN3 to be differentially expressed in severe vs. mild endometriosi40. CELF2 was reported as a putative tumor suppressor gene in colon cance41. IL17RA plays a pathogenic role in many inflammatory and autoimmune diseases. Bunch et al. (2011) reported that the expression of PGRMC1 is significantly decreased in the endometrium of women with endometriosi42. Fang et al. (2011) suggested that abnormal HRH4 expression plays a role in the progression of colorectal cancer43.
Lastly, the finding of many cancer and tumor associated genes using GenomeForest approach is consistent with our previous findings using other machine learning classifier44. Several studies investigated the relationship of endometriosis with cancer. Sato et al. (2000) and Thomas et al. (2000) have found some cancer associated mutations in endometriotic lesion44,45 and significant shared genetic correlation in both endometrial cancer and endometriosi46. Some other studies have found that endometriosis patients are at a higher risk of developing several malignancies: ovarian cancer, breast cancer, renal cancer, thyroid cancer and brain tumo47. Both cancer and endometriosis have some similar characteristics: metastasis, angiogenesis and resistance to apoptosis. However, only endometriosis is considered to be a benign condition. More genomic studies are needed to investigate the association of endometriosis with cancer.
In summary, this study demonstrated that GenomeForest, an ensemble machine learning classifier, is a robust and reliable approach for classifying endometriosis using transcriptomics or methylomics data. We concluded that an appropriate GenomeForest diagnostic pipeline for endometriosis should use (a) either transcriptomics or methylomics data, (b) vNorm for protein-coding genes, qNorm for all genes and either qNorm or vNorm for the methylomics data, (c) chromosomal partitioning with ensemble of decision trees for greatest increase in classification performance, (d) no differential analysis is necessary for feature reduction, and (e) F1 score for both ranking of the individual models and generating the composite score. The conclusion was made based on the use case of endometriosis classification in this study. Further study is needed to generalize the results across multiple disease classification cases. Also, development of prediction classification models for endometriosis and other diseases using integrated multi-omics data would be an interesting investigation.
Data Availability
The datasets generated for this study can be found in the Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134052; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134056.
Acknowledgement
The biological samples were collected in collaboration with the University of Missouri and Boone Hospital, Columbia, MO, and University of California, San Francisco, CA. Some tissue samples were provided by the NIH SCCPIR Human Endometrial Tissue Bank and DNA Bank at UCSF (NIH grant HD055764-06). We are thankful to Stacy Syrcle, Sarah Crowder, Danny J. Schust, and Breton Barrier for help with sample collection. This work was supported by NIEHS R21 ES020039, University of Missouri Institute for Clinical and Translational Science, NIH/National Center for Advancing Translational Sciences (NCATS) grant UL1TR002345, and University of Missouri Research Council.
Figures & Tables
References
- 1.David Adamson G, Kennedy S, Hummelshoj L. Creating solutions in endometriosis: Global collaboration through the World Endometriosis Research Foundation. J Endometr. 2010;2(1):3–6. doi:10.5301/JE.2010.4631. [Google Scholar]
- 2.Selçuk I, Bozdağ G. Recurrence of endometriosis; risk factors, mechanisms and biomarkers; review of the literature. J Turkish Ger Gynecol Assoc. 2013;14(2):98–103. doi: 10.5152/jtgga.2013.52385. doi:10.5152/jtgga.2013.52385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeung P, Sinervo K, Winer W, Albee RB. Complete laparoscopic excision of endometriosis in teenagers: is postoperative hormonal suppression necessary? Fertil Steril. 2011;95(6):1909–1912.e1. doi: 10.1016/j.fertnstert.2011.02.037. doi:10.1016/j.fertnstert.2011.02.037. [DOI] [PubMed] [Google Scholar]
- 4.Burkett D, Horwitz J, Kennedy V, Murphy D, Graziano S, Kenton K. Assessing current trends in resident hysterectomy training. Female Pelvic Med Reconstr Surg. 2011;17(5):210–214. doi: 10.1097/SPV.0b013e3182309a22. doi:10.1097/SPV.0b013e3182309a22. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, Outley J, Botteman M, Spalding J, Simon JA, Pashos CL. Economic burden of endometriosis. Fertil Steril. 2006;86(6):1561–1572. doi: 10.1016/j.fertnstert.2006.06.015. doi:10.1016/j.fertnstert.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Dunselman GAJ, Vermeulen N, Becker C, et al. ESHRE guideline: management of women with endometriosis. Hum Reprod. 2014;29(3):400–412. doi: 10.1093/humrep/det457. doi:10.1093/humrep/det457. [DOI] [PubMed] [Google Scholar]
- 7.Parasar P, Ozcan P, Terry KL. Endometriosis: Epidemiology, Diagnosis and Clinical Management. Curr Obstet Gynecol Rep. 2017;6(1):34–41. doi: 10.1007/s13669-017-0187-1. doi:10.1007/s13669-017-0187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal SK, Chapron C, Giudice LC, et al. Clinical diagnosis of endometriosis: a call to action. Am J Obstet Gynecol. 2019;220(4):354.e1–354.e12. doi: 10.1016/j.ajog.2018.12.039. doi:10.1016/j.ajog.2018.12.039. [DOI] [PubMed] [Google Scholar]
- 9.Lee B, Du H, Taylor HS. Experimental murine endometriosis induces DNA methylation and altered gene expression in eutopic endometrium. Biol Reprod. 2009;80(1):79–85. doi: 10.1095/biolreprod.108.070391. doi:10.1095/biolreprod.108.070391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarca AL, Carey VJ, Chen X, Romero R, Drăghici S. Machine Learning and Its Applications to Biology. PLoS Comput Biol. 2007;3(6):e116. doi: 10.1371/journal.pcbi.0030116. doi:10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015;16(6):321–332. doi: 10.1038/nrg3920. doi:10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GTEx Consortium Gte. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. doi:10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirooznia M, Yang JY, Yang MQ, Deng Y. A comparative study of different machine learning methods on microarray gene expression data. BMC Genomics. 2008;9(Suppl 1(Suppl 1)):S13. doi: 10.1186/1471-2164-9-S1-S13. doi:10.1186/1471-2164-9-S1- S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13(10):705–719. doi: 10.1038/nrg3273. doi:10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 15.Johnson NT, Dhroso A, Hughes KJ, Korkin D. Biological classification with RNA-seq data: Can alternatively spliced transcript expression enhance machine learning classifiers? RNA. 2018;24(9):1119–1132. doi: 10.1261/rna.062802.117. doi:10.1261/rna.062802.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue Q, Lin Z, Cheng Y-H, et al. Promoter methylation regulates estrogen receptor 2 in human endometrium and endometriosis. Biol Reprod. 2007;77(4):681–687. doi: 10.1095/biolreprod.107.061804. doi:10.1095/biolreprod.107.061804. [DOI] [PubMed] [Google Scholar]
- 17.Akter S, Xu D, Nagel SC, Joshi T. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; 2018. A Data Mining Approach for Biomarker Discovery Using Transcriptomics in Endometriosis; pp. 969–972. doi:10.1109/BIBM.2018.8621150. [Google Scholar]
- 18.Akter S, Xu D, Nagel SC, et al. Machine Learning Classifiers for Endometriosis Using Transcriptomics and Methylomics Data. Front Genet. 2019;10:766. doi: 10.3389/fgene.2019.00766. doi:10.3389/FGENE.2019.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. doi:10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. http://www.ncbi.nlm.nih.gov/pubmed/12538238 . [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57:289–300. doi:10.2307/2346101. [Google Scholar]
- 22.Quinlan JR. C4. 5: Programs for Machine Learning. Morgan Kaufmann; 1993. [Google Scholar]
- 23.Data Mining Tools See5 and C5.0. 2015. http://www.rulequest.com/see5-info.html. Accessed November 21.
- 24.Rinaudo P, Boudah S, Junot C, Thévenot EA. biosigner: A New Method for the Discovery of Significant Molecular Signatures from Omics Data. Front Mol Biosci. 2016;3:26. doi: 10.3389/fmolb.2016.00026. doi:10.3389/fmolb.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aburjania Z, Jang S, Whitt J, Jaskula-Stzul R, Chen H, Rose JB. The Role of Notch3 in Cancer. Oncologist. 2018;23(8):900–911. doi: 10.1634/theoncologist.2017-0677. doi:10.1634/theoncologist.2017-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braune E-B, Seshire A, Lendahl U. Notch and Wnt Dysregulation and Its Relevance for Breast Cancer and Tumor Initiation. Biomedicines. 2018;6(4) doi: 10.3390/biomedicines6040101. doi:10.3390/biomedicines6040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H-Y, Wang Y, Lan H, Zhang Y-X. Expression of Notch receptors and their ligands in pancreatic ductal adenocarcinoma. Exp Ther Med. 2018;16(1):53–60. doi: 10.3892/etm.2018.6172. doi:10.3892/etm.2018.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su T, Yang X, Deng J-H, et al. Evodiamine, a Novel NO TCH3 Methylation Stimulator, Significantly Suppresses Lung Carcinogenesis in Vitro and in Vivo. Front Pharmacol. 2018;9:434. doi: 10.3389/fphar.2018.00434. doi:10.3389/fphar.2018.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsuhashi Y, Horiuchi A, Miyamoto T, Kashima H, Suzuki A, Shiozawa T. Prognostic significance of Notch signalling molecules and their involvement in the invasiveness of endometrial carcinoma cells. Histopathology. 2012;60(5):826–837. doi: 10.1111/j.1365-2559.2011.04158.x. doi:10.1111/j.1365-2559.2011.04158.x. [DOI] [PubMed] [Google Scholar]
- 30.González-Foruria I, Santulli P, Chouzenoux S, Carmona F, Chapron C, Batteux F. Dysregulation of the ADAM17/Notch signalling pathways in endometriosis: from oxidative stress to fibrosis. MHR Basic Sci Reprod Med. 2017;23(7):488–499. doi: 10.1093/molehr/gax028. doi:10.1093/molehr/gax028. [DOI] [PubMed] [Google Scholar]
- 31.Su R-W, Strug MR, Joshi NR, et al. Decreased Notch pathway signaling in the endometrium of women with endometriosis impairs decidualization. J Clin Endocrinol Metab. 2015;100(3):E433–42. doi: 10.1210/jc.2014-3720. doi:10.1210/jc.2014-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trimarchi MP, Yan P, Groden J, Bundschuh R, Goodfellow PJ. Identification of endometrial cancer methylation features using combined methylation analysis methods. PLoS One. 2017;12(3):e0173242. doi: 10.1371/journal.pone.0173242. doi:10.1371/journal.pone.0173242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Mara TA, Zhao M, Spurdle AB. Meta-analysis of gene expression studies in endometrial cancer identifies gene expression profiles associated with aggressive disease and patient outcome. Sci Rep. 2016;6:36677. doi: 10.1038/srep36677. doi:10.1038/srep36677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Feng Y, Sun Z, et al. [Expression of ZBTB8A in gastric cancer and its clinical significance] Zhonghua Wei Chang Wai Ke Za Zhi. 2013;16(12):1199–1202. [PubMed] [Google Scholar]
- 35.Aghajanova L, Giudice LC. Molecular Evidence for Differences in Endometrium in Severe Versus Mild Endometriosis. Reprod Sci. 2011;18(3):229–251. doi: 10.1177/1933719110386241. doi:10.1177/1933719110386241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baek J-H, Lee J, Yun HS, et al. Kinesin light chain-4 depletion induces apoptosis of radioresistant cancer cells by mitochondrial dysfunction via calcium ion influx. Cell Death Dis. 2018;9(5):496. doi: 10.1038/s41419-018-0549-2. doi:10.1038/s41419-018-0549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu K, Song X, Chen Z, Qin C, He Y, Zhan W. XRCC2 Promotes Colorectal Cancer Cell Growth, Regulates Cell Cycle Progression, and Apoptosis. Medicine (Baltimore) 2014;93(28):e294. doi: 10.1097/MD.0000000000000294. doi:10.1097/MD.0000000000000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park DJ, Lesueur F, Nguyen-Dumont T, et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90(4):734–739. doi: 10.1016/j.ajhg.2012.02.027. doi:10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Fan Y, Liu B, Qi X, Guo Z, Li L. Med19 promotes breast cancer cell proliferation by regulating CBFA2T3/HEB expression. Breast Cancer. 2017;24(3):433–441. doi: 10.1007/s12282-016-0722-3. doi:10.1007/s12282-016-0722-3. [DOI] [PubMed] [Google Scholar]
- 40.Tamaresis JS, Irwin JC, Goldfien GA, et al. Molecular Classification of Endometriosis and Disease Stage Using High-Dimensional Genomic Data. Endocrinology. 2014;155(12):4986–4999. doi: 10.1210/en.2014-1490. doi:10.1210/en.2014-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramalingam S, Ramamoorthy P, Subramaniam D, Anant S. Reduced Expression of RNA Binding Protein CELF2, a Putative Tumor Suppressor Gene in Colon Cancer. Immunogastroenterology. 2012;1(1):27–33. doi: 10.7178/ig.1.1.7. doi:10.7178/ig.1.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunch K, Tinnemore D, Huff S, Hoffer Z, Burney R, Stallings J. PGRMC1 And PGRMC2 Expression is Significantly Decreased in the Endometrium of Women With Endometriosis. Fertil Steril. 2011;95(4):S10–S11. doi:10.1016/j.fertnstert.2011.01.061. [Google Scholar]
- 43.Fang Z, Yao W, Xiong Y, et al. Attenuated expression of HRH4 in colorectal carcinomas: a potential influence on tumor growth and progression. BMC Cancer. 2011;11(1):195. doi: 10.1186/1471-2407-11-195. doi:10.1186/1471-2407-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato N, Tsunoda H, Nishida M, et al. Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res. 2000;60(24):7052–7056. http://www.ncbi.nlm.nih.gov/pubmed/11156411 . [PubMed] [Google Scholar]
- 45.Thomas EJ, Campbell IG. Molecular genetic defects in endometriosis. Gynecol Obstet Invest. 2000;50(Suppl 1):44–50. doi: 10.1159/000052878. doi:10.1159/000052878. [DOI] [PubMed] [Google Scholar]
- 46.Painter JN, O’Mara TA, Morris AP, et al. Genetic overlap between endometriosis and endometrial cancer: evidence from cross-disease genetic correlation and GWAS meta-analyses. Cancer Med. 2018;7(5):1978–1987. doi: 10.1002/cam4.1445. doi:10.1002/cam4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melin A, Sparén P, Bergqvist A. The risk of cancer and the role of parity among women with endometriosis. Hum Reprod. 2007;22(11):3021–3026. doi: 10.1093/humrep/dem209. doi:10.1093/humrep/dem209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134052; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134056.