Abstract
Healthcare analytics is impeded by a lack of machine learning (ML) model generalizability, the ability of a model to predict accurately on varied data sources not included in the model’s training dataset. We leveraged free-text laboratory data from a Health Information Exchange network to evaluate ML generalization using Notifiable Condition Detection (NCD) for public health surveillance as a use case. We 1) built ML models for detecting syphilis, salmonella, and histoplasmosis; 2) evaluated generalizability of these models across data from holdout lab systems, and; 3) explored factors that influence weak model generalizability. Models for predicting each disease reported considerable accuracy. However, they demonstrated poor generalizability across data from holdout lab systems being tested. Our evaluation determined that weak generalization was influenced by variant syntactic nature of free-text datasets across each lab system. Results highlight the need for actionable methodology to generalize ML solutions for healthcare analytics.
Keywords: Machine learning, Generalizability, Notifiable condition detection, Public health surveillance
Introduction
Rapid advances in Artificial Intelligence (AI), uptake of health information infrastructure and the ever-increasing availability of electronic health datasets present considerable potential to leverage machine learning solutions to address medical and population health needs. In contrast to more archaic rule-based systems, machine learning approaches (a) do not rely on labor intensive human-driven rule development1, (b) learns multiple ‘overlapping rules’ to classify each instance, thereby facilitating increased classification robustness2, and (c) are capable of more complex classification tasks that cannot be effectively automated by rule-based approaches3. Machine learning has led to state- of-the-art results on a number of health challenges such as cancer case detection4, prediction of in-hospital mortality, unplanned readmissions5, need of treatment for depression6, and access to social risk and needs7.
The impact of robust decision models will only uphold expectations if they demonstrate external validity by generating accurate predictions across a variety of diverse healthcare systems8. A machine learning model that cannot accurately predict outcomes for a population that is different to the original population it was trained on suffers from weak generalization9,10. Machine learning researchers have long sought to develop generalizable algorithms11. However, efforts to demonstrate generalizability for health analytics has been limited12,13. Replication of published machine learning models often show substantially different conclusions on optimal modeling techniques. Several attempts to replicate existing efforts have led to results significantly in the opposite direction of the original study11. Thus, implementers seeking to deploy successful machine learning solutions across various sites must often expend considerable time and resources to ‘re-invent the wheel’ using facility-specific datasets.
Generalizability is impacted by the cost and effort of obtaining diverse datasets for decision model training. However, there is little methodological knowledge on the causes of weak generalizability in healthcare , nor the value of leveraging varied datasets from multiple sources for better generalizability. Better assessment of weak generalizability across varied patient populations and datasets could significantly improve our understanding of the generalization challenge.
Objectives
Increasing interest in Notifiable Condition Detection (NCD) for public health surveillance presents a suitable use case for our efforts. Notifiable conditions are diseases of public health concern that must be reported to government authorities by law14. However, accurate, automatic identification of notifiable diseases using existing patient data are an ongoing challenge. Previous work has demonstrated the ability to leverage machine learning for detecting notifiable diseases such as Hepatitis C, Hepatitis B, Chlamydia, and Gonorrhea15 using free-text laboratory datasets extracted from an Health Information Exchange (HIE), a network of interconnected health information systems16, example laboratory report messages can be found in appendix D. Prior efforts have demonstrated the ability to identify cancer cases using plaintext laboratory reports and machine learning approaches for case reporting to public health registries4,17 Neither study on machine learning in public health reporting evaluated the model architecture’s ability to generalize. Given the state of the existing literature, we sought to; a) train decision models capable of identifying three notifiable conditions using test and train data sampled independently of their source, b) assess generalizability of these decision models by exploring how well they perform on previously unseen data sources from lab systems excluded from the training process, and c) evaluate potential causes of weak generalization.
Materials and methods
Data extraction and preparation.
The Indiana Network for patient care (INPC)18 served as the data source for our study. One of the largest, continuously operated statewide Health Information Exchanges (HIE) in the US, the INPC covers 32 health systems, >100 hospitals, a multitude of laboratory systems and over 40,000 providers spread across Indiana19. We processed all electronic laboratory reports sent to the INPC from various participating lab systems during the years 2016 to 2017 to identify reports related to Syphilis, Salmonella, and Histoplasmosis. Each report was available in Health level Seven version 2 (HL7 v2) format20. Thus, relevant reports were identified by use of Logical Observation Identifiers Names and Codes (LOINC)21 codes related to each disease as determined by the Public Health Information Network Vocabulary Access and Distribution System (PHINVADS)22 and supplemented by a keyword search (appendix A). We created a gold standard for supervised learning by manually reviewing each of the identified reports and tagging them as either positive or negative for each disease under test.
Objective 1 – Decision Models for Notifiable Condition Detection (Overall model training method)
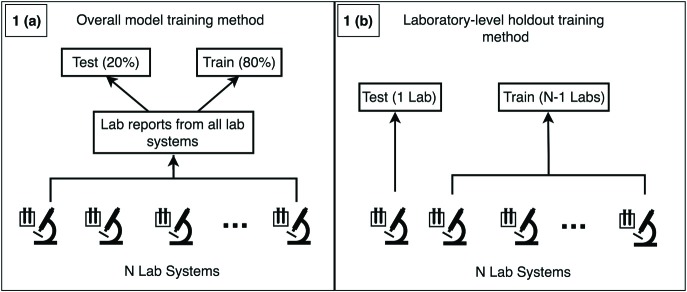
We vectorized reports for each disease using a bag-of-words approach23. HL7 V2 OBX5 (observation) and NTE (note) entries of each report were combined into a single string and tokenized by splitting using white space. We selected tokens to be included in the feature vectors by pooling all tokens for each disease and selecting tokens with a frequency > 5. We randomly split the data for each disease into 80%-20% train-test splits. We used the scikit-learn python package24 to train Random Forest25 classification algorithms using 20 max random features and 1000 trees as parameters to train models to predict if a report is positive or negative for the corresponding disease under test (figure 1.a). Random forest was selected due to previous success in machine learning tasks addressing various healthcare challenges.17 We chose 1000 trees due to acceptable train time and performance trade off and 20 random features because it is within the range of √m to log2(m+1) where m is the number of features that demonstrate optimal performance.25 We evaluated machine learning performance by using holdout test datasets to calculate precision, recall, F1-Score (harmonic mean between precision and recall), and Area Under the ROC Curve (ROC-AUC) for each disease.
Figure 1.
Overall model training method (1.a) vs. the laboratory-level holdout method for generalizability assessment (1.b).
Objective 2 – Generalizability assessment (Laboratory-level holdout method)
To evaluate the generalizability of decision models across data from previously unsampled lab systems, we used the following methods. We linked laboratory reports on the three diseases to lab systems they originated from. Next, we used a frequency threshold to filter out reports from lab systems with the lowest reporting rates for each disease. For each selected lab system x and disease under evaluation, we iteratively used all reports not associated with the lab system x as the train dataset and all reports associated with lab system x as the test dataset. We refer to this process as the ‘laboratory-level holdout’ method ( (figure 1.b). To evaluate the performance of models using the laboratory- holdout methods, we measured the performance of each model’s prediction on its corresponding test set using precision, recall, F1-Score, and ROC-AUC scores.
(Figure 1 contrasts the overall model training method (1.a) vs. the laboratory-level holdout method for generalizability assessment (1.b).
Objective 3 – Explore Causes of weak Generalizability
We investigated the relative distribution of features across reports submitted from each lab system to quantitatively assess differences in report content. This enabled us to determine what factors contributed to performance variations in models trained using the overall and laboratory-holdout methods. We hypothesized that clustering feature vectors extracted from each lab system would highlight relationships between feature vectors and the corresponding lab system they were extracted from. However, clustering feature vectors from all lab systems was untenable as the distribution of messages from N institutions distributed among N clusters would result in (N-1) x (N-1) degrees of freedom. This would invalidate the normality assumption for all counts, thereby making it impossible to effectively apply chi-squared analysis to determine relationships between lab systems and cluster on report feature vectors.26 Instead, we sought to identify all possible combinations of two lab systems (lab system pairs) for each disease, cluster data from these pairs into two clusters, and assess the relationship between lab system pair and feature vector for statistical analysis.
We selected the same subset of lab systems identified in objective 2 for this assessment. Feature vectors for all lab system pairs were analyzed using the K-Means algorithm27 with K=2. Chi-squared test for independence was used to determine whether the clusters of feature vectors were independent from the lab system of origin.
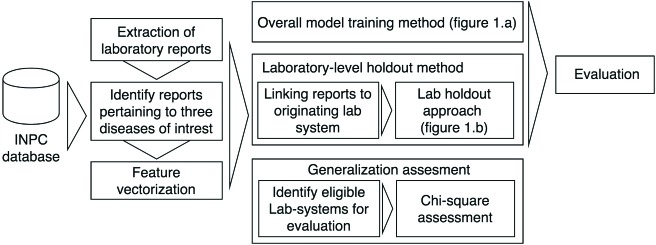
Figure 2 presents an overview of our research methods.
Figure 2.
Flowchart presenting the complete study approach, from data exaction to evaluation of each objective.
Results
Data extraction and preparation
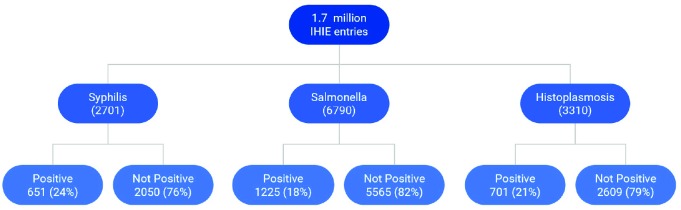
We evaluated a total of 1.7 million laboratory reports collected by the INPC between 2016-2017. Figure 3 presents the number of reports per disease, together with results of the manual review (% positive or negative/indeterminate for each disease). Disease prevalence (% of positive cases) ranged from 18% to 82%. Vectorizing reports for each disease generated feature vectors of lengths 940, 3446, and 1634 for Syphilis, Salmonella, and Histoplasmosis respectively.
Figure 3.
Prevalence of each disease across overall lab report set
Overall model training method
Table 1 presents the precision, recall, F1-Score, and ROC-AUC for each disease, as calculated using the overall model training method. We note that all performance metrics for each disease were relatively high despite our use of basic supervised learning approaches without any optimization. We verified the robustness of thse results by 10-fold cross- validation and found the minimum and maximum F1-Scores for all overall models to be within ±0.05 of the presented leave-one-out F1-scores. As such, we continue to use the leave-one-out performance results in this paper for ease of exposition.
Table 1.
Performance of overall models
| Disease | # of positive reports / # total reports (Prevalence %) | Precision | Recall | F1-Score | ROC-AUC |
|---|---|---|---|---|---|
| Syphilis | 133/541 (24.6%) | 0.89 | 0.91 | 0.90 | 99.22% |
| Salmonella | 223/1358 (16.4%) | 0.97 | 0.95 | 0.96 | 99.91% |
| Histoplasmosis | 143/662 (21.6%) | 0.88 | 0.96 | 0.92 | 99.18% |
Laboratory-level holdout method for generalizability assessment
Based on the distribution of lab reports across each lab system, we identified 29 reports as the optimal minimum for inclusion of a lab system for the laboratory-level holdout method. This cutoff was chosen to account for a break in the frequency of messages across laboratory systems in the Syphilis dataset, which was our most data-constrained disease dataset. We identified (8, 10, 12) labs with >= 29 reports for Syphilis, Salmonella, and Histoplasmosis, respectively. Table 2 shows the minimum, maximum, and median values for each performance metric. A comprehensive list of performance metrics for each laboratory-level holdout test is presented in appendix B.
Table 2.
Summary of holdout model performances for each disease
| Performance metric | Syphilis | Salmonella | Histoplasmosis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Median | Min | Max | Median | Min | Max | Median | |
| Precision | 0.24 | 1 | 0.97 | 0 | 1 | 0.97 | 0.66 | 1 | 0.925 |
| Recall | 0.11 | 0.95 | 0.655 | 0.14 | 1 | 0.73 | 0.18 | 1 | 0.935 |
| F1Score | 0.2 | 0.97 | 0.76 | 0.01 | 0.99 | 0.715 | 0.29 | 1 | 0.925 |
| ROC-AUC | 66.67% | 99.11% | 98.02% | 48.27% | 100% | 99.56% | 72.34% | 100% | 99.32% |
| Frequency Pos | 5 | 156 | 44.5 | 2 | 228 | 44.5 | 12 | 224 | 18.5 |
| Frequency Neg | 10 | 958 | 25 | 154 | 1031 | 415.5 | 16 | 1086 | 22.5 |
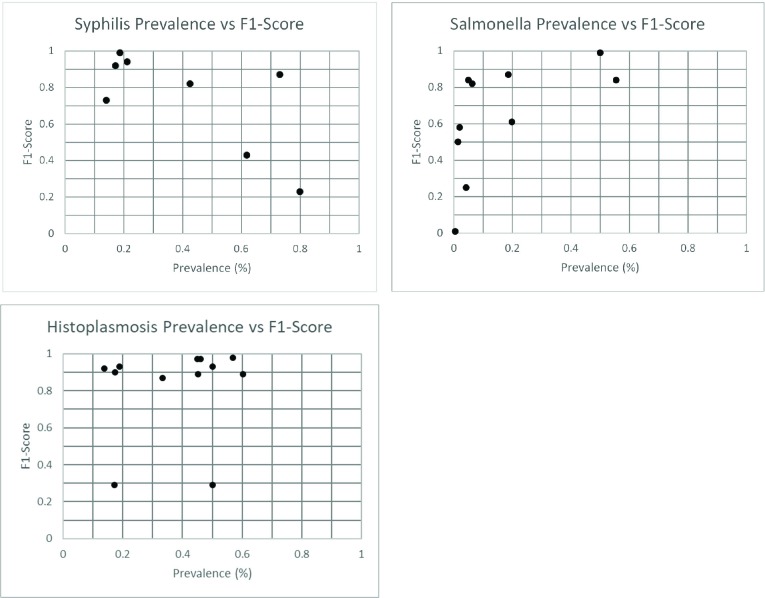
To investigate the role of disease prevalence on varying performance metrics reported across each laboratory-level holdout assessment, we plotted disease prevalence vs. F1-score for each disease under study (figure 4). We note that F1-scores may vary significantly across systems that reported similar prevalence.
Figure 4.
prevalence vs. F1-Score for each laboratorylevel holdout assessment.
Assessment of report variations across various lab systems
We identified (38, 32, 35) lab pairs for Syphilis, Salmonella, and Histoplasmosis, respectively. However, not all unique pairs clustered by K-Means were suitable for chi-squared analysis. Table 3 describes all pairs suitable for chi-squared analysis (valid lab pairs), the number of pairs that failed to show a statistically significant relationship between the feature vector cluster and lab system pair at the 99% confidence level (non-significant pairs), number of unique lab systems comprised the pairs of lab systems that failed to show a significant relationship (unique non-significant labs), and the average proportion of reports belonging to the cluster with fewer messages (average lesser cluster size). Each pair was clustered into two groups, so if the average lesser cluster size is P, then the average greater cluster size is (1-P)%.
Table 3.
Variations in lab reports across various lab systems
| Disease | Valid lab pairs | Non- Significant Pairs (%) | Unique Non- Significant Locations | Average Lesser Cluster Size |
|---|---|---|---|---|
| Syphilis | 12 | 0 (0%) | 0 | 35.45% |
| Salmonella | 183 | 8 (4%) | 5 | 26.17% |
| Histoplasmosis | 33 | 8 (24%) | 6 | 31.34% |
Discussion
Despite widespread acknowledgement of its value28, generalization of decision models for healthcare is rarely performed8. We found that decision models that deliver strong performance metrics using data from multiple systems still fail to generalize to data from other holdout systems. The approach we adopted to investigate model generalizability represents a simple method readily transferable to other datasets and use cases.
Models trained using the overall training method performed well with F1-Scores >= 0.9 for all three diseases. However, the laboratory-system holdout method demonstrated highly variable performance when models were tested on holdout datasets. These variations in holdout model performance are much greater than can be accounted for by random variation in model training as evidenced by 10-fold cross validation F1-scores of the overall models. The median F1- score of 0.925 for Histoplasmosis indicated a substantial number of models maintained high performance. However, F1-scores for Syphilis and Salmonella were 0.76 and 0.715, demonstrating that a majority of models exhibited substantial performance degradation when generalizing. Additionally, all three diseases reported minimum F1-scores of < 0.3. The uniformly poor minimum scores indicate the high likelihood of a model failing to generalize to certain institutions.
Comparison of prevalence vs. F1-Score for each laboratory-level holdout assessment (figure 4), as well as other performance metrics from each lab system (appendix B) demonstrate that variations in model performance does not mimic differences of data sources at a macroscopic level. Prevalence vs. F1-Score plots (figure 4) do not show a clear relationship between F1-score and the positive-negative balance of the holdout dataset. Many lab systems with roughly equivalent prevalence reported considerable differences in machine learning performance. This suggests that differences in individual report characteristics are a likely primary contributor to performance variability instead of overall characteristics of the holdout dataset. Clustering evaluation provided quantitative support for the difference in report characteristics across lab systems. The overwhelming number of lab system pairs showing a significant relationship between report institution and cluster suggest that differing message structure is a likely cause for poor generalizability. The average proportion of data in the lesser cluster shows that the difference in clusters is not a few outlying vectors, but that each cluster corresponds to a significant number of lab messages. Both salmonella and histoplasmosis datasets contained eight pairs of lab systems that failed to show a significant relationship between lab systems and clustering of messages. However, the eight salmonella pairs contained only five unique lab systems, and the eight histoplasmosis pairs contained only six unique lab systems. This affirms the consistency of clustering procedure, as it would be expected that if messages from lab system A overlaps with B and B overlaps with C, then A and C are more likely to overlap. These checks indicate that the clustering analysis correctly detects substantial differences in lab messages across holdout datasets.
Our results provide important evidence that performance of a machine learning model on a singular dataset does not imply the model will generalize well to an external dataset. We also provide empirical evidence for an intuitive explanation that this phenomenon is caused by different characteristics of the individual reports such as vocabulary and sentence structure as opposed to the size or positive-negative balance of the overall datasets. One might expect that a machine learning algorithm might perform poorly on messages containing unfamiliar words and phrases or combinations thereof. From a mathematical perspective, the clustering experiment emphasizes that the machine learning algorithm is often forced to extrapolate when predicting on unseen data sources, which is known to be concerning in any statistical model. Even though these results may be intuitive, they highlight the need for a clear path towards implementing machine models in real healthcare settings. Currently, papers that exemplify the potential of machine learning in healthcare focus almost solely on performance on a homogeneous dataset, leading to uncertainty of model generalizability and the pathway to implementing the model across real healthcare settings unclear.
We identified several limitations in our study. We leveraged laboratory reports on specific illness from a large HIE network. Thus, these results may not be applicable to other healthcare use-cases or datasets. Further, we used bag-of- words as our sole vectorization procedure. The generalizability and clustering results are likely dependent on the vectorization procedure used. Thus, it is unclear how use of other common preprocessing methods such as negation, word vectors or use of context would influence model performance. Additionally, we were unable to identify specific lab report characteristics of a lab system that caused poor generalizability. Our current experiments were limited to holding out one lab system at a time. However, multiple lab systems could be held out in a combinatorial approach to gain more robust results.
Future research will evaluate the feasibility of more inclusive HIE participation for analytics. When healthcare systems contribute to the training dataset, then generalizability is more likely. Determining the feasibility of enabling healthcare systems to train models on their own data could be a solution for similar reasons. Improvements to our understanding of model generalizability in the healthcare setting could also guide other solutions. Possibly there could be a representative dataset with a minimal number of contributing institutions. Discovering how to identify a minimal set of data sources to build a generalizable model is still unknown. Error analysis of messages misclassified by the developed holdout models will be a crucial component of determining factors of generalizability. Alternatively, a degree of data standardization could improve observed generalizability. Finally, techniques that improve model generalizability would alleviate some of the need for the above solutions. Our results indicate that suboptimal models fail to accommodate new words missing from the training data set. Normalizing data could minimize the impact of this issue, for example, by using a medical or word vectors to map similar words to words to similar vectors, so that a feature vector containing unseen words will still have a similar distribution to vectors in the train dataset.
Conclusion
We demonstrate that studies showing highly performant machine learning models for public health analytical tasks cannot be assumed to perform well when applied to data not sampled by the model’s train dataset. Our clustering results provide evidence that differences in data representation among data sources account for this poor generalization. These results highlight need to consider more inclusive training pathways for machine learning models in healthcare. It is currently unknown the degree to which many published ML models in the healthcare domain are generalizable. Administrative or technical support structures would likely be required to implement these models at other institutions. Considering model generalizability when designing future algorithms and evaluating model generalizability could reduce the need for external support structures and ease difficulty in achieving practical application of machine learning in healthcare.
Appendix A. List of Keywords and LOINC codes used to identify HL7 reports pertaining to each diseases.
Syphilis
LOINC Codes: PHVS_LabTestName_Syphilis Version 9. OID: 2.16.840.1.114222.4.11.4212 Additional Keyword(s): None
Salmonella
LOINC Codes: PHVS_LabTestName_Salmonellosis Version 7. OID: 2.16.840.1.114222.4.11.4198 Additional Keyword(s): “salmonella”
Histoplasmosis
LOINC Codes*: PHVS_LabTestName_Histoplasmosis Version 6. OID: 2.16.840.1.114222.4.11.4163 Additional Keyword(s): “histoplas”
*LOINC code 24647-0 was removed from histoplasmosis list as this code corresponded to test showing past histoplasmosis infection
Appendix B. Results of the Laboratory-holdout assessment per each notifiable condition. Note that laboratory names have been masked using identifiers.
| Syphilis | |||||
|---|---|---|---|---|---|
| Institution # | # of reports | Disease prevalence (%) | Precision | Recall | F1-Score |
| 1 | 1114 | 156 (14%) | 0.24 | 0.58 | 0.34 |
| 2 | 703 | 131 (18.63%) | 0.98 | 0.95 | 0.97 |
| 3 | 222 | 47 (21.17%) | 0.9 | 0.79 | 0.84 |
| 4 | 114 | 91 (79.82%) | 1 | 0.11 | 0.2 |
| 5 | 68 | 42 (61.76%) | 1 | 0.19 | 0.32 |
| 6 | 40 | 17 (42.5%) | 0.86 | 0.71 | 0.77 |
| 7 | 37 | 27 (72.97%) | 0.96 | 0.85 | 0.9 |
| 8 | 29 | 5 (17.24%) | 1 | 0.6 | 0.75 |
| Salmonella | |||||
| Institution # | # of reports | Pos-Support | Precision | Recall | F1-Score |
| 1 | 1099 | 68 (6.19%) | 0.98 | 0.71 | 0.82 |
| 2 | 807 | 17 (2.11%) | 1 | 0.41 | 0.58 |
| 3 | 620 | 9 (1.45%) | 1 | 0.33 | 0.5 |
| 4 | 507 | 94 (18.54%) | 0.84 | 0.9 | 0.87 |
| 5 | 503 | 21 (4.17%) | 1 | 0.14 | 0.25 |
| 6 | 420 | 2 (0.48%) | 0 | 1 | 0.01 |
| 7 | 411 | 228 (55.47%) | 0.96 | 0.75 | 0.84 |
| 8 | 363 | 72 (19.83%) | 0.89 | 0.46 | 0.61 |
| 9 | 342 | 17 (4.97%) | 0.93 | 0.76 | 0.84 |
| 10 | 308 | 154 (50%) | 0.98 | 1 | 0.99 |
| Histoplasmosis | |||||
| Institution # | # of reports | Pos-Support | Precision | Recall | F1-Score |
| 1 | 1310 | 224 (17.1%) | 0.66 | 0.18 | 0.29 |
| 2 | 1244 | 215 (17.28%) | 0.88 | 0.93 | 0.9 |
| 3 | 94 | 13 (13.83%) | 0.92 | 0.92 | 0.92 |
| 4 | 74 | 14 (18.92%) | 0.88 | 1 | 0.93 |
| 5 | 53 | 32 (60.38%) | 0.93 | 0.84 | 0.89 |
| 6 | 44 | 25 (56.82%) | 1 | 0.96 | 0.98 |
| 7 | 42 | 21 (50%) | 0.88 | 1 | 0.93 |
| 8 | 42 | 19 (45.24%) | 0.94 | 0.84 | 0.89 |
| 9 | 40 | 18 (45%) | 1 | 0.94 | 0.97 |
| 10 | 39 | 18 (46.15%) | 1 | 0.94 | 0.97 |
| 11 | 36 | 12 (33.33%) | 0.91 | 0.83 | 0.87 |
| 12 | 32 | 16 (50%) | 0.66 | 0.18 | 0.29 |
Appendix C. Table containing the minimum, median, and maximum number of whitespace delimited tokens contained in laboratory reports of each disease dataset.
| Minimum | Median | Maximum | |
| Syphilis | 4 | 26 | 815 |
| Salmonella | 4 | 45 | 7055 |
| Histoplasmosis | 4 | 103 | 1151 |
Appendix D. Two example messages corresponding to a positive and negative salmonella laboratory report. Note the lack of consistent structure across messages. Protected health information has been redacted from the messages.
Salmonella (Positive case):
gram stain of blood culturevial indicates presence ofgram negative bacillus. direct mass spectrometry testing performed on positive blood culture bottle indicatespresence of salmonella species. additional testing including susceptibility when appropriate to follow. identifications performed by maldi tofmass spectrometry were developed and performance characteristics were determined by pcl alverno hammond in. salmonella species identified by maldi tof mass spectrometry. sent to illinois state department of public health.
gram stain of blood culturevial indicates presence of gram negative bacillus. culture in progress. identifications performed by maldi tofmass spectrometry were developed and performance characteristics were determined by pcl alverno hammond in. salmonella species identified by maldi tof mass spectrometry.
Salmonella (Negative case):
normal gi flora present preliminary report <date> at <time> no enteric pathogens isolated stool screened for salmonella shigella staphylococcusaureus campylobacter and sorbitol negative e. colifinal report <date> at <time>
Figures & Table
References
- 1.Zheng T, Xie W, Xu L, et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. International Journal of Medical Informatics. 2017;97:120–7. doi: 10.1016/j.ijmedinf.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yala A, Barzilay R, Salama L, et al. Using machine learning to parse breast pathology reports. Breast Cancer Research and Treatment. 2017;161(2):203–11. doi: 10.1007/s10549-016-4035-1. [DOI] [PubMed] [Google Scholar]
- 3.Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH. Advances in electronic phenotyping: From rule-based definitions to machine learning models. Annual Review of Biomedical Data Science. 2018;1(1):53–68. doi: 10.1146/annurev-biodatasci-080917-013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasthurirathne SN, Dixon BE, Gichoya J, et al. Toward better public health reporting using existing off the shelf approaches: The value of medical dictionaries in automated cancer detection using plaintext medical data. Journal of biomedical informatics. 2017;69:160–76. doi: 10.1016/j.jbi.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine. 2018;1(1):18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasthurirathne SN, Biondich PG, Grannis SJ, Purkayastha S, Vest JR, Jones JF . Identification of patients in need of advanced care for depression using data extracted from a statewide health information exchange: A machine learning approach. Journal of medical Internet research. 2019;21(7):e13809. doi: 10.2196/13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasthurirathne SN, Vest JR, Menachemi N, Halverson PK, Grannis SJ. Assessing the capacity of social determinants of health data to augment predictive models identifying patients in need of wraparound social services. Journal of the American Medical Informatics Association. 2017;25(1):47–53. doi: 10.1093/jamia/ocx130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moons KG, Kengne AP, Grobbee DE, et al. Risk prediction models: Ii. External validation, model updating, and impact assessment. Heart. 2012;98(9):691–8. doi: 10.1136/heartjnl-2011-301247. [DOI] [PubMed] [Google Scholar]
- 9.Witten IH, Frank E, Hall MA, Pal CJ. Data mining: Practical machine learning tools and techniques: Morgan Kaufmann. 2016 [Google Scholar]
- 10.Domingos P. A few useful things to know about machine learning. Communications of the ACM. 2012;55(10):78–87. [Google Scholar]
- 11.Gardner J, Yang Y, Baker R, Brooks C. Enabling end-to-end machine learning replicability: A case study in educational data mining. ArXiv e-prints [Internet] 2018. Jun 01, 2018. Available from: https://ui.adsabs.harvard.edu/#abs/2018arXiv180605208G.
- 12.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association. 2012;19(e1):e162-e9. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohn S, Wang Y, Wi C-I, et al. Clinical documentation variations and nlp system portability: A case study in asthma birth cohorts across institutions. Journal of the American Medical Informatics Association. 2017;25(3):353–9. doi: 10.1093/jamia/ocx138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon BE, Grannis SJ, Revere D. Measuring the impact of a health information exchange intervention on provider-based notifiable disease reporting using mixed methods: A study protocol. BMC medical informatics and decision making. 2013;13(1):121. doi: 10.1186/1472-6947-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kochmann M, Wang J, Dixon BE, Kasthurirathne SN, Grannis SJ. AMIA; 2017. Evaluation of text mining methods to support reporting public health notifiable diseases using real-world clinical data. [Google Scholar]
- 16.Dixon BE. What is health information exchange? In: Dixon BE, editor. Health information exchange: Navigating and managing a network of health information systems. Waltham, MA: Academic Press; 2016. pp. 3–20. [Google Scholar]
- 17.Kasthurirathne SN, Dixon BE, Gichoya J, et al. Toward better public health reporting using existing off the shelf approaches: A comparison of alternative cancer detection approaches using plaintext medical data and non-dictionary based feature selection. Journal of biomedical informatics. 2016;60:145–52. doi: 10.1016/j.jbi.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 18.McDonald CJ, Overhage JM, Barnes M, et al. The indiana network for patient care: A working local health information infrastructure. 2005;24(5):1214–20. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 19.Kasthurirathne SN, Grannis SJ. Machine learning approaches to identify nicknames from a statewide health information exchange2019: AMIA Informatics summit 2019 Conference Proceedings. [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez DM, Blobel B. Architectural approaches for hl7-based health information systems implementation. Methods of information in medicine. 2010;49(02):196–204. doi: 10.3414/ME09-02-0012. [DOI] [PubMed] [Google Scholar]
- 21.McDonald CJ, Huff SM, Suico JG, et al. Loinc, a universal standard for identifying laboratory observations: A 5-year update. Clinical chemistry. 2003;49(4):624–33. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 22.Ganesan S. Public health information network vocabulary access distribution system (phin vads): Public health vocabulary standards development, distribution and implementation. 2013 [Google Scholar]
- 23.Joulin A, Grave E, Bojanowski P, Mikolov T. Bag of tricks for efficient text classification. arXiv preprint arXiv:160701759. 2016 [Google Scholar]
- 24.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in python. Journal of machine learning research. 2011;12(Oct):2825–30. [Google Scholar]
- 25.Breiman L. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 26.McHugh ML. The chi-square test of independence. Biochemia medica: Biochemia medica. 2013;23(2):143–9. doi: 10.11613/BM.2013.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain AK. Data clustering: 50 years beyond k-means. Pattern recognition letters. 2010;31(8):651–66. [Google Scholar]
- 28.Doshi-Velez F, Kim B. Considerations for evaluation and generalization in interpretable machine learning. Explainable and interpretable models in computer vision and machine learning. Springer; 2018. pp. 3–17. [Google Scholar]