Abstract
The Department of Veteran’s Affairs (VA) archives one of the largest corpora of clinical notes in their corporate data warehouse as unstructured text data. Unstructured text easily supports keyword searches and regular expressions. Often these simple searches do not adequately support the complex searches that need to be performed on notes. For example, a researcher may want all notes with a Duke Treadmill Score of less than five or people that smoke more than one pack per day. Range queries like this and more can be supported by modelling text as semi-structured documents. In this paper, we implement a scalable machine learning pipeline that models plain medical text as useful semi-structured documents. We improve on existing models and achieve an F1-score of 0.912 and scale our methods to the entire VA corpus.
Introduction
The Veteran’s Health Administration (VHA) is America’s largest integrated health care provider. The VHA provides care to United States Veterans at over 1,250 health care facilities nationwide. Currently, the VHA uses the Veterans Health Information Systems and Technology Architecture (VistA) system to house the electronic health records (EHR) of more than nine million veterans. VistA hosts structured data, images, and unstructured text.
Text Integration Utilities (TIU) is one of the VistA software packages. The TIU package allows healthcare staff to document patients’ care processes as a medical note. The TIU package manages clinical notes creation, editing and search. Clinicians use TIU to enter discharge summaries, progress notes, and other clinical documents. Healthcare facility managers also use TIU to search for documents. TIU also provides users with the ability to sign documents.
The content among TIU notes varies widely. Content varies depending on clinician, facility, and the type of care the patient received. Additionally, content for a specific type of care varies over time. TIU manages this change by creating a hierarchy of document types. All documents inherit from a base document type, and then notes are specialized into discharge summaries, progress notes, etc.
The structure in a newly created TIU note is determined by what attributes that document inherited from the document type hierarchy. Notes often have precompiled templates with boilerplate sentences, text fields, check boxes, and section headings. Clinicians fill out the appropriate fields while giving care to the patient. Clinicians are able to modify documents by adding custom fields or typing additional information.
There is no standard hierarchy of document attributes. Each deployment of the TIU package allows documents to be configured differently. The result is that a document at facility A may record a patient’s weight in a field named weight while another document at facility B records weight in a field called wt. So there can be a wide variety of documents across all the VistA systems.
The Department of Veterans Affairs corporate data warehouse (CDW) stores data from across the enterprise. Extract, transform, and load (ETL) scripts are responsible for moving data from the 130+ different VistA systems into the appropriate enclave in the CDW. Inside the CDW enclaves, most data is stored in traditional relational databases. The clinical documents from TIU are stored in the research enclave and loaded into a single text column of a fact Table 1.
Table 1:
Comparing different sets of features on the testing data
| l1-regularization | l2-regularization | |||||
|---|---|---|---|---|---|---|
| Recall | Precision | F1-score | Precision | Recall | F1-score | |
| aDAI Features | 0.898 | 0.874 | 0.886 | 0.900 | 0.878 | 0.889 |
| xJSONize Features | 0.892 | 0.890 | 0.891 | 0.921 | 0.902 | 0.912 |
These data are extremely valuable to the medical research community because they contain information not captured in the structured portions of the EHR. Yet these data are difficult to use due to the unstructured nature of the text columns. Text columns lend themselves well to inverted indices, keyword searches and look ups but fail to support complex queries on fields within the document. For example, it is reasonable for a researcher to ask for all of the notes where a clinician wrote diabetes in a patient’s past medical history. If the notes are stored as unstructured text, then that user must write a custom application to find the answer to their query. We propose a machine learning approach to model unstructured text to support more complex queries.
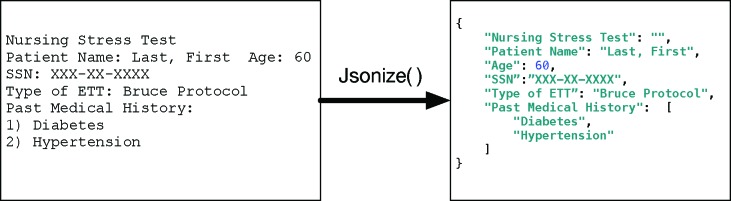
In this paper, we introduce JSONize, a scalable machine learning pipeline that models medical text as scehma-free JSON documents. JSON is a human readable text format for expressing structured data with support for many programming languages 2. A JSON document is a collection of fields and values in a structured format3. Figure 1 shows an example of a clinical document modelled as unstructured text on the left and modelled as a semi-structured JSON document on the right.
Figure 1:
Using JSONize to model a clinical document (left) to a JSON document (right).
Related Work
JSONize is not a clinical information extraction system4. Information extraction deals with modelling unstructured or semi-structured data as structured data5. JSONize models text as semi-structured documents. This distinction separates the JSONize approach from other approaches geared toward information extraction6,7. JSONize makes the simple observation that TIU notes have enough structure to be modelled, stored and queried as semi-structured documents.
JSONize builds on previous approaches to the section identification problem. In a survey of approaches to section identification, Pomares et al. categorize current approaches as either rule-based, purely machine learning or machine-learning- rules hybrids 8. Pomares et al. also compared approaches based on a number of factors like the language the note was written in, narrative type, and performance metrics. The survey shows that conditional random fields out-perform other machine learning approaches while maintaining the ability to generalize. Pomares et al. notes that rule-based approaches lack the ability to generalize to notes from different healthcare facilities and authors.
One of the requirements of our system is to process 20 years worth of clinical documents from 130 VistA systems by hundreds of different authors. Hence we choose to use a token based approach with conditional random fields (CRF) similar to the one introduced by Dai et al.9, which is demonstrated to work well with publicly available data sets. Dai et al. use MedPost to tokenize the medical text. Then Dai et al. generate and normalize multiple features based on word, affix, orthography, lexicon, semantic, and layout. Then Dai et al. train and test a linear-chain CRF model on a publicly available data set achieving an F1-score of 0.942.
Methods
We propose a two-step approach to modelling medical text as a JSON document. First, we solve the section identification problem8. Then we follow up with a rule-based post-processing module that provides structure to the annotated sections. To address the size of our data, we scale out using Apache Spark.
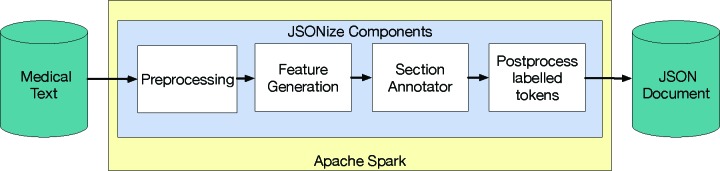
We approached the section identification problem using a method adapted from Dai et al.9. First, we preprocess each document using data set specific techniques that clean the data. Next, we tokenize the documents and generate features. Then we use a binary classifier to classify each token as either a section header or not a section header.
Preprocessing
Prior to processing notes we replace all unicode code point characters less than 32 and greater than 126 with a newline. This fixes errors caused by optical character recognition and accidental insertion of binary data10.
Feature Generation
The feature generation module generates the features that are needed by the CRF classifier for section annotation. We begin with the Apache cTakes implementation of the Penn TreeBank tokenizer to tokenize documents 11. This converts each note into a series of T tokens. For each token, we compute the same features as described in Dai et al.9 and use the SecTag vocabulary for the lexicon features 12. We make two changes in this feature set:
We remove the semantic features that are intended to disambiguate abbreviations. Abbreviations are common throughout medical text and difficult to disambiguate even when context is present. For example, the abbreviation pt appears in approximately 10% of the documents in the corpus and can mean a variety of things depending on the context. In a corpus of billions of documents over a 20 year time period, the amount of abbreviations in the corpus is so vast that the simple semantic features of Dai et al.9 did not make sense.
We change the behavior of the newline feature to be the count of newlines before a token, instead of just checking for an empty line before a token.
We implement the Dai et al. features using Python and denote the resulting feature vector for token t ∈ {1, ..., T} by a t.
There is no standard definition of a section in a medical note. Denny et al. define a section header as a segment that includes words that provide context for the encapsulated text, but whose words themselves do not add specific clinical information 13. This means that each token is classified as a section header or not a section header, which we denote as {key, value}, respectively. We use Denny et al.’s definition of a section header but remove the constraint that a section header cannot add clinical information. We interpret a segment as a phrase not part of a grammatically correct sentence. Consider the sentence, ”The patient’s HR is XX, and the patient’s BP is XX.” We would not label a section header. Consider the string ”Vital Signs: HR XX BP XX”. We would label all of VitalSigns, HR and BP as sections headers (keys). In this regard we deviate from Dai et al. and label sections and subsections as keys.
The a t features model topmost sections well. A topmost section is a section that cannot be viewed as further describing a previous section. Topmost sections are usually sections like Allergies, Medications, and Vital Signs. Rather than only modelling topmost sections, we model all sections in the note and offer the following improvements to the a t features. After the removal of the semantic features from feature change 1 above and the way newlines are calculated from feature change 2, the resulting set of features is called b t.
We add a layout feature, c t, a count of white space preceding a token. The original layout features only account for newline characters. In the VA data set, tabs and spaces can also signal the beginning of a new section. For example, consider Figure 1. The clinical document on the left uses two spaces before the Age : section starts. The c t feature is intended to extend all sections and not just topmost sections.
The output from the feature generation module for a single note with T tokens is a set of T feature vectors x t = (b t, c t), where t = 1, ..., T.
Section Annotation
The section annotation module uses the features from the previous module to classify tokens as a section header or not. CRFs are well suited to classification tasks with a large number of interrelated features. Linear-chain CRFs are probabilistic sequence models for computing the conditional probability p(Y |X) of a label sequence Y = {y 1, ..., y T}, in our case a binary sequence coding {key, value}, given input sequence X = {x 1, ..., x T}. A linear-chain CRF model for token t is given by,
| (1) |
where Z(x) is an instance specific normalization term such that the sum of all possible sequence labels sum to 1, is a set of real-valued feature functions, and λk is areal-valued weight of the feature function. The number of feature functions, K, is given by the number of possible feature vector combinations, where each feature function is coded by a one-hot (dummy variable) 14.
In this paper, we used CRFSuite’s implementation of linear-chain conditional random fields 15. We compared l1-regularization and l2-regularization during training, and we tuned the regularization parameters using grid search on k-fold cross validation on the training data.
Postprocessing
Our post-processing module uses a rule-based approach to organize labelled tokens into a JSON document. JSON documents are built on two structures: objects and arrays. Objects are a collection of field-value pairs. Arrays are collections of values. A value can be a string, number, boolean, null, array, or another object. There are extensions to JSON that support additional data types such as binary data. For this paper we do not consider binary data.
Postprocessing begins by pairing fields with their respective values. Fields are formed by merging all consecutive tokens labelled as a section header. Values are formed by merging together all consecutive tokens labelled as not a section header. Fields are paired to the first value after the last token in the field. In Figure 1, the tokens Last,,, and First were all labelled as not section headers. Since those tokens are consecutive and labelled as not section headers, they were grouped into a single value. The field Patient Name precedes them in the text so together they form a field-value pair.
The field-value pairs form the basis for the JSON document. All field-value pairs that are not part of a subsection are inserted in the JSON document. The current implementation of this module creates subsections for unordered lists and numbered lists, otherwise afield-value pair is not considered a subsection. An unordered list is a field that begins with a hyphen. An ordered list is a field that begins with a Roman numeral, numeric with or without surrounding punctuation, or single letter with punctuation. In Figure 3 impression is section heading, and the different ordered lists are subsections of the impression section.
Figure 3:
Examples of Ordered Lists in Medical Notes.
Apache Spark
Apache Spark is a distributed computing framework 16. The Spark application programming interface supports a map-reduce programming paradigm. A core component of Spark is the DataFrame abstraction which is a data set that is horizontally distributed into a number of partitions. Operations on DataFrames are broken up into tasks that can be executed in parallel and distributed to workers. These workers can efficiently and reliably work in parallel across many nodes.
In addition to fault tolerant computation across multiple nodes, Spark provides parallel input/output libraries based on the Apache Parquet file format 17. Spark allows easy use of the parquet file format through the DataFrame application programming interface. Parquet allows us to store clinical documents efficiently and retrieve them quickly.
For this pipeline, we configured Apache Spark with two executors per node. Each executor had 55 GB of memory and 16 cores. We ran the master and driver on the same node. We used the local disks for spill directories and wrote the final outputs to a parallel file system.
Results
As of the writing of this paper, the Oak Ridge National Laboratory (ORNL) version of the VA corpus does not update via the ETL process. The ORNL version was last updated in October 2016. This version contains 3.4 billion TIU notes. We exported these notes out of SQL Server and stored the notes on a parallel file system. Due to security constraints, we implemented a Kerberos friendly Java Database Connectivity wrapper for Apache Spark to connect to SQL Server. After the notes were extracted from SQL Server. the notes were stored in parquet format with gzip compression. The total size of all 3.4 billion notes is 1.9 TB.
Generating the CRF Model
We randomly sampled 1,000 documents. We manually labelled all sections in 515 documents. The number of distinct tokens labelled was 154,148. The number of tokens annotated as sections were 25,332. We used our own annotation software for this task. After labelling we preprocessed all of the documents and generated features.
We split the labelled documents into training data and testing data. Our training data was 309 documents, and the test data was 206 documents. We tuned the regularization parameters on the training data using grid search and k-fold cross validation. We used five folds and searched the range between 0.02 and 2.00 in equal increments of 0.02 for the l1- and l2- regularization parameters. After choosing the parameters we trained a CRF model on the training data. Then we evaluated the model based on precision, recall, and F1-score. We performed this process for the a DAI features and for the x JSONize features (see Table 1). We found that the F1-score for the model using x JSONize was 0.912 and the F1-score for the model using a DAI was .889.
Finally, we trained a CRF model using all of the labelled data with the features and parameters that worked best on the test data. We stored the resulting model in a pickle file. This model was used by the section annotation module.
Running the Entire Corpus
We converted all 3.4 billion clinical documents to JSON documents using the JSONize pipeline. This pipeline was implemented in Python and Scala using 20 nodes of the Apache Spark cluster. The entire pipeline finished in 103 hours. 23 hours were spent in preprocessing and feature generation modules. The section annotator used 75 hours, and the postprocessing module took 5 hours. After the post-processing module was completed, we stored the final output in parquet files with one column which was a JSON document.
Final Analysis
After running JSONize on the entire VA corpus, we reviewed the output by counting the frequency of fields in the JSON documents. We went through each document and counted the number of times each field appears. Note that we did not review arrays. The total number of unique fields was 220,868,034. There are 36,247 fields that have a frequency greater than 100,000. Table 2 lists the top 50 fields by frequency count. There are fields related to medications, allergies, vital signs, and Subjective Objective Assessment Plan (SOAP) notes.
Table 2:
The top 50 most frequent fields from the 3.4 billion JSON documents
| Field | Frequency (millions) | Field | Frequency (millions) |
|---|---|---|---|
| sig | 597 | medications | 139 |
| active outpatient medications | 397 | o | 137 |
| comment | 364 | a | 136 |
| refills | 360 | level of understanding | 132 |
| qty | 357 | if | 132 |
| comments | 339 | chief complaint | 132 |
| last | 317 | heent | 126 |
| expr | 315 | location | 126 |
| plan | 311 | blood pressure | 126 |
| issu | 304 | vital signs | 124 |
| other | 303 | resp | 119 |
| allergies | 283 | skin | 115 |
| pulse | 271 | abdomen | 115 |
| date | 271 | bmi | 112 |
| pain | 251 | labs | 112 |
| weight | 245 | lungs | 111 |
| p | 215 | provider | 110 |
| assessment | 189 | eval | 108 |
| bp | 172 | objective | 109 |
| temp | 161 | neck | 105 |
| height | 161 | active non-va medications | 102 |
| age | 157 | activity | 100 |
| diagnosis | 150 | respiration | 97 |
| s | 149 | neuro | 97 |
| temperature | 140 | os | 96 |
Medication related keys appeared frequently. The TIU notes often contain a list of current medications, expired medications, and active medications prescribed by a doctor outside the VA. Many medication lists provide the drug name, usage information sig, the amount qty, the number of refills refills, when the medication was prescribed issu, when the medication was last filled last, and when the medication expires expr.
SOAP is a common method for structuring a patient’s note. In the EHR, these fields are often abbreviated to just s, o, a, and p. The token subjective only appears in 4% of notes, and in an overwhelming majority of the documents that contain the token subjective, it is only used once. JSONize made 64% of the uses of the word subjective a field. The word subjective probably does not have many uses in the EHR outside of SOAP notes. The words assessment and plan are common in prose and also used as section headings. Table 3 summaries these numbers and provides field frequency, word frequency, and document frequency.
Table 3:
This table gives field frequency, word frequency and the document frequency of selected words from the 3.4 billion JSON documents. All units are in millions.
| Field | Field Frequency | Word Frequency | Document Frequency |
|---|---|---|---|
| subjective | 96 | 150 | 144 |
| objective | 109 | 192 | 149 |
| assessment | 189 | 1,478 | 771 |
| plan | 312 | 1,463 | 900 |
| s | 150 | 2,061 | 906 |
| o | 137 | 1,094 | 623 |
| a | 137 | 7,836 | 1,832 |
| p | 215 | 1,123 | 674 |
Discussion
We were able to model 3.4 billion TIU notes as JSON documents. Our method is scalable and reasonable to compute on big medical data. The resulting JSON documents can support complex queries not easily performed on unstructured notes.
We used this data in an initial use case for improving existing tobacco and alcohol use phenotypes. We were able to query the most frequent fields that record tobacco and alcohol habits. We queried fields such as tobacco, tobacco use, alcohol, alcohol use, and etoh. We were able to combine the structured data on smoking habits with the smoking data that we found in the medical text. We found that often the medical text provides higher resolution of tobacco use than the structured data. For example, notes can record how many packs per day are smoked.
We acknowledge some of the limitations of our paper. We plan to label more documents so that we have more training and test data. Also, the postprocessing module needs more logic to correctly model all of the different ways that notes can be recorded. For example, occasionally keys can follow the value in medical text. Lastly, we plan to evaluate deep learning approaches in the section annotation module.
Conclusion
We implemented a machine learning pipeline that transformed clinical documents into JSON documents. This pipeline uses state-of-the-art techniques and machine learning features. We evaluated our CRF model using a labelled data set from the VA corpus of TIU Notes and achieved an F1-score of 0.912. This is 0.023 higher than a leading state-of-the-art method using CRFs.
We scaled our technique to process 3.4 billion clinical documents. Our entire pipeline finished in 103 hours using a 20 node Apache Spark cluster. After we modelled all the documents as JSON documents, we counted the occurrences of fields in the JSON documents. For SOAP notes, we compared the field frequency with word frequency and document frequency.
Acknowledgments
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award #MVP000. This publication does not represent the views of the Department of Veterans Affairs or the United States Government.
This manuscript has been in part co-authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725, and under a joint program with the Department of Veterans Affairs under the Million Veteran Project Computational Health Analytics for Medical Precision to Improve Outcomes Now. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
This research used resources of the Knowledge Discovery Infrastructure at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725.
Figures & Table
Figure 2:
The components of JSONize.
References
- [1].Price LE, Shea KD, Gephart SM. The Veterans Affairs’s Corporate Data Warehouse: Uses and Implications for Nursing Research and Practice. Nursing administration quarterly. 2015;39(4):311–8. doi: 10.1097/NAQ.0000000000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].ECMA International. The JSON Data Interchange Syntax. 2017 Available from: http://www.ecma-international.org/publications/files/ECMA-ST/ECMA-404.pdf. [Google Scholar]
- [3].MongoDB. The MongoDB Manual. 2019 Available from: https://docs.mongodb.com/manual/ [Google Scholar]
- [4].Wang Y, Wang L, Rastegar-Mojarad M, Moon S, Shen F, Afzal N, et al. Clinical information extraction applications: A literature review. Journal of Biomedical Informatics. 2018;77:34 – 49. doi: 10.1016/j.jbi.2017.11.011. Available from: http://www.sciencedirect.com/science/article/pii/S1532046417302563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sarawagi S. Information Extraction. Foundations and Trends in Databases. 2008;1(3):261–377. Available from: http://dx.doi.org/10.1561/1900000003. [Google Scholar]
- [6].Finch DK, McCart JA, Luther SL. TagLine: Information Extraction for Semi-Structured Text in Medical Progress Notes. AMIA Annu Symp Proc. 2014;2014:534–543. [PMC free article] [PubMed] [Google Scholar]
- [7].Tran LT, Divita G, Redd A, Carter ME, Samore M, Gundlapalli AV. Scaling Out and Evaluation of OBSecAn, an Automated Section Annotator for Semi-Structured Clinical Documents, on a Large VA Clinical Corpus. AMIA Annu Symp Proc. 2015;2015:1204–1213. [PMC free article] [PubMed] [Google Scholar]
- [8].Pomares-Quimbaya A, Kreuzthaler M, Schulz S. Current approaches to identify sections within clinical narratives from electronic health records: a systematic review. BMC Medical Research Methodology. 2019 Jul;19(1):155. doi: 10.1186/s12874-019-0792-y. Available from: https://doi.org/10.1186/s12874-019-0792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dai HJ, Syed-Abdul S, Chen CW, Wu CC. Recognition and Evaluation of Clinical Section Headings in Clinical Documents Using Token-Based Formulation with Conditional Random Fields. BioMed Research International. 2015 doi: 10.1155/2015/873012. Available from: http://dx.doi.org/10.1155/2015/873012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zeng QT, Redd D, Divita G, Jarad S, Brandt C, Nebeker J. Characterizing Clinical Text and Sublan-guage: A Case Study of the VA Clinical. Journal of Health and Medical Informatics. 2011 Available from: http://dx.doi.org/10.4172/2157-7420.S3-001. [Google Scholar]
- [11].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010 09;17(5):507–513. doi: 10.1136/jamia.2009.001560. Available from: https://doi.org/10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Denny JC, Spickard A, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a Method to Identify and Categorize Section Headers in Clinical Documents. Journal of the American Medical Informatics Association. 2009 11;16(6):806–815. doi: 10.1197/jamia.M3037. Available from: https://doi.org/10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Denny J, A Miller R, B Johnson K, Spickard A. Development and Evaluation of a Clinical Note Section Header Terminology; AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2008. pp. 156–60. 2008. [PMC free article] [PubMed] [Google Scholar]
- [14].Sutton C, McCallum A. An Introduction to Conditional Random Fields. Found Trends Mach Learn. 2012 apr;4;(4):267–373. Available from: http://dx.doi.org/10.1561/2200000013. [Google Scholar]
- [15].Okazaki N. CRFsuite: a fast implementation of Conditional Random Fields (CRFs) 2007 Available from: http://www.chokkan.org/software/crfsuite/ [Google Scholar]
- [16].Zaharia M, Chowdhury M, Franklin MJ, Shenker S, Stoica I. Spark: Cluster Computing with Working Sets; Proceedings of the 2Nd USENIX Conference on Hot Topics in Cloud Computing. HotCloud’10; Berkeley, CA, USA; 2010. pp. 10–10. USENIX Association. Available from: http://dl.acm.org/citation.cfm?id=1863103.1863113. [Google Scholar]
- [17].Apache Software Foundation. Apache Parquet. 2019 Available from: https://parquet.apache.org/ [Google Scholar]