Abstract
Half a million people die every year from smoking-related issues across the United States. It is essential to identify individuals who are tobacco-dependent in order to implement preventive measures. In this study, we investigate the effectiveness of deep learning models to extract smoking status of patients from clinical progress notes. A Natural Language Processing (NLP) Pipeline was built that cleans the progress notes prior to processing by three deep neural networks: a CNN, a unidirectional LSTM, and a bidirectional LSTM. Each of these models was trained with a pre- trained or a post-trained word embedding layer. Three traditional machine learning models were also employed to compare against the neural networks. Each model has generated both binary and multi-class label classification. Our results showed that the CNN model with a pre-trained embedding layer performed the best for both binary and multi- class label classification.
Introduction
Tobacco use is a primary cause of many afflictions ranging from coronary heart disease to lung cancer. Approximately 500,000 people die every year from smoking-related issues in the United States [1], and 50% of people who smoke die of smoking-related complications. Cigarette smoking is the number one cause of lung cancer, contributing to about 90% of lung cancer related deaths [2]. Therefore, it is of utmost urgency to employ preventive measures such as smoking cessation and correspondingly, reduce smoking and tobacco products consumption. Identification of individuals who smoke will enable providers to implement methods to treat tobacco dependence. However, accurate detection of smokers is problematic due to clinic workflows and data quality issues.
The Patient Past Medical History (PMS) includes questions for identifying smoking status in clinical settings, however data quality is limited by inconsistencies. First, PMS is often misleading due to the poor wording used during the interview [3]. Second, survey administration procedures are not standardized across organizations [3]. Smoking status can be extracted from unstructured sources like Electronic Health Records (EHRs) and progress notes [4] as clinicians record patients' use of tobacco in their progress notes [5]. However, a major obstacle to parsing through progress notes is their unstructured nature [6]. Hence, extracting relevant information requires going through progress notes manually [7].
Natural language processing (NLP) has advanced to make unstructured texts more accessible and consumable [8-9]. NLP techniques transform free text into extracted concepts that a machine can better identify. This allows machine learning to perform a variety of tasks such as classifying and adjusting intensive care risks by identifying procedure and diagnosis [10], detecting heart failure criteria [11], identifying adverse drug effects [12-13], detecting the status of autism spectrum disorder [15] or asthma [14], and estimating the activity of rheumatoid arthritis [16].
Many of the first algorithms introduced to extract information used rule-based approaches and traditional machine learning methods. Traditional machine learning methods include logistic regression, random forest, naive-bayes, and most notablely, support vector machine (SVM). Of particular relevance to this paper is the Mayo Clinic NLP System developed to address the i2b2 Smoking Status Discovery Challenge [17]. In this system, three levels of classification were implemented, the first two of which were based on making decisions from a rule-based perspective. The third level of classification employed a SVM with manually selected temporal resolution words and date indications as the features [18]. Although, the system was fairly accurate in classifying patients on their smoking status, desired improvements were identified. Many of these improvements were rooted in the need to manually select features for
the machine learning algorithm. This led to certain key features not being measured or non-relevant features being over analyzed [17].
Deep learning techniques have effectively captured long-range dependencies through deep hierarchical feature construction [19]. These techniques require a lot of computational power and huge stores of training data. With the ever-increasing abundance of patient data, applying deep learning models to health records and achieving comparable results have become realistic. A number of publications have applied deep learning to EHR data in order to perform clinical informatics tasks. These publications indicate that deep learning techniques often perform better than traditional machine learning methods and do not require as thorough preprocessing or feature engineering [20-21]. Furthermore, deep learning produces higher accuracy in a myriad of applications. Therefore, our hypothesis is that smoking status can be extracted from EHR with better accuracy through the use of deep neural networks.
In this paper, we investigate the application of three different deep learning or Deep Neural Network (DNN) models on EHR data in order to extract patient smoking status. The primary aim of this study is to identify whether deep learning has the potential to identify smoking status of patients better than parsing past medical history or traditional machine learning models. Three traditional machine learning models were developed and trained to serve as a benchmark. We looked at both binary classification of status (Smoker vs. Non-smoker) as well as multi-class classification (Current Smoker vs. Former Smoker vs. Non-smoker). Before entering the data through these models, the text within each of the progress notes were cleaned through an NLP pipeline. In addition, two sets of word embeddings were created for the vocabulary present in the progress notes, one pre-trained, the other trained on the progress notes themselves.
Methods
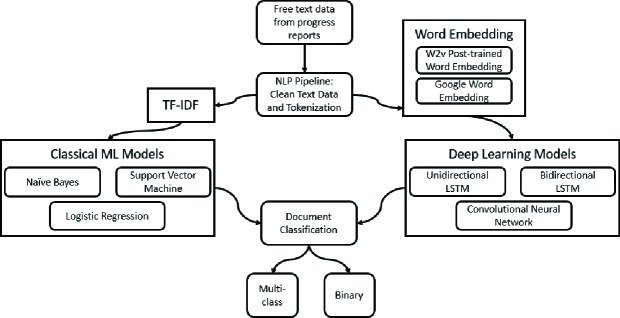
A generalized workflow used to process text and classify documents is shown in Figure 1. The DEMON-Isilon, a High Performance Computing Cluster at Wake Forest University Health Sciences was used to train the deep-learning models. The DEMON’s centralized storage system provides over 190 TB of storage, 1,358 2.6GHz Intel CPU cores, and 4,992 706MHz Nvidia GPU cores, and is useful for running complex models.
Figure 1.
General Workflow of Text Processing and Document Classification for this study.
Establishing Labels:
This study was approved by the Wake Forest Health Sciences IRB. A total of 6,298 de-identified progress notes belonging to 781 patients were obtained from the Translational Data Warehouse. These progress notes were compiled together based on a unique patient ID. Henceforth, we will refer to these compiled notes as patient notes. For each patient, smoking status labels were extracted from the Patient Social History in Epic electronic health records (EHR). Because of the errors that are present in social histories, labels for each progress note were cleaned through a rule- based algorithm. All 781 patients were given two labels, one for the binary DNN models, another for the multi-class DNN models. Binary labels were 'Smoker' and 'Never Smoker'. Multi-class labels were 'Current Smoker', 'Former Smoker', and 'Never Smoker'.
Preprocessing Text:
Prior to processing by the machine learning models, the patient notes were cleaned via a natural language processing (NLP) pipeline. The NLP pipeline contains several features applied to clean the text. These features were created through the use of nltk, regex, and autocorrect python libraries. Figure 2 depicts the flow in which these features were applied to the text.
Figure 2.
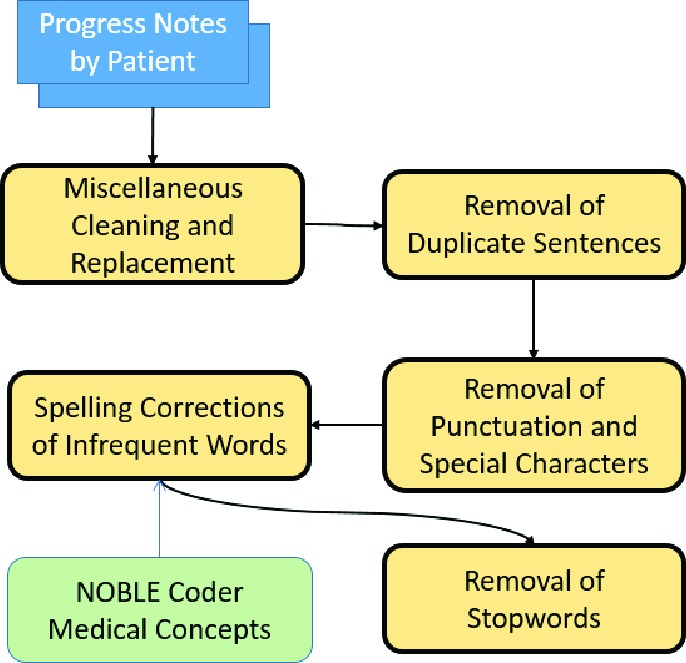
Order of steps taken to preprocess patient notes
I. Miscellaneous Cleaning:
Strings in the text were standardized to fit a more common English Lexicon. This involves replacing certain strings of text with certain other characters that were better processable as well as removing contractions and cleaning abbreviations. For example, a replacement would include changing all instances of "e - mail" to "email".
II. Removal of Repeat Sentences:
Physicians often copy sections of the progress notes from previous visits into the new progress notes [22]. It was decided that such copied lines provided no new information. To increase performance and efficiency when training machine learning models, verbatim sentences in each patient note were removed, leaving only the first instance of a sentence in the patient note.
III. Removal of Punctuation:
Punctuation was not necessary for our algorithm to capture the semantic meaning inside the progress notes [10]. Hence, for more efficient processing, punctuation and special characters were removed.
IV. Correction of Spelling:
To embed the words present in the progress notes, it was essential for the words to be recognized by the pre-trained corpus. Moreover, it was desirable for the post-trained corpus to recognize two words as the same despite any slight spelling inaccuracies. Correspondingly, we decided to correct the spelling of the most infrequent words: words that appear only once throughout the text of each patient note due to the fact that many misspelled words would be in the infrequent list. In order to distinguish the biomedical term that appears only once and a misspelled word, we utilized the NOBLE Coder, which autocodes free text with concepts from NCIt controlled terminology [23]. The Noble Coder result allowed us to identify whether an infrequent word is a medical concept, consequently it would not be subject to autocorrection. Once this identification was complete, any words that were in the infrequent list, but not a medical concept, were autocorrected.
V. Removal of Stopwords:
As employed by many natural language processing techniques, our pipeline has removed the most common stop words
(e.g. "a", "the", and "it" etc.). These words don't present much semantic meaning to a sentence so their removal greatly improves the efficiency of later algorithms [7].
Word Embeddings:
Once the text was cleaned, two different word embeddings were created: pre-trained and post-embedding. Word embeddings can capture the semantic meaning of words by converting them into numeric vectors [24]. Vectors of semantically similar words would be closer to each other. Word embedding has been applied in many biomedical named entity recognition (NER) tasks [25-26].
Before creating the embeddings, the data sequences (patient notes) lengths were truncated to ~100,000 tokens. This greatly improved efficiency of word embedding training as well as the eventual deep learning model training. The first embedding uses a word2vec model pre-trained from the Google news corpus. This word vector model contains approximately three million three-hundred dimensional vectors. The second embedding was word2vec model created specifically from the patient notes, which we will refer to as post-trained. The latter word2vec model theoretically captures more of the complex medical terminology that is present in the patient notes. Moreover, word2vec is more capable of capturing the semantic relationships between words within each progress note. In order to control any variables with dimensionality, we set the length of post-trained word2vec word vectors to be three hundred. The gensim library from Python was employed to process both these word embeddings.
Traditional Machine Learning Methods:
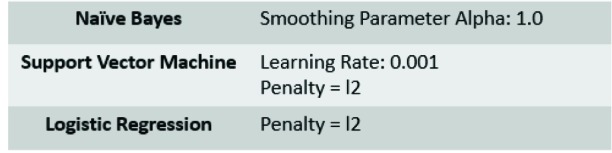
Three traditional machine learning methods were created in this study to serve as a comparison for model performance of deep learning. These models are the Naive Bayes (NB) [27], Support Vector Machine (SVM) [28], and Logistic Regression (LR) [29]. We have generated the term frequency–inverse document frequency (TF-IDF) vectors [30] on the processed text using unigrams with a minimum document frequency of 1, and a maximum document frequency of 100%. Parameters for the algorithms are shown in Table 1.
Table 1.
Hyperparameters for Traditional machine learning models
 |
Deep Learning Models:
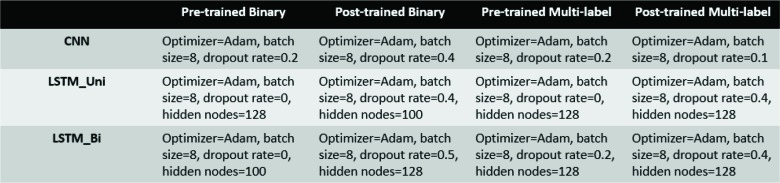
We have used three deep learning models to classify patients based on smoking status: a unidirectional Long short- term memory (LSTM) model, a bidirectional LSTM model, and a Convolutional Neural Network (CNN) model. The specifications for each of the deep learning models are listed below in Table 2.
Recurrent Neural Networks:
Recurrent Neural Networks (RNN) are neural networks that attempt to model time dependencies and sequential events by adding additional weights to the network, creating cycles in the network [26]. There are many variations of RNN. Especially relevant to this study is the long short-term memory (LSTM) network which was built to handle the gradient vanishing problem that often occurs in a standard RNN [31]. DeepCare, a network that predicts future medical outcomes using electronic health records, was created using an LSTM model [32]. In this current study, both a unidirectional LSTM (LSTM_Uni) and a bidirectional LSTM (LSTM_Bi) model were created to extract smoking status from patient notes.
CNN:
Convolutional neural networks have been applied successfully in many previous image processing and text processing studies [32]. CNNs work by modeling hierarchical complicated patterns using smaller and simpler patterns [33]. Convolutional layers along with the max-pooling layer allows models to learn useful word representations while also making the model more computationally efficient.
Hyperparameter Optimization:
The grid-search technique was performed to optimize hyperparameters in the deep learning models. Parameters that were tuned include learning rate, dropout, and number of hidden layers.
Model Setup and Evaluation:
To be consistent between the different deep learning models, we split the data into 0.66/0.33 train/test datasets. There were no overlapping patients between the training and testing set for each model. Five-fold cross-validation was performed for each model in order to produce more accurate metrics. For DNNs that classified patient notes based on binary labels, binary cross-entropy was used as the loss function. For DNNs classifying patient notes based on multi- categorical labels, categorical cross-entropy was used as the loss function. For both RNN and CNN algorithms, early- stopping was implemented. In other words, the model stopped training if the loss function does not improve for 2 epochs on the validation dataset. Four evaluation metrics were used to compare the performance of the models: accuracy, precision, recall, and F1 score.
Open Source Platforms:
High-level NN API Keras (https://keras.io/) using Tensorflow (https://github.com/tensorflow/tensorflow) as a backend was used to set up the neural network structures. Word embedding was performed using Gensim (https://radimrehurek.com/gensim/). One of the embedding techniques used in this investigation employed Google News Vectors ((https://github.com/mmihaltz/word2vec-GoogleNews-vectors). Noble Coder (http://noble- tools.dbmi.pitt.edu/) was used to extract medical concepts for token comparison in the NLP pipeline. Deep learning models were created using the Keras python library. Code for this study can be found at https://github.com/criw- git/NLP_Smoking_Extraction.
Results
Due to the stochastic nature of machine learning algorithms, each model was repeated 10 times. We calculated the average metrics and corresponding standard deviations. The performance metrics for each model is shown below in Table 3 and Table 4. Performance metrics are further elaborated upon in Figures 3-6.
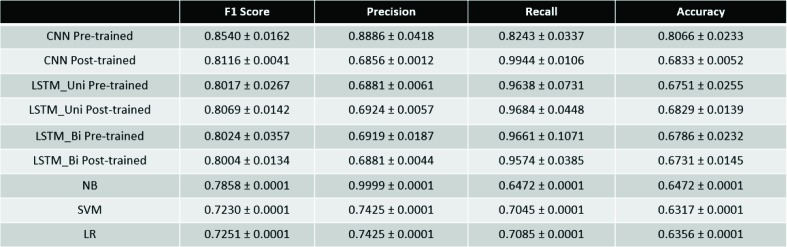
Table 3.
Performance Metrics for Binary Classification Tasks
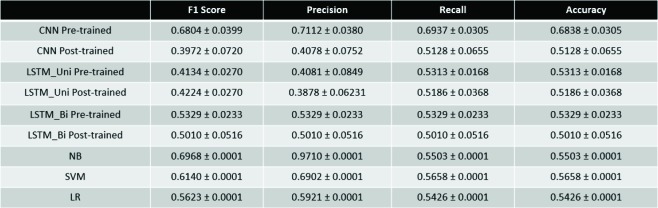
Table 4.
Performance Metrics for Multi-class Classification Tasks
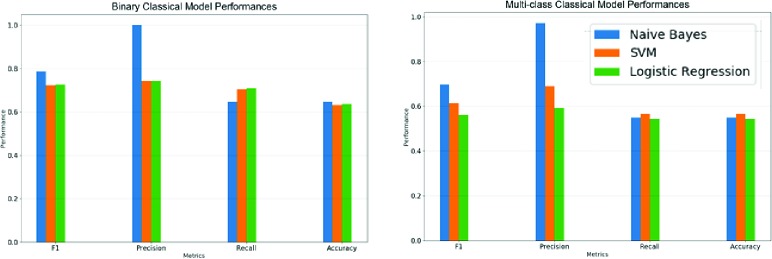
Figure 3.
Performance metrics for traditional machine learning models.
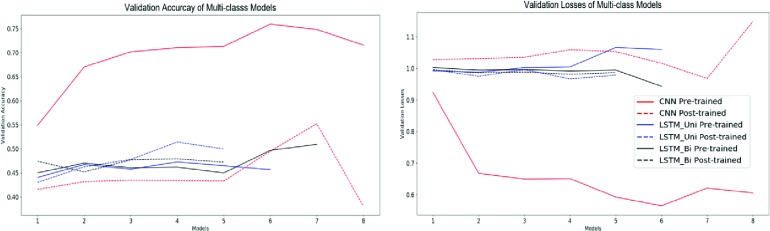
Figure 6.
Validation accuracies and losses for multi-class classification deep learning models.
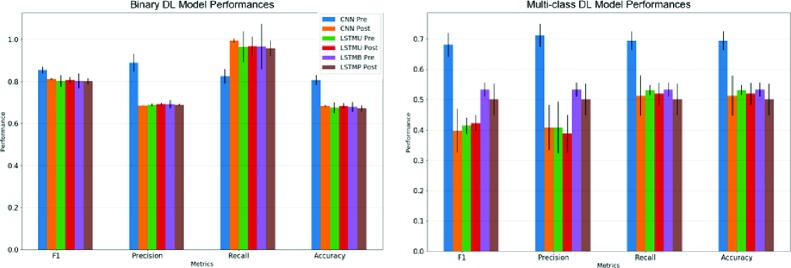
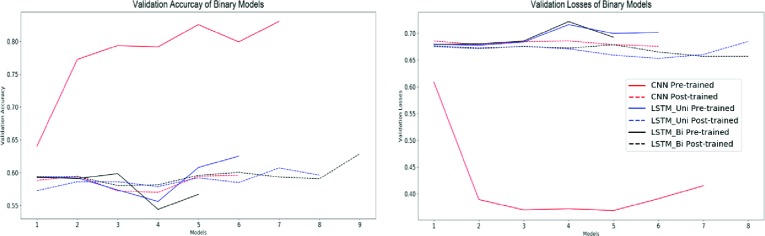
Because the tasks carried out by the models were classification-based, accuracy and F1 score will be the principal metrics we use to evaluate these models. For binary classification, the CNN with pre-trained word embeddings performed the best both in accuracy (0.8066) and F1 score (0.8540). It should also be noted that all the binary classification DNNs outperformed all of the traditional learning methods in both accuracy and F1 score.
For multi-class classification, the CNN with pre-trained embedding performed the best in terms of accuracy (0.6838). However, the Naive-Bayes model provided a comparable F1 Score (0.6968) to the CNN Pre-trained model (0.6804).
Discussion
In this study, we have successfully applied a natural language pipeline coupled with deep learning methods to extract smoking status from clinical progress notes. When looking specifically at the deep learning models, the CNN model with pre-trained word embeddings demonstrated significantly better performance than any of the models both in binary label classification and multi-class label classification.
There may be multiple reasons for this outcome. CNN models are able to learn text input by detecting patterns of multiple sizes. In essence, this learning mechanism allows CNNs to extract local and position-invariant features well [34]. RNNs, like LSTMs, make use of sequential data where the current step has a relationship with a previous one [26]. RNNs works well in applications where sequential information is especially important. For tasks which are more classification based, like the ones performed in this study, CNNs often perform better and provide a more efficient model structure. On the other hand, RNNs are better used in machine translation applications or in cases where the model is to predict what comes next in a certain sequence.
Another reason for the performance result may be the limited data. For this study, we only had access to 781 patient notes. Initially, the idea of not compiling separate progress notes into one patient note was considered. However, not
all progress notes had associated smoking status labels. The only accessible labels were patient-level which is why we compiled all of one person's progress notes into a patient note (one data sample input). There are many effects of this lack of data. First, the post-trained word2vec models were significantly worse than the pre-trained word embedding models. The post-trained model learns the meaning of words based on the context of those words within each progress note. As such, larger numbers of progress notes would allow for a superior post-trained word2vec model and embedding. The lack of data prevented the post-trained word2vec model from learning to its best ability and therefore, the pre-trained word2vec model was able to perform better. Second, the lack of data may have affected the learning potential of the models. As deep learning models take much more data to train [20], we speculate that both the CNN and RNN models were impacted by the deficit of data but the RNN models were especially impaired. he results demonstrate that all of the deep learning models, with the exception of the CNN pre-trained model, hit a certain maximum in terms of performance. This is particularly true for the binary classification models where metrics closed out at an F1 Score of 0.80 and an accuracy of 0.68. We hypothesize that this may be due to the DNNs hitting a ceiling in their learning capacity. Essentially, they are unable to learn further information with the amount of data provided to them [35]. We believe that given more data, these models would be able to overcome this ceiling.
When comparing the traditional and deep learning methods against each other, it seems that DNNs outperformed traditional methods in binary classification tasks. In multi-class classification tasks, all of the models provided more or less similar accuracy scores save for the CNN Pre-trained model, which outperformed all of the models. However, the traditional models did provide significantly better F1 scores than many of the deep learning models. We suspect that this may be due to the lack of data. Given more data, we presume that DNNs will be able to outperform traditional methods in multi-class classification as well.
Our goal in this study was to implement an automated system for clinical progress note classification using deep learning. Moving forward, we plan to further investigate the capabilities of the NLP and DNN analysis pipelines given a larger data set. We will also look to see if this pipeline can be used to perform other classification tasks. Another possibility for future studies is to generalize this pipeline to read data from multiple research facilities and multiple resources. Not only would this increase the number of samples for models to train on, but it would also allow for a more robust pipeline.
Conclusion
Automated NLP coupled with deep learning was demonstrated to have potential in document classification tasks for unstructured clinical progress notes. It is important that this field be widely investigated in order to develop better ways of identifying target patients.
Acknowledgements
We thank Roshan Kumar Subramanian for providing us aid on several steps throughout the project. We also thank Meijian Guan for providing a template model for the deep learning algorithms. We would also like to thank Brian Ostasiewski for providing administrative and data support. The work is partially supported by the Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197) and by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (UL1TR001420).
Figures & Table
Figure 4.
Performance metrics for deep learning models. For reference, LSTMU is the unidirectional LSTM model whereas LSTMB is the bidirectional LSTM model
Figure 5.
Validation accuracies and losses for binary classification deep learning models.
Table 2.
Hyperparameters for deep learning models
References
- 1.Lariscy J. T. SSM - population health. 2019. Smoking-attributable mortality by cause of death in the United States: An indirect approach; p. 100349. doi:10.1016/j.ssmph.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health. 2014. U.S. Department of Health and Human Services.The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. [Google Scholar]
- 3.Agency for Healthcare Research and Quality. Data Sources for Health Care Quality Measures [Internet]. Content last reviewed July 2018. Available From: https://www.ahrq.gov/talkingquality/measures/understand/index.html. [Google Scholar]
- 4.Geisler BP, Schuur JD, Pallin DJ. Estimates of Electronic Medical Records in U.S. Emergency Departments. PLoS ONE. 2010;5(2) doi: 10.1371/journal.pone.0009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polubriaginof F. PMC US National Library of Medicine. 2018. Collecting Smoking Status in Electronic Health Records; pp. 1392–400. [PMC free article] [PubMed] [Google Scholar]
- 6.Charles D. 2008-2014. Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals. 2015. [cited 2019 Jun 14] Available From: https://www.healthit.gov/sites/default/files/databrief/2014HospitalAdoptionDataBrief.pdf. [Google Scholar]
- 7.Sebastiani F. Machine learning in automated text categorization. ACM Computing Surveys (CSUR) 2002;31(1):1–47. [Google Scholar]
- 8.Liao KP, Ananthakrishnan AN, Kumar V. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One. 2015;10(8):e0136651. doi: 10.1371/journal.pone.0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCoy TH, Castro VM, Cagan A. Sentiment measured in hospital discharge notes is associated with readmission and mortality risk: an electronic health record study. PLoS One. 2015;10(8):e0136341. doi: 10.1371/journal.pone.0136341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marafino BJ, Davies JM, Bardach NS, Dean ML, Dudley RA. N-Gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. J Am Med Inform Assoc. 2014;21(5):871–5. doi: 10.1136/amiajnl-2014-002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd RJ, Steinhubl SR, Sun J. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int J Med Inform. 2014;83(12):983–92. doi: 10.1016/j.ijmedinf.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi- corpus training. J Biomed Inform. 2015;53:196–207. doi: 10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S. Text Mining for Adverse Drug Events: the promise, challenges, and state of the art. Drug Saf. 2014;37(10):777–90. doi: 10.1007/s40264-014-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ST W, Juhn YJ, Sohn S, Liu H. Patient-level temporal aggregation for text-based asthma status ascertainment. J Am Med Inform Assoc. 2014;21(5):876–84. doi: 10.1136/amiajnl-2013-002463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J. Autism Spectrum disorder detection from semi-structured and unstructured medical data. EURASIP J Bioinforma Syst Biol. 2017;3:1–9. doi: 10.1186/s13637-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, Karlson EW, Canhao H, Miller TA, Dligach D, Chen PJ. Automatic prediction of rheumatoid arthritis disease activity from the electronic medical records. PLoS One. 2013;8(8):e69932–10. doi: 10.1371/journal.pone.0069932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn S, Savova G. K. AMIA ... Annual Symposium proceedings. AMIA Symposium. 2009. (2009). Mayo clinic smoking status classification system: extensions and improvements; pp. 619–623. [PMC free article] [PubMed] [Google Scholar]
- 18.Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Computation. 2001;13(3):637–49. [Google Scholar]
- 19.Goodfellow I, Bengio Y, Courville A. MIT Press. 2016. Deep learning. [Google Scholar]
- 20.Domingos P. “A few useful things to know about machine learning,” Communications of the ACM. 2012;55(10):78–87. [Google Scholar]
- 21.Edward Choi, Mohammad Taha Bahadori, Andy Schuetz, Walter F Stewart, Jimeng Sun. 2016. Doctor ai: Predicting clinical events via recurrent neural networks; In Machine Learning for Healthcare Conference; pp. 301–318. [PMC free article] [PubMed] [Google Scholar]
- 22.Thielke S, Hammond K, Helbig S. Copying and pasting of examinations within the electronic medical record. Int J Med Inform. 2007;76(1):S122–S128. doi: 10.1016/j.ijmedinf.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Tseytlin E, Mitchell K, Legowski E, Corrigan J, Chavan G, Jacobson R. S. (2016). NOBLE - Flexible concept recognition for large-scale biomedical natural language processing. BMC bioinformatics 17. 32 doi: 10.1186/s12859-015-0871-y. doi:10.1186/s12859-015-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengio Y, Ducharme R, Vincent P. A neural probabilistic language model. J Mach Learn Res 2003. 3:1137–55. [Google Scholar]
- 25.Liu S, Tang B, Chen Q. Effects of semantic features on machine learning-based drug name recognition systems: word embeddings vs. manually constructed dictionaries. Information 2015. 6(4):848–65. [Google Scholar]
- 26.Tang B, Cao H, Wang X. Evaluating word representation feature in biomedical named entity recognition tasks. Biomed Res Int 2014. 2014;1 doi: 10.1155/2014/240403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazmierska J, Malicki J. Application of the Naı v̈ e Bayesian classifier to optimize treatment decisions. Radiother Oncol 2008. 86(2):211–6. doi: 10.1016/j.radonc.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Joachims T. Machine learning: ECML-98. Berlin, Heidelberg: Springer. 1998. Text categorization with support vector machines: learning with many relevant features; pp. 137–42. doi:10.1007/BFb0026683. [Google Scholar]
- 29.Speech and Language Processing. https://web.stanford.edu/jurafsky/slp3/ (accessed June 18 2018) [Google Scholar]
- 30.Salton G, Wong A, Yang CS. A vector space model for automatic indexing. Commun ACM. 1975;18(11):613–20. [Google Scholar]
- 31.Hochreiter S, Bengio Y, Frasconi P. Wiley-IEEE Press. 2001. Gradient flow in recurrent nets: The difficulty of learning long- term dependencies. [Google Scholar]
- 32.Liu S, Tang B, Chen Q. Comput Math Methods Med 2016. 2016. Drug-drug interaction extraction via convolutional neural networks; p. 6918381. doi:10.1155/2016/6918381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collobert R, Weston J. New York, NY: ACM. 2008. A unified architecture for natural language processing: deep neural networks with multitask learning. In: Proceedings of the 25th International Conference on Machine Learning; pp. 160–7. [Google Scholar]
- 34.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM 2017. 60(6):84–90. [Google Scholar]
- 35.Roncancio H, Hernandes AC, Becker M. IEEE Workshop on Robot Vision (WORV) 2013. Ceiling analysis of pedestrian recognition pipeline for an autonomous car application 2013. [Google Scholar]