Abstract
A growing quantity of health data is being stored in Electronic Health Records (EHR). The free-text section of these clinical notes contains important patient and treatment information for research but also contains Personally Identifiable Information (PII), which cannot be freely shared within the research community without compromising patient confidentiality and privacy rights. Significant work has been invested in investigating automated approaches to text de-identification, the process of removing or redacting PII. Few studies have examined the performance of existing de-identification pipelines in a controlled comparative analysis. In this study, we use publicly available corpora to analyze speed and accuracy differences between three de-identification systems that can be run off-the-shelf: Amazon Comprehend Medical PHId, Clinacuity’s CliniDeID, and the National Library of Medicine’s Scrubber. No single system dominated all the compared metrics. NLM Scrubber was the fastest while CliniDeID generally had the highest accuracy.
Introduction
Electronic health records (EHR) data is increasingly being used to support research in the translation research commu-nity1. A significant amount of important information is trapped in free-text format in clinical notes2,3. However, these text notes often contain patient names, dates of services, and other types of personally identifiable information (PII), which imparts a significant burden associated with the risk to patient confidentiality on the utility of these important sources of data. In the United States, the Health Insurance Portability and Accountability Act (HIPAA) provides data privacy and security provisions for safeguarding the confidentiality of patients4. Moreover, based on the Common Rule, in order to use patient data for research, researchers are required to obtain either an informed consent from patients or a waiver of informed consent from an institutional Internal Review Board (IRB)5. Therefore, automated de-identification may alleviate the regulatory burden associated with the use of text data for research. A significant amount of work has been invested in investigating various approaches for de-identification6.
However, few studies have examined the performance of existing text de-identification software in a controlled comparative analysis7. In this study, we use publicly available corpora to analyze speed and accuracy differences between three de-identification systems that can be run off-the-shelf. The three systems were chosen to reflect a diversity of approaches in currently maintained systems that can be run without any additional training or configuration.
Methods
First, we discuss the annotations of interest that will be the focus of our accuracy metrics. Second, we provide a brief overview of the reference corpora used to determine system accuracy. Third, we describe the three text de-identification systems to be evaluated. Fourth, we explain the tool used to generate the accuracy metrics.
The HIPAA Privacy Rule “Safe Harbor” method lists eighteen categories of identifiers or PII that a complete de-identification system must target. Because we are dealing with text data exclusively, we can put aside imaging and audio-related categories. The remaining PII categories applicable to text are: names, geographic subdivisions, dates, telephone & fax numbers, vehicle identifiers, serial numbers, device identifiers, email addresses, URLs, social security numbers, IP addresses, medical record numbers, account numbers, certificate numbers, license numbers, and any other unique identifying number, characteristic, or code.
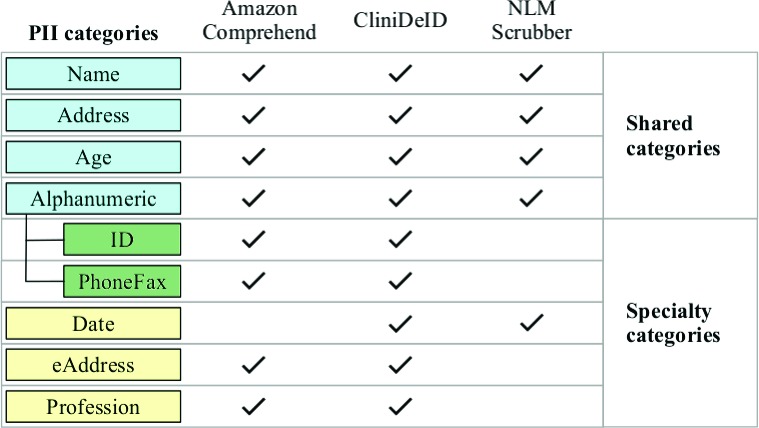
Not all of these categories are addressed by the systems we evaluated. Even those categories addressed by all systems are not equally addressed. As such, we have pared down the possible categories for evaluation into two sets: shared categories (i.e., those targeted by all three systems) and specialty categories (i.e., those addressed by two of the three systems). Figure 1 contains a visual summary of those two sets and what underlying categories compose them.
Figure 1:
PII categories coverage
The ADDRESS category includes standard mailing address components such as street, city, state, country, and postal code, even though the Safe Harbor method only includes components smaller than a state. Other geographic locations, like named geographical features, also fall under this category.
The AGE category comes in two flavors due to the specificity of the HIPAA Safe Harbor method. Technically, only ages over 89 are considered PII. The NLM Scrubber targets precisely this class. Many analyses (including Amazon Comprehend, CliniDeID, and both reference corpora) actually treat all ages as PII. Potentially, this definitional gap between AGE 90+ and ALL AGES could inequitably hurt NLM Scrubber’s scores as compared to the other systems. The evidence does not support this possibility, as addressed in the results section.
The ALPHANUMERIC category is a composition of the more commonly separated PHONEFAX category and all the general identifier categories (e.g., social security numbers and device IDs), which tend to be alphanumeric strings. Amazon Comprehend and CliniDeID treat PHONEFAX as separate from the other identifiers but NLM Scrubber merges them all. As such, we considered evaluation at the composite level to be more equitable. For completeness, we split PHONEFAX from other ID’s when evaluating specialty types for Amazon Comprehend and CliniDeID.
Finally, the NAME category also required special consideration. The Safe Harbor method only requires names of patients, relatives, employers, or household members to be de-identified. The two reference corpora include patient and provider names. The three de-identification systems tag patient, provider, and other names (e.g., relatives of the patient). In the output of Amazon Comprehend and NLM Scrubber, we cannot differentiate between the name subtypes. As such, we evaluated all systems at the name level rather than filtering by subtype. We should expect to see a slight inflation of false positive counts across all three systems as a consequence.
With respect to specialty categories, ID and PHONEFAX were treated individually for Amazon Comprehend and CliniDeID. The DATE category is annotated by CliniDeID and NLM Scrubber but not by Amazon Comprehend1. Specifically, calendar dates like “June "9th” are tagged as PII but not times, seasons, etc. The EADDRESS category includes electronic addresses like email addresses and URLs, which Amazon Comprehend and CliniDeID tag but not NLM Scrubber. The PROFESSION category includes job titles and is tagged by Amazon Comprehend and CliniDeID but not by NLM Scrubber. Further, each system was also evaluated against their own individual maximum possible set of categories out of all the above mentioned types.
While we report individual category accuracy metrics, we also evaluated each system as if all annotations belonged to a single supertype of PII. This analysis is separate from micro- and macro-averaging individual category results. Finding PII in a note but tagging it as the wrong category will lower the micro- and macro-average scores but will still count as a true positive instance of the PII supertype.
All evaluations were done with respect to two publicly available corpora: the 2014 and 2016 i2b2 de-identification challenge corpora8,9. These corpora include more annotated PII categories than the shared and specialty categories, such as health care units and organization names (both interpreted as geographic subdivisions). The 2014 corpus is split into 790 training notes and 514 test notes. The 2016 corpus is split into 200 development notes, 600 training notes, and 400 test notes. A note in the 2016 corpus is longer than one in the 2014 corpus with an average of 1,863 words per note versus 617 words per note, respectively. For consistency purposes, all corpora were processed as plain text files with one note per file. Timing results are aggregated across the training and testing documents. Performance scores are reported for both the training and testing splits. All de-identification systems were tested out-of-the-box.
Amazon Comprehend Medical2 is a cloud-based natural language processing (NLP) system. We have focused only on their detect phi service endpoint (also called PHId), which can be run via API calls or remotely on an AWS instance. Our testing leveraged a simple Python script to read in a note, post it to the service endpoint, and then convert the returned output to the brat standoff format10. The server region, which determines the physical location of the servers that data is processed on, is the only additional parameter providing during these API calls. The service was rate-limited to at most 20k characters per note. Thirty five notes exceeded this limit and were not analyzed. For production purposes, additional logic could be constructed to split notes into overlapping tiles of text to still maintain all context clues to PII while keeping under the rate-limit.
Clinacuity’s CliniDeID3 is available for Linux, macOS, and Windows. The on-premises version can be run using a graphical user-interface (GUI) or via the command-line. The test results reported below used the CentOS command-line version 1.5.0. CliniDeID offers several levels of de-identification. We selected the level ‘Beyond HIPAA Safe Harbor’ to align the coverage of categories such as names with other systems (e.g., so provider names were also tagged). Timing results did not include resynthesizing the PII detected (i.e., consistently replacing PII with realistic random surrogates), although that option was available. We ran our evaluations using a special testing license.
The National Library of Medicine’s Scrubber4 is the oldest of the three systems and freely available online for Linux and Windows. The most recent version (v.19.0403L) included both a command-line and GUI version. The default system output is a redacted version of the original note. The only parameters we provided were the input and output directories. Additional parameters, such as a file of whitelisted terms to never de-identify (“preserved terms”), were not used. The classic opening line of Moby Dick is converted to “Call me [NAME].” Because our evaluation tool (described in more detail below) relied on annotation offsets noted in the reference corpus for scoring, we needed to map the NLM Scrubber’s output file to a character offset anchored annotation format. Namely, the string “Ishmael” covers the ninth to fifteenth characters in the original. The bracketed redaction “[NAME]” covers the ninth to twenty-second characters, which both fails to match the original offsets and bumps out the offsets of all future annotations. To solve this problem, we wrote a script to convert NLM Scrubber’s output to brat standoff format. The script attempts to line up the preceding and following context around all bracketed forms to anchor the redaction in the original note. Consecutive redactions (e.g., the “[NAME]\n[ADDRESS]” format used on envelopes) were merged. That is, because we could not reliably determine the character boundary between the annotations, we associated both annotations with the full span of both, effectively creating two fully-overlapping annotations. This decision forced all evaluations to be run as ‘partial matching’, as described below, so as to not unfairly count overlapping annotations against the system. Roughly speaking, as long as a system output PII annotation text span at least partially overlaps a reference PII annotation span in the same category, it is considered a true positive.
Performance evaluation was done using ETUDE11 (Evaluation Tool for Unstructured Data and Extractions), a freely available open source tool5, which was developed and maintained by the first author. All scoring was done using the command-line engine to facilitate scripting. A GUI is also available6. The ETUDE engine parses each of the reference and system documents based on corpus specific configuration files. Using separate configuration files per corpus allows each to be in a different format (e.g., brat vs. inline XML) with different schemata (e.g., the engine is responsible for binning CliniDeID’s patient and provider names into a single category for scoring). We used the partial matching flag for evaluating annotation alignment. That is, as long as two annotations at least partially overlap their spans and match categories, it is considered a true positive. Using the exact match flag would inequitably hurt systems when they differ from the reference standard by punctuation (e.g., “Ishmael” vs. “Ishmael.”) or when the system aggressively splits annotations (e.g., “[Moby Dick]” vs. “[Moby] [Dick]”). Metrics include True Positives (TP), False Positives (FP), False Negatives (FN), Precision (i.e., positive predictive value), Recall (i.e., sensitivity), and F1 (i.e., an F-measure with a β value of one).
Results
No single system dominated all the compared metrics. Before we delve into the particulars of speed tests and annotation evaluation, we need to look deeper at the nlm2brat.py Python script performance summarized in Table 1. As described above, nlm2brat.py (available on-line7) is responsible for converting the output of the NLM Scrubber to brat standoff format. The latter of these formats can be scored using ETUDE. The NLM Scrubber generates five tags (with the normalized category type in parentheses): ADDRESS (ADDRESS), AGE90+ (AGE), ALPHANUME-RICID (ALPHANUMERIC), DATE (DATE), PERSONALNAME (NAME). The second column in Table 1 shows the total counts of each tag across the training and test splits in both corpora. The third column shows the counts of tags present in the brat files generated by the script. Note that all values are identical, indicating that all tags were correctly found. Finding a tag is a necessary but not sufficient condition for extracting the character offsets for said tag. The fourth column shows the counts of all tags found but not anchored by character offsets. These unanchored tags are predominantly the first of three consecutive tags separated by nothing or limited whitespace. The final column reports the total percentage of successfully anchored tags. The high percentages here (99.7+% for all types) allows us to feel confident that the tag extraction and format conversion has not introduced significant noise.
Table 1:
Annotation Counts for the Five Primary NLM Scrubber Annotation Types at Each Stage of Processing
| Type | NLM Scrubber Annotations | nlm2brat.py Annotations | Anchorless Annotations | % Anchored Annotations Out of All Annotations |
|---|---|---|---|---|
| ADDRESS | 4304 | 4304 | 10 | 0.9977 |
| AGE90+ | 119 | 119 | 0 | 1.0000 |
| ALPHANUMERICID | 10297 | 10297 | 5 | 0.9995 |
| DATE | 9980 | 9980 | 10 | 0.9990 |
| PERSONALNAME | 32372 | 32372 | 63 | 0.9982 |
Table 2 summarizes our speed test results. The NLM Scrubber was indisputably the fastest of the three systems. As per the second column, the time taken to process 10k characters is roughly half as long as for Amazon Comprehend and an order of magnitude faster than for CliniDeID. Likewise, the characters processed per second (column three) is roughly
Table 2:
Key Speed Test Characteristics of the Three Systems
| Tool | Secs / 10k Char | Chars / Sec | Total Notes | Secs / Note | Notes / Sec |
|---|---|---|---|---|---|
| Amazon Comprehend | 32.38 | 308.88 | 2269 | 0.57 | 1.75 |
| CliniDeID | 189.61 | 52.74 | 2304 | 3.39 | 0.29 |
| NLM Scrubber | 13.69 | 730.61 | 2300 | 0.24 | 4.09 |
twice that of Amazon Comprehend and, again, an order of magnitude more than for CliniDeID. As noted in column four, NLM Scrubber failed to process four notes. This failure was due to the presence of non-ASCII characters and should not bias our results. In contrast, the thirty-five notes not processed by Amazon Comprehend were the longest notes. The failure here has the potential to slightly increase the real average speed per note for this tool. As a final quirk of the timing evaluation, the CliniDeID and the NLM Scrubber tests were run on a different class of computer from the Amazon Comprehend tests. Amazon Comprehend is always run on an arbitrarily large cloud server in AWS. In contrast, the other two systems were run on a development server at MUSC with four dual core 2.66 GHz processors and 32 GB of RAM.
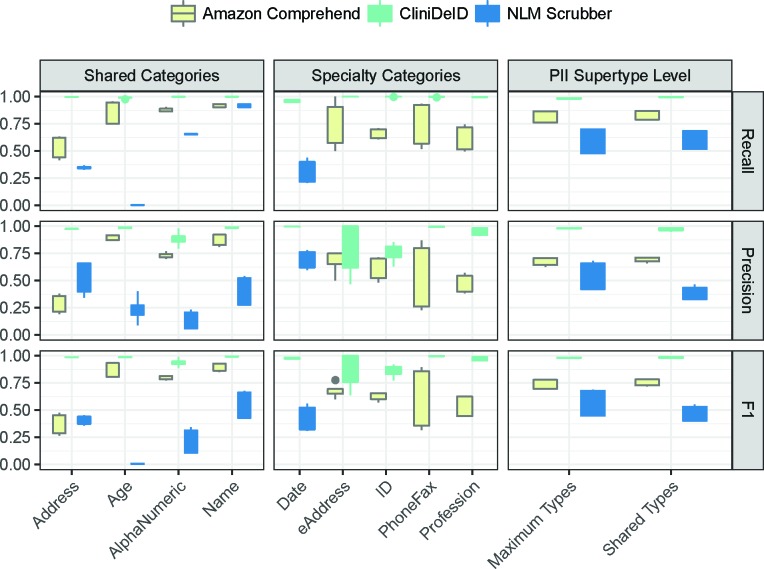
Figure 2 presents a visual overview of the relative performance metrics for our tools. The complete data underlying these graphs is presented in Tables 3–8. The box-and-whisker plot highlights the full range of performance across the training and test splits for both corpora. That is, four individual data points are summarized by the box-and-whisker. To see the specific performance on the test split for each corpus, refer to the tables at the end.
Figure 2:
Accuracy for shared categories (left), specialty categories (center), and at the PII supertype level (right)
At a high level, across categories, CliniDeID reliably outperformed the other two systems. ADDRESS is the only category for which Amazon Comprehend did not clearly beat NLM Scrubber. The two systems are roughly equivalent in terms of F1 with Amazon Comprehend having a higher recall score and NLM Scrubber having a higher precision.
More specifically, Amazon Comprehend seemed to have the largest performance gap around ADDRESS. The range for AGE recall is also fairly wide compared to the other categories. The very low raw number of EADDRESS annotations makes it difficult to draw any real conclusions about performance. PHONEFAX performance is surprisingly fragile with a wide range for a category that, intuitively, should be fairly consistent. Digging in to Table 6, we can see a very high false positive rate for this category. Generally, this tool has better recall than precision.
Table 6:
Complete Table of Amazon Comprehend Specialty Category Performance Results
| Training | Testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eAddress | 3 | 1 | 3 | 0.7500 | 0.5000 | 0.6000 | 1 | 1 | 0 | 0.5000 | 1.0000 | 0.6667 |
| IDs | 535 | 227 | 343 | 0.7021 | 0.6093 | 0.6524 | 387 | 156 | 238 | 0.7127 | 0.6192 | 0.6627 |
| PhoneFax | 164 | 550 | 152 | 0.2297 | 0.5190 | 0.3184 | 127 | 341 | 90 | 0.2714 | 0.5853 | 0.3708 |
| Profession | 174 | 151 | 60 | 0.5354 | 0.7436 | 0.6225 | 127 | 95 | 52 | 0.5721 | 0.7095 | 0.6334 |
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| eAddress | 3 | 1 | 2 | 0.7500 | 0.6000 | 0.6667 | 7 | 3 | 1 | 0.7000 | 0.8750 | 0.7778 |
| IDs | 30 | 32 | 13 | 0.4839 | 0.6977 | 0.5714 | 22 | 19 | 9 | 0.5366 | 0.7097 | 0.6111 |
| PhoneFax | 126 | 19 | 11 | 0.8690 | 0.9197 | 0.8936 | 100 | 29 | 7 | 0.7752 | 0.9346 | 0.8475 |
| Profession | 706 | 1141 | 652 | 0.3822 | 0.5199 | 0.4406 | 478 | 707 | 483 | 0.4034 | 0.4974 | 0.4455 |
For CliniDeID, EADDRESS and ID are the most difficult categories. All other categories’ performance metrics are reliably in the upper nineties. If anything, recall tends to be slightly higher than precision across categories.
For NLM Scrubber, AGE is not reliably tagged. We would expect higher recall than precision if the performance metrics were dominated by the mismatch between the reference standard, which tags all ages, and the system, which only tags ages 90+. In fact, we find higher precision than recall, which implies the system is more conservative in its tagging of spans as AGE. Manual inspection of errors indicates that many of the spans incorrectly annotated as NAME are in fact organizations or hospitals. For instance, “Hollings Cancer Center” is annotated as “[PERSONALNAME] Cancer Center”. Likewise, the low precision for ALPHANUMERIC stems from dates and other numbers being tagged erroneously. For instance, “Living with father since [ALPHANUMERICID]” is actually a date.
Discussion
The rightmost graph in Figure 2 provides a good holistic view of the systems. Amazon Comprehend performance is generally acceptable. CliniDeID performs at near ceiling. NLM Scrubber, despite poor performance on several shared categories, generally performs better than average and only slightly worse than Amazon Comprehend when taking frequency of annotation types into account.
In prioritizing patient privacy, users should prioritize recall over precision. In that light, we should be more concerned with NLM Scrubber’s performance on ADDRESS tags than on ALPHANUMERIC tags even though the F-measure for the latter is the higher of the two categories because its recall is also the lower of the two. In a similar vein, the type of note to be de-identified should also play into decision-making. For instance, NLM Scrubber’s relatively low AGE category performance is less important for a pediatric dataset, which is unlikely to have many ages mentioned over the required threshold. Likewise, Amazon Comprehend should be safer to use on pathology reports, which are unlikely to include many instances of ADDRESS, than on history and physical notes, which are more likely to include references to previous residences or care facilities. The lack of comprehensive coverage of all PII categories by NLM Scrubber and Amazon Comprehend also restricts the viable distribution options for notes processed by these systems. That is, notes processed by CliniDeID will have more types of PII de-identified than those processed by the other two systems. This wider range of de-identified information means these notes can be shared with a broader audience without leaking PII. For instance, releasing notes with the original professions and electronic addresses, which NLM Scrubber does not annotate, still intact may only be advisable for more circumscribed distribution.
All three systems require a minimum amount of basic engineering work to encapsulate them in a regularly run pipeline (e.g., hourly or daily). Even batch processing, as we did in our evaluation, likely requires reformatting notes from their source format (e.g., dumping them from a database into flat files) and output file parsing and/or reformatting. CliniDeID has the widest range of output options.
The gaps in category coverage of NLM Scrubber and Amazon Comprehend may be acceptable or may need to be supplemented by a secondary system. As mentioned above, for instance, combining the detect entities and the detect phi service endpoints would extend Amazon Comprehend’s category coverage. This doubling of the pipeline would, of course, double the direct cost for using Amazon Comprehend. NLM Scrubber could be run in tandem with another de-dentification system with better performance on AGE or ADDRESS categories. It could also be run in series with a filter engine to reduce the high false positive rate in those categories with high recall but low precision. Along these same lines, de-identification could need to be integrated into a more comprehensive processing pipeline that included annotation or extraction beyond PII categories. Amazon Comprehend is the most flexible for integration into larger pipelines through the use of API calls. On the other hand, these calls require notes containing PII to be sent to an off-site server. In contrast, CliniDeID and NLM Scrubber can be run fully locally.
Coverage, ease of integration, cost, and output formats are three key components we have only lightly touched on due to the wide range of context-dependent contingencies tied to these factors.
Conclusion
While this evaluation has strived to be thorough, a wide range of additional evaluations need to be run to better understand the full landscape of text de-identification tools. For instance, speed evaluations were conducted under a simplistic analysis of one machine type that is not otherwise under any significant system load. We still need to investigate the impact of document size on processing speed. Average note size from three different silos in our own Research Data Warehouse range from 375 to 1900 characters. Are speed increases linear or exponential with character count? Testing additional systems will also improve our understanding of the software landscape. However, expanding the evaluation to include more systems is a non-trivial task. For instance, MIST12, the MITRE Identification Scrubber Toolkit8, only ships with a de-identification engine and not the model necessary to run the engine, which must be trained on local data. On the other end of the spectrum, Microsoft’s Presidio9 includes all necessary pre-trained models but has a relatively complex installation procedure.
The next two classes of extension relate to improved evaluation tooling rather than additional test conditions. The nlm2brat.py script we developed can be extended in functionality. We documented the impact of three consecutive redacted spans in a row on conversion. Performance here could be improved to guarantee all redacted spans are anchored. Smarter anchoring algorithms could be tested to see if we can more reliably find exact spans for the redacted annotation rather than needing to merge adjacent redactions. Other output formats could also be added to the script (e.g., inline XML files or CAS XMI files for UIMA compatibility). Finally, the core issue in de-identification surrounds leaking PII. We evaluated performance at the coarse level of the annotation. Finer-grained analyses at the token and character levels will help us understand exactly how much and what type of PII is most likely to be leaked by the various systems.
While no perfect solution exists for text de-identification, the ideal tool for your particular application will need to integrate and balance a wide range of constraints.
Acknowledgements
We would like to thank SM for the test license, Gary Underwood & Andrew Trice for help understanding the output formats for scoring, and Jean Craig for pulling numbers to help ground our system estimates. This project was supported, in part, by the National Center for Advancing Translational Sciences of the National Institutes of Health under Grant Number UL1 TR001450 and the SmartState Endowment for Translational Biomedical Informatics. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Amazon provides a separate detect entities service endpoint that does annotate full dates (i.e., month, day, and year together), days of the week, months, and times. However, because it is annotated by a separate service on a separate endpoint from the other PII categories, we would need to run every note through two systems for full coverage, which is beyond our intent of evaluating simple, off-the-shelf systems.
Competing Interests Statement
PH and JO have no competing interests to declare. SM is associated with Clinacuity. To avoid potential conflicts of interest, SM had no involvement or influence on running the text de-identification systems and analyzing their output.
Figures & Table
Table 3:
Complete Table of Amazon Comprehend Shared Category Performance Results
| Training | Testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Address | 501 | 2095 | 700 | 0.1930 | 0.4172 | 0.2639 | 380 | 1351 | 463 | 0.2195 | 0.4508 | 0.2953 |
| Age | 1160 | 99 | 70 | 0.9214 | 0.9431 | 0.9321 | 730 | 67 | 34 | 0.9159 | 0.9555 | 0.9353 |
| AlphaN. | 1031 | 445 | 163 | 0.6985 | 0.8635 | 0.7723 | 728 | 283 | 114 | 0.7201 | 0.8646 | 0.7858 |
| Name | 3924 | 349 | 265 | 0.9183 | 0.9367 | 0.9274 | 2593 | 187 | 198 | 0.9327 | 0.9291 | 0.9309 |
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Address | 1598 | 2951 | 986 | 0.3513 | 0.6184 | 0.4481 | 1007 | 1643 | 580 | 0.3800 | 0.6345 | 0.4753 |
| Age | 2586 | 378 | 844 | 0.8725 | 0.7539 | 0.8089 | 1641 | 255 | 569 | 0.8655 | 0.7425 | 0.7993 |
| AlphaN. | 159 | 48 | 21 | 0.7681 | 0.8833 | 0.8217 | 125 | 45 | 13 | 0.7353 | 0.9058 | 0.8117 |
| Name | 3034 | 721 | 337 | 0.8080 | 0.9000 | 0.8515 | 1990 | 400 | 212 | 0.8326 | 0.9037 | 0.8667 |
Table 4:
Complete Table of CliniDeID Shared Category Performance Results
| Training | Testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Address | 1197 | 26 | 5 | 0.9787 | 0.9958 | 0.9872 | 840 | 25 | 3 | 0.9711 | 0.9964 | 0.9836 |
| Age | 1224 | 30 | 9 | 0.9761 | 0.9927 | 0.9843 | 758 | 13 | 6 | 0.9831 | 0.9921 | 0.9876 |
| AlphaN. | 1196 | 307 | 2 | 0.7957 | 0.9983 | 0.8856 | 839 | 110 | 3 | 0.8841 | 0.9964 | 0.9369 |
| Name | 4189 | 46 | 3 | 0.9891 | 0.9993 | 0.9942 | 2788 | 45 | 3 | 0.9841 | 0.9989 | 0.9915 |
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Address | 2784 | 65 | 7 | 0.9772 | 0.9975 | 0.9872 | 577 | 15 | 1 | 0.9747 | 0.9983 | 0.9863 |
| Age | 3605 | 38 | 32 | 0.9896 | 0.9912 | 0.9904 | 752 | 6 | 17 | 0.9921 | 0.9779 | 0.9849 |
| AlphaN. | 191 | 27 | 0 | 0.8761 | 1.0000 | 0.9340 | 46 | 1 | 0 | 0.9787 | 1.0000 | 0.9892 |
| Name | 3660 | 66 | 6 | 0.9823 | 0.9984 | 0.9903 | 757 | 4 | 0 | 0.9947 | 1.0000 | 0.9974 |
Table 5:
Complete Table of NLM Scrubber Shared Category Performance Results
| Training | Testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Address | 444 | 845 | 758 | 0.3445 | 0.3694 | 0.3565 | 294 | 416 | 549 | 0.4141 | 0.3488 | 0.3786 |
| Age | 5 | 50 | 1228 | 0.0909 | 0.0041 | 0.0078 | 6 | 20 | 758 | 0.2308 | 0.0079 | 0.0152 |
| AlphaN. | 795 | 3210 | 403 | 0.1985 | 0.6636 | 0.3056 | 547 | 1807 | 295 | 0.2324 | 0.6496 | 0.3423 |
| Name | 3794 | 3523 | 398 | 0.5185 | 0.9051 | 0.6593 | 2501 | 2111 | 290 | 0.5423 | 0.8961 | 0.6757 |
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Address | 953 | 484 | 1814 | 0.6632 | 0.3444 | 0.4534 | 566 | 292 | 1162 | 0.6597 | 0.3275 | 0.4377 |
| Age | 5 | 18 | 3604 | 0.2174 | 0.0014 | 0.0028 | 6 | 9 | 2348 | 0.4000 | 0.0025 | 0.0051 |
| AlphaN. | 124 | 2145 | 66 | 0.0546 | 0.6526 | 0.1009 | 99 | 1562 | 52 | 0.0596 | 0.6556 | 0.1093 |
| Name | 3376 | 8880 | 265 | 0.2755 | 0.9272 | 0.4247 | 2258 | 5866 | 146 | 0.2779 | 0.9393 | 0.4290 |
Table 7:
Complete Table of CliniDeID Specialty Category Performance Results
| Training | Testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Date | 7263 | 35 | 232 | 0.9952 | 0.9690 | 0.9820 | 4838 | 18 | 142 | 0.9963 | 0.9715 | 0.9837 |
| eAddress | 6 | 0 | 0 | 1.0000 | 1.0000 | 1.0000 | 1 | 0 | 0 | 1.0000 | 1.0000 | 1.0000 |
| IDs | 880 | 303 | 1 | 0.7439 | 0.9989 | 0.8527 | 625 | 107 | 0 | 0.8538 | 1.0000 | 0.9211 |
| PhoneFax | 317 | 30 | 0.9906 | 1.0000 | 0.9953 | 216 | 1 | 1 | 0.9954 | 0.9954 | 0.9954 | |
| Profession | 233 | 2 | 1 | 0.9915 | 0.9957 | 0.9936 | 178 | 4 | 1 | 0.9780 | 0.9944 | 0.9861 |
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Date | 5390 | 24 | 333 | 0.9956 | 0.9418 | 0.9679 | 1066 | 5 | 53 | 0.9953 | 0.9526 | 0.9735 |
| eAddress | 7 | 8 | 0 | 0.4667 | 1.0000 | 0.6364 | 2 | 1 | 0 | 0.6667 | 1.0000 | 0.8000 |
| IDs | 44 | 26 | 0 | 0.6286 | 1.0000 | 0.7719 | 4 | 1 | 0 | 0.8000 | 1.0000 | 0.8889 |
| PhoneFax | 147 | 1 | 0 | 0.9932 | 1.0000 | 0.9966 | 42 | 0 | 0 | 1.0000 | 1.0000 | 1.0000 |
| Profession | 1464 | 144 | 7 | 0.9104 | 0.9952 | 0.9510 | 355 | 30 | 1 | 0.9221 | 0.9972 | 0.9582 |
Table 8:
Complete Table of NLM Scrubber Specialty Category Performance Results
| Training | Testing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Date | 2906 | 935 | 4589 | 0.7566 | 0.3877 | 0.5127 | 2177 | 617 | 2803 | 0.7792 | 0.4371 | 0.5601 |
| 2016 | TP | FP | FN | Prec | Rec | F1 | TP | FP | FN | Prec | Rec | F1 |
| Date | 1257 | 745 | 4421 | 0.6279 | 0.2214 | 0.3273 | 795 | 538 | 3026 | 0.5964 | 0.2081 | 0.3085 |
References
- 1.Obeid JS, Beskow LM, Rape M, Gouripeddi R, Black RA, Cimino JJ, et al. A survey of practices for the use of electronic health records to support research recruitment. Journal of Clinical and Translational Science. 2017;1(4):246–252. doi: 10.1017/cts.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearbook of medical informatics. 2008:128–44. [PubMed] [Google Scholar]
- 3.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association. 2014 11;21(2):221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.HIPAA Privacy Rule, 45 CFR Part 160, Part 164(A,E). U.S. Department of Health and Humans Services. 2002 [Google Scholar]
- 5.Federal Policy for the Protection of Human Subjects (’Common Rule’) [Internet] 2009 [cited 2018-1120]; Available from: https://www.hhs.gov/ohrp/regulations-and-policy/regulations/common-rule/index.html. [Google Scholar]
- 6.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010;10(70) doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrandez O, South BR, Shen S, Friedlin FJ, Samore MH, Meystre SM. Evaluating current automatic de-identification methods with Veteran’s health administration clinical documents. BMC Medical Research Methodology. 2012;12(1):109. doi: 10.1186/1471-2288-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. Journal of Biomedical Informatics. 2015 Dec:S11–9. doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stubbs A, Filannino M, Uzuner Ö. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID shared tasks Track 1. Journal of Biomedical Informatics. 2017 Nov;75:S4–S18. doi: 10.1016/j.jbi.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenetorp P, Pyysalo S, Topié G, Ohta T, Ananiadou S, Tsujii J. brat: a web-based tool for NLP-assisted text annotation; Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics. Association for Computational Linguistics; 2012. pp. 102–107. [Google Scholar]
- 11.Heider PM, Accetta JK, Meystre SM. ETUDE for Easy and Efficient NLP Application Evaluation; Presented at the AMIA NLP-WG Pre-Symposium; San Francisco, CA, USA; 2018. [Google Scholar]
- 12.Wellner B, Huyck M, Mardis S, Aberdeen J, Morgan A, Peshkin L, et al. Rapidly Retargetable Approaches to De-identification in Medical Records. Journal of the American Medical Informatics Association. 2007 Sep;14(5):564–573. doi: 10.1197/jamia.M2435. [DOI] [PMC free article] [PubMed] [Google Scholar]