Abstract
Mapping local terminologies to standardized terminologies facilitates secondary use of electronic health records (EHR). Penn Medicine comprises multiple hospitals and facilities within the Philadelphia Metropolitan area providing services from primary to quaternary care. Our Penn Medicine (PennMed) data include medications collected during both inpatient and outpatient encounters at multiple facilities. Our goal was to map 941,198 unique medication terms to RxNorm, a standardized drug nomenclature from the National Library of Medicine (NLM). We chose three popular tools for mapping: NLM’s RxMix and RxNav-in-a-Box, OHDSI’s Usagi and Mayo Clinic’s MedXN. We manually reviewed 400 mappings obtained from each tool and evaluated their performance for drug name, strength, form, and route. RxMix performed the best with an F1 score of 90% for drug name versus Usagi’s 82% and MedXN’s 74%. We discuss the strengths and limitations of each method and tips for other institutions seeking to map a local terminology to RxNorm.
1. Introduction
1.1. Standardization of Medication Nomenclature and RxNorm
Data interoperability is a concern for effective communication of health information. Standard medication nomenclature can promote patient safety and facilitate secondary use of clinical medication data. The National Library of Medicine (NLM) produces RxNorm, a standardized nomenclature for clinical drugs1,2. RxNorm provides standard names for clinical drugs for human use, both branded and generic, and their links to active ingredients, drug components, and other related brand names. The purpose of RxNorm is to allow health systems to communicate drug-related information efficiently and clearly. In the latest release of the RxNorm dataset includes about 54,000 distinct active drug names (RxNorm August 2019 Release).
1.2. Mapping Local Medication Terms to RxNorm
Manually mapping local terms to RxNorm concepts is a time-consuming process and requires a significant amount of work by domain experts3. Developing a tool to automate this process requires time and efforts to validate the results when the tool is applied in different contexts. Using publicly available tools can shorten the process to map local terminologies to RxNorm. However, it is important to decide which is the most appropriate tool for the job. In this paper, we describe the process in which we found the most appropriate medication-mapping tool for a master list of medications from our institution at Penn Medicine (PennMed).
2. Methods
2.1. Tools for Mapping Terms to RxNorm
2.1.1. RxNorm API and RxMix
The NLM has developed publicly available drug Application Programming Interfaces (APIs) through Simple Object Access Protocol (SOAP) and Representational State Transfer (RESTful) web services for users with programming skills to integrate into applications4. Bodenreider and Peters developed RxMix to enable users who do not have programming skills to benefit from NLM’s API resources4. RxMix is a web-based interface enabling users to create queries with combined functions of the RxNorm, RxTerms, NDF-RT, RxClass, and Interaction APIs (which can be accessed via https://mor.nlm.nih.gov/RxMix/). Users can build a workflow of API functions to execute batch queries against the multiple APIs. An interactive mode allows users to test queries on a single input value, before executing in batch mode. RxMix offers output in Extensible Markup Language (XML), JavaScript Object Notation (JSON), or text format.
From the RxNorm API, we used the function getApproximateMatch (REST) or getApproximateTerm (SOAP) to normalize medication strings to RxNorm5. The normalization approach used is described in more detail here6. A drug-centric token matching approach is used in the approximate matching method. Drug names are identified against a drug name list for matches, along with several other methods: token splitting, drug name expansion, and spelling correction. Extracted candidate strings containing the drug are scored to determine the closeness to the user string. Tokens of the candidate string are compared to tokens of the input string and similarity is determined by calculating the Jaccard’s coefficient.
Users can also install RxNav-in-a-Box to query the RxNorm RESTful API locally7. RxNav-in-a-Box uses Docker to create two container images (Tomcat container and MariaDB container). However, the user is responsible to keep the system up to date with the latest version, which is generated on a monthly basis. In addition, the spelling suggestions resource is not included in RxNav-in-a-Box, and therefore this will have an effect on the Approximate Term function. No output will be given for a query of a misspelled medication, where normally the RxNorm API will give an output with a comment, stating the spelling suggestion for the token.
For the purpose of this paper, we reviewed outputs from the RxMix application and RxNav-in-a-Box. RxMix accesses the RxNorm SOAP web services, while RxNav-in-a-Box provides a local version of RxNorm REST web services. We used Docker Desktop Community Edition (version 2.0.0.3) for the RxNav-in-a-box (downloaded June 2019) two container images: Tomcat and MariaDB. We fetched JSON data from the local REST API in RStudio (Version 1.2.1335).
2.1.2. MedXN
Sohn et al. (2014) developed an open source Medication Extraction and Normalization (MedXN) system to extract medication information from clinical notes and normalize it to the most appropriate RxNorm concept unique identifier (RXCUI). MedXN uses Apache’s Unstructured Information Management Architecture (UIMA) in a serialized hierarchal annotation flow. MedXN extracts the mediation from text, parses out the attributes, and composes the medication and attribute association. The attributes include the following: dosage, strength, frequency, form, and route. Next, the MedXN system converts this information to the Rxnorm Standard and maps it to the corresponding RXCUI. The output will have two RXCUI: the DrugRxCUI (i.e. sulfaSALAzine [RXCUI = 9524]), and the NormRxCUI with the medication attributes (i.e. Sulfasalazine 500 MG Oral Tablet [AZULFIDINE] RXCUI = 208437). The MedXN system was developed on the ‘Current Medication’ section in clinical notes, which varies from unstructured narratives to lists of medications8.
2.1.3. Usagi
The Observational Health Data Sciences and Informatics (OHDSI) program is a multi-stakeholder, interdisciplinary collaborative that grew out of the Observational Medical Outcomes Partnership (OMOP). The OMOP Common Data Model (CDM) is a standardization approach that allows for the systematic analysis of disparate observational databases9. Usagi is a java application that creates mappings from source coding systems into standards terminologies stored in the OMOP Vocabulary10. Standard vocabularies for use in OHDSI tools are available for download through the Athena download site (http://athena.ohdsi.org). Usagi uses Apache Lucene free text search engine and provides a score for each match, with 1.00 being a 100% match10.
2.2. Filtering of Drug Names and Linking Synonymous Drugs
Our Penn Medicine (PennMed) data include medications collected during both inpatient and outpatient encounters at multiple hospitals and facilities. Because of this, we have medication strings sourced from several applications and contexts. The medication list used in this paper also includes medications that in some cases were last ordered between 1980s through 2019.
We started by filtering the medication strings to prepare them for the matching tools. Some strings had medications along with other information that would be unnecessary for the matching process. While MedXN was created to find medications and their attributes within a clinical note, Usagi and RxNorm API require cleaner data. For instance, the score given by the RxNorm API depends upon the amount of words in the string. In the original string, there may be attributes that could be important clinically, but would lower the accuracy of the matching algorithm, as illustrated in Table 1. Instructions mentioned after the medication are outside the scope of RxNorm, which does not encompass frequency and duration in their clinical drug concepts. MedXN was developed to capture these attributes, but having extraneous information in the medication string would likely increase error for tools not developed for extraction from clinical notes.
Table 1.
Difference in RxMix Approximate Match output with and without clinical information.
| Input | Score | RXCUI Match | STR match | |
|---|---|---|---|---|
| Original | ZOMETA EVERY THREE MONTHS AS PREVIOUSLY DIRECTED | 14 | 18346 | AS |
| Cleaned | ZOMETA | 100 | 285143 | Zometa |
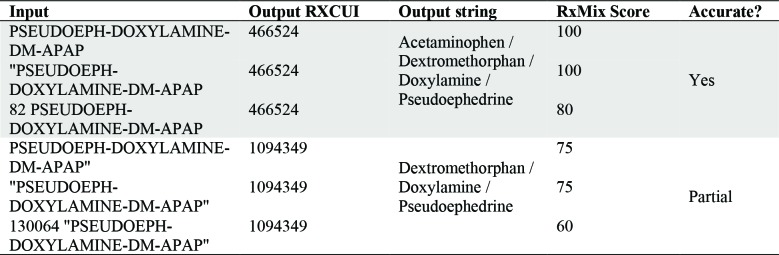
Quotes around the string, and other added characters can have an effect the score (Table 2). If the input string contains four drugs: acetaminophen (APAP), dextromethorphan (DM), doxylamine, and pseudoephedrine (PSEUDOEPH). For a query in the RxNorm API, an input with PSEUDOEPH-DOXYLAMINE-DM-APAP” resulted in the partially correct output (RXCUI = 1094349), but “PSEUDOEPH-DOXYLAMINE-DM-APAP gave the correct output (RXCUI = 466524). Punctuation and erroneous characters can greatly affect the results. The partially correct output is missing one of the four drugs from the input string: acetaminophen.
Table 2.
RxMix Output RXCUI, output string, RxMix Score, and accuracy for variations of the same input.
Before and during the filtration process, we used the interactive mode of RxMix. This helped to see what aspects of strings should be filtered out. See Table 3 for specific examples of filtered terms. The descriptions of the topics are not extensive, as there are several variations within each topic. Because the list contained a large amount of strings, not all unnecessary strings could be removed without significant effort. By the point of the process at which we linked synonymous strings, any unnecessary strings caught after are still included in the list due to time constraints.
Table 3.
Examples of filtered terms from local medication list.
In RStudio, we filtered out non-drug related strings using regular expressions in base R11. For each of the strings, we created a new ID number and set the strings to all uppercase. We set each non-drug string ID equal to zero, to be removed later. A record of the original IDs and the variants was kept. See Figure 1 for details of the filtering process and the size of the list at each stage. Non-drug strings pertained to other orders and notes, such as work notes, instructions, procedures, lab orders, health services, and medical equipment orders. We also filtered out drug strings of cancelled orders, or notes to change the above order. Sections of drug strings with sensitive information were removed, such as patient names, date of birth, dates, time, location, and health practitioner names. Common medical prescription abbreviations and general medical terms were referenced to filter non-drug strings and unnecessary information from drug strings 12–14. In order to prevent filtering actual drug strings from the list, the filtering regex was completed manually, overlooking which strings were filtered out by each function.
Figure 1.

Filtering process of master medication list.
The last stage of the filtration process was completed by removing all spaces from strings and removing duplicates. This decreased the size of the list significantly, since many duplicates included different practitioner names or instructions within the string. We filtered out names using personal pronouns, possessive adjectives, possessive pronouns, and common names15–17.
2.3. Manual Review of Results
A random sampling of 400 medication ID strings from the cleaned version of the medication list was extracted into a comma-separated values (CSV) and text file. The sample list was manually reviewed for possible patient information and categorized as a medication or non-medication related string. Not all non-medication strings were filtered from the list. 38 of the 400 strings were determined as non-medication strings. For example, “FETAL MONITORING FOR MULTIPLES”
indicates a health service, but has no mention of medications. In RxMix, the workflow was set to the RxNorm:getApproximateMatch function, the sample text file was loaded for input, and the output was selected to text. RxMix automatically uses the most recent RxNorm release. For the MedXN 1.0.1 collection processing engine (CPE) GUI, the sample set was processed with the default settings to run the matches. This MedXN version is from December 2013 and the RxNorm version is limited to that year. For Usagi, we downloaded the latest relase of the vocabulary from OHDSI’s Athena and built the index in Usagi from this vocabulary (https://github.com/OHDSI/Athena). The sample set CSV file was imported into Usagi (v.1.2.7, downloaded 4/3/2019). We filtered by vocabulary (RxNorm), and by domain (drug). The first suggested match (the highest scored match) was approved for the entire list.
The output for each tool was imported into RStudio to convert into a CSV file. Each output was compared to the original sample list, to determine medication strings that were not matched by the tool. The RxMix output was specifically limited to one output per medication id—specifically, the first candidate string for each string. For ease of comparison, the SAB=RXNORM normalized drug names and term types were merged with the output by RXCUI and RxNorm atom unique identifier (RXAUI) from the RXNCONSO data file 1. Next, the output was converted to CSV and manually reviewed.
Each medication string was reviewed manually, rather than against a gold standard because there can be several RXCUI for a medication, depending on the granularity. The output was reviewed upon five variables: medication, drug, strength, form, and route. Refer to Table 4 for more detail on the definitions of these variables.
Table 4.
Variables for manual review and their definitions.
| Type | Definition |
|---|---|
| Medication | Is this a medication string? |
| Drug | The medication or ingredient |
| Strength | The strength of the medication (eg, ‘800 mg’) |
| Form | The physical form in which the drug is specified to be administered (eg capsule, tablet) |
| Route | The route of administration (eg, intravenous, oral, etc) |
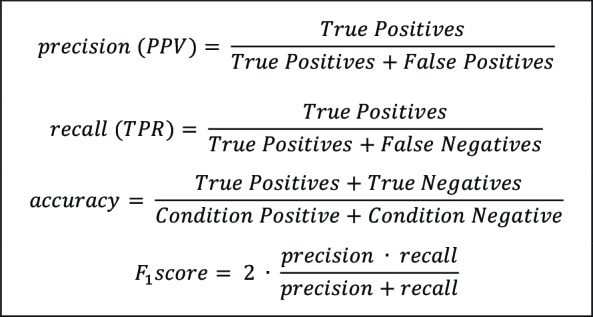
The medication variable refers to whether the string pertained to a medication or a non-medication string, which would be determined as yes (Y) or no (N). Each output that was determined to be a medication string was then classified as true positive (TP), false positive (FP), false negative (FN), true negative (TN), or partial (PART) for the following attributes: drug, strength, form, and route. TP means the match was correct, TN means there was correctly no match, FP means there was an incorrect match, FN means there was incorrectly no match, and partial indicates there was a component of the drug missing. To compute the manual review, binary classification was used. 1 indicates a positive class, and 0 indicates a negative class. The first digit indicates the predicted value, and the second indicates the actual value: TP (11), FP (10), FN (01), and TN (00). These values were analyzed in RStudio to create confusion tables, violin plots, and to calculate precision, recall, accuracy, and F1 score. See Table 5 for the formulas for each of these measures. In addition, we calculated the analysis of means in RStudio, using the ANOM package to understand significance of score and performance18.
Table 5.
Precision, recall, accuracy and F1 score formulas.
 |
See Figure 2 for two examples of the manual review of the output strings. The first example demonstrates a match where the drug is the same and the form is the same between the input and output. However, the strength changes from 250 mcg to 150 mcg in the output and is therefore inaccurate. In RxNorm there is no RXCUI for 250 mcg Synthroid tablets, meaning this strength is not included in RxNorm. For the means of this analysis, this is classified as not applicable for the strength attribute. If the strength was included in RxNorm, it would be classified as false positive. The attributes for this example would be as follows: TP for drug, NA for strength, TP for form, and TN for route. In the second example from Figure 2, both the input and output contain semantically equivalent variations of normal saline. The output string incorrectly matches hydromorphone to the string, causing this to be reviewed as a partially correct output for the drug variable. Overall, if a component of the string is within the RxNorm vocabulary, then the attribute is applicable. If the drug, strength, form or route is not within RxNorm, then it is not expected and is labeled NA.
Figure 2.
Comparison of input and output strings for manual review.
After analyzing the results from the matching tools with the sample set, we decided to use the RxNav-in-a-Box to query the full medications list.
Lastly, we conducted a manual review of two subsets of the filtered medication list: inpatient and outpatient. The inpatient and outpatient delivery medication lists consist of medications received at any time point between 2006- 2017 by women who eventually delivered a baby at Penn Medicine. These lists include the distinct count of medical record number (MRN) of each string. The top 100 of the distinct MRN count were manually reviewed for precision, recall, accuracy and F1 score.
3. Results
3.1. Medication Matching Tool Queries
The time to run each medication-matching tool varied. For the 400 medications in our evaluation sample, the tools took the following amount of time: MedXN ran just under 1 second, Usagi ran in approximately 1 minute, the local RxNorm query ran in approximately 90 seconds, and RxMix sent the output via email in under 2 minutes. RxMix, local RxNorm API, and Usagi each give a score for every output (MedXN did not provide a score).
The sample set included 362 medication strings and 38 non-medication strings. See Table 6 for the number of matches per tool and the percentages from the total of 400 strings. MedXN has the highest number of unmatched strings, leaving only 225 strings with matches. Of those 225 matched strings, 31 were successfully matched to a more specific RXCUI (NormRxCUI). The local API query has a higher number of unmatched strings compared to the RxMix output due to the lack of spelling suggestions. For this query, an additional 34 strings (8.50%) left unmatched. Usagi matched 100 percent of the strings.
Table 6.
Comparison of Matches
 |
In Figure 3, all of the tools performed well on the medication subdomain. MedXN consistently demonstrated precise outputs for all of the subdomains, however the lower recall scores (Table 7) resulted in lower F1 scores for especially medication, form and route domains. The Usagi results indicate lower precision scores than the other tools reviewed, but the high recall leads to overall higher F1 scores for all domains. As expected, both variations of the RxNorm API perform comparably.
Figure 3.
Plots of Precision and F1 scores for each tool by subdomain.
Table 7.
Precision (P), Recall (R), Accuracy (A), F1 score, and match score p-values (p) for TP and FP classifications for each subdomain (drug, strength, form, route) by each tool.
As expected, the RxMix output and Local RxNorm API query performed similarly (Table 7). MedXN had great precision for drug, strength, and form, but the low recall contributed to a lower F1 score for those variables. Usagi
overall had poor precision, but the F1 score shows to be higher due to the high recall for drug, form and route. The p values for each tool across the domains indicate that score and performance are related. FP output scores of form and route are not significant for RxMix, and the FP output scores were not shown to be significant for the local RxNorm API query.
In Figure 4, each row displays the violin plot for the drug, strength, form, and route, showing the density of true positives and false positives among the scores. For each tool, a lower output score will indicate a poor match (Figure 4). False drug matches tend to have lower scores with a higher density of scores from 20-30, or 0.2-0.3 respectively. Strength FP matches sit higher around 40, or 0.4, and have higher scores with errors than drug FP outputs. From and route FP matches generally sit low, but the contrast is not as great as those illustrated on the drug and strength violin plots.
Figure 4.

Violin plots of False Positives and Scores. Each row of the figure indicates the medication matching tool: RxMix, Local RxNorm API, and Usagi, respectively. The distribution of scores for the drug and strength.
3.2. RxNav-in-a-Box for local RxNorm API query of filtered medication list
The query took 17 hours and 18 minutes to complete and matched 445,695 (90.20%) of the 494,146 strings in the filtered medication list, leaving 48,451 (9.80%) unmatched to RXCUI. In the output, 360,894 (80.97%) strings have a score greater than or equal to 30, 230,868 (51.80%) strings have a score greater than or equal to 50, and 66,638 (15%) have a score greater than or equal to 80.
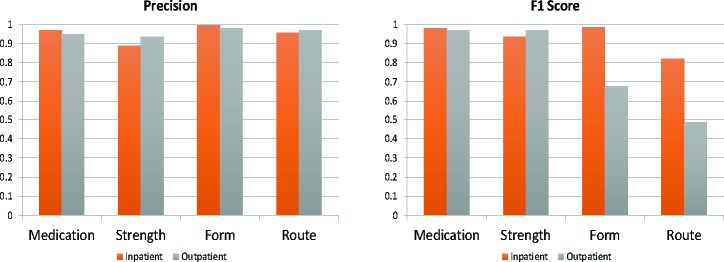
Shown in Figure 5, the outpatient and inpatient queries performed comparably for precision across all subdomains. However, as shown in Table 8, the outpatient query review indicates lower F1 scores on the form and route medication attributes, while the F1 score for inpatient falls lower for only the route subdomain. The inpatient and outpatient queries perform well on drug and strength attributes of the strings.
Figure 5.
Precision and F1 scores by subdomain for inpatient and outpatient subsets
Table 8.
Precision (P), Recall (R), Accuracy (A), and F1 score for each variable inpatient and outpatient
 |
The outpatient form and route attribute F1 scores show the lowest performance in comparison to the F1 scores in Table 7, due to low recall. Additionally, these attributes also show low accuracy scores for the outpatient subset.
4. Discussion
4.1. Strengths and Weaknesses
Each of the tools have been developed for different use cases to map medication strings to a standardized format. The approximate match function from RxNorm API was developed to normalize medication strings to RxNorm and therefore performs well for our review. MedXN is meant to extract medication information from clinical notes. This may have led to poorer performance on our sample. While MedXN matched over half of the sample to a general medication RXCUI, only 31 (~14%) of those generally matched were also given a more specific RXCUI as the result of parsing and normalizing the input string. While this tool is shown to perform well on the medication section of clinical notes, that performance did not generalize as well to our medication list. The Usagi tool was developed to accommodate for several vocabularies within the scope of the CDM, and not just for RxNorm. Usagi’s scope and purpose is to map to the CDM, and it is assumed that each match should be reviewed and approved. Because the matching algorithm is not specific to the RxNorm nomenclature, its holds back the performance for matching to RxNorm. This tool is more useful to mapping a shorter, perhaps higher-level list to the CDM.
The tools tended overlap for strings that have matches that could not be classified as true positives. None of the tools correctly matched strings with the drug morphine sulphate abbreviated as “MSO4” in the input string. Abbreviations of dextrose (e.g. “D50”, “D5W”) also led to incorrect matches across all tools. Strings with combination drugs (e.g. “CHLORPHEN-DIPHENHYD-PE-APAP PO MISC”) tended to only partially match the drugs in the string. Across all tools, strings with exact brand names of cold/flu medications resulted in false positive drug matches.
4.2. Importance of Input Strings for Accurate Output
There are several reasons that the tools would point to the wrong match, or provide no match for an input string. While the tools overall perform well, even an unnecessary character can cause an error in the output. This makes the tools highly sensitive to the quality of the input strings. In addition, the medication list included several strings of concepts not captured within the scope of RxNorm, these include: non-therapeutic radiopharmaceuticals, bulk powders, contrast media, food, dietary supplements, and medical devices1. Our list also includes several spelling errors. The source of the medication strings, such as the health information system, will have an effect on the overall quality of the strings. The software’s own local terminology may have local abbreviations and formats that a medication matching tool may not necessarily capture appropriately. Several strings included strengths outside of RxNorm. It may be an entry with dose and strength combined into the amount of the drug given to the patient, a strength not included in RxNorm (i.e. foreign drug), or simply a typo.
The outpatient and inpatient PennMed subsets are from any time point between 2006 and 2017, during which it is more likely for the data to be in a more standardized format. The complete filtered medication list and the sample set are from approximately the 1980’s to 2019, in which such a time frame may have changes in the medication records and a lack on standardization in the earlier years. Also, we reviewed the most used medication strings in the outpatient and inpatient subsets (again they are more likely to be in a standardized format). Because of this, the inpatient and outpatient queries performed better for drug and strength attributes in review than the queries from the sample set. In the inpatient medication query, over half of the medication strings matched with a score of 100. Moreover, the outpatient query shows low accuracy and F1 scores. This may be due to a number of reasons: more standardized nomenclature in the inpatient setting, and more out-of-scope strings in the outpatient list. In addition, the outpatient data was obtained from 1,048 distinct clinics within PennMed as described previously19, and data may have been captured differently across clinics.
Cleaning and filtering the medication list before using a matching tool should help improve the tool’s performance. In the case for Usagi, it may be required to have strings parsed, standardized, and merged before using the tool. Processing through the list, such as removing information out of the scope of RxNorm, and removing sensitive information should be part of the process before using a matching tool. It is important to note that when there is a possibility of sensitive information in the medication list, that the list should not be submitted on a query to another server. Therefore, it would be more appropriate to use a locally installable API, such as the RxNav-in-a-Box.
4.3. Abbreviations and Acronyms in Medication Strings
The NLM lists abbreviations and acronyms used in the RxNorm normalization process20. The user may be able to refer to this list to find acronyms and abbreviations in their list that will not be matched properly. For instance, ‘MSO4’ (morphine sulfate) is not listed as an abbreviation, and therefore the RxNorm API does not perform well and fails to match it the corresponding RXCUI. In this case, MedXN has more flexibility because a user with knowledge of UIMA can customize patterns in the system. Medication strings from older data will likely include outdated acronyms and abbreviations. For example, MSO4 (morphine sulphate) and MgSO4 (magnesium sulfate) are abbreviations that should be spelled out since they are likely to become confused for one another. Several of these ambiguous practices are included on recommendation lists to promote safe medication practice. While these tools have been developed to accommodate such terms, they can be limited to the terms captured in their data.
4.4. What Can We Learn About Using Publicly Available Medication Mapping Tools?
Because the input string quality is very important to the quality of the output, the user must consider the amount of pre-processing necessary. The effort of data preparation is time-consuming, as described earlier during the filtering process. For example, it would be a significant effort to extract medications from clinical notes in order to prepare the data for use with the RxNorm API. Perhaps in this case it would be useful to use MedXN, and then used the parsed data to capture RXCUI of higher granularity for outputs without specific NormRXCUI.
The amount of time a tool takes to run its algorithm should be considered. While MedXN quickly matched our sample set, a significant portion were unmatched to RXCUI. Moreover, a local query depends on the server in use. Several standard vocabularies are available from Athena for use in Usagi, but an index of multiple vocabularies significantly slows down the algorithm. An index of just the RxNorm vocabulary runs in approximately one minute, while a complete collection of vocabulary runs in approximately 4 minutes. The file format can also have an effect on the run time. Usagi performs better with a CSV file input in comparison to a text file format, which took approximately ten minutes to run. MedXN also favors a CSV format file and took just under one second to process compared to the three seconds for a text file. RxMix requires a text file format for batch jobs, while MedXN and the local RxNorm query are more flexible.
We observe that publicly available state-of-the-art medication matching tools for RxNorm standardization perform well overall (>70% F1 score across all three tools). Scores help to indicate where a user should set a threshold to increase the accuracy and precision of the output matches. Unfortunately, scores were not provided among MedXN output, making this type of filtering impossible. For tools that provide a score, having cleaned input greatly improves the score and quality of the output. This is especially important concerning longer strings and strings with more than one drug. Users may test a sample and determine an appropriate score threshold for their use case. Clear description of the algorithm used in publicly available tools can also help users understand and provide appropriate error handling.
4.5. Limitations and Future Work
There are several limitations of our work. Our manual review was conducted by an informaticist (LD) and not a pharmacologist and therefore certain nuances with regards to specific drug combos and their equivalence or lack of equivalence may be less then ideal. In addition, several strings in the filtered list still contained terminologies outside of the scope of RxNorm, such as medical devices and dietary supplements. This could have an effect on the overall analysis, while the tools could have performed better without these unnecessary strings. The 400-string sample was selected randomly, however no methods were used to ensure a representative sampling was obtained. In result, the sampling is not necessarily representative of the filtered master medication list. Lastly, MedXN’s performance may be underestimated due to depending on a 2013 RxNorm release.
Future work includes further classifying input strings into drug types to understand matching performance based upon class. It may also include comparing input strings across years, investigating how the strengths of the tools may be combined, and assessing newer tools as they become available. Further work could use lenient metrics when reviewing the performance of these tools, to have assess for partially correct matches for the medication attributes.
5. Conclusion
In conclusion, we performed a comparative analysis of three state-of-the-art medication-mapping tools—RxMix, Usagi, and MedXN - for converting local medication terminologies to RxNorm using non-standardized medication strings as input. Using filtering, we were able to clean a set of 941,198 unique medication terms from PennMed down to 494,146 (52.5% of original size). This filtering greatly reduced the efforts of applying any of the three tools, especially with regards to time intensiveness. Our analysis shows that all of the tools performed reasonably well (>70% F1 score). RxMix performed the best with an F1 score of 90% for drug name versus Usagi’s 82% and MedXN’s 74%. Our findings also indicate the importance of understanding the local medication terminology, and understanding the development context of the matching tools in consideration (e.g., clinical notes, medication terminology) because this can alter the performance of the tool for a given task. Our comparative results of these three popular and state-of-the-art tools for mapping non-standardized medication strings to RxNorm can help direct others efforts to choose an appropriate method for their use case.
Acknowledgments
We thank the Perelman School of Medicine at the University of Pennsylvania for generous funds to support this project. We also thank Nebojsa Mirkovic, Mark Miller, Sunil Thomas and Selah Lynch for feedback on medication terminologies within the Penn Data Store and Penn Data Analytics Center.
References
- 1.U.S. National Library of Medicine. RxNorm overview [Internet] Available from: https://www.nlm.nih.gov/research/umls/rxnorm/overview.html. [Google Scholar]
- 2.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc [Internet] 2011;18(4):441–8. doi: 10.1136/amiajnl-2011-000116. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21515544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saitwal H, Qing D, Jones S, Bernstam E V., Chute CG, Johnson TR. Cross-terminology mapping challenges: A demonstration using medication terminological systems. J Biomed Inform [Internet] 2012 Aug 1;45(4):613–25. doi: 10.1016/j.jbi.2012.06.005. Available from: https://www.sciencedirect.com/science/article/pii/S1532046412000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenreider O, Peters L. RxMix – Use of NLM drug APIs by non-programmers. 2017:2291. [Google Scholar]
- 5.U.S. National Library of Medicine. Approximate matching in the RxNorm API [Internet] 2015 Available from: https://rxnav.nlm.nih.gov/RxNormApproxMatch.html. [Google Scholar]
- 6.Peters L, Kapusnik-Uner JE, Bodenreider O. Methods for managing variation in clinical drug names. AMIA Annu Symp Proc [Internet] 2010;2010:637. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3041346/pdf/amia-2010_sympproc_0637.pdf. [PMC free article] [PubMed] [Google Scholar]
- 7.Peters L, Rice R, Bodenreider O. RxNav-in-a-Box – A locally-installable version of RxNav and related APIs. 2018;2102 [Google Scholar]
- 8.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: An open source medication extraction and normalization tool for clinical text. J Am Med Informatics Assoc. 2014;21(5):858–65. doi: 10.1136/amiajnl-2013-002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Informatics Assoc [Internet] 2012 Jan 1;19(1):54–60. doi: 10.1136/amiajnl-2011-000376. Available from: https://academic.oup.com/jamia/article-lookup/doi/10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.OHDSI/Usagi [Internet] Available from: https://github.com/OHDSI/Usagi. [Google Scholar]
- 11.Kopacka I. Basic regular expressions in R: Cheat sheet [Internet] 2016 Available from: https://www.rstudio.com/wp-content/uploads/2016/09/RegExCheatsheet.pdf. [Google Scholar]
- 12.Common medical abbreviations [Internet] 2002 Available from: http://dhss.alaska.gov/dph/Emergency/Documents/ems/assets/Downloads/Common_Med_Abbrev.pdf. [Google Scholar]
- 13.List of abbreviations used in medical prescriptions [Internet] Available from: https://en.wikipedia.org/wiki/List_of_abbreviations_used_in_medical_prescriptions. [Google Scholar]
- 14.American Speech-Language Hearing Association. Common medical abbreviations [Internet] Available from: https://www.asha.org/uploadedfiles/slp/healthcare/medicalabbreviations.pdf. [Google Scholar]
- 15.100 most popular american last names [Internet] Available from: https://www.rong-chang.com/namesdict/100_last_names.htm. [Google Scholar]
- 16.Butler R. Most common last names for blacks in the U.S. [Internet] Available from: https://names.mongabay.com/data/black.html. [Google Scholar]
- 17.Spitzer Y. Most common jewish names [Internet] 2012 Available from: https://yannayspitzer.net/2012/07/24/most-common-jewish-names/ [Google Scholar]
- 18.Pallmann P, Hothorn LA. Analysis of means: A generalized approach using R. J Appl Stat. 2016;43(8):1541–60. [Google Scholar]
- 19.Boland MR, Alur-Gupta S, Levine L, Gabriel P, Gonzalez-Hernandez G. Disease associations depend on visit type: results from a visit-wide association study. BioData Min [Internet] 2019 Dec 11;12(1):15. doi: 10.1186/s13040-019-0203-2. Available from: https://biodatamining.biomedcentral.com/articles/10.1186/s13040-019-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. National Library of Medicine. Abbreviations and acronyms in RxNorm normalization [Internet] Available from: https://rxnav.nlm.nih.gov/Abbreviations.html. [Google Scholar]