Abstract
Laboratory tests are a common aspect of clinical care and are the primary source of clinical genomic data. However, most laboratories use PDF documents to store and exchange the results of these tests. This locks the data into a static format and leaves the results only human-readable. The ordering clinician uses the results, but after that the information is unlikely to be used again. Future use would require a clinician to know that the test was performed, know where to find the PDF report, and take the time to open it and determine relevance to that future scenario. New computational standards such as SMART on FHIR and CDS Hooks present opportunities to better utilize these results, both physically upon receipt and asynchronously in future clinical encounters for that patient.
Full app available at https://github.com/mwatkin8/FHIR-Lab-Reports-App.
Demo available at http://hematite.genetics.utah.edu/FHIR-Lab-Reports/.
Introduction
Genetic laboratory tests are becoming increasingly common in clinical care. One study found that during the period of 2014–17 there were approximately 75,000 genetic tests on the market with about ten new tests entering the market daily.1 A growing subset of these tests are pharmacogenomic in nature. Although many of the promises of precision medicine via clinical genomic data are years away from realistic utility, pharmacogenomics2 is an area which has shown early promise.3, 4 Typically, these tests detect the presence of one or more variants which have been associated with deficiencies in the metabolism of a type of drug. The ordering clinician then uses this information to tailor a more appropriate medication regimen for the patient. Once the statuses of these variants are known, they are compiled into a report to be sent back to the ordering clinician. Inclusion of external test validation, recommendations, and other statistics into that report are specific to each laboratory as part of their business model. The actual transmission of the report typically involves it being compiled as a PDF and sent via email or other secure messaging system. Once received, the report is clicked open, viewed, and closed. The report is then likely saved to the patient’s record for future reference.
This system of reporting results and exchanging genomic data is flawed and represents a waste of resources.5, 6 Locking the data into a static PDF means that the data is no longer computable7 and is only beneficial when physically read by a clinician or other provider. This means that potential medical knowledge is likely to be buried in the patient’s health record in a form that is not accessible to computational clinical decision support systems. This could lead to preventable adverse effects, the possibility of re-ordering tests to confirm findings that were previously tested for, and an inability to reanalyze any genomic results included.8, 9 However, if the results were formatted in a standardized way and returned electronically, they could be utilized by asynchronous clinical decision support systems for the rest of the patient’s life.11, 12 Such a system could detect important reminders for clinicians which are based on a laboratory test that may have been performed several years earlier. Due to the specific nature of genetic tests and results, it is likely that the relevant implications would only surface periodically during the patient’s lifetime.13 It is highly improbable to expect a clinician to consult a PDF report for a test performed years previously (whose link is probably buried in the patient’s medical record) when such sporadic conditions do arise. The FHIR Lab Reports application has been designed as a solution to these various problems and presents one way of using new standards to better utilize the results of laboratory tests. The laboratory test which serves as a use-case for this application is pharmacogenomic. This was done deliberately because the results of pharmacogenomic tests are relevant not only when the ordering clinician first views the test results but at all later times when that certain drug class is being considered for the patient. Despite this specific use-case, the framework explained here can be applied to any type of laboratory test. The project code can be found at https://github.com/mwatkin8/FHIR-Lab-Reports-App.
Background
Several standards have been actively developed in recent years to address these issues. While these standards are being spread throughout the medical informatics community, there is a constant need for implementation projects to inform the iterative process of refining and improving them.
SMART on FHIR
FHIR (Fast Healthcare Interoperability Resources) is an HL7 data standard which has become very popular over the past few years.14 It is designed to divide a clinical scenario into distinct entities called resources. These resources are designed by HL7 workgroups and are now in their fourth version (R4).15 Each resource represents a clinical entity and clearly outlines how to store data related to that entity (e.g., the birthday of a patient in the Patient resource, or the date of a patient visit in the Encounter resource). These resources are then strung together according to defined profiles. This ensures uniform implementation of the standard. In instances where the base resources do not provide a specific data field needed for a certain scenario, extensions can be defined and published. FHIR is a very scalable clinical standard which can be adopted even while more complex entities are still being debated and formulated by working groups. This is an important characteristic which has played a major role in its widespread adoption. FHIR is being implemented by stakeholders across the spectrum of healthcare. These include EHR vendors such as Epic, Cerner, and AllScripts; commercial payers such as Anthem, the BlueCross BlueShield Association, Humana, and UnitedHealthcare; and technology companies such as Amazon, Google, Microsoft, IBM, and Oracle.16 SMART on FHIR17 is a design specification for clinical decision support (CDS) applications which use data formatted in the FHIR standard. It dictates how to combine four important components of software development to produce a standardized computer application. The four aspects of SMART on FHIR applications are: clean, structured data (FHIR), scopes and permissions (OAuth2), simple sign-in (OpenID Connect), and lightweight UI integration (HTML5). Each of these represent a solution to a slew of interoperability issues that have prevented the widespread adoption of CDS applications in the past.
CDS Hooks
Another aspect which will become increasingly important as the volume of CDS applications begins to rise is the ability to access them when needed and ignore them otherwise. While CDS applications promise increased quality in care, injudicious triggering criteria can lead to over-alerting and clogged workflows. CDS Hooks is an HL7 standard which will “hook” relevant CDS services into the EHR to provide services only when relevant. There are eight hooks in the current specification but the most commonly used are the patient-view hook (activated when a user opens a patient’s record) and the order-select hook (activated when a user selects an order such as a test or medication). Once activated, a hook queries the data server to access “pre-fetch” information. This information confirms whether or not to use the hook to pull a certain CDS service into the health system for use. Once hooked, the CDS service (hosted externally) returns a response card directly to the EHR which contains decision support for the clinician and can include suggestions, warnings, or even links to relevant SMART on FHIR applications18. Previous work has explored the use of the CDS Hooks specification in delivering pharmacogenomic recommendations and has shown it to be a promising approach.19
FHIR Genomics
The FHIR genomics team has produced a new implementation guide, soon to be released, which details the process of representing genomic lab reports, in all of their variety, as FHIR version R4 resources.20 Earlier work done by the team resulted in a 2015 publication of a SMART on FHIR genomic application which demonstrated how those new genomic profiles were to be implemented. With the new FHIR R4 General Genomics Reporting Implementation Guide (version 1.0.0), representing four years of progress since that earlier application, a new implementation project to provide similar feedback is timely. This was a driving factor in the development of the FHIR Lab Reports application. The profiles in the guide provide a glimpse at the complexity and breadth of genomic test reporting data types. They range from basic types such as variant, genotype, haplotype, etc., to specific types such as sequence phase relationship, inherited disease pathogenicity, somatic prognostic implication, etc. Having the data points strictly profiled as FHIR resources enables interoperable approaches to reclassification, exchange, and additional genomic services which would otherwise be very difficult.9, 21, 22
Methods
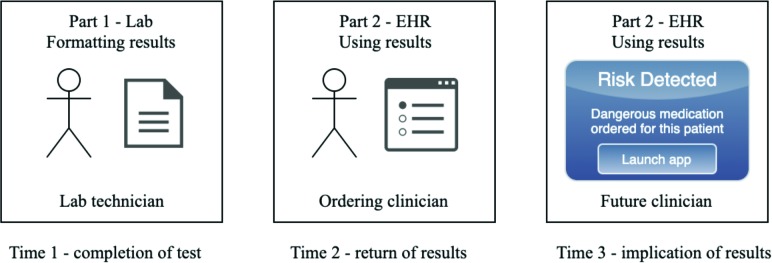
One of the major contributions of the FHIR Lab Reports application is to demonstrate the duality which an approach needs to adequately address the issues explained previously. In other words, the solution must involve both generating and consuming the data. The data must be structured as appropriately-formed FHIR resources by the laboratory if it is to be utilized by a SMART on FHIR application within the EHR. Also, the results must be accessed not only on receipt of the report, but at all relevant future times. Thus, this application demonstrates how those two different parts should operate at three main time periods, as explained in Figure 2.
Figure 2.
Graphical representation of how, and when, this application is intended to be used.
Part 1—Lab
The use-case for this prototype is a test selected from the ARUP Genetics Test Menu. Most of the tests therein include example PDF reports of the different possible results. The test selected for this project was a Statin Sensitivity test23 which reports the presence or absence of a variant of the SLCO1B1 gene. The main functionality of the first part of the FHIR Lab Reports application is to create resources for the clinical scenario in which the test was ordered, as well as subsequent related clinical scenarios such as a blood draw or other consultation relating to the test.
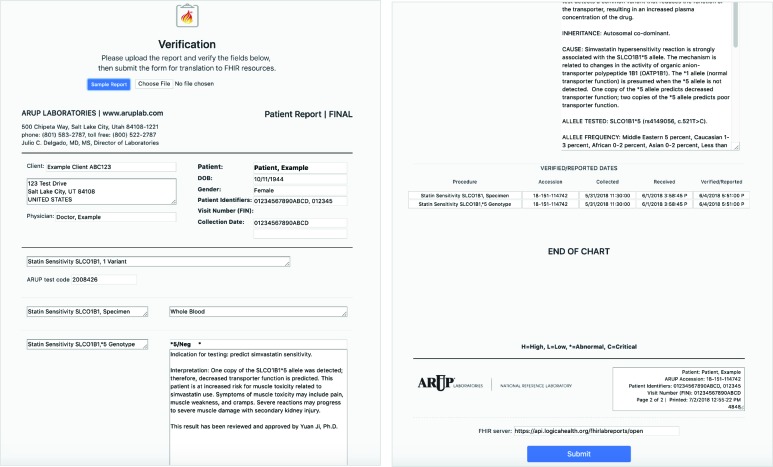
Implementation of this part of the application in a live laboratory setting would involve intercepting the data before the generation of the PDF report. The data concepts could then be mapped reliably to the appropriate fields of the FHIR resources. However, since the inner file systems and workflows of laboratories are proprietary, this application uses the publicly available example PDF report as input data and performs custom parsing to create the FHIR resources. Parsing a PDF document is not a scalable solution because the text is largely unstructured and must be extracted through regular expressions specific to each test. This approach is used only to allow relevant and realistic test data to be made available for this proof-of-concept application. As a result of this limitation, a verification step was included to allow the user to make sure that the data from the uploaded PDF has been parsed as expected. This verification step is an HTML version of the test report, shown in Figure 3.
Figure 3.
Report upload and verification screen of the FHIR Lab Reports application.
This has been made to resemble the PDF so that the user can confirm that the data are parsed into the appropriate fields before they are used to generate the FHIR resources. The user can directly move or edit any misplaced data before submitting. Because this application is not in a live production setting, a field for a FHIR data server URL has also been included. On submission, the data is extracted from those user-verified fields to generate the twenty FHIR resources needed to describe the full scenario. These resources are pushed to the specified FHIR data server. The chain of events and their corresponding resource types relating to this test are described in Table 1.
Table 1.
Resources created by the FHIR Lab Reports application for the results of a Statin Sensitivity test.
| Event in the clinical scenario | FHIR resource(s)24 created |
|---|---|
| The patient sits down with a clinician at a clinic. | Patient, Practitioner, Encounter, Location |
| The clinician orders the genetic test and the blood sample needed for the test. | ServiceRequest (test), ServiceRequest (blood draw) |
| The patient sits down with the in-clinic phlebotomist. | Practitioner, Encounter |
| The phlebotomist performs the blood draw. | Procedure, Specimen |
| The blood sample is sent to the lab technician at ARUP. | Practitioner, Location |
| The lab technician runs the test. | Procedure |
| A report is made of the findings. | DiagnosticReport, Observation (interpretation), Observation (implication), Observation (genotype), Observation (variant) |
| A CDS service is defined to be hooked if the implications ever arise. | PlanDefinition |
| A CDS Hooks card is created to be returned if implications ever arise. | RequestGroup |
The FHIR Genomics working group implementation guide was used to create the four observation resources used to represent the findings of the test. Specific FHIR profiles have been made for each of those observations.25–28 Explanations of each of these profiles are included in Table 2. Similarly, FHIR profiles guided the creation of the two resources which are used to provide the CDS Hooks functionality of the application.29, 30 There is variability in the types of results returned by the many pharmacogenomic tests available. Our use-case was chosen because its results include a variant, an indicated medication, and a genotype. These data points are less complex than other test results, but are common among pharmacogenomic tests. More complex results are possible and their modeling as FHIR resources would be dictated by the implementation guide.
Table 2.
Explanations of genomic test result resource profiles and examples from the test report.
| Profile | Explanation | Example from test report |
|---|---|---|
| Observation (obs-overall) | Overall Genomic Interpretation: a high-level summary observation of the entire report | One copy of the SLCO1B1*5 allele was detected; therefore, decreased transporter function is predicted. |
| Observation (obs-implication) | Genetic Implication: These represent observations about the patient based on the genetic test results. | This patient is at increased risk for muscle toxicity related to simvastatin use…(snipped for brevity). |
| Observation (obs-genotype) | Genotype: combinations of genetic variations that together are associated with a particular phenotype | *5/Neg * |
| Observation (obs-variant) | Variant: differences between parts of specimen sequences and the equivalent portions of reference sequence(s). | c.521T<C |
Part 2—EHR
Once the test results are translated to FHIR resources and pushed to the appropriate data server, they are available to be consumed by the SMART on FHIR part of the application within the EHR. This is anticipated to happen at two different time periods. The first is upon completion of the test and the transmission of the results. This completion will be communicated to the ordering clinician who can use the application to view the results. The second is any future scenario where the findings of the test have direct implications for the care of the patient. This is achieved through the two CDS Hooks resources created during part 1.
The first is a PlanDefinition resource which defines the application itself as a CDS service available to respond to hook events with response cards if appropriate. This is not linked to the patient but is global to the health system and is not duplicated for a new patient if already found in the data server. It defines the hook events supported by the application as well as the pre-fetch information it requires. This resource would be consumed by the health system in order to register the application as an available CDS Hooks service.
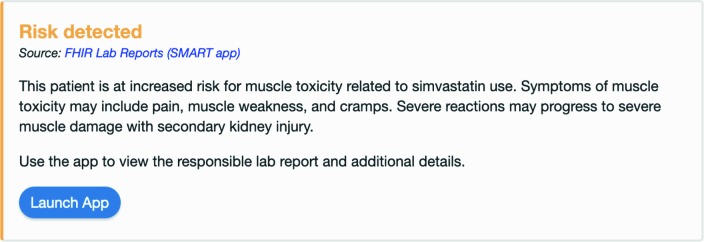
The second is a RequestGroup resource which defines the response card to be returned for the patient if a hook event triggers the application. This is specific to the patient. It is also unique to the test which was used to create it. Since each test will have different implications, each test will have one or more response cards which will ensure that those implications are delivered to clinicians when appropriate. For example, a positive result to our example Statin Sensitivity test means that the patient should not be prescribed Simvastatin as part of any medication regimen. To ensure this, the app will return a response card if Simvastatin is ever the medication associated with an order-select hook which is invoked during a clinical scenario where that patient is the subject. The response card contains the implication directly from the test result as well as a link to launch the FHIR Lab Reports application. The card would be displayed within the EHR to the clinician who is about to order Simvastatin for the patient.
Through CDS Hooks, the result of the test is made available not only to the ordering clinician, but asynchronously for the rest of the patient’s life. This ensures that all future clinicians will be warned against ordering Simvastatin for that patient without those same clinicians needing to locate the original PDF for a test which they may or may not know was ever performed for the patient.
Results
The application has four main views, as shown in Figures 5-8. Each of these views are the result of the design considerations mentioned previously and can be viewed and demoed at http://hematite.genetics.utah.edu/FHIR-Lab-Reports/. Each test result associated with the patient is shown in a list in order of the date it was performed. A search bar is provided for finding a test by name rather than by date. When the app is used by the ordering clinician to view the result, it will be at the top of the list since it was the likely most recent performed. The clinician will also be familiar with the name of the test and will know which one to select since they are the ones who ordered it.
Figure 5.
The “Report Details” view of the FHIR Lab Reports application.
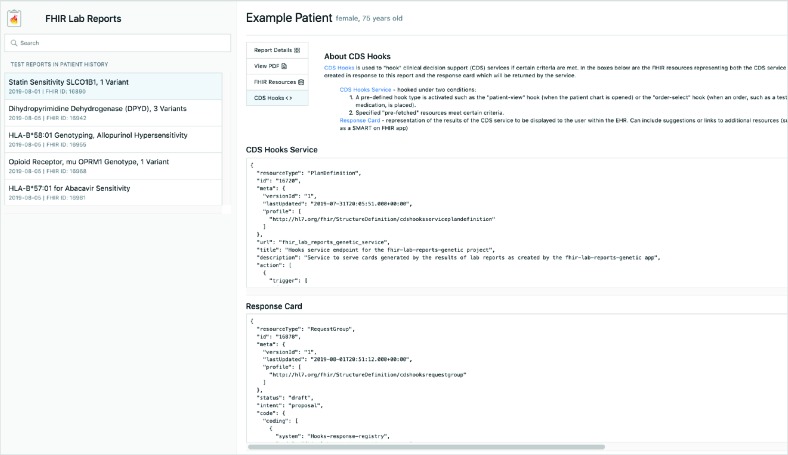
Figure 8.
The “CDS Hooks” view of the FHIR Lab Reports application.
If the application is launched from a CDS Hooks response card, the associated test is placed at the top of the list and automatically selected for that clinician who won’t be familiar with the test and who will only know what the card told them.
Discussion
As clinicians increasingly rely on CDS applications to perform services and ease their cognitive load, efforts must be made to foster their understanding of underlying standards and technologies being used. This application demonstrates how to provide those kinds of educational elements can be incorporated. Each of the views includes combinations of educational elements and CDS functionality. While this is a proof-of-concept project, similar applications could be made which include more robust combinations of popular CDS services, knowledge-retrieval tools, and educational elements. Applications built for live implementation should prioritize clinician feedback in the design process to ensure ease of use and simplicity in the display and available tools.31, 32
A summary view is displayed when a test is selected. It provides the same information as is in the PDF of the test result. However, since it is not a static display, it can be enhanced with knowledge-retrieval CDS services. This test example contains links to relevant medication and genetic knowledge bases. This allows clinicians who may have little prior exposure to the jargon and formats of clinical genomics to be able to link directly to educational resources, allowing them to provide more informed care for the patient.
The PDF is still a valuable source of information. In part 1 of the application, the PDF is stored as a base64 string in the DiagnosticReport FHIR resource. This means that future clinicians don’t need to track down the link to the original PDF in the patient’s medical history. The application is able to pull it right back out of the resource and render it when needed. It is important that the current methods of laboratories returning results are not completely abandoned. PDFs have value in their own right and represent various business services built up by the laboratory. Making this original report more easily accessible and usable will foster adoption by the laboratory, helping them see these kinds of applications as complementary rather than competing services.
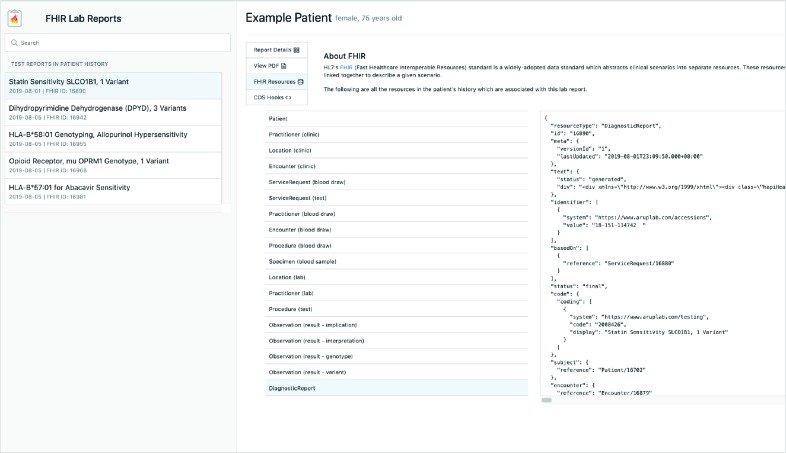
Because FHIR is a relatively new data standard, a view was included that gives a very brief overview of FHIR and allows the clinician to view all the resources which were created in part 1 of the application. This helps them get a better understanding of FHIR and how it works. However, a more valuable contribution is to be able to quickly view the original ordering clinician, the location of the clinic where the test was ordered, who the phlebotomist was, who the lab technician was, etc. Every piece of relevant clinical data being linked and made available through FHIR.
Alerting clinicians directly within the EHR should be a closely-guarded privilege. This view gives a brief introduction to CDS Hooks and explains how the response card was generated. This is a step toward helping clinicians understand where the alert came from and develop trust that the application won’t send unnecessary alerts in the future.
Conclusion
Laboratory tests are filled with useful information. By standardizing their content and using SMART on FHIR applications to display their results, various other types of CDS services can be included. By enabling CDS Hooks, these results can be better utilized at future points in the care of the patient rather than just read upon receipt of the test result. This is likely to result in reduced adverse events, reduced ordering of redundant laboratory tests, and increased cost savings to participating health systems. The FHIR Lab Reports application demonstrates how to combine these standards and technology into a useful innovation.
While it is still in the prototype stage, collaboration with participating laboratories can lead to more practical insight about the translation of test results to FHIR resources before the final PDF is generated. This mapping process will be unique to each laboratory depending on the intermediate file types being used and on how where the relevant pieces of information are being pulled from for the final report. However, if this mapping process were completed, the SMART on FHIR EHR-facing part of the application wouldn’t change because the underlying FHIR data structure would not change. These efforts of a laboratory to implement the FHIR standard into their existing systems would be a significant and complex challenge but would result in a more robust business model where interoperability is a strong component. This and additional future reference implementations will hopefully demonstrate to laboratories that the resources required to utilize the FHIR standard would be well worth the effort.
Acknowledgements
This work was supported by the National Institute of Health: R01HG008628 to KE, and NLM T15-LM007124 training predoctoral slot to MW.
Figures & Table
Figure 1.
Example PDF report for the results of a pharmacogenomic laboratory test, taken from the ARUP Genetics Test Menu.10
Figure 4.
Response card generated from the test result which is displayed when Simvastatin is ordered for the patient.
Figure 6.
The “View PDF” view of the FHIR Lab Reports application.
Figure 7.
The “FHIR Resources” view of the FHIR Lab Reports application.
References
- 1.Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic Test Availability And Spending: Where Are We Now? Where Are We Going? Health affairs (Project Hope) 2018;37(5):710–716. doi: 10.1377/hlthaff.2017.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. https://ghr.nlm.nih.gov/primer/genomicresearch/pharmacogenomics.
- 3.Ji Y, Skierka JM, Blommel JH, Moore BE, VanCuyk DL, Bruflat JK, et al. Preemptive Pharmacogenomic Testing for Precision Medicine. The Journal of Molecular Diagnostics. 2016 May;18(3):438–445. doi: 10.1016/j.jmoldx.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross T, Daniel J. Overview of pharmacogenomic testing in clinical practice. The mental health clinician. 2018 Sep;8(5):235–241. doi: 10.9740/mhc.2018.09.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knoppers BM. Framework for responsible sharing of genomic and health-related data. The HUGO journal. 2014 Dec;8(1):3. doi: 10.1186/s11568-014-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Global Alliance for Genomics and Health. A federated ecosystem for sharing genomic, clinical data. Science. 2016 Jun;352(6291):1278–1280. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 7.Lubin IM, Aziz N, Babb LJ, Ballinger D, Bisht H, Church DM, et al. Principles and Recommendations for Standardizing the Use of the Next-Generation Sequencing Variant File in Clinical Settings. The Journal of Molecular Diagnostics. 2017;19(3):417–426. doi: 10.1016/j.jmoldx.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu P, Meng L, Normand EA, Xia F, Song X, Ghazi A, et al. Reanalysis of Clinical Exome Sequencing Data. New England Journal of Medicine. 2019 Jun;380(25):2478–2480. doi: 10.1056/NEJMc1812033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genetics in Medicine. 2017 Feb;19(2):209–214. doi: 10.1038/gim.2016.88. [DOI] [PubMed] [Google Scholar]
- 10. https://www.aruplab.com/genetics/tests.
- 11.Masys DR, Jarvik GP, Abernethy NF, Anderson NR, Papanicolaou GJ, Paltoo DN, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. Journal of Biomedical Informatics. 2012 Jun;45(3):419–422. doi: 10.1016/j.jbi.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch BM, Eilbeck K, Fiol GD, Meyer LJ, Kawamoto K. Technical desiderata for the integration of genomic data with clinical decision support. Journal of Biomedical Informatics. 2014 Oct;51:3–7. doi: 10.1016/j.jbi.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nature Reviews Genetics. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. https://www.healthit.gov/buzz-blog/interoperability/heat-wave-the-u-s-is-poised-to-catch-fhir-in-2019.
- 15. https://www.hl7.org/fhir/overview.html.
- 16. https://healthitanalytics.com/features/fhir-is-blazing-a-path-to-patient-centered-data-driven-healthcare.
- 17. https://docs.smarthealthit.org/
- 18. https://cds-hooks.org/specification/1.0/
- 19.Dolin RH, Boxwala A, Shalaby J. A Pharmacogenomics Clinical Decision Support Service Based on FHIR and CDS Hooks. Methods of Information in Medicine. 2018 Dec;57(S 02):e115–e123. doi: 10.1055/s-0038-1676466. [DOI] [PubMed] [Google Scholar]
- 20. http://build.fhir.org/ig/HL7/genomics-reporting/general.html.
- 21.Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, et al. Prevalence of Variant Reclassification Following Hereditary Cancer Genetic Testing. JAMA. 2018 Sep;320(12):1266. doi: 10.1001/jama.2018.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaminathan R, Huang Y, Astbury C, Fitzgerald-Butt S, Miller K, Cole J, et al. Clinical exome sequencing reports: current informatics practice and future opportunities. Journal of the American Medical Informatics Association. 2017 Nov 1;24(6):1184–1191. doi: 10.1093/jamia/ocx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. http://ltd.aruplab.com/tests/pub/2008426.
- 24. https://www.hl7.org/fhir/resourcelist.html.
- 25. http://build.fhir.org/ig/HL7/genomics-reporting/obs-overall.html.
- 26. http://build.fhir.org/ig/HL7/genomics-reporting/obs-implication.html.
- 27. http://build.fhir.org/ig/HL7/genomics-reporting/obs-genotype.html.
- 28. http://build.fhir.org/ig/HL7/genomics-reporting/obs-variant.html.
- 29. http://hl7.org/fhir/cdshooksserviceplandefinition.html.
- 30. http://hl7.org/fhir/cdshooksrequestgroup.html.
- 31.Shirts BH, Salama JS, Aronson SJ, Chung WK, Gray SW, Hindorff LA, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. Journal of the American Medical Informatics Association. 2015 Jul;:ocv065. doi: 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cutting E, Banchero M, Beitelshees AL, Cimino JJ, Fiol GD, Gurses AP, et al. User-centered design of multi-gene sequencing panel reports for clinicians. Journal of Biomedical Informatics. 2016 Oct;63:1–10. doi: 10.1016/j.jbi.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]