Abstract
POTE is a primate-specific gene family that encodes cancer testis antigens that contain three domains, although the proteins vary greatly in size. The amino-terminal domain is novel and has three cysteine-rich domains of 37 amino acids. The second and third domains are rich in ankyrin repeats and spectrin-like helices respectively. In humans, 13 highly homologous paralogs are dispersed among eight chromosomes. Some members of the POTE gene family have an actin insertion at the carboxyl end of the protein. The expression of the POTE gene in normal adult tissues is restricted, but several POTE paralogs are frequently expressed in many cancers including breast, prostate, and lung cancers. We show here that POTE is expressed in several human embryonic stem (ES) cell lines. We found that UC06, WA01 and ES03 cell lines express mainly a POTE-2γ transcript but ES02 and ES04 cell lines predominantly express POTE-2α. The WA09 cell line expressed both POTE-2γ and POTE-2α. There is no detectable POTE gene expression in fetal tissues (ages 16–36 weeks). The POTE paralogs that are expressed in ES cells may have a specific function during lineage-spe-cific differentiation of ES cells.
INTRODUCTION
In our effort to identify new targets for the immunotherapy of prostate cancer, we have identified a new gene family, POTE, that is expressed in normal prostate, testis, ovary, and placenta [1–3]. There are at least 13 paralogs of the POTE genes in the human genome and these are dispersed among eight different chromosomes [4,5]. They are POTE-2α, POTE-2β, POTE-2γ, and POTE-2δ, all of which are on chromosome 2; POTE-8 is on chromosome 8; POTE-14α and POTE-14β are on chromosome 14; POTE-15 is on chromosome 15; POTE-18 is on chromosome 18; POTE-21 is on chromosome 21; and POTE-22 is on chromosome 22. POTE paralogs are only found in humans and other primates. There are no obvious orthologs of POTE gene in other species; however, a distant ancestral gene has been identified in rodents [5]. Recently, we reported that there is an actin retroposon insertion at the carboxyl terminus of one of the ancestral POTE paralogs that occurred before the divergence of Old World monkeys and apes [6]. The resulting gene has duplicated and diverged into several POTE paralogs. These encode a new chimeric protein that contains both POTE and actin in the same molecule [6].
Human embryonic stem (hES) cells derived from the embryoblast are pluripotent cells that maintain their ability to self-renew and are a potential source of differentiated cells for a variety of therapeutic uses. Identification of molecular components that contribute to the pluripotency of hES cells provides valuable information for both developmental biology as well as stem cell research [7].
Because POTE is expressed in testis and ovary, we decided to extend our analysis and investigate the expression of POTE paralogs in undifferentiated hES cells. We have investigated the relative expression of POTE paralogs in six commonly used hES cell lines. Our results show that either POTE-2α or POTE-2γ is predominantly expressed in undifferentiated ES cell line.
MATERIALS AND METHODS
Primers
Primers used in this study are listed as follows: T444, 5′ -CAA TGC CAG GAA GAT GAA TGT GCG-3′; T445, 5′-TCT CTG GCC GTC TGT CCA GAT AGA T-3′; actin, forward, 5′-GCA TGG GTC AGA AGG AT-3′, reverse, (5′-CCA ATG GTG ATG ACC TG-3′). Primers were synthesized in Lof-strand Labs Limited (Gaithersburg, MD).
Northern analysis
Total RNAs were isolated from the WA01 ES cell line using Trizol reagent (Invitrogen, Carlsbad, CA). About 2.0 μg of mRNAs (purchased from Clontech Laboratories Inc., Mountain View, CA) from testis and brain and 20 μg of total RNA from the WA01 cell line were run on an agarose gel and transferred to a nylon membrane. The 1.2-kb probe was generated by PCR and labeled with 32P using the random priming extension method. The probe location is shown in Fig.1. Membranes were prehybridized for more than 2 h in hybridization buffer followed by addition of denatured probe and incubation for additional 12 h. Membranes were washed two times for 20 min each in 2 × SSC and 0.1% sodium dodecyl sulfate (SDS) at room temperature and then washed two times for 20 min each in 0.1 × SSC and 0.1% SDS at 60°C. The membranes were then subjected to autoradiography.
FIG. 1.
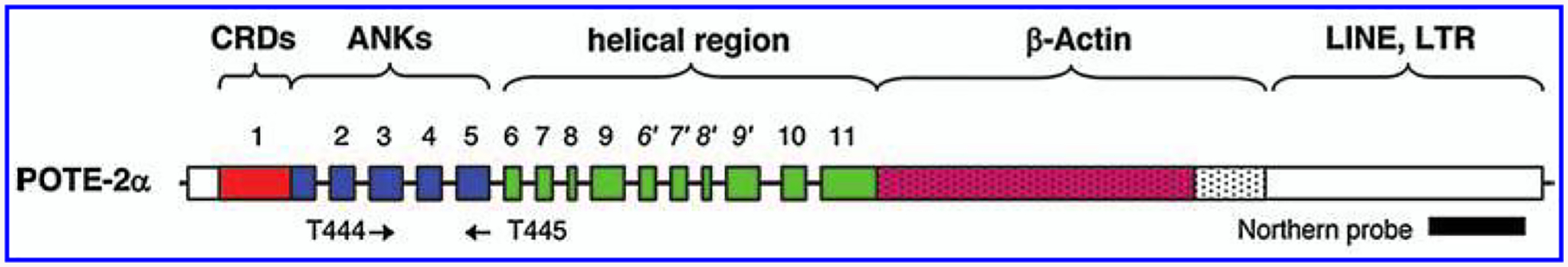
Schematic showing the coding exon and the predicted protein domains of POTE-2α. Primer binding sites (arrows) and the region of probe used for northern analysis are also shown. CRDs, Cysteine-rich domains; ANKs, ankyrin repeats.
5-Aza-2′ -deoxycytidine treatment
Cells were maintained in growth medium containing 5-aza-2′ -deoxycytidine (DAC) at 1 mM for 48 h, with exchange of fresh medium containing DAC every 12 h. After the 48 h of exposure, the DAC-containing medium was removed and replaced with fresh medium without DAC. The cells were harvested for RNA 48 h after the DAC exposure was complete.
Expression of POTE by RT-PCR analysis
Total RNAs from different ES cell lines were isolated using TRIzol reagent (Invitrogen). First-strand cDNA was prepared from the isolated total RNA using the First Strand cDNA synthesis kit (Amersham, Piscataway, NJ) following the manufacturer’s instructions. PCR was performed on cDNA from different RNA samples following the manufacturer’s instructions using the error-proof polymerase, pfu Turbo (Stratagene, La Jolla, CA). PCR-ready first-strand cDNA from human fetal tissues were purchased from Clontech Laboratories Inc. The PCR conditions used were as follows: initial denaturation at 94°C for 35 sec, 30 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 1 min, and elongation at 72°C for 2 min. The T444 and T445 primers described above were expected to generate a 386-bp DNA fragment (Fig. 1).
Cloning and analysis of PCR product
The PCR product obtained from the RT-PCR reaction was purified from a low-melting-point agarose gel using gel extraction kit (Qiagen, Alameda, CA) and cloned into the pCR4Blunt-TOPO vector (Invitrogen). Positive colonies from each plate were selected randomly and plasmids were isolated from each colony. Plasmids with the correct insert were sequenced and analyzed. To determine the origin of the transcripts, we devised an in-house BLAST Web server specialized for identification of POTE paralogs, as described earlier [8]. Briefly, we collected exon sequences of all POTE paralogs from the human genomic sequence. The sequences excluding the primer binding regions of the PCR- amplified fragment, were prepared from each paralog and converted to a database that could be run against BLAST. The identification of a clone is based on its BLAST score against the standard POTE sequences: The paralog giving the best score is assigned to the clone.
Analysis of the POTE gene cluster on chromosome 2
We defined the boundary of the POTE gene cluster on chromosome 2 with a BLAT search of the human genome assembly by using a POTE cDNA sequence as a query at the UCSC Human Genome Browser Database (http://genome.ucsc.edu). Each of the five POTE gene transcriptional units was identified. Segmental duplication was assessed by a dot-plot analysis using the LBDOT program (downloaded from http://www.lynnon.com/dotplot.html; Lynnon Corporation, Vaudreuil-Dorion, Quebec, Canada).
RESULTS
Expression of POTE in HES cells
We reported earlier that POTE paralogs are expressed in a wide variety of cancers including prostate, colon, lung, breast, ovary and pancreas, but the expression of the POTE paralogs in normal tissues is restricted to testis, prostate, ovary, and placenta [2,8]. Because of the expression of POTE in ovary and testis, we were prompted to examine POTE expression in hES cells by RT-PCR analysis. As shown in Fig. 2A, a specific band of 386 bp was obtained in WA01 and WA09 lanes when RT-PCR was performed using the T444 and T445 primers, indicating POTE is expressed in both WA01 and WA09 ES cells. To test the quality of the generated cDNA, separate PCR analyses were performed with primers specific to actin.
FIG. 2.
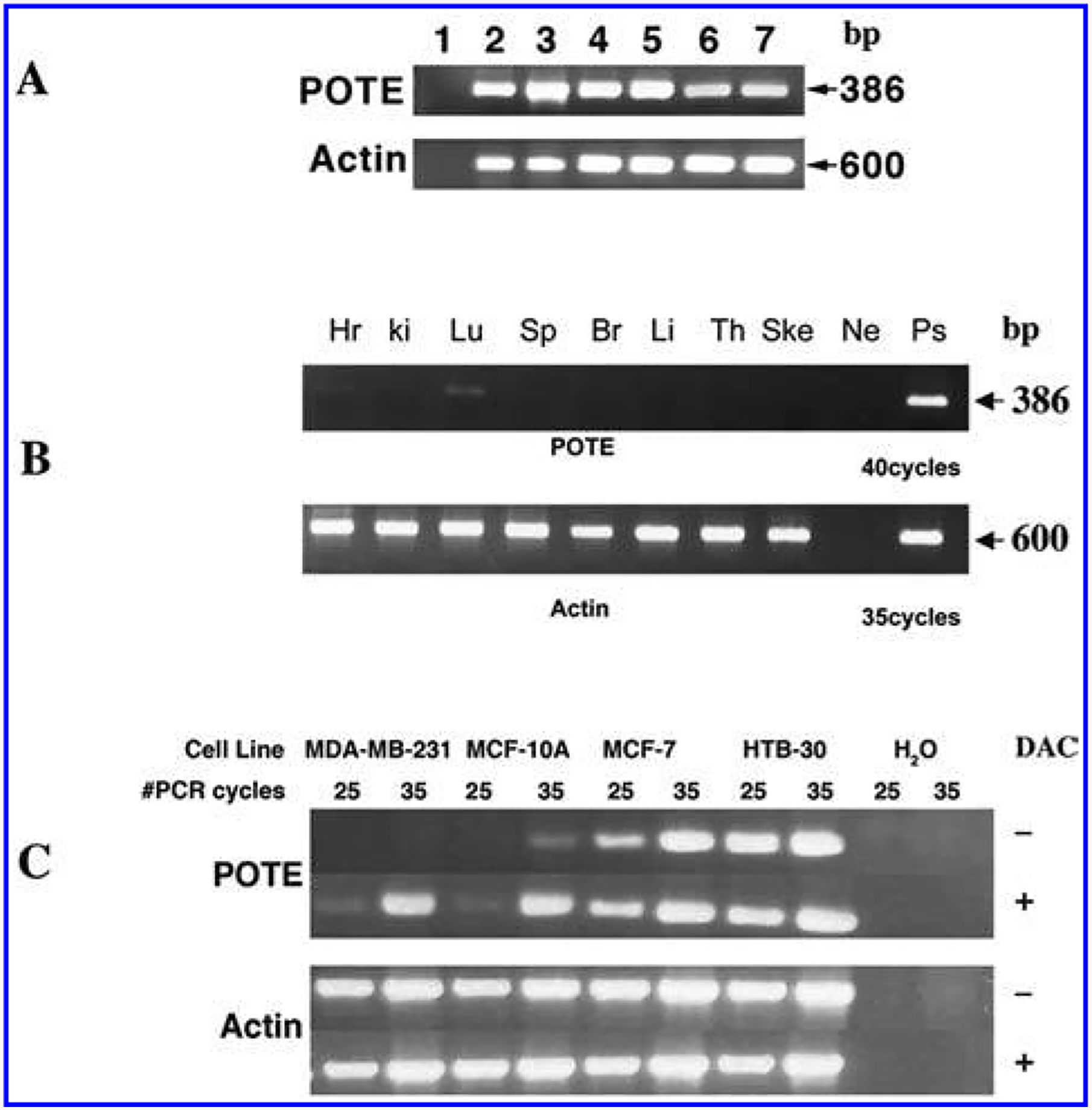
RT-PCR analysis of POTE expression. (A) Expression of POTE in six different ES cell lines. Lane 1, Negative control; lane 2, ES02; lane 3, ES03; lane 4, ES04; lane 5, UC06; lane 6, WA01; lane 7, WA09. (B) POTE expression in human fetal tissues: Hr, heart; ki, kidney; Lu, lung; Sp, spleen; Br, brain; Li, liver; Th, thymus; Ske, skeletal muscle; Ne, negative control; Ps, positive control. (C) POTE expression in breast cancer cell lines after DAC treatment. The expected size of the POTE PCR fragment is 386 bp. Amplification of actin cDNA was used as internal control and the expected size of the actin PCR product is 600 bp.
To determine the size of the POTE transcript expressed in hES cells, total RNA from the WA01 cell line was analyzed by northern blot hybridization using a probe 3′ end of POTE transcript [6]. A specific band of about 7.5 kb in size was detected in WA01 lane, suggesting that the POTE transcript in this cell line is 7.5 kb in size (Fig. 3). As expected, there is a strong but smeary signal in testis, which is known to express many POTE paralogs. There is no detectable band in the POTE-negative brain RNA sample.
FIG. 3.
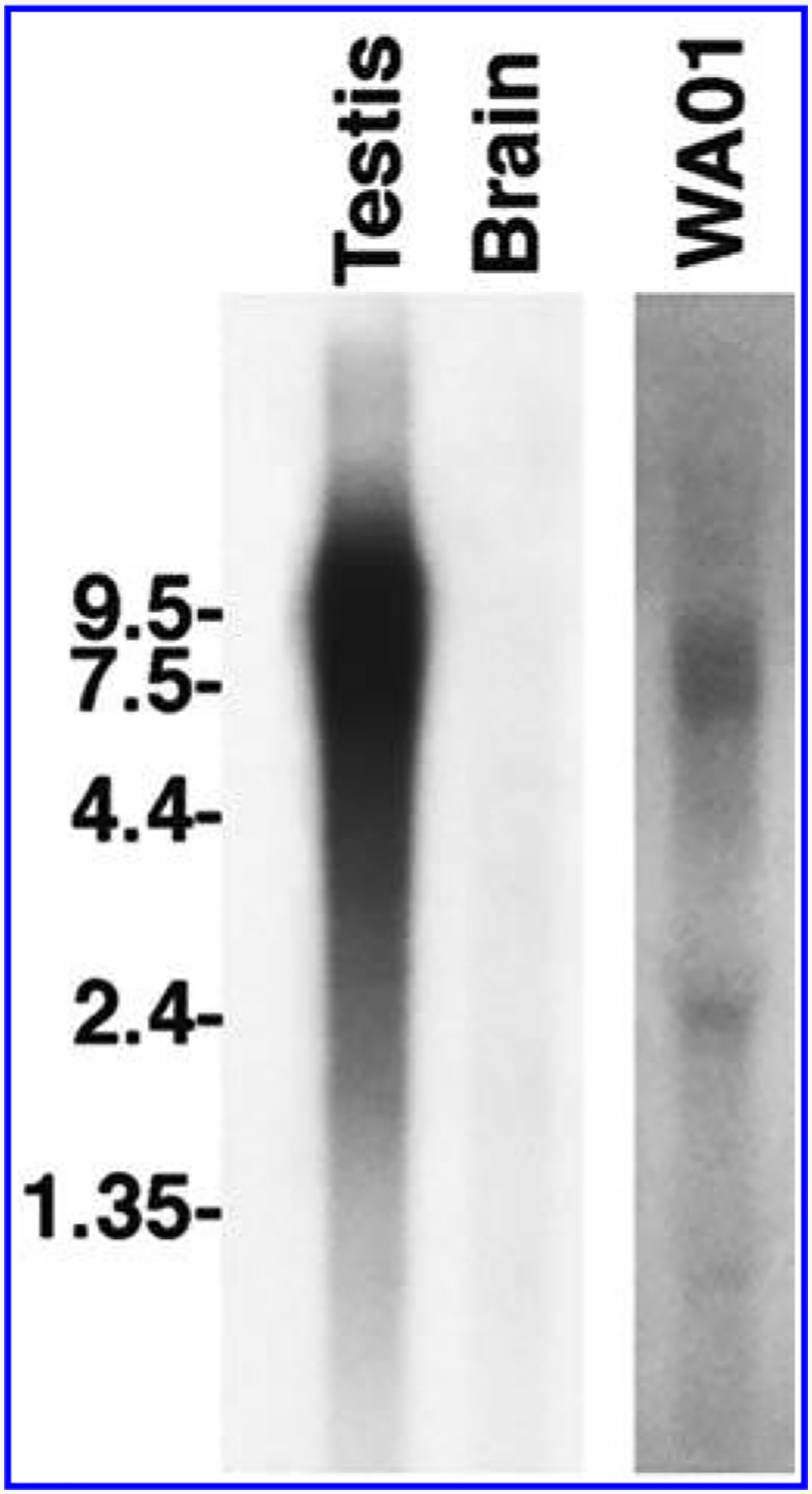
Analysis of POTE transcript in WA01 ES cell line. A specific band of about 7.5 kb in size was detected in the WA01 line. In testis the signal is some what diffuse, over 6.5–9.0 kb in size, and there are no detectable bands in brain.
We then obtained total RNA for four other ES cell lines—ES02, ES03, ES04, and UC06 from different sources—and performed RT-PCR analysis as described above. In all cases a specific 386-bp fragment of the POTE gene was detected (Fig. 2A), indicating that POTE is expressed in all ES cell lines.
POTE expression in human fetal tissue
To investigate if POTE is expressed in tissues during fetal development, we performed RT-PCR analysis with cDNAs prepared from fetal brain, lung, liver, kidney, heart, spleen, thymus, and skeletal muscle. Each tissue cDNA represents RNA pooled from 10–38 spontaneously aborted fetuses. There is no detectable POTE expression in any of these tissues after 35 cycles of PCR (data not shown). However, as shown in Fig. 2B, there is a very weak band detected in lung after 40 cycles of PCR, indicating POTE expression is turned off during fetal development of these tissues.
POTE expression is induced by the demethylating agent DAC
To determine if POTE expression is regulated by methylation of the gene, we analyzed the promoter region of the POTE gene family using the CpG Island Searcher program (http://www.uscnorris.com/cpgisland2/cpg.aspx). As shown in Fig. 4, all POTE members are associated with CpG islands at the 5′ region of the gene, suggesting POTE expression might be regulated by methylation of the gene. We treated two POTE-negative cell lines (MCF-10A and HTB30) and two POTE-positive cell lines (MCF7 and MDA-MB-231) with DAC and performed a semiquantitative RT-PCR analysis to test this hypothesis. As shown in Fig. 2C, DAC treatment significantly induces POTE expression in MDA-MB-231 and MCF10A cell lines. The induction of POTE expression is modest in MCF7 cells, but there is no induction in HTB 30 cells. There is no difference in actin expression between the treated and untreated groups in any cell line. This result suggests that POTE expression is regulated by the methylation status of the DNA. However, more direct methods, such as bisulfite sequencing or pyrosequencing of individual CpG dinucleotides in the POTE promoter will be required to confirm that a hypomethylation state is the primary mechanism by which POTE expression is regulated.
FIG. 4.
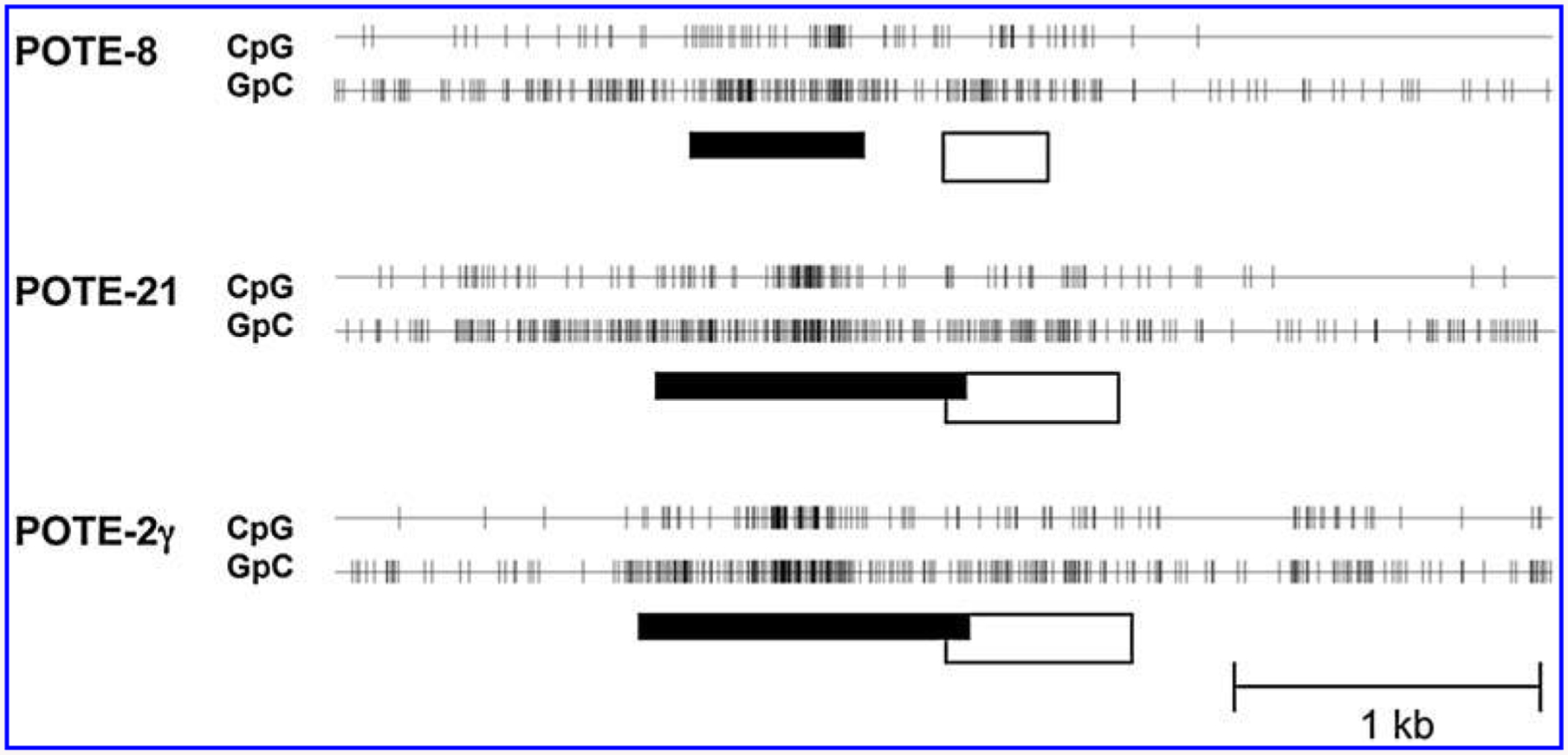
CpG islands of representative POTE genes. The CpG Island Searcher program predicted CpG islands ranging from 556 to 1,218 bp in all of the POTE gene loci. The locations of CpG or GpC dinucleotides in representative POTE paralog member from each group are shown. (White box) Exon 1 of each POTE gene; (vertical lines) CpG or GpC dinucleotides; (black box) putative CpG island where the frequency of CpG dinucleotide is elevated.
Selective POTE paralogs are expressed in ES cells
Previously, we reported that the relative expression of POTE paralogs in cancer and in normal tissues varied. We found that POTE-2α, POTE-2β, POTE-2γ, and POTE-22 are the major paralogs expressed in cancers, but the expression of POTE paralogs in normal tissues is more diverse [8]. To determine the relative expression of POTE paralogs in different ES cell lines, we cloned the PCR amplified product from each cell line and sequenced randomly selected individual clones. As shown in Table 1, 100% of the 10 clones analyzed from the ES02 cell line are transcribed from POTE-2α. In contrast, all 10 clones analyzed from ES03 are transcribed from POTE-2γ. Similarly, all clones analyzed from cell lines WA01 and UC06 are transcribed from the POTE-2γ paralog. In case of cell line ES04, 90% of the analyzed clones are transcribed from POTE-2α, and only 10% of the clones are from POTE-2β. We found expression of three POTE paralogs in cell line WA09; 53% of the 27 clones analyzed are from POTE-2α, 43% are from POTE-2γ, and only 4% are transcribed from POTE-2β. This result shows that there is very selective POTE paralog expression in ES cells, which is in contrast to the expression profile of POTE paralogs in adult tissues and in different cancer specimens.
TABLE 1.
Relative Expression of POTE Paralogs in ES Cells
| ES cell line | Clones analyzed | POTE-2α (%) | POTE-2β (%) |
POTE-2γ (%) |
|---|---|---|---|---|
| ES02 | 10 | 100 | ||
| ES03 | 10 | 100 | ||
| ES04 | 10 | 90 | 10 | |
| WA01a | 10 | 100 | ||
| WA01b | 9 | 100 | ||
| WA09a | 16 | 88 | 12 | |
| WA09b | 11 | 18 | 9 | 73 |
| UC06 | 30 | 100 |
Expression of POTE paralogs not observed is not listed here.
RNA provided by James Thomson (Wisconsin National Primate Center).
RNA provided by Dan S. Kaufman (University of Minnesota).
DISCUSSION
We reported earlier that the POTE gene is only expressed in a few normal adult tissues, including testis, but has a wider expression in many cancers, indicating POTE is a new member of cancer testis (CT) antigens [8]. CT antigens are usually located on the X chromosome, but some CT antigens have been discovered to be encoded from other chromosomes. POTE paralogs are dispersed on eight different chromosomes, and none of the members are located on the X chromosome. To date, more than 40 CT antigens have been identified and have been reported to be overexpressed in many cancers as well as in human mesenchymal stem cells [9,10]. We show here for the first time that POTE, a new member of the CT antigen family, is expressed in undifferentiated ES cell lines. We have tested six commonly used ES cell lines from different laboratories with different passage numbers. POTE is expressed in all six cell lines tested. Among all POTE paralogs, only POTE-2α and POTE-2γ are predominantly expressed in ES cell lines.
The expression pattern of POTE paralogs in different cell types may be influenced by their chromosomal location and chromosome evolution during human evolution. All POTE genes, except those on chromosome 2, are found within pericentromeric regions [2,5]. The pericentromeric regions generally exhibit low transcriptional activity [11]. During human evolution, two ancestral ape chromosomes were fused into one to generate the present human chromosome 2 [12,13]. One of the two ancestral centromeres was located at 2q21.2, and the POTE gene cluster at 2q21.1 is located in the pericentromeric region of this ancient cen tromere (Fig. 5). The degeneration of this centromere probably allowed the POTE-2 genes to escape, at least partially, from the pericentromeric repression [14]. We do not know why some POTE paralogs escape the pericentromeric repression and are expressed in cancer and germ cells [8], but it has been reported that epigenetic mechanisms control the expression of CT antigens [15] and that DNA methylation is deregulated in cancer cells. The results shown in Fig.2C support the view that promoter demethylation is one mechanism by which POTE expression may be induced in cancer.
FIG. 5.
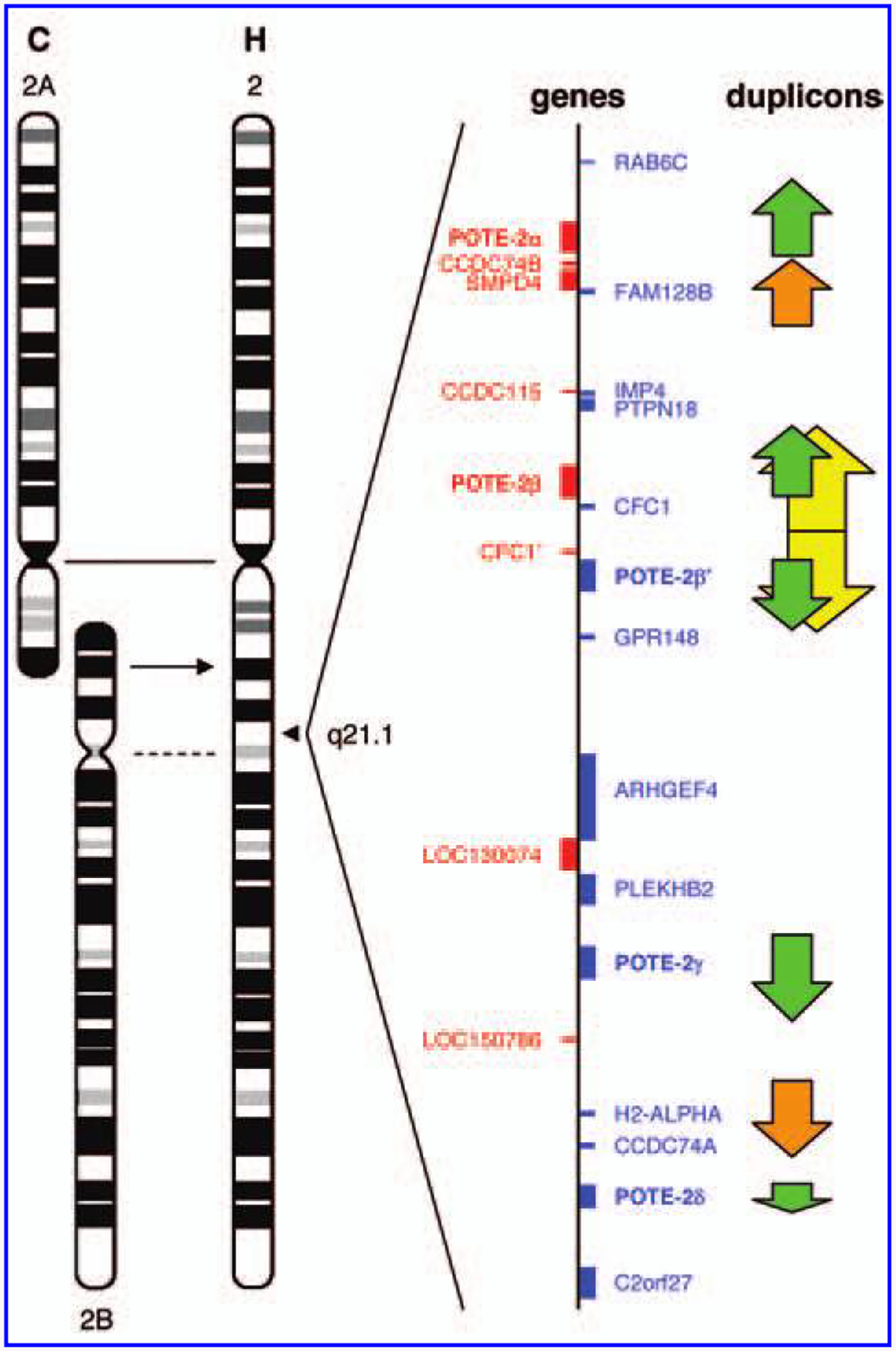
A POTE gene cluster on the human chromosome 2. The human chromosome 2 (H) is a fusion of two ancestral chromosomes equivalent to the chimpanzee (C) chromosomes 2A and 2B. The centromere of chromosome 2A is retained (solid line) in the human chromosome 2 whereas that of chromosome 2B has been degenerated (dotted line). A POTE gene cluster is located in an ancestral pericentromeric region (2q21.1), which contains a total of five POTE genes. Genes in the POTE cluster are shown in blue (transcribed downward) and in red (transcribed upward). A dot-plot analysis reveals duplicons (arrows at the right) in this region. Homologous duplicons are depicted using the same color. The duplicons in green contain POTE genes. The palindromic region in yellow has been recently generated by a tandem duplication event.
Interestingly, there is selective expression of either POTE-2α or POTE-2γ in a given ES cell line. Both ES02 and ES04 cell lines express mostly POTE-2α, whereas in ES03, WA01, and UC06 cell lines POTE-2γ is the only paralog that is expressed. The only cell line that expresses both POTE-2α and POTE-2γ to a similar extent is WA09. The relevance and the mechanism behind this selective POTE paralog expression is not clear. One possibility is that the POTE-2α and POTE-2γ genes are regulated selectively by factors that determine the pluropotency of ES cells and that the level of expression of these paralogs depends on the differentiation state of the cell line. Each of the POTE genes, including those on chromosome 2, is embedded in larger duplicons (Fig. 5). Therefore, POTE-2α, POTE-2β, and POTE-2γ genes, which encode similar proteins, probably share similar regulatory elements. Previously we showed that the POTE genes and POTE proteins have been evolving rapidly [2]. Out of 1,075 amino acids in the coding region, POTE-2α and POTE-2β show a 35-residue difference; POTE-2α and POTE-2γ show a 15-residue difference; and between POTE-2β and POTE-2γ there is a 32-residue difference (Fig. 6). Protein divergence and selective adaptation may drive changes in the regulatory element of the corresponding gene. Various transacting molecules may intensify the subtle differences in the regulatory elements of the genes exposed to diverse cellular microenvironments [16,17].
FIG. 6.
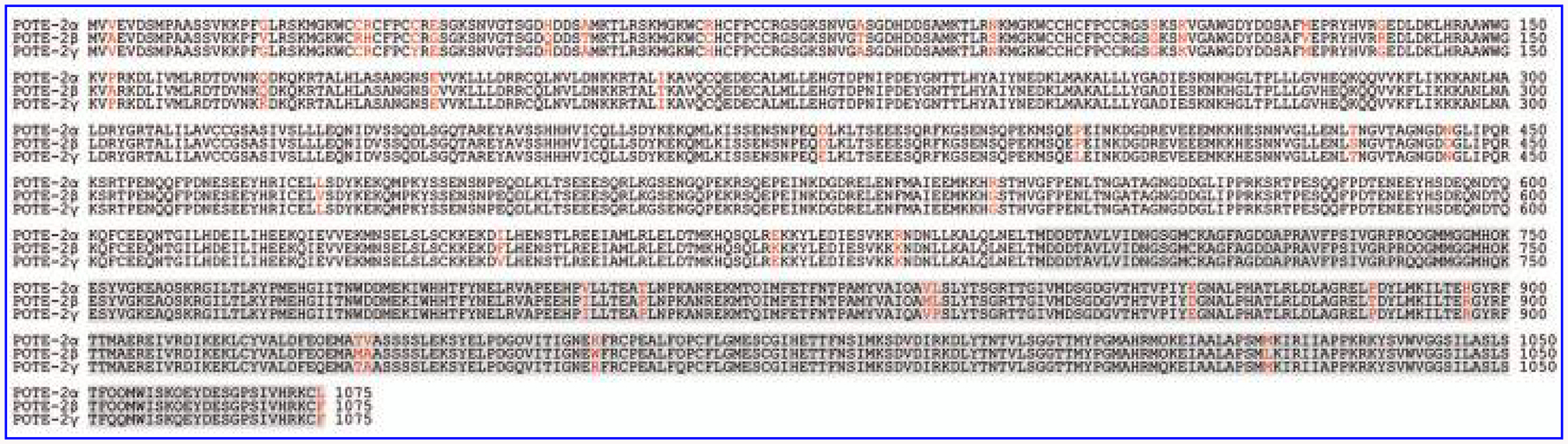
Amino acid sequence alignment of POTE-2α, POTE-2β, and POTE-2γ. The protein sequences were deduced from composite cDNA sequences derived from the human genome sequence. The positions that are not conserved in the three proteins are highlighted in red. Residues in gray indicate β-actin module.
Another interesting finding is that POTE paralogs in adult tissue have restricted expression, but the relative expression of different paralogs in those tissues is somewhat diverse. On the other hand, the POTE paralogs that are expressed in many cancers, and the relative expression of POTE paralogs in cancers is restricted to POTE-2α and POTE-2γ, which is very similar to the expression profile in ES cells. Studies from several groups suggest the existence of a cancer stem cell, which may drive the growth and metastasis of many types of cancers [18]. It will be interesting to investigate if POTE is expressed in those cells and whether it could be a potential marker for “cancer stem cells.”
ACKNOWLEDGMENTS
We thank Dr. B.J. Trask (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing ideograms and Dr. James Thomson (Wisconsin National Primate Center) for providing RNA samples from WA01 and WA09 cell lines; Dawn Walker for reviewing the manuscript; and Anna Mazzuca for editorial assistance. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
REFERENCES
- 1.Vasmatzis G, Essand M, Brinkmann U, Lee B and Pastan I I. (1998). Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc Natl Acad Sci USA 95:300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bera TK, Zimonjic DB, Popescu NC, Sathyanarayana BK, Kumar V, Lee B and Pastan I. (2002). POTE, a highly homologous gene family located on numerous chromosomes and expressed in prostate, ovary, testis, placenta, and prostate cancer. Proc Natl Acad Sci USA 99:16975–16980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bera TK, Egland KA, Lee BK and Pastan I. (2005). Identification of novel cancer target antigens utilizing EST and genome sequence databases In: Cancer Drug and Discovery and Development: The Oncogenomics Handbook. LaRochelle WJ and Shimkets RA, eds. Humana Press, New Jersey, pp 31–42. [Google Scholar]
- 4.Bera TK, Huynh N, Maeda H, Sathyanarayana BK, Lee B and Pastan I. (2004). Five POTE paralogs and their splice variants are expressed in human prostate and encode proteins of different lengths. Gene 337:45–53. [DOI] [PubMed] [Google Scholar]
- 5.Hahn Y, Bera TK, Pastan IH and Lee B. (2006). Duplication and extensive remodeling shaped POTE family genes encoding proteins containing ankyrin repeat and coiled coil domains. Gene 366:238–245. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Ise T, Ha D, Saint Fluer A, Hahn Y, Liu XF, Nagata S, Lee B, Bera TK and Pastan I. (2006). Evolution and expression of chimeric POTE-actin genes in the human genome. Proc Natl Acad Sci USA 103:17885–17890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Shin S, Zeng X, Zhan M, Gonzalez R, Mueller FJ, Schwartz CM, Xue HP, Li H, Baker SC, Chudin W, Barker DL, McDaniel TK, Oeser S, Loring JF, Mattson MP and Rao MS. (2006). Genome wide profiling of human embryonic stem cells (hESCs), their derivatives and embryonal carcinoma cells to develop base profiles of U.S. Federal government approved hESC lines. BMC Dev Biol 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bera TK, Saint-Fleur A, Lee Y, Kydd A, Hahn Y, Popescu NC, Zimonjic DB, Lee B and Pastan I. (2006). POTE paralogs are induced and differentially expressed in many cancers. Cancer Res 66:52–56. [DOI] [PubMed] [Google Scholar]
- 9.Costa FF, Le Blanc K and Brodlin B. (2006). Concise Review: cancer/testis antigens, stem cells, and cancer. Stem Cells 25:707–711. [DOI] [PubMed] [Google Scholar]
- 10.Simpson AJG, Caballero OL, Jungbluth A, Chen YT and Old LJ. (2005). Cancer/testis antigens, gametogenesis and cancer. Nature Rev Cancer 5:615–625. [DOI] [PubMed] [Google Scholar]
- 11.She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C, Bailey JA, Sahinalp C, Rocchi M, Haussler D, Wilson RK, Miller W, Schwartz S and Eichler EE. (2004). The structure and evolution of centromeric transition regions within the human genome. Nature 430:857–864. [DOI] [PubMed] [Google Scholar]
- 12.Yunis JJ and Prakash O. (1982). The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530. [DOI] [PubMed] [Google Scholar]
- 13.Fan Y, Linardopoulou E, Friedman C, Williams E and Trask BJ. (2002). Genomic structure and evolution of the ancestral chromosome fusion site in 2q13–2q14.1 and paralogous regions on other human chromosomes. Genome Res 12:1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson M. (2003). Duplicate, decouple, disperse: the evolutionary transience of human centromeric regions. Curr Opin Genet Dev 13:629–635. [DOI] [PubMed] [Google Scholar]
- 15.Kimmins S and Sassone-Corsi. (2005). Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix MJC, Seftor EA, Seftor REB, Kasemeier-Kulesa J, Kulesa PM and Postovit LM. (2007). Reprogramming metastatic tumour cells with embryonic microenvironments. Nature Rev Cancer 7:246–255. [DOI] [PubMed] [Google Scholar]
- 17.Postovit LM, Seftor EA, Seftor REB and Hendrix MJC. (2005). A three-dimensional model to study the epigenetic effects induced by the microenvironment of human em bryonic stem cells. Stem Cells 24:501–505. [DOI] [PubMed] [Google Scholar]
- 18.Clarke MF and Fuller M. (2006). Stem cells and cancer: Two faces of eve. Cell 124:1111–1115. [DOI] [PubMed] [Google Scholar]


