Abstract
The inhibition of soluble epoxide hydrolase (sEH) is a promising therapeutic approach to treat inflammation and other disorders. In our present investigation on searching for sEH inhibitors from traditional Chinese medicines, we found that Alisma orientale displayed inhibition of sEH. We constructed a small library of protostane-type triterpenoids (1–25) isolated from A. orientale, and screened their inhibitory activities. Alismanin B (1), 11-deoxy-25-anhydro alisol E (4), 11-deoxy alisol B (5), and 25-O-ethyl alisol A (15) displayed concentration-dependently inhibitory activities against sEH with IC50 values from 3.40 ± 0.57 μM to 9.57 ± 0.88 μM. 11-Deoxy-25-anhydro alisol E (4) and 11-deoxy alisol B (5) were defined as mixed-type competitive inhibitors with Ki values of 12.6 and 3.48 μM, respectively, based on the result of inhibition kinetics. The potential interaction mechanism of 11-deoxy alisol B (5) with sEH was analyzed by molecular docking and molecular dynamics, revealing that amino acid residues Trp336 and Tyr466 were vital for its inhibitory activity.
Keywords: Soluble epoxide hydrolase, Protostane-type triterpenoids, Inhibition kinetics, Molecular dynamics stimulation
1. Introduction
Soluble epoxide hydrolase (sEH, E.C. 3.3.2.10) is a bifunctional enzyme of the α/β hydrolase family. It possesses a C-terminal hydrolase activity and an N-terminal phosphatase activity [1,2]. The C-terminal epoxide hydrolase can hydrolyze a variety of epoxides [3–5], whereas the N-terminal lipid phosphatase acts on 12-phosphonooxy-octadec-9Z-enoic acid, erythro-9,10-phosphonooxy-hydroxy-octadecanoic acid, threo-9,10-phosphonooxy-hydroxy-octadecanoic acid, and 12-phosphonooxy-octadec-9E-enoic acid [3–5]. The C-terminal function of sEH has received the most attention since it can hydrolyze epoxides of arachidonic acid (AA) epoxy eicosatrienoic acids (EETs) via opening of the epoxide into a vicinal diol termed dihydroxy eicosatrienoic acids (DHETs) [6,7]. The inhibition of sEH is a method of elevating the concentration of EETs, EETs and other epoxy fatty acids (EpFA) have many effects including glycogen synthase kinase-3 beta (GSK3β) and nuclear factor kappa B (NF-κB) inhibition. Inhibition of sEH exerts anti-inflammatory effects in Parkinson’s disease and cardiovascular diseases [6,8–14]. Therefore, sEH is considered as a promising therapeutic approach for resolution of inflammation [15–17].
Recently, a collection of sEH inhibitors with varied skeleton, including ureas, amides, triterpenoids, and flavones, have come to light [18–23], which stimulated medicinal and natural product chemists to develop and discover more efficacious sEH inhibitors with excellent specificity and high safety profile [24–26]. To date, the most powerful of sEH inhibitors are ureas, such as N-[1-(1-oxopropyl)-4-piperidinyl]-N’-[4-(trifluoromethoxy)phenyl]urea (TPPU) and 12-[[(tricyclo [3.3.1.13,7]dec-1-ylamino)carbonyl]amino]-dodecanoic acid (AUDA), that can competitively bind to the epoxide binding in the catalytic site – Asp335, Tyr466, and Tyr383 [27–31]. Traditional Chinese medicines are the important natural resources for medicine [32–35], which provides possibility of searching for the new generation of sEH inhibitors. This also raises the possibility of finding valuable natural supplements, meanwhile, Alisma orientale displayed the potential of inhibition on sEH, therefore, we collected twenty-five protostane-type triterpenoids (1–25, Fig. 1) from A. orientale and screened their inhibitory activities. 11-Deoxy alisol B (5) showed potent inhibitory activity against sEH, which stimulated us to further investigate its inhibition behavior and mechanism of action by molecular docking and molecular dynamics.
Fig. 1.

The structures of twenty-five protostane-type triterpenoids.
2. Results and discussion
2.1. sEH inhibitory effects
The mammalian microsomal has been well studied in xenobiotic metabolism. The sEH can degrade some xenobiotics or foreign compounds but it is best known for degrading biologically active, 1,2-disubstituted epoxides of fatty acids [19,23,28]. Some of potential sEH inhibitors have been isolated from traditional Chinese medicines or synthesized by chemists [15,19–23], including capsaicin, dihydrocapsaicin, piperlonguminine, dihydropiperlonguminine, futoamide, and urea analogues, stimulating us to screen the sEH inhibitors from natural products. We screened the inhibitory activities of twenty-five protostane-type triterpenoids (1–25) against sEH based on the method of sEH-mediated fluorogenic substrate PHOME hydrolysis [16,23]. As shown in the heatmap Fig. 2A and Table 1, compounds 1, 3–6, and 15 possessed potential inhibition on sEH activity (> 70%) at 50 μM, especially compound 5 (> 95%). Compounds 11, 13, 16, 18, 19, 24, and 25 could inhibit more than 50% sEH activity at the final concentration of 50 μM. In order to quantify the inhibitory potentials of triterpenoids on sEH, the relationship between inhibitor concentrations and inhibitory potential were plotted. Compounds 1, 4, 5, and 15 could dose-dependently inhibit sEH activity, and their IC50 values were 7.15 ± 0.59, 5.94 ± 0.54, 3.40 ± 0.57, and 9.57 ± 0.88 μM, respectively (Fig. 2), whereas compounds 3, 6, 11, 13, 16, 18, 19, 24, and 25 displayed moderate inhibitory effects with IC50 values ranging from 14.3 ± 1.1 μM to 36.3 ± 4.0 μM.
Fig. 2.

(A) Compounds 1–25 (A1-A5, 1–5; B1-B5, 6–10; C1-C5, 11–15; D1-D5, 16–20; E1-E5, 21–25) displayed inhibitory activities against sEH at 50 μM; (B-E) Compounds 1, 4, 5, and 15 dose-dependently inhibited sEH activities in vitro.
Table 1.
Inhibitory activities of compounds 1–25 against sEH.
| Compounds | Inhibition of compounds against sEH | |
|---|---|---|
| 50 μM (%) | IC50 (μM) | |
| 1 | 75.31 ± 2.42 | 7.15 ± 1.09 |
| 2 | 27.33 ± 2.81 | >50 |
| 3 | 78.54 ± 1.86 | 14.8 ± 1.1 |
| 4 | 71.21 ± 1.56 | 5.94 ± 0.54 |
| 5 | 95.23 ± 1.24 | 3.40 ± 0.57 |
| 6 | 71.6 ± 2.12 | 14.6 ± 1.1 |
| 7 | 24.45 ± 1.03 | >50 |
| 8 | 44.42 ± 1.43 | >50 |
| 9 | 37.63 ± 0.86 | >50 |
| 10 | 18.64 ± 0.45 | >50 |
| 11 | 65.62 ± 1.65 | 36.3 ± 4.0 |
| 12 | 44.97 ± 0.95 | >50 |
| 13 | 66.84 ± 2.09 | 22.1 ± 1.4 |
| 14 | 18.30 ± 0.35 | >50 |
| 15 | 78.66 ± 2.19 | 9.57 ± 0.88 |
| 16 | 58.56 ± 1.31 | 21.2 ± 0.5 |
| 17 | 49.44 ± 1.15 | >50 |
| 18 | 56.93 ± 1.26 | 31.5 ± 1.4 |
| 19 | 58.34 ± 1.31 | 22.2 ± 1.0 |
| 20 | 17.63 ± 0.56 | >50 |
| 21 | 39.33 ± 0.92 | >50 |
| 22 | 17.96 ± 0.69 | >50 |
| 23 | 16.27 ± 0.72 | >50 |
| 24 | 59.45 ± 1.53 | 14.6 ± 1.6 |
| 25 | 56.98 ± 1.86 | 21.4 ± 1.7 |
| TPPUa | -b | 53 nM |
Positive control.
No tested.
The structures of triterpenoids 1–25 and IC50 values were used to analyze their structure-activity relationships. Comparison of IC50 values of compounds 2 with 15 and 12 with 18 revealed that the alkylation of C-25 hydroxy group was in favor of inhibitory activity. The IC50 value of compound 14 was more than 50 μM, whereas compound 13 displayed significant inhibitory activity with an IC50 value of 22.1 μM, which indicated that the C-11 hydroxy substitution negatively affected its inhibitory activity. An analogous case was also observed for compounds 5, 9, and 19, compound 5 showed significant inhibitory activity (IC50 = 3.40 μM), whereas compounds 9 and 19 did not displayed significantly inhibitory activities. The dehydration of OH-25 in compound 14 led to the produce of compound 16, and the latter displayed more inhibitory activity than the former, revealing that the Δ25(26) olefinic bond was essential to inhibitory activity. The similar result was also observed for compounds 2 (IC50 > 50 μM) and 4 (IC50 = 5.94 μM). Compared with inhibitory efficiency of compounds 2 with 8 and compounds 13 with 22 suggested that the hydrolyzation of C-7 and C-12 went against inhibitory activity. Additionally, the Δ25(26) olefinic bond and 16-oxo group operated to the disadvantage of inhibitory activity via an analysis of inhibitory activities of compounds 4 and 12.
2.2. Inhibition mechanism of compounds 4 and 5
The potent inhibitory activity of compounds 4 and 5 stimulated us to study their inhibition behaviors. Inhibition kinetics on compounds 4 and 5 were performed to assign the inhibition type which was competitive, mixed competitive, noncompetitive, or uncompetitive type on the basis of Michealis-Menten and Lineweaver-Burk plots (Fig. 3). Lineweaver-Burk plots clearly demonstrated that compounds 4 and 5 functioned as mixed-type competitive inhibitors against sEH in Fig. 3C and D showing that a series of lines were intersected at the second quadrant, and the inhibition constants (Ki) were defined as 12.6 and 3.48 μM (Fig. 3E and F and Table 2), respectively.
Fig. 3.
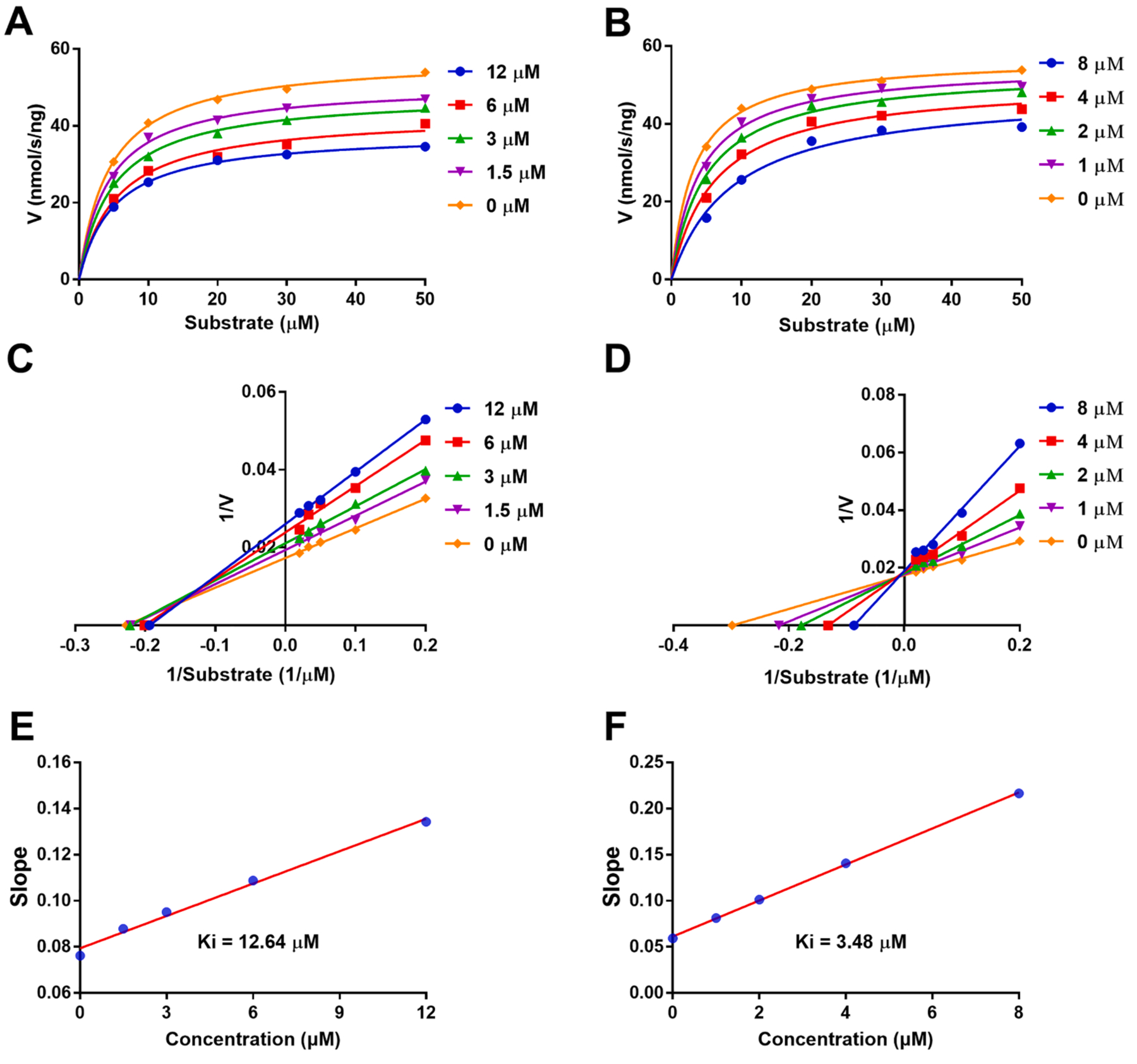
(A-B) Michealis-Menten plots of compounds 4 and 5 against sEH. (C-D) Lineweaver-Burk plots of compounds 4 and 5 against sEH. (E-F) Slope plots of compounds 4 and 5 against sEH.
Table 2.
Kinetic parameters of compounds 4 and 5 against sEH.
| Compounds | Inhibition type | Ki (μM) | Goodness of fit (r) |
|---|---|---|---|
| 4 | mixed-type | 12.6 | 0.9988 |
| 5 | mixed-type | 3.48 | 0.9974 |
2.3. Interaction analysis
The binding sites of compounds 4 and 5 with sEH were predicted by molecular docking on the basis of their inhibition kinetic results. Compounds 4 and 5 could bond to the catalytic cavity of sEH, and their lowest binding energies were −36.45 and −40.52 kcal/mol (Fig. 4 and Table 3), respectively. Compound 4 interacted with amino acid residues Asp335, Tyr383, and Tyr466 with the distances of 2.80, 2.59, and 2.68 Å through three hydrogen bonds, respectively. Compound 5 possessed two hydrogen bonds between the hydroxy group at C-23 and amino acid residues Trp336 and Tyr466 at the distances of 3.01 and 2.95 Å, respectively. Furthermore, compounds 4 and 5 also had hydrophobic interactions with other surrounding amino acid residues (Asp335, Met339, Thr343, Thr360, Ile363, Ala365, Pro371, Ile375, Phe381, Thr383, Gln384, Met469, Trp473, Val498, Leu499, and His524) of the catalytic pocket [21,23,26,27]. According to the above-mentioned results, the hydroxy group of C-23 and amino acid residue Tyr466 were critical for inhibitory activities of protostane-type triterpenoids against sEH.
Fig. 4.

(A-B) Compounds 4 (A) and 5 (B) could be bonded into the catalytic cavity of sEH. (C-D) 2D interactions of compounds 4 (C) and 5 (D) with sEH. The green dotted lines expressed hydrogen bonds.
Table 3.
Interaction information of compounds 4 and 5 with sEH.
| Compounds | Interaction amino acids | Hydrogen bonds | Lowest binding energy (kcal/mol) |
|---|---|---|---|
| 4 | Asp335, Trp336, Met339, Thr360, Ile363, Pro371, Ile375, Phe381,Tyr383, Gln384, Tyr466, Val498, Leu499, His524 | Asp335, Tyr383, Tyr466 | −36.45 |
| 5 | Asp335, Trp336, Met339, Tyr343, Thr360, Ala365, Ile363, Pro371, Tyr383, Gln384, Met469, Tyr466, Trp473 | Trp336, Tyr466 | −40.52 |
2.4. Molecular dynamics
Because compound 5 showed potent inhibitory activity against sEH with Ki value of 3.48 μM and interacted with key amino acid residue Trp366 and Tyr466, therefore, we used Gromacs to investigate the molecular dynamics (MD) of compound 5 with sEH. The potential energy of the complex of sEH with compound 5 was held at about −2.16 × 106 kJ/mol during the simulation (Fig. 5). RMSD, a parameter to access the stability of the protein-ligand system [19,20,22], was also calculated, and the backbone RMSD of compound 5 was represented at a distance of about 3.5 Å during 10 ns of MD simulation (Fig. 5). RMSF of residues was acquired to figure out regions exhibiting higher flexibility with a distance of approximately 2.0 Å in MD simulation of compound 5 (Fig. 5). The number of hydrogen bonds and their distances throughout the MD simulation trajectory were determined using the gmx hbond and gmx distance tools in Gromacs. As shown in Fig. 5, compound 5 interacted with sEH via 1–3 hydrogen bonds during 10 ns of MD stimulation. The docking result indicated that amino acid residues Trp336 and Tyr466 could form hydrogen bonds with compound 5, therefore, we analyzed the level of the interactions of compound 5 with the above-mentioned residues. In MD simulation of compound 5, amino acid residue Trp336 was held at a distance of about 3.0 Å with the C-23 hydroxy group, amino acid residue Tyr466 was held at a distance of about 3.25 Å with the C-23 hydroxy group, which suggested that amino acid residues Trp336 and Tyr466 were vital for the inhibition of compound 5 against sEH. The above-mentioned result further supported the molecular docking result of compound 5. This finding revealed that compound 5 could be regarded as a potential sEH inhibitor.
Fig. 5.

The RMSD (A), RMSF (B), potential energy (C), and hydrogen bond number (D) of compound 5 with sEH. The distance of compound 5 with amino acid residues Trp336 (E) and Tyr466 (F).
3. Conclusions
In summary, we screened the inhibitory potentials of twenty-five protostane-type triterpenoids against sEH, and compounds 1, 4, 5, and 15 showed inhibition on sEH activity with IC50 values from 3.40 ± 0.57 μM to 9.57 ± 0.88 μM. Compounds 4 and 5 were assigned as mixed-type competitive inhibitors with Ki values of 12.6 and 3.48 μM, respectively. Molecular docking and molecular dynamics were used to analyze the potential interaction mechanism of compound 5 with sEH, suggesting that amino acid residues Trp336 and Tyr466 were vital for the inhibitory activity of compound 5.
4. Material and methods
4.1. General experimental procedures
sEH protein was provided by the laboratory of Bruce D. Hammock in Department of Entomology and Nematology, UC Davis Comprehensive Cancer Center, University of California. Alismanin B (1), alisol A (2), alismanin D (3), 11-deoxy-25-anhydro alisol E (4), 11-deoxy alisol B (5), alismanin F (6), alismanol O (7), 7α,12α-dihydroxyalisol A (8), alisol B 23-acetate (9), 7α-hydroxy-24-oxo alisol A (10), 16-oxo-11-anhydro alisol A 23,24-acetonide (11), alismanol B (12), 11-deoxy alisol F (13), alisol F (14), 25-O-ethyl alisol A (15), 25-anhydro alisol F (16), alismanin E (17), 25-ethoxy-16-oxo-11-anhydro alisol A (18), 20-hydroxy alisol C (19), 25-methoxy alisol F 23-acetate (20), 11-deoxy-12β-hydroxy alisol F 24-acetate (21), 11-deoxy-12β-hydroxy alisol F (22), 1-deoxy-12β-hydroxy-16β-methoxy alisol A 24-acetate (23), alisol G (23, 24)-diol acetonide (24), and alisol A 24-acetate (25) were isolated from A. orientale by the authors [35], their structures were identified by NMR and MS spectra, and their purities were more than 98% analyzed by HPLC.
4.2. sEH inhibitory activity
The inhibitory activities of compounds 1–25 against sEH were screened according to sEH-meditated probe substrate PHOME (Cayman Chemical, Ann Arbor, MI) hydrolysis [16,18]. Compounds 1–25 were added into the standard incubation system (200 μL), including sEH (0.1 ng/mL), BisTris-HCl buffer (pH 7.4, 25 mM), and probe PHOME (10 μM). The reaction was performed for 20 min at 37 °C, then the fluorescence signal at 465 nm was recorded. TPPU (Cayman Chemical, Ann Arbor, MI) was used as a positive control [23].
4.3. Inhibitory kinetic analysis
The inhibition kinetics of compounds 4 and 5 were performed by using Michealis-Menten analyses [23]. Their inhibition types (competitive inhibition, noncompetitive inhibition, uncompetitive inhibition, or mixed-type inhibition) were identified according to Lineweaver-Burk plots, and their inhibition constants (Ki) were calculated by slope plots and four inhibition kinetics models [Eq. (1): Competitive inhibition; Eq. (2): Noncompetitive inhibition; Eq. (3): Uncompetitive inhibition; Eq. (4): Mixed-type inhibition].
| (1) |
| (2) |
| (3) |
| (4) |
V, reaction rate; Vmax, maximum rate; Km, Michaelis constant, Ki, inhibition constant; S, the concentrations of substrate; I, the concentrations of inhibitor.
4.4. Molecular docking
The interactions of compounds 4 and 5 with sEH (PDB ID: 4OCZ) were analyzed by molecular docking according to the previous method [23]. The 3D structure of compounds 4 and 5 were optimized at CHARMm force field by Discovery Studio software (BIOVIA Inc., San Diego, USA). The interactions of compounds 4 and 5 with sEH were analyzed at CHARMm force field using the CDOCKER protocol of Discovery Studio [35,36].
4.5. Molecular dynamics
Molecular dynamics (MD) simulation of compound 5 with sEH was calculated by GROMACS using the AMBER99SB force field [19,20,22]. The restrained electrostatic potential charges of ligand were calculated at B3LYP/6–31+G (d, p) level by the Gaussian 09 program. Then, the starting topology of the ligand was generated by antechamber and ACPYPE programs. The complex of compound 5 with sEH was placed centrally in a rectangular box with a size of 1.2 nm. After the assembly, the steepest descent energy minimization was applied to relax the system. Then, NVT and NPT were used to equilibrate the system at the constant temperature of 300 K and the constant pressure of 1 bar, respectively. MD interaction of compound 5 and sEH was calculated at 300 K and 1 bar pressure for 10 ns after equilibration. The trajectory file was analyzed by GROMACS to obtain the root mean square deviation (RMSD), root mean square fluctuations (RMSF). The total number of intermolecular hydrogen bonds (HB) formed between compound 5 and sEH and HB distribution for the system.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81703769), Revolutionizing Innovative, Visionary Environmental Health Research Program of the National Institutes of Environmental Health Sciences (R35 ES030443-01), Superfund Basic Research Program of the National Institutes of Environmental Health Sciences (P42 ES04699), and Dalian Young Star of Science and Technology.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Beetham JK, Tian T, Hammock BD, cDNA cloning and expression of a soluble epoxide hydrolase from human liver, Arch. Biochem. Biophys 305 (1) (1993) 197–201. [DOI] [PubMed] [Google Scholar]
- [2].Grant DF, Storms DH, Hammock BD, Molecular cloning and expression of murine liver soluble epoxide hydrolase, J. Biol. Chem 268 (23) (1993) 17628–17633. [PubMed] [Google Scholar]
- [3].Decker M, Adamska M, Cronin A, Di Giallonardo F, Burgener J, Marowsky A, Falck JR, Morisseau C, Hammock BD, Gruzdev A, Zeldin DC, Arand M, EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides, J. Lipid Res 53 (10) (2012) 2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Newman JW, Morisseau C, Harris TR, Hammock BD, The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity, P. Natl. Acad. Sci. USA 100 (4) (2003) 1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Przybyla-Zawislak BD, Srivastava PK, Vazquez-Matias J, Mohrenweiser HW, Maxwell JE, Hammock BD, Bradbury JA, Enayetallah AE, Zeldin DC, Grant DF, Polymorphisms in human soluble epoxide hydrolase, Heterocycles 64 (2) (2003) 482–490. [DOI] [PubMed] [Google Scholar]
- [6].Wagner KM, McReynolds CB, Schmidt WK, Hammock BD, Soluble epoxide hydrolase as a therapeutic target for pain, inflammatory and neurodegenerative diseases, Pharmacol. Ther 180 (2017) 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ren Q, Ma M, Yang J, Nonaka R, Yamaguchi A, Ishikawa K, Kobayashi K, Murayama S, Hwang SH, Saiki S, Akamatsu W, Hattori N, Hammock BD, Hashimoto K, Soluble epoxide hydrolase plays a key role in the pathogenesis of Parkinson’s disease, P. Natl. Acad. Sci. USA 115 (25) (2018) E5815–E5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhong LJ, Zhou JT, Wang DW, Zou XJ, Lou YX, Liu D, Yang B, Zhu Y, Li XX, Proteomics and bioinformatics analysis of mouse hypothalamic neurogenesis with or without EPHX2 gene deletion, Int. J. Clin. Exp. Pathol 8 (10) (2015) 12634–12645. [PMC free article] [PubMed] [Google Scholar]
- [9].Schunck WH, Konkel A, Fischer R, Weylandt KH, Therapeutic potential of omega-3 fatty acid-derived epoxyeicosanoids in cardiovascular and inflammatory diseases, Pharmacol. Ther 183 (2018) 177–204. [DOI] [PubMed] [Google Scholar]
- [10].Ostermann AI, Reutzel M, Hartung N, Franke N, Kutzner L, Schoenfeld K, Weylandt KH, Eckert GP, Schebb NH, A diet rich in omega-3 fatty acids enhances expression of soluble epoxide hydrolase in murine brain, Prostaglandins Other Lipid Mediat. 133 (2017) 79–87. [DOI] [PubMed] [Google Scholar]
- [11].Deng BQ, Luo Y, Kang X, Li CB, Morisseau C, Yang J, Lee KSS, Huang J, Hu DY, Wu MY, Peng A, Hammock BD, Liu JY, Epoxide metabolites of arachidonate and docosahexaenoate function conversely in acute kidney injury involved in GSK3 beta signaling, P. Natl. Acad. Sci. USA 114 (47) (2017) 12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ren Q, Soluble epoxide hydrolase inhibitor: a novel potential therapeutic or prophylactic drug for psychiatric disorders, Front. Pharmacol 10 (2019) 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang WC, Yang J, Zhang JA, Wang YX, Hwang SH, Qi WP, Wan DB, Kim D, Sun J, Sanidad KZ, Yang HX, Park Y, Liu JY, Zhao XF, Zheng XH, Liu ZH, Hammock BD, Zhang GD, Lipidomic profiling reveals soluble epoxide hydrolase as a therapeutic target of obesity-induced colonic inflammation, P. Natl. Acad. Sci. USA 115 (20) (2018) 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xu DY, Davis BB, Wang ZH, Zhao SP, Wasti B, Liu ZL, Li N, Morisseau C, Chiamvimonvat N, Hammock BD, A potent soluble epoxide hydrolase inhibitor, t-AUCB, acts through PPAR gamma to modulate the function of endothelial progenitor cells from patients with acute myocardial infarction, Int. J. Cardiol 167 (4) (2013) 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Eldrup AB, Soleymanzadeh F, Farrow NA, Kukulka A, De Lombaert S, Optimization of piperidyl-ureas as inhibitors of soluble epoxide hydrolase, Bioorg. Med. Chem. Lett 20 (2) (2010) 571–575. [DOI] [PubMed] [Google Scholar]
- [16].Wolf NM, Morisseau C, Jones PD, Hock B, Hammock BD, Development of a high-throughput screen for soluble epoxide hydrolase inhibition, Anal. Biochem 355 (1) (2006) 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xie WS, Tang XY, Lu Q, Ames RS, Ratcliffe SJ, Li H, Development of a high throughput cell-based assay for soluble epoxide hydrolase using BacMam technology, Mol. Biotechnol 45 (3) (2010) 207–217. [DOI] [PubMed] [Google Scholar]
- [18].Burmistrov V, Morisseau C, Harris TR, Butov G, Hammock BD, Effects of adamantane alterations on soluble epoxide hydrolase inhibition potency, physical properties and metabolic stability, Bioorg. Chem 76 (2018) 510–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kim JH, Cho IS, Ryu J, Lee JS, Kang JS, Kang SY, Kim YH, In vitro and in silico investigation of anthocyanin derivatives as soluble epoxide hydrolase inhibitors, Int. J. Biol. Macromol 112 (2018) 961–967. [DOI] [PubMed] [Google Scholar]
- [20].Kim JH, Jo YD, Jin CH, Isolation of soluble epoxide hydrolase inhibitor of capsaicin analogs from Capsicum chinense Jacq. cv. Habanero, Int. J. Biol. Macromol 135 (2019) 1202–1207. [DOI] [PubMed] [Google Scholar]
- [21].Lukin A, Kramer J, Hartmann M, Weizel L, Hernandez-Olmos V, Falahati K, Burghardt I, Kalinchenkova N, Bagnyukova D, Zhurilo N, Rautio J, Forsberg M, Ihalainen J, Auriola S, Leppanen J, Konstantinov I, Pogoryelov D, Proschak E, Dar’in D, Krasavin M, Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase, Bioorg. Chem 80 (2018) 655–667. [DOI] [PubMed] [Google Scholar]
- [22].Thao NP, Kim JH, Luyen BTT, Dat NT, Kim YH, In silico investigation of cycloartane triterpene derivatives from Cimicifuga dahurica (Turcz.) Maxim. roots for the development of potent soluble epoxide hydrolase inhibitors, Int. J. Biol. Macromol 98 (2017) 526–534. [DOI] [PubMed] [Google Scholar]
- [23].Liu ZB, Sun CP, Xu JX, Morisseau C, Hammock BD, Qiu F, Phytochemical constituents from Scutellaria baicalensis in soluble epoxide hydrolase inhibition: kinetics and interaction mechanism merged with simulations, Int. J. Biol. Macromol 133 (2019) 1187–1193. [DOI] [PubMed] [Google Scholar]
- [24].Kim IH, Heirtzler FR, Morisseau C, Nishi K, Tsai HJ, Hammock BD, Optimization of amide-based inhibitors of soluble epoxide hydrolase with improved water solubility, J. Med. Chem 48 (10) (2005) 3621–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Manickam M, Pillaiyar T, Boggu P, Venkateswararao E, Jalani HB, Kim ND, Lee SK, Jeon JS, Kim SK, Jung SH, Discovery of enantioselectivity of urea inhibitors of soluble epoxide hydrolase, Eur. J. Med. Chem 117 (2016) 113–124. [DOI] [PubMed] [Google Scholar]
- [26].McElroy NR, Jurs PC, Morisseau C, Hammock BD, QSAR and classification of murine and human soluble epoxide hydrolase inhibition by urea-like compounds, J. Med. Chem 46 (6) (2003) 1066–1080. [DOI] [PubMed] [Google Scholar]
- [27].Lee KSS, Liu JY, Wagner KM, Pakhomova S, Dong H, Morisseau C, Fu SH, Yang J, Wang P, Ulu A, Mate CA, Nguyen LV, Hwang SH, Edin ML, Mara AA, Wulff H, Newcomer ME, Zeldin DC, Hammock BD, Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy, J. Med. Chem 57 (16) (2014) 7016–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hiesinger K, Kramer JS, Achenbach J, Moser D, Weber J, Wittmann SK, Morisseau C, Angioni C, Geisslinger G, Kahnt AS, Kaiser A, Proschak A, Steinhilber D, Pogoryelov D, Wagner K, Hammock BD, Proschak E, Computer-aided selective optimization of side activities of talinolol, Acs Med. Chem. Lett 10 (6) (2019) 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Oster L, Tapani S, Xue YF, Kack H, Successful generation of structural information for fragment-based drug discovery, Drug Discov. Today 20 (9) (2015) 1104–1111. [DOI] [PubMed] [Google Scholar]
- [30].Pilger J, Mazur A, Monecke P, Schreuder H, Elshorst B, Bartoschek S, Langer T, Schiffer A, Krimm I, Wegstroth M, Lee D, Hessler G, Wendt KU, Becker S, Griesinger C, A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles, Angew. Chem. Int. Edit 54 (22) (2015) 6511–6515. [DOI] [PubMed] [Google Scholar]
- [31].Takai K, Chiyo N, Nakajima T, Nariai T, Ishikawa C, Nakatani S, Ikeno A, Yamamoto S, Sone T, Three-dimensional rational approach to the discovery of potent substituted cyclopropyl urea soluble epoxide hydrolase inhibitors, Bioorg. Med. Chem. Lett 25 (8) (2015) 1705–1708. [DOI] [PubMed] [Google Scholar]
- [32].Liu TT, Huo XK, Tian XG, Liang JH, Yi J, Zhang XY, Zhang S, Feng L, Ning J, Zhang BJ, Sun CP, Ma XC, Demethylbellidifolin isolated from Swertia bimaculate against human carboxylesterase 2: Kinetics and interaction mechanism merged with docking simulations, Bioorg. Chem 90 (2019) 103101. [DOI] [PubMed] [Google Scholar]
- [33].Wang C, Huo XK, Luan ZL, Cao F, Tian XG, Zhao XY, Sun CP, Feng L, Ning J, Zhang BJ, Ma XC, Alismanin A, a triterpenoid with a C34 skeleton from Alisma orientale as a natural agonist of human pregnane X receptor, Org. Lett 19 (20) (2017) 5645–5648. [DOI] [PubMed] [Google Scholar]
- [34].Yi J, Bai R, An Y, Liu TT, Liang JH, Tian XG, Huo XK, Feng L, Ning J, Sun CP, Ma XC, Zhang HL, A natural inhibitor from Alisma orientale against human carboxylesterase 2: kinetics, circular dichroism spectroscopic analysis, and docking simulation, Int. J. Biol. Macromol 133 (2019) 184–189. [DOI] [PubMed] [Google Scholar]
- [35].Luan ZL, Huo XK, Dong PP, Tian XG, Sun CP, Lv X, Feng L, Ning J, Wang C, Zhang BJ, Ma XC, Highly potent non-steroidal FXR agonists protostane-type triterpenoids: structure-activity relationship and mechanism, Eur. J. Med. Chem 182 (2019) 111652. [DOI] [PubMed] [Google Scholar]
- [36].Liang JH, Luan ZL, Tian XG, Zhao WY, Wang YL, Sun CP, Huo XK, Deng S, Zhang BJ, Zhang ZJ, Ma XC, Uncarialins A-I, monoterpenoid indole alkaloids from Uncaria rhynchophylla as natural agonists of the 5-HT1A receptor, J. Nat. Prod 82 (12) (2019) 3302–3310. [DOI] [PubMed] [Google Scholar]


