SUMMARY
Lung injury activates specialized adult epithelial progenitors to regenerate the epithelium. Depending on the extent of injury, both remaining alveolar type II cells (AEC2s) and distal airway stem/progenitors mobilize to cover denuded alveoli and restore normal barriers. The major source of airway stem/progenitors other than basal-like cells remains uncertain. Here, we define a distinct subpopulation (~5%) of club-like LNEPs marked by high H2-K1 expression critical for alveolar repair. Quiescent H2-K1high cells account for virtually all in vitro regenerative activity of airway lineages. After bleomycin injury H2-K1 cells expand and differentiate in vivo to alveolar lineages. However injured H2-K1 cells eventually develop impaired self-renewal with features of senescence, limiting complete repair. Normal H2-K1high cells transplanted into injured lungs differentiate into alveolar cells and rescue lung function. These findings indicate that small subpopulations of specialized stem/progenitors are required for effective lung regeneration and are a potential therapeutic adjunct after major lung injury.
Keywords: Adult lung epithelial stem/progenitor cells, Single cell transcriptomics, Quiescent progenitors, dedifferentiation, Injury and regeneration, Bleomycin injury, In vivo transplantation
eTOC Blurb
Through single-cell transcriptomic profiling of distal lung airway epithelium Kathiriya et al. reveal rare progenitors that activate and mobilize after injury to help re-establish alveolar lining and barrier function. The findings favor expansion of pre-existing stem cells over widespread de-differentiation of mature cells as a major pathway of lung regeneration.
INTRODUCTION
Injury to adult lung elicits activation of various cell types to repair the epithelium, depending on the type and severity of the injury (Chen, 2017; Hogan et al., 2014). In the lung, almost all cell types can re-enter the cell cycle and display some degree of motility to promote barrier reconstitution. Yet, small populations of stem/progenitor cells with multipotency are known to expand after major injury to either conducting airways or the alveoli. In the injured lung parenchyma, both remaining type II cells (AEC2s) and stem/progenitors emanating from distal airways contribute importantly to alveolar repair (Barkauskas et al., 2013; Kumar et al., 2011; McQualter, 2019; Zacharias et al., 2018; Zheng et al., 2017). Bronchioalveolar Stem Cells (BASCs), rare p63+ cells, and Sca-1+ cells have all been reported as distal airway stem/progenitor cells capable of migration into alveoli (Kim et al., 2005; Liu et al., 2019; McQualter, 2019; Vaughan et al., 2015). In addition, relying on the use of lineage tracing using the club cell marker Secretoglobin 1A1 (Scgb1a1), dedifferentiation or transdifferentiation of club cells as the source of stem/progenitor cell population in the distal lung has been reported (McConnell et al., 2016; McQualter, 2019; Rawlins et al., 2009). Indeed, under some conditions, Scgb1a1-traced cells are observed to dedifferentiate to basal cells that then expand and promote repair (Tata et al., 2013), raising the possibility that dedifferentiation of mature lung cell types is a major pathway of epithelial stem/progenitor cell expansion, analogous to similar paradigms in other species (Brawley and Matunis, 2004; Brockes and Kumar, 2002; Kai and Spradling, 2004; Kragl et al., 2009). However, it has been observed that mature lineage tracers such as Sox2 or Scgb1a1 label a more heterogeneous group of cells than previously appreciated, raising the possibility that small populations of stem/progenitors could be hiding under the tracing umbrella of a mature lineage. Therefore, it remains unclear whether fully mature cells de-differentiate and repopulate the epithelium as a major paradigm or, somewhat like adult muscle (Relaix and Zammit, 2012), there exists a specialized stem/progenitor population in the distal lung airways with preferential ability to expand, migrate, and differentiate towards other mature cell types in the event of major injury.
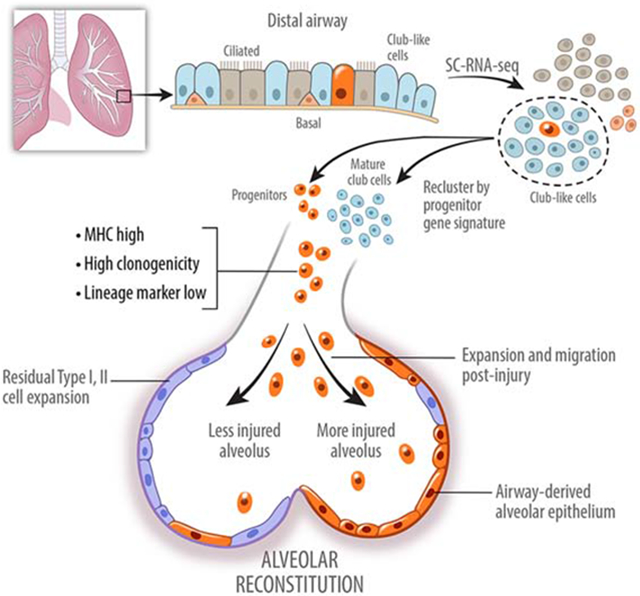
We observe that Scgb1a1-lineage tracing marks at least three types of mature lung epithelial cells in an uninjured adult murine lung as well as unstained cells of uncertain origin or function (Figure 3). Therefore, to understand the source of regenerative activity of an airway epithelium, we performed single cell transcriptomic analysis of club-like cells searching for cells enriched with a progenitor gene signature. We report identification of a specialized progenitor population that is labeled by mature lineages (Sox2 and Scgb1a1) but remain quiescent in a normal and uninjured murine lung. These cells are not fully differentiated and are not bronchioalveolar stem cells (BASCs) by the virtue of their in situ localization and lack of surfactant protein C expression, or p63+ cells previously reported to expand after viral injury (Vaughan et al., 2015; Yang et al., 2018). We demonstrate that these specialized progenitor cells are characterized by high levels of antigen-presenting genes, can be isolated by high expression of MHC class I marker H2-K1, and account for in vitro and in vivo regenerative activity of the lineage labeled airway epithelium and aid in functional recovery of injured mice. Along with the rare p63+ stem/progenitors previously observed mobilizing during influenza infection, these p63neg/H2-K1high cells comprise a family of stem/progenitors previously termed lineage negative epithelial progenitors (LNEPs) that become remarkably mobilized after injury. These findings support the paradigm that pre-existing but quiescent specialized stem/progenitor cells are the major pathway for epithelial regeneration of lungs after major injury.
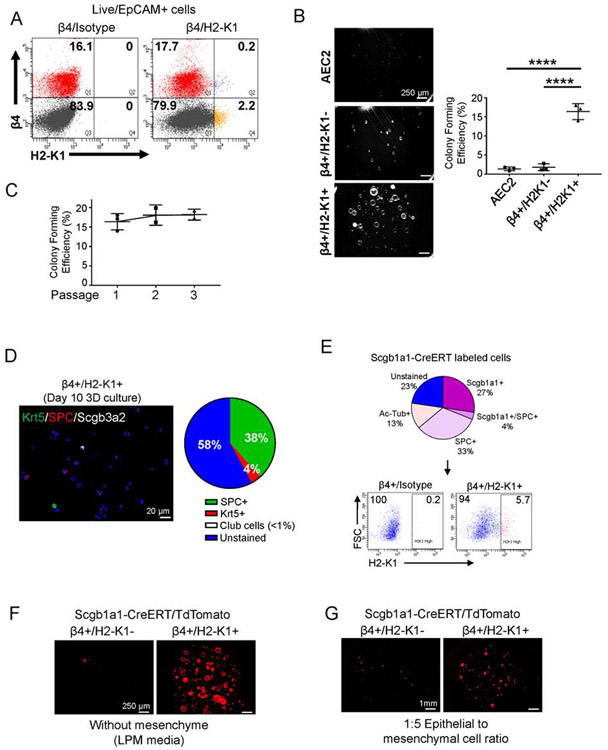
Figure 3: H2-K1high cells account for the in vitro regenerative activities of airway epithelium.
(A) High H2-K1 expression identifies a small fraction (~3%) of EpCAM+/β4+ cells via flow cytometry. Flow plot representative of >10 independent experiments.
(B) β4+/H2-K1high cells have the highest colony forming activity in vitro in mesenchyme free 3-dimensional culture with lung progenitor media (LPM). Lung progenitor media has key growth factors typically secreted from mesenchyme (Figure S3). Each data point represents a biological replicate. Data are presented as mean ± SD. **** p < 0.0001 (One-way ANOVA followed by Tukey’s multiple comparisons test).
(C) β4+/H2-K1high cells maintain their colony forming efficiency for at least three passages in culture. Each data point represents a biological replicate.
(D) At the end of first passage, β4+/H2-K1high colonies can differentiate towards either alveolar fate (SPC+) or airway fate (Krt5+ basal and Scgb3a2+ club cells).
(E) Scgb1a1-CreERT lineage labeled at least three major populations in the lung epithelium as determined by cytospin analysis. Various cell types labeled by Scgb1a1-CreERT included β4+/H2-K1high cells, which represented ~6% of total lineage labeled airway cells by Scgb1a1-CreERT.
(F, G) Scgb1a1-CreERT lineage labeled H2-K1high cells also account for all colony forming cells in (F) mesenchyme-free or (G) mesenchyme-dependent 3D culture conditions. β4+/H1-K1high cells represent the cells with progenitor activity ascribed to lineage labeled club cells.
See also Figure S3.
RESULTS
Identification of subpopulations of airway epithelial cells with progenitor potential.
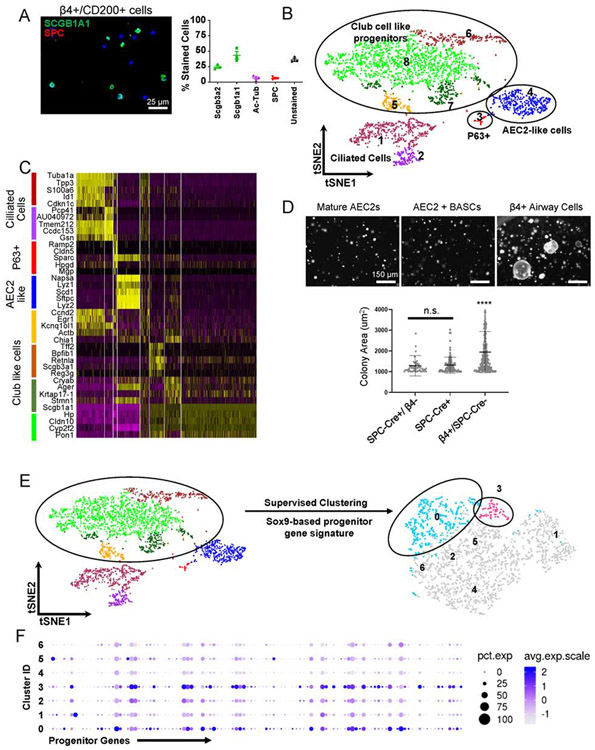
Although Scgb1a1-creERT lineage trace labels cells with regenerative potential in vivo and has been used extensively to isolate cells that function as progenitors in in vitro organoids, the actual cells of origin of these progenitor population(s) remain unclear. That uncharacterized epithelial stem/progenitor cells may exist among mature airway epithelial cells is suggested by immunostaining of epithelial cells isolated by flow sorting on the basis of expression of integrin β4 (β4), which marks airway epithelial cells, and CD200, which excludes mature ciliated cells (Vaughan et al., 2015). Using this strategy, we determined that ~60% of the isolated cells stain for either club cell specific protein (SCGB1A1), the ciliated cell marker Ac-Tubulin, or SPC while ~40% of the cells did not stain for any mature lineage markers of the airway epithelium (Figure 1A). To identify unknown cell populations within these cell types, we then performed single cell transcriptomics analysis of EpCAM+/β4+/CD200+ cells. t-distributed stochastic neighbor embedding (tSNE) plot of single cell identified 8 distinct clusters (Figure 1B, C). As expected, we identified a small cluster of rare distal p63+ epithelial cells (cluster 3) previously shown to be activated by influenza infection (Kumar et al., 2011; Vaughan et al., 2015; Zuo et al., 2015) and a subpopulation of cells with a highly similar transcriptomes to mature AEC2s (cluster 4). Another source of alveolar AEC2s is the BASCs residing at the bronchioalveolar duct junction (BADJ) (Kim et al., 2005; Liu et al., 2019; Salwig et al., 2019). The BASCs can be traced with both a club cell lineage marker Scgb1a1− and SPC-CreERT2 (Figure S1A) (Liu et al., 2019). To assess the in vitro clonogenicity of mature AEC2s, the BASCs, and other β4+ airway cells as a measure of in vivo stem/progenitor potential, we established a 3-dimensional organoid culture system as previously described (Barkauskas et al., 2013; Zacharias et al., 2018). We isolated SPC-CreERT2+/β4− cells (mature AEC2s), SPC-CreERT2+ cells (mature AEC2s and BASCs), and the β4+/SPC-CreERT2− cells (remaining airway epithelial cells) and co-cultured with mesenchymal cells as described previously and analyzed colony size on Day 14 (Figure 1D, Figure S1B). These co-cultures established that SPC-Cre-neg airway cells held the most in vitro regenerative potential among the lung epithelial cells giving rise to SPC+/RAGE1+ alveolar colonies (Figure 1D; Figure S1C). These findings imply that a yet to be identified subpopulation of epithelial cells possesses robust clonogenic and stem/progenitor potential, which we will focus on in the subsequent experiments.
Figure 1: Identification of non-BASC airway cell population with progenitor-like features.
(A) Cytospin analysis of EpCAM+/ITGB4+/CD200+ cell population enriched for epithelial progenitors identified ~45% cells stained with clara cell specific protein Scgb1a1, ~7% of cells were stained with Ac-Tubulin and Surfactant Protein C (SPC) each while ~40% of cells were unstained for mature lineage markers. ~25% of Scgb3a2+ cells were subpopulation of Scgb1a1+ cells. Scale bar = 25 μm.
(B) Single cell transcriptomic sequencing of progenitor enriched cell population was performed on 10X Genomics platform. t-distributed stochastic neighbor embedding (tSNE) plot of single cells displays 8 distinct clusters.
(C) Heatmap shows five top upregulated genes for each of the eight clusters identified through single cell mRNA sequencing of β4+/CD200+ cells. Four major types of cells were identified: Ciliated cells, p63+ cells, AEC2-like cells, and four clusters of club-like cells – all with high expression of Scgb1a1 and Cyp2f2 (club cell markers).
(D) non-BASC airway epithelial cells (β4pos/SPC-CreERT2-neg) cells in 3-dimensional co-culture with freshly sorted mesenchyme (1:5 ratio) are the primary sources of largest colonies (n=3 replicates were analyzed). Scale bar = 150 μm. Area mean ± SD.**** p < 0.0001. Brown-Forsythe and Welch ANOVA tests followed by Holm-Sidak’s multiple comparisons test.
(E) To identify minor population within the club cell like clusters with high progenitor activity, we performed supervised clustering using a Sox9-based progenitor gene signature of the bud tip cells from a developing mouse lung (Ostrin et al., 2018).
(F) Progenitor genes used to re-cluster club like cells were enriched in clusters 0 and 3. See also Figures S1.
Identification of H2-K1high cells as stem/progenitor cells.
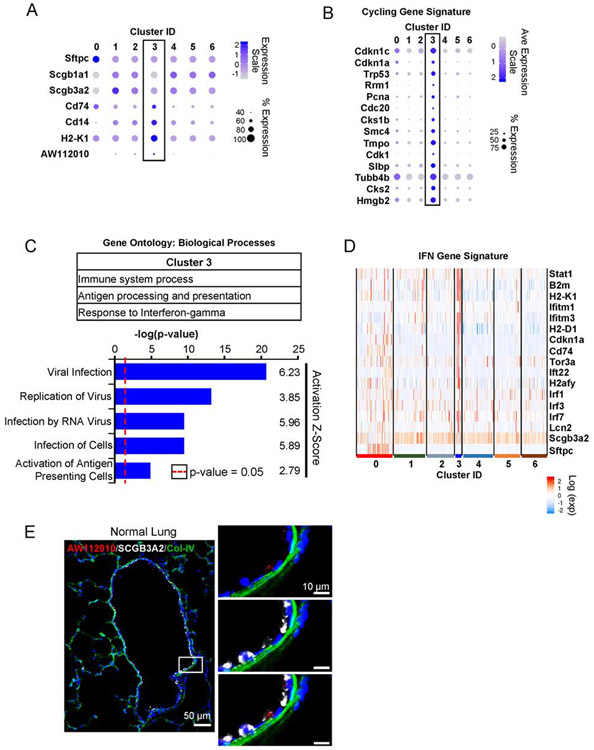
To search for additional stem/progenitor cells within the club-like cells we re-clustered the transcriptomes of these cells using a Sox9 gene signature assigned to embryonic epithelial bud tip cells with known stem cell function (Figure 1E) (Ostrin et al., 2018). This re-clustering identified two cell populations (clusters 0 and 3) that were enriched in the Sox9-progenitor signature (Figure 1F). Cells in cluster 3 are marked by lower expression of mature lineage markers such as Sftpc and Scgb1a1 and higher expression of several MHC class I and class II genes, including H2-K1 in comparison to remaining clusters (Figure 2A, S2A). Interestingly, cells in cluster 3 have elevated expression of several genes involved in cell cycle progression while simultaneously expressing key cell cycle inhibitors Cdkn1a (p21) and Cdkn1c (p57) (Figure 2B), that could explain their lack of proliferation and maintenance in quiescence in vivo. Because of the low levels of mature lineage transcripts and high levels of cell cycling genes of cluster 3 compared with all other cell clusters, we subsequently focused on characterizing cluster 3. Cells in cluster 3 are highlighted by activation of host-defense related signaling pathways (Figure 2C, D). Cells in cluster 3 also express a number of genes associated with viral infection, which suggests a susceptibility to viral infection (Figure 2C). Indeed, a recent study suggested that influenza injury specifically targets β4+/CD200+ distal airway progenitors, which includes cells in Cluster3 (Gaur et al., 2011; Quantius et al., 2016). Consistent with activation of host-defense related signaling pathways, these cells have increased expression of interferon-regulated genes (Figure 2D). Importantly, Cluster 3 cells are marked by a significantly high expression of lncRNA AW112010 (Figure 2A, Figure S2A, B). Therefore, we performed in situ hybridization for the lncRNA to identify cells of cluster 3 in uninjured mouse lungs. AW112010high/Scgb3a2low cells were localized to the distal airway epithelium but there was no signal in Scgba3a2+ club cells or at the bronchioalveolar duct junctions (Figure 2E; Figure S2C), further confirming that AW112010high cells are not BASCs. Sca-1 (Ly6a), another marker attributed to bronchioalveolar stem cells (BASC) (Kim et al., 2005), is expressed at varying levels in multiple clusters with some Ly6ahigh cells residing in cluster 3 (Figure S2D). Finally, ~3% of the β4pos cells express AW112010 (Figure S2E), while cells in cluster 3 represent ~2.2% of sequenced cells. The difference in the frequency could be due to insufficient sequencing depth which would fail to pick up all AW112010 expressing cells. In summary, cells in cluster 3 marked by elevated expression of AW112010 and several immune response genes are identified as non-BASC airway epithelial progenitor-like cells, many of which also express Sca-1.
Figure 2: Host defense-related signaling pathways highlight cells in progenitor cluster.
(A) Cluster 3 is negative or low for lineage markers of mature club (Scgb1a1, Scgb3a2) and alveolar epithelial type 2 cells (Sftpc). The cells in this cluster are marked by enhanced expression of surface markers Cd14, Cd74, H2-K1, and lncRNA AW112010.
(B) Cells in cluster 3 are also enriched in genes associated with cell cycle in addition to cyclin inhibitors Cdkn1a and Cdkn1c.
(C) Gene ontology analysis and pathway analysis identifies processes related with host-defense signaling pathways. Signaling pathways required for viral infection and replication are predicted to be activated in cells of cluster 3. Activation scores > 2 indicate a significant activation of a given pathway.
(D) Consistent with activated host-defense pathway, cells in cluster 3 are highlighted by enhanced interferon regulated genes.
(E) In situ staining for AW112010 lncRNA, which is expressed in H2-K1high progenitor cluster, localizes the progenitor cells in the airway. AW112010 positive cells (red) with low or no detectable expression of club cell marker Scgb3a2 (white). The image is a composite image of multiple images taken at 20X and stitched together (See STAR Methods).
See also Figures S2.
H2-K1high cells account for in vitro regenerative activities of airway epithelium.
Expression of H2-K1, an MHC class I antigen, was significantly increased in cells of cluster 3. Flow cytometry analysis showed that high expression of H2-K1 marked ~3% of all airway epithelium (marked by β4 expression) (Figure 3A), and matched with the frequency of AW112010-expressing cells. Using our earlier established 2-D culturing conditions (Chapman et al., 2011), we observed that β4+/H2-K1high cells represented the primary colony forming cells (Figure S3A). To assess the clonogenicity of H2-K1high cells, we established a culture system to test whether H2-K1high airway epithelial cells represent the airway colony forming epithelial cells (see Methods). We cultured cells in a mesenchyme-free 3D organoid assay in lung progenitor media (Nichane et al., 2017). This media has several growth factors that are thought to be secreted by the mesenchyme in an epithelial/mesenchymal 3D organoid condition (Figure S3B). Under these conditions, β4+/H2-K1high cells represented virtually all colony forming cells within an uninjured lung epithelium (Figure 3B). The high colony forming efficiency of these cells was maintained for at least three passages (Figure 3C). To determine the differentiation into either airway or alveolar cells, we stained either whole colonies or cytospins prepared from cultured cells. Analysis of cytospins revealed that these cells either differentiated towards SPC-expressing AEC2s (38%) or cytokeratin 5 (Krt5) expressing basal cells (4%) while Scgb3a2+ club cells were rarely observed (<1%) (Figure 3D). Likewise, whole organoids were exclusively either basal (Krt5+) or alveolar (SPC+/RAGE1+) (Figure S3C), indicating majority of the H2-K1high cells formed alveolar colonies with few cells forming airway basal colonies. To assess whether H2-K1high cells clonally expanded, we mixed H2-K1high cells from either a wild-type or a UBC-GFP mouse, which constitutively expressed eGFP (Schaefer et al., 2001). The colony mixing experiment demonstrated that 99% of 422 organoids were either GFP+ or GFP-, indicating clonal expansion of H2-K1high progenitors (Figure S3D).
Several studies have made use of Scgb1a1-CreERT lineage to label and isolate “club progenitor cells” to study their regenerative capacity in organoid assays (Lee et al., 2017; McConnell et al., 2016; Rawlins et al., 2009; Zheng et al., 2012; Zheng et al., 2017). Immunostaining of Scgb1a1-CreERT-traced cells identified at least three mature epithelial populations including SPC+, Scgb1a1+, Ac-Tub+, and a fraction of traced cells had undetectable levels of mature lineage markers (Figure 3E). Therefore, we tested whether H2-K1high cells are labeled by Scgb1a1-CreERT. Indeed, our analysis identified that ~6% of Scgb1a1-CreERT labeled airway cells (β4+) were H2-K1high, which accounted for the colony forming cells of Scgb1a1-lineage in mesenchyme-free or previously described mesenchyme-dependent conditions (Figure 3E-G). In summary, these experiments identified a subpopulation of cells that by lineage tracing appear indistinguishable from club cells, but in fact have distinguishing features and represent the colony forming cells within the club-like cells under 2D (Chapman et al., 2011), 3D with mesenchyme (Barkauskas et al., 2013; Lee et al., 2017; McConnell et al., 2016; Zacharias et al., 2018), and 3D without mesenchyme (Nichane et al., 2017) culture conditions.
β4+/H2-K1high cells expand after injury and differentiate toward alveolar lineages.
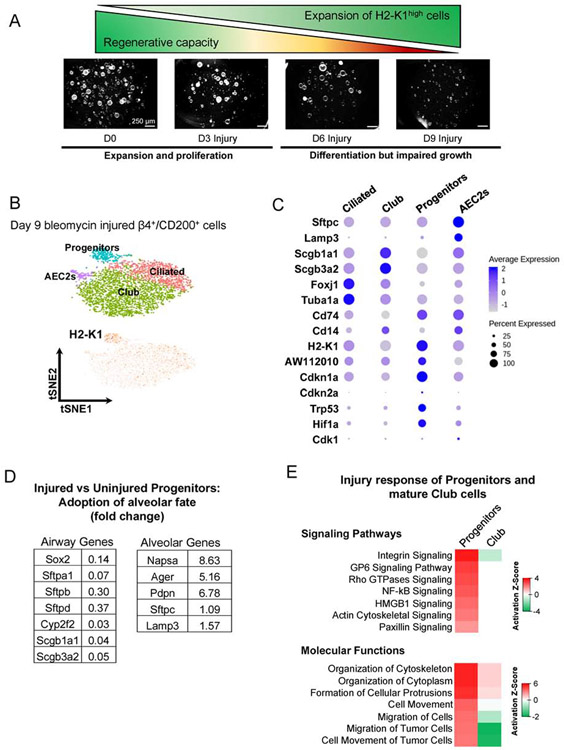
To determine whether H2-K1high cells respond to injury, we injured mice with bleomycin and assessed their response. Although H2-K1high cells in uninjured lungs are quiescent and do not proliferate (0% Edu+ cells in H2-K1high vs 1.7% Edu+ cells in H2-K1low fraction of airway cells), injury preferentially elicits a proliferative response in H2-K1high cells (11% Edu+ cells in H2-K1high vs 2% Edu+ cells in H2-K1low fraction) (Figure S4A, B). This is reflected in expansion of H2-K1high cells and formation of large colonies during in vitro culture from days 3-6 (Figure 4A, S4C). By day 9 almost all of the colonies were smaller, suggesting possible differentiation. Analysis of single cell transcriptomes of β4pos/CD200pos at 9 days post injury and subsequent supervised clustering of these cells confirmed a larger H2-K1high cluster when compared to uninjured H2-K1high cluster (Figure 4B, S4D). Injured H2-K1high cells maintained lower levels of mature lineage markers such as Sftpc, Scgb1a1, and Scgb3a2 in comparison to other injured mature cells (Figure 4C). However, when compared with uninjured/quiescent H2-K1high progenitors, the injured progenitors had decreased expression of multiple airway genes whereas several alveolar genes including Napsa, Ager, and Podoplanin (Pdpn) were increased, indicating that the injured H2-K1high progenitors have adapted a more alveolar fate (Figure 4D). Injured H2-K1high cells also had upregulation of signaling pathways associated with cellular mobility whereas mature club cells failed to mobilize after injury, as judged by the transcriptional profiles (Figure 4E), consistent with lack of proliferative response in β4pos/H2-K1low club cells (Figure S4A, B). Finally, bleomycin injury elicited preferential increased expression of Cdkn1a, Cdkn2a, Trp53, and Hif1a genes in H2-K1high cells that, along with differentiation, could be expected to reduce colony size (Figure 4C). Overall, these findings reveal an injury-induced H2-K1high initial expansion, increased migration, and differentiation towards alveolar fate.
Figure 4: β4+/H2-K1high cells expand after injury and acquire senescence-associated markers during fibrotic phase of bleomycin injury.
(A) Fraction of β4+/H2-K1high cells increases preferentially immediately after injury as determined by flow cytometry at 3 and 6 days after bleomycin injury. These expanded H2-K1high cells gradually lose their ability to form large colonies in vitro at days 3, 6, and 9 post bleomycin injury. (data representative of n=2 independent experiments from n=3 mice each).
(B, C) Single cell RNA-seq was performed on EpCAM+β4+/CD200+ cell population 9 days after injury followed by supervised clustering using the Sox9-based gene signature of progenitor cells (Figure 1D), which identified H2-K1high cell cluster (labeled as Progenitors). Cells in this cluster expressed low levels of mature lineage markers such as Surfactant Protein C and secretoglobins (Scgb1a1 and Scgb3a2), as expected. These cells also express senescence associated markers such as Cdkn1a, Cdkn2a, Hif1a, and Trp53.
(D) Injured β4+/H2-K1high cells (d9 post injury) have increased expression of alveolar genes and decreased expression of airway genes when compared with uninjured β4+/H2-K1high progenitor cells, suggesting acquisition of alveolar fate.
(E) Pathway analysis of injured β4+/H2-K1high cells in comparison with uninjured progenitor cells identify upregulation of signaling pathways associated with cellular movement while mature club cells from injured lungs remain unresponsive to the injury when compared to uninjured club cells.
See also Figure S4.
β4+/H2-K1high cells differentiate into alveolar cells in vivo.
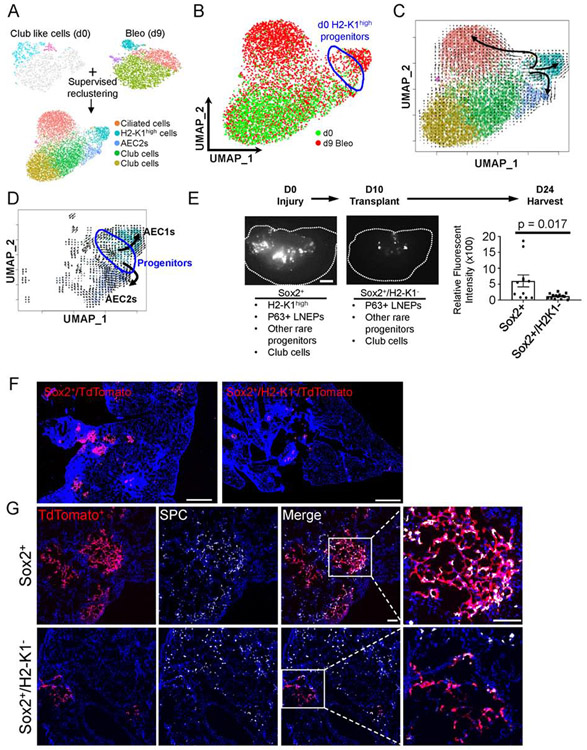
Our findings have revealed that within the large group of club-like cells, a minor population of H2-K1high cells express a progenitor gene signature and account for the in vitro clonogenic activity commonly attributed to club cells. These cells expand following bleomycin injury as judged by their larger population size in sc-RNA-seq data at day 9 (Figure 4B). To further explore the apparent expansion and differentiation response of quiescent H2-K1high cells after injury we compared the transcriptomes of all club-like cells, including mature club cells and immature H2-K1high cells, before and after injury. We merged uninjured club like cells (Figure 1E) including H2-K1high progenitors with injured β4+/CD200+ cells also containing the progenitors as well as any cells potentially derived from H2-K1high cells (Figure 4B) followed by supervised re-clustering again using the progenitor gene signature (Figure 5A). Subsequently, we identified four large populations – ciliated cells, club cells, AEC2s, and H2-K1high cells (Figure 5A; Figure S5A). Uninjured H2-K1high progenitors lie within the H2-K1high cluster next to a group of Ager+/Pdpn+ cells and Sftpc+ cells (Figure 5B; S5A), which indicated a potential lineage relationship between uninjured H2-K1high cells and alveolar cells appearing after injury. To test a relationship between H2-K1high and other mature cells, we performed RNA velocity analysis, which computationally predicts the future of cells based on a ratio of spliced to unspliced mRNA levels (La Manno et al., 2018). RNA velocity analysis predicted three distinct trajectories (Figure 5C). To probe specific lineage relationship between d0 H2-K1high progenitors and alveolar cells, we sub-clustered cells that included uninjured and injured H2-K1high cells, AEC1s, and AEC2s based on cell-type specific markers (Figure S5A) followed by calculation of RNA velocity. This approach identified a lineage trajectory from naïve progenitors to Ager+/Pdpn+ AEC1s and naïve progenitors to Sftpc+AEC2s (Figure 5D, S5A). Similarly, trajectories towards club cells and ciliated cells were also identified (Figure S5B). The conclusion that H2-K1high cells expand and contribute to the restoration of multiple airway and alveolar lineages in vivo after bleomycin injury was recently validated by an independent bioinformatic analysis that used high resolution temporal single cell sequencing combined with RNA velocity (Strunz et al., 2019). This study identified an injury-dependent expansion of a “club cell subset” marked by expression of several MHC class I and class II genes, consistent with the interferon signature of our uninjured H2-K1high cells (Figure 2D, S2A), that ultimately gives rise to AEC1s and AEC2s during alveolar repair. Markers used to identify the “club cell subset” by Strunz et al are also highly expressed in injured H2-K1high cells (Figure S5C), confirming that the H2-K1high progenitors described here and those identified by Strunz et. al. are likely identical and indeed expand during injury resolution to give rise to alveolar cells.
Figure 5: β4+/H2-K1high cells can differentiate into alveolar cells in vivo.
(A) To understand lineage relationship between H2-K1high progenitors from uninjured lungs and alveolar, club, and expanded H2-K1high cells post injury, all uninjured club like cells (including H2-K1high progenitors) were merged with bleomycin injured β4+/CD200+ cells followed by supervised clustering based on progenitor genes as used in Figure 1D. Four cell types are identified: H2-K1high cells, Foxj1+ ciliated cells, Scgb1a1+ club cells, and Sftpc+ AEC2s (See Figure S5).
(B) UMAP embedded merged objects shows either d0 (uninjured; green) or d9 bleomycin injured cells (red). Uninjured H2-K1high (d0) progenitors are highlighted by blue circle.
(C) RNA velocity was calculated to predict lineage differentiation of the merged objects and plotted onto the UMAP embedding. The RNA velocity shows three distinct lineages emanating from H2-K1high cells: H2-K1high cells to Sftpc+ AEC2s, H2-K1high cells to Foxj1+ ciliated cells, and H2-K1high cells into further H2-K1+ cells, some of which express AEC1 markers (Ager, Pdpn; Figure S5).
(D) Sub-setting AEC1s, AEC2s, and Progenitors followed by re-calculation of RNA velocity identified a trajectory from uninjured progenitors to AEC1s and AEC2s. Uninjured H2-K1high (d0) progenitors are highlighted by blue circle.
(E) 75,000 freshly sorted Sox2-lineage labeled airway cells containing ~4% (or ~3000) H2-K1high cells or 75,000 freshly sorted Sox2-lineage labeled cells depleted of H2-K1high cells were transplanted 10 days after mice were injured with bleomycin. Scale bar = 1mm. Dotted line outlines an individual lobe. Sox2-lineage label excludes BASCs and includes H2-K1high progenitors, P63+ LNEPs, other rare airway progenitors, and mature club cells. Whereas Sox2+/H2-K1low includes all of those progenitors except the H2-K1high progenitors. Total fluorescent intensity corresponding to transplanted cells was captured and quantified. 2-3 lobes/mouse were imaged from n=4 mice in each condition. Each data point represents quantification of fluorescent intensity from an individual lobe. Mann-Whitney test was used to determine significance. p-value < 0.05 is considered significant. Data are presented as mean ± SD.
(F) A representative section of a mouse lobe transplanted with either Sox2-labeled or Sox2-labeled/H2-K1neg cells is visualized showing limited engraftment of Sox2-labeled/H2-K1neg cells. Image represents a composite of image of multiple images captured at 10X and stitched together (see STAR Methods). Scale bar = 1mm.
(G) Sox2-labeled cells, which include H2-K1high progenitors, have engrafted in injured lung and express pro-Surfactant Protein C (pro-SPC), indicative of their differentiation towards AEC2 fate. Image represents a composite of image of multiple images captured at 20X and stitched together (see STAR Methods). Scale bar = 200 μm.
To test whether the implied trajectories of H2-K1high cells after injury (Figure 5C) indeed occur in vivo, we performed transplantation assays to determine the capacity of β4+/H2-K1high cells to engraft and differentiate towards alveolar cells. We used Sox2-CreERT2/tdTomato mice to label all airway epithelial cells including the H2-K1high progenitor cells. Selection of Sox2-labeled cells excludes all alveolar cells and BASCs based on their lack of Sox2 expression (Liu et al., 2019). We transplanted either total Sox2-traced airway epithelial cells (inclusive of β4+/H2-K1high progenitors) or Sox2-traced/H2-K1low cells, which would include mature club cells, rare p63+distal basal cells, and other rare airway progenitors but exclude the H2-K1high progenitors. Analysis of injured lungs 24 days post injury revealed robust engraftment of the Sox2+/H2-K1high population (Figure 5E, F). Consistent with our RNA velocity analysis, we found differentiation of engrafted H2-K1high progenitors into both AEC1s and AEC2s in bleomycin injured lungs (Figure 5G, S6A). Likewise, consistent with relatively rare differentiation into Krt5+ or Scgb1a1+ airway cells in vitro, we found rather infrequent regions of transplanted cells differentiating into Krt5+ or Scgb1a1+ airway cells post injury in vivo (Figure S6B, C). In summary, our analysis reveals the ability of H2-K1high progenitors to meaningfully contribute to the regeneration of multiple cell types during bleomycin injury.
Expanded progenitor cells differentiate in vivo and aid in functional recovery of injured mice.
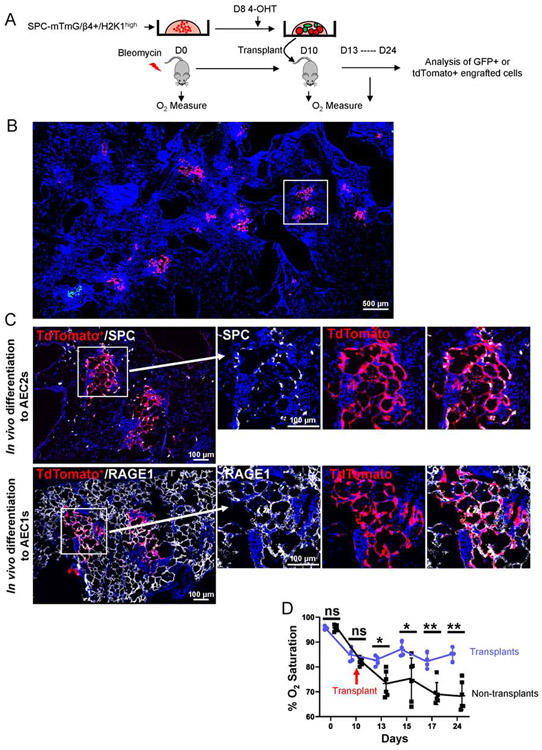
We tested whether purified H2-K1high cells expanded in vitro can engraft and aid in recovery of injured mice. We isolated and cultured progenitor cells from an unlabeled SPC-CreERT2 mouse for 10 days and labeled SPC+ cells during the last 48 hours of culture using 4-hydroxytamoxifen (Figure 6A). We then transplanted all cultured cells in bleomycin injured mice and analyzed engrafted regions and their effect on oxygenation (Figure 6A). Although 38% of the cultured cells differentiate into SPC+ cells (Figure 3D), only ~15% of the engrafted regions were GFP+, indicating that ~85% of the engrafted regions were tdTomato+ and coming from undifferentiated H2-K1high progenitors (Figure 6B), consistent with a recent study showing the need for a significantly higher number of mature AEC2s for effective engraftment (Weiner AI, 2019). In part, the higher engraftment efficiency of H2-K1high cells likely reflects the previously reported higher proliferative activity of sox2+ airway cells migrating into injured alveolar compartments relative to residual endogenous AEC2s (Xi et al., 2017). Many of the unlabeled, and therefore undifferentiated, progenitors differentiated into SPC-expressing AEC2s and RAGE1+ squamous AEC1s covering large areas of alveolar walls, indicating their capacity for alveolar regeneration in vivo (Figure 6C). Accordingly, transplantation of H2-K1high progenitor cells significantly improved oxygen saturation levels and promoted functional recovery of injured mice as early as 3 days post-transplant (Figure 6D).
Figure 6: Expanded progenitor cells differentiate in vivo and aid in functional recovery of injury mice.
(A) Schematic experimental design. Freshly sorted progenitor cells from unlabeled SPC-CreERT2 mice were cultured in mesenchyme-free 3D conditions with LPM for 10 days. 100μM 4-Hydroxytamoxifen (4-OHT) was added in culture for the last 48 hours to label SPC expressing cells, which accounted for ~38% of all cells (see Figure 3D). Wild-type mice were injured with bleomycin on D0. Either 250,000 cells or saline were transplanted in injured mice at 10 days post injury and lungs were harvested on 24 days after initial injury. Oxygen saturation was measured at indicated time-points.
(B) A representative section of a mouse lobe transplanted with either SPC-expressing (GFP+) or SPC-negative (tdTomato+) cells shows engraftment of tdTomato+ cells accounts for >90% of total engraftment areas. Boxed region is enlarged in C. Image represents a composite of image of multiple images captured at 10X and stitched together (see STAR Methods).
(C) tdTomato+ cells, upon engraftment, differentiate into pro-SPC+ AEC2s and RAGE1+ AEC1s (same engrafted region from sequential section is shown). Image represents a composite of image of multiple images captured at 10X and stitched together (see STAR Methods).
(D) Oxygen measure at day 0 (day of the injury) and starting day 10 post injury (day of the transplants) indicate improved oxygenation in mice that received transplant (n=4 for transplant and n=5 for non-transplant mice). Each data point represents oxygen saturation level of one mouse at the indicated time point. Mann-Whitney test was used to determine significance. * p < 0.05, **p < 0.01. Data are presented as mean ± SD.
DISCUSSION
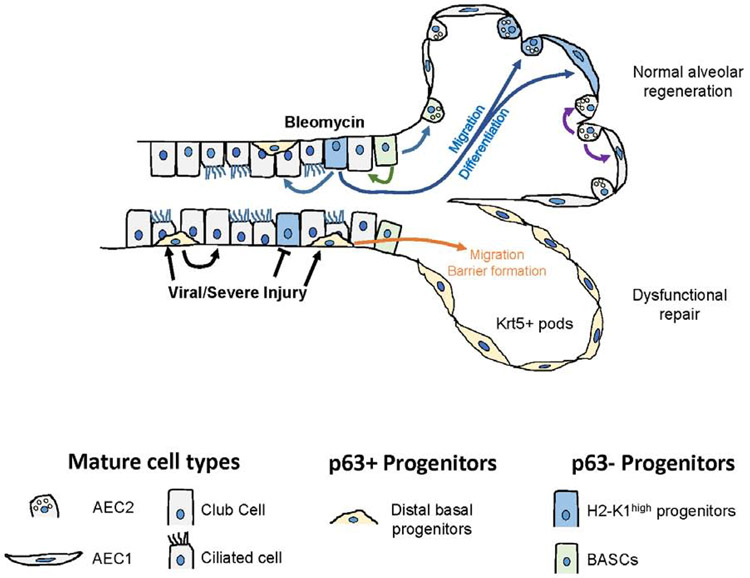
Repair and regeneration of the lung after significant injury involves expansion and migration of both residual AECs within alveoli and airway derived stem/progenitor cells. In the mouse, after bleomycin injury approximately half of the mature AEC2s present in the recovered lungs derive from distal airways or bronchioalveolar junctions (Barkauskas et al., 2013; Chapman et al., 2011). Compared with proliferating AEC2s in alveolar sacs, distal airway cells, once mobilized, are in a favorable position to migrate and cover large areas of denuded or heavily damaged alveolar epithelium and then restore normal lining. But the nature of the stem/progenitors mobilized after injury has remained uncertain. Rare p63+ basal like cells, Sca1+ cells or BASCs, and Scgb1a1-traced cells, which are thought to be club cells, are all reported to expand and mobilize after injury to repair alveolar epithelium (Kim et al., 2005; Liu et al., 2019; McQualter, 2019; McQualter et al., 2010; Vaughan et al., 2015; Zuo et al., 2015). Moreover, club cells in the conducting airway are reported to dedifferentiate into a basal stem cell population that can then expand and differentiate into a number of mature cell types, raising the possibility that club cell de-differentiation is a pathway of regeneration in the injured lung (McQualter, 2019; Tata et al., 2013), although the extent to which this regenerative pathway is utilized during physiologically relevant injury remains uncertain. The findings reported here that a small, quiescent subpopulation of SCA-1 expressing, Scgb1a1-traced stem/progenitor cells without mature protein lineage markers and distinct from either p63+ cells or BASCs account for virtually all of the in vitro clonogenic activity of distal airway cells and significantly contribute to in vivo regenerative responses can clarify some of the current uncertainties. First, the findings point to the promiscuous nature of existing lineage tracers that allow small subpopulations of relatively immature and specialized progenitor cells to hide among transcriptionally similar mature cell types. Similarly, a small fraction of AEC2s in murine and human lungs, but not all AEC2s, was recently shown to contribute to alveolar repair (Nabhan et al., 2018; Zacharias et al., 2018). Secondly, recent studies have revealed that BASCs do not express SCA-1 (Salwig et al., 2019), a biomarker originally used to isolate BASCs (Kim et al., 2005). Instead the H2-K1high population identified here appears to be the major airway subpopulation expressing surface SCA-1 (Figure S2) and are likely isolated by flow strategies employing SCA-1 antibodies. An additional surprising finding is that H2-K1high cells show high levels of genes supporting viral replication, which is consistent with our data showing high susceptibility to H1N1 PR8 viral injury (Figure S7). This could explain their missing in action in influenza models of lung injury (Vaughan et al., 2015; Yang et al., 2018). Airway p63+ basal cells preferentially expand after PR8 influenza viral infection whereas the β4+CD200+ airway cells containing H2-K1high cells are preferentially targeted and lose their regenerative activity (Figure S7) and (Buchweitz et al., 2007; Quantius et al., 2016; Tata et al., 2013). In this case, the lung is left with p63+ basal-like cells as their only recourse to mobilize airway cells in a desperate attempt at alveolar regeneration. By contrast, in the bleomycin model studied here the H2-K1high subpopulation appears to be the major airway-derived cell supporting type II and type I cell differentiation and alveolar regeneration. Overall, these findings support the paradigm that pre-existing specialized stem/progenitor cells preferentially mobilize following injury to repopulate the alveolar epithelium and the particular type of responder is strongly influenced by the nature of the injury (Summarized schematically in Figure 7).
Figure 7: Injury-specific mobilization of distal airway stem/progenitor cells.
Distinct airway progenitors activate and expand depending on the type of injury. Newly identified H2-K1high progenitors are transcriptionally highly similar to mature club cells but have unique regenerative characteristics that are identified through single cell mRNA-seq. These cells are preferentially targeted during viral injury, which leaves p63+ distal basal cells as the primary effectors of alveolar injury resolution. In bleomycin injury, p63neg H2-K1high progenitors, along with BASCs, are mobilized to promote regeneration of normal alveolar epithelium.
See also Figure S7.
As expected by their in vitro growth and differentiation potential, an expanded population of H2-K1high appears following bleomycin injury. But are these actually derived from the pre-existing H2-K1high cells? Recent developments in the application of computational biology techniques to cellular dynamics using time-based sampling allows us to predict single cell lineage changes over time (Kester and van Oudenaarden, 2018). We used RNA velocity to predict the lineage relationship between preexisting normal H2-K1high cells and the H2-K1high cells that we observed at d9 of bleomycin injury. This analysis predicted an expansion trajectory of uninjured H2-K1high, but not mature club cells, into not only more H2-K1high cells but also several mature lineages including AEC2s, AEC1-like cells, and ciliated cells (Figure 5, Figure S5). Interestingly, the lineage prediction depicts appreciable expansion of H2-K1high cells into more H2-K1high cells and differentiation into AEC1s, with seemingly less robust differentiation into AEC2s. One caveat is that differentiation toward AEC2s results in dramatic loss of integrin β4 (Vaughan et al, 2015) which would bias against their recovery by the flow cytometry used to isolate the populations for scRNA-seq analysis and potentially underweight the RNA velocity trajectory toward these cells. Moreover, we have not considered trans-differentiation to ciliated cells in experiments here that focus on alveolar injury and repair. But we validated the RNA velocity predictions by direct transplantation of uninjured H2-K1high cells into bleomycin-injured lungs after which we observed expansion of the H2-K1high population and differentiation to both AEC2s and AEC1-like cells. Robust differentiation of transplanted H2-K1high cells into AEC2s in vivo underscores the high potential of these progenitors to restore the AEC2 population. Interestingly, a recent independent bioinformatic analysis of bleomycin-injured lungs in mice made strikingly similar predictions and also described a core source of stem/progenitors within the “club cell” population characterized by high expression of immune response genes, sharing many features with H2-K1high cells (Strunz et al., 2019). The importance of high levels of immune-response genes to the function of these stem/progenitor cells is completely unclear and remains to be defined. Nonetheless, collectively these findings strongly support a lineage relationship between the reservoir of normal airway H2-K1high cells and expansion of airway-derived stem/progenitors into the alveolar compartment after bleomycin injury.
The progressive accumulation of senescent epithelial stem/progenitors in mouse and human fibrotic disorders is widely believed to limit the repair and regeneration (Campisi, 2016; Hamsanathan et al., 2019). Indeed, we observed the acquisition of biomarkers associated with senescence in the H2-K1high population (Figure 4), suggesting that while these progenitors expand and contribute to alveolar regeneration, their numbers may be insufficient to fully restore normal function in the bleomycin fibrosis model. To explore this issue we undertook transplantation of normal H2-K1high cells into bleomycin-injured mouse lungs at a time (day10) when the endogenous cells had already largely lost their renewal potential. We observed robust engraftment and differentiation to AEC2s and RAGE1 expressing AEC1s, consistent with our in silico lineage prediction. This result establishes that normal stem/progenitors can productively expand in the harsh environment of injured lungs. Moreover, using 10 day culture-expanded H2-K1high cells, we achieved both extensive engraftment and improved lung oxygenation. Analysis of the engrafted regions also signified that immature progenitors were more efficient at engrafting and differentiating into AEC2s in vivo than mature AEC2s derived from the same progenitors in vitro, as expected. Surprisingly, the oxygenation levels improved as early as 3 days post transplantation. We could not determine whether the improved oxygenation as early as day 3 could be completely explained by formation of new alveolar barriers by transplanted cells (prior to terminal differentiation), or possibly also indirect effects on the local inflammatory process that could attenuate enhanced permeability and improve gas exchange. These aspects of early regeneration will require further studies focused on reversal of hypoxia by cell-based therapies. Nonetheless, these findings point to the possibility of future cell-based adjunctive therapy for severe lung injury and the need for further characterization of the most robust transplantable progenitors in humans.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further requests for resources and reagents should be directed to and will be fulfilled by the first author Harold A Chapman (hal.chapman@ucsf.edu). There were no new unique reagents generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal models and lineage labeling
All animals were housed and treated according to the procedures approved by the Institutional Animal Care and Use Committee of UCSF and all animal experiments were done in compliance with ethical guidelines and the approved protocols. All mice were housed in humidity- and temperature-controlled rooms on a 12 h light-dark cycle with free access to food and water. Sox2-CreERT2 (Arnold et al., 2011), Scgb1a1-CreERT (Scgb1a1tm1(cre/ERT)Blh) (Rawlins et al., 2009), SPC-CreERT2 (Sftpctm1(cre/ERT2,rtTA)Hap) (Chapman et al., 2011), Ai14-tdTomato (Madisen et al., 2010), and mTmG (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo) (Muzumdar et al., 2007) mice were previously described. For lineage tracing, mice were treated with 0.25mg/g dose of tamoxifen (intraperitoneal injection). Sox2-CreERT2 mice received three doses, SPC-CreERT2 mice received four doses, and Scgb1a1-CreERT mice received five doses of tamoxifen in one dose/alternate day protocol. Mice between 6-10 weeks of age of both sexes in equal ratios were used.
METHOD DETAILS
Bleomycin Injury
All animal studies utilized a minimum of 4 mice per group. Mice were injured with intra-tracheal instillation of bleomycin (1.5 U/kg body weight; Sigma Aldrich, Cat# B5507) by oral aspiration.
Mice were weighed twice a week and sacrificed at day 24 post injury for histopathological analysis
Edu Incorporation
50mg/kg body weight of 5-Ethynyl-2′-deoxyuridine (Edu) in PBS was delivered to mice via intraperitoneal injection every 24 hours starting immediately after bleomycin or PBS instillation (D0) for three days total. Mice were sacrificed at least 2 hours after the last dose of Edu. Lungs of mice were fixed and embedded in OCT followed by staining for Edu using the Click-iT EdU Alexa Fluor 647 imaging kit.
Transplantation of progenitor cells
Freshly sorted 75,000 EpCAM+/Sox2-labeled or EpCAM+/Sox2-labeled/H2-K1low cells were transplanted at 10 days after bleomycin injury. For pulse oximetry experiments, 250,000 cultured cells (10 days in culture) were transplanted in mice at 10 days after injury. Lungs were harvested 24 days post injury to determine level of engraftment.
Pulse oximetry
Arterial oxygen saturation was measured from non-anesthetized mice using the MOUSEOX Pulse Oximeter system (Starr Life Science). Mice were shaved prior to bleomycin injury and measures were taken on Day 0 (day of the injury), followed by day 10 (day of the transplantation) followed by alternate days. 5 measurements/second were taken for at least 4 minutes per mouse after establishing the first successful reading. All measurements were then analyzed for associated error code and error-free (code = 0) measurements were then averaged for each time point for each mouse. At least 4 mice per group were used.
Mouse lung epithelial cell prep and flow cytometry
Single cell suspension of mouse lungs was created as previously described (Vaughan et al., 2015). For FACS analysis, single cell preparations were incubated with the following antibodies for 30-45 minutes at 4°C in DMEM without phenol red) + 2% FBS: Rat anti-mouse CD45 (1:200, BD 30-F11), rat anti-mouse EpCAM (1:500, Biolegend, G8.8), rat anti-mouse integrin β4 (1:75, BD, 346-11a), rat anti-mouse CD200 (1:100, Biolegend, OX-90), mouse anti-mouse H2-K1 (1:100, Biolegend, 34-1-2S). Cells were then washed twice with 1X PBS. Sorting and subsequent analyses were performed on BD FACS Aria cytometers.
RNA In Situ Hybridization
Paraformaldehyde-fixed OCT embedded lung sections were processed for RNA in situ hybridization for AW112010 (Cat# 487351) using the RNAScope Multiplex Fluorescent Reagent Kit v2 (Cat# 323100, Advanced Cell Diagnostics) according to the manufacturer’s protocol (ACDBio, California, USA).
Cell Culture
Primary cells were cultured in either 3-dimensional system or on top of matrigel (Corning #CB-40230A) in a 2-dimensional system. 3D culture with mesenchymal cells: Cells were cultured with mesenchyme in 3-D inserts as described before (Barkauskas et al., 2013). Briefly, 5000 epithelial cells were mixed with 30,000 freshly sorted CD45neg/CD31neg/EpCAMneg mesenchymal cells in 1:1 matrigel:media in transwell inserts in a 24-well plate. Matrigel was allowed to solidify for 20 minutes at 37°C and 500μL Small Airway Basal Medium + MTEC media (You et al., 2002) was supplemented in the lower chamber. Mesenchyme-free 3D culture: Cells were cultured in mesenchyme-free 3D culture conditions as described before (Nichane et al., 2017), 500 or 1000 epithelial cells were mixed in 45μL growth factor reduced matrigel and a 40μL drop was placed in the center of a well in a 24-well plate. Matrigel was allowed to solidify at 37°C followed by addition of 500μL of lung progenitor media (LPM) (See Figure S3). Cells were imaged or harvested 10 days after setting up the culture. 2D culture: Cells were cultured on top of matrigel as described before (Chapman et al., 2011) with Small Airway Growth Media (Lonza, #CC-3118). Upon plating cells, rock inhibitor (Y27632 Santa Cruz Biotechnology, #sc-281642; 10μM) was added for the first 48 hours followed by addition of KGF (Peprotech, # AF-100-19; 10ng/mL).
Immunofluorescence analysis
Lung tissues were processed after euthanasia as described before (Xi et al., 2017). Briefly, OCT/PFA (94% OCT, 2% PFA) -inflated lungs were fixed with 4% PFA for 1 hour at room temperature and subsequently embedded in OCT. Colonies in 3D matrigel with mesenchyme-dependent or –free conditions were fixed with 4% PFA and embedded in OCT. 7 μm sections were cut on a cryostat. Sections were incubated for three 10 minute intervals with 1 mg/ml sodium borohydride (Sigma) in PBS to reduce aldehyde-induced background fluorescence. Slides were subsequently blocked ≥ 45 minutes in blocking buffer (1X PBS + 1% bovine serum albumin (Affymetrix), 5% nonimmune horse serum (UCSF Cell Culture Facility), 0.1% Triton X-100 (Sigma), and 0.02% sodium azide (Sigma). Primary antibodies listed below, diluted in blocking buffer, were then applied and incubated overnight at 4°C. Slides were washed three times with PBS + 0.1% Tween 20, and incubated with secondary antibodies (typically Alexa Fluor conjugates, Life Sciences) at a 1:1000 dilution in blocking buffer for 45-60 minutes. Finally, slides were again washed, incubated with 1μM DAPI for 5 min, and mounted using Prolong Gold (Life Sciences). The following antibodies were used: rabbit anti-proSPC (1:3000; Millipore, AB3786), goat anti-Scgb1a1 (1:10,000; a gift from Barry Stripp), goat anti-Scgb3a2 (1:500, R&D, AF3465), Rabbit anti-Collagen IV (1:1,000; Abcam, ab6586), chicken anti-Krt5 (1:1000; Covance, SIG-3475), mouse anti-acetylated tubulin (1:500; Sigma, 6-11B-1), rabbit anti-phospho histone H3 (1:500; Millipore, 06-570), and rat anti-RAGE1 (1:250; R&D, MAB1179). Images were captured using Zeiss Imager M1 and analyzed using AxioVision 4.8.2 (Zeiss, Germany). Where indicated, multiple images at 20X were captured using the “MosaiX” function and stitched together using “Tile Stitch” function in the AxioVision software.
Single cell transcriptomics and subsequent analysis
Single cell sequencing was performed on a 10X Chromium instrument (10X Genomics, Pleasanton, CA) at the Institute of Human Genetics (UCSF, San Francisco, CA). Cells were isolated via flow cytometry according to the experimental design. Cells were washed twice and finally re-suspended in 1X PBS supplemented with 0.04% BSA at 1000 cells/μL. Single cells were loaded on a Chromium Controller instrument (10X Genomics) that generated single-cell Gel Bead-In-EMulsions (GEMs). Libraries were prepared by performing reverse transcription on Bio-Rad C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA), following which, GEMs were harvested to amplify cDNA also using Bio-Rad C1000 Touch Thermal Cycler. SPRIselect (Beckman Coulter, Brea, CA) was used to select for amplified cDNA. Indexed sequencing libraries were constructed using the Chromium Single-Cell 3’ library kit (10X Genomics) and sequenced on NovaSeq 6000 (Illumina) with the following parameters: Read 1 (26 cycles), Read 2 (98 cycles), and i7 index (8 cycles) to obtain a sequencing depth of ~100,000 read/cell. Reads were aligned to mouse genome (mm10) and quantified using Cell Range Single-Cell Software Suite. Secondary analysis was performed using R package (Seurat v2.0; Butler et al, 2018). Dimensionality reduction was performed based on t-Distributed Stochastic Neighbor Embedding (t-SNE) or Uniform Manifold Approximation and Projection (UMAP) and clusters were determined to identify unique cell populations. Differential gene expression analysis was performed using “FindMarkers” function in Seurat package. Differentially expressed genes were then used for network and pathway analysis using Ingenuity Pathway Analysis (IPA, QIAGEN, www.qiagen.com/ingenuity). Comparison analysis was performed using IPA by first preparing differentially expressed genes of normal vs injured progenitor and club cells. These differentially expressed genes were then used to perform pathway analysis to identify differentially expressed pathways between uninjured and injured cell types. Z-scores of these pathways were then compared between progenitor and club cells. Uninjured club like progenitors and injured β4pos/CD200pos transcriptomes were integrated using the standard integration workflow of Seurat v3 (Stuart et al., 2019). Briefly, common anchors between were identified between the two datasets using FindIntegrationAnchors() function followed by integrating the two datasets using these anchors with IntegrateData() function. Subsequently, supervised clustering was performed on the integrated dataset using the Sox9 associated progenitor genes (Ostrin et al., 2018) to identify clusters of AEC2s, H2-K1high, ciliated and club cells. RNA velocity was then calculated on the integrated object using Velocyto package in R (La Manno et al., 2018) and RNA velocity in SeuratWrappers package in R.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analyses are reported where applicable in the figure legends. Statistical analyses were performed in GraphPad Prism 8. As shown in figure legends, data in graphs are shown as mean ± SD (at least n=3). All animal studies had at least n=4 mice in each group. For quantification of engraftment in Figure 5, 2-3 lobes from a total of 4 mice/group were imaged and fluorescence was quantified in Fiji distribution of ImageJ. Mann Whitney test was used to calculate significant difference. Statistical significance of differences between three or more groups in Figure 1D was calculated by Brown-Forsythe and Welch ANOVA tests followed by Holm-Sidak’s multiple comparisons test and in Figure 3B was calculated by one-way ANOVA followed by Tukey’s multiple comparison tests. The researchers were not blinded to the experimental groups. No sample-size estimation was performed for experiment in Figure 6. No data points were excluded from the analyses performed in this study.
DATA AND CODE AVAILABITLITY
All raw data generated in this manuscript are hosted at NCBI Gene Expression Omnibus (GEO) with the following accession numbers: GSE129937 and GSE130077.
Supplementary Material
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-pro-SPC (1:3000) | Millipore | Cat# AB3786, RRID:AB_91588 |
| Chicken anti-KRT5 (1:1000) | Covance | Cat# SIG-3475-100, RRID:AB_10720202 |
| Mouse anti-Acetylated Tubulin (1:500) | Sigma-Aldrich | Cat# T7451, RRID:AB_609894 |
| Rabbit anti-Histone H3 Phospho (1:500) | Millipore | Cat# 06-570, RRID:AB_310177 |
| Rat anti-RAGE1 (1:250) | R&D Systems | Cat# MAB1179, RRID:AB_2289349 |
| Goat anti-Scgb3a2 (1:500) | R&D Systems | Cat# AF3465, RRID:AB_2183550 |
| Goat anti-Scgb1a1 (1:1000) | Millipore | Cat# ABS1673 |
| Rabbit anti-Collagen IV | Abcam | Cat# ab6586, RRID:AB_305584 |
| Secondary antibodies raised in Donkey included FITC-, Cy3, or Cy5-conjugated | ThermoFisher | Cat#A32787; Cat#A32849; Cat#A32766; Cat#A32790; Cat#A10042; Cat#A-11055 |
| Donkey anti-Chicken IgY | Sigma-Aldrich | Cat# AP194SA6 |
| DAPI (4’,6’-diamidino-2-phenylindole) | ThermoFisher | Cat# D1306; RRID:AB_2629482 |
| APC Rat anti-mouse CD45 | BD Biosciences | Cat# 550539; RRID:AB_2174426 |
| BV421 Rat anti-Mouse CD326 | BD Biosciences | Cat# 563214; RRID:AB_2738073 |
| PE anti-mouse CD104 | BioLegend | Cat# 123610; RRID:AB_2563544 |
| APC anti-mouse H-2K1 | BioLegend | Cat# 114714; RRID:AB_2734174 |
| APC anti-mouse CD200 (OX2) antibody | BioLegend | Cat# 123810, RRID:AB_10900447 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Mouse FGF-10 | R&D Systems | Cat# 6224-FG |
| Recombinant Mouse FGF-9 | R&D Systems | Cat# 7399-F9 |
| Recombinant Mouse EGF | R&D Systems | Cat# 2028-EG-200 |
| CHIR99021 | Cayman Chemical | Cat# 13122 |
| BIRB796 | Tocris | Cat# 5989 |
| A83-081 | Tocris | Cat# 2939 |
| Y-27832 dihydrochloride | Sigma Aldrich | Cat# Y0503 |
| Insulin, Human | Sigma Aldrich | Cat# 11376497001 |
| Heparin Sodium Salt | Sigma Aldrich | Cat# H3149 |
| Recombinant Human Transferrin | Peprotech | Cat# 10-366 |
| KGF | Peprotech | Cat# 100-19 |
| SABM Basal Media | Lonza | Cat# CC-3119 |
| Advanced DMEM/F-12 | Invitrogen | Cat# 12634010 |
| Tamoxifen | Sigma | Cat# T5648 |
| Corn Oil | Fisher Scientific | Cat# S25271 |
| 5-Ethynyl-2′-deoxyuridine | Santa Cruz | Cat# sc-284628 |
| Growth Factor Reduced Matrigel | Corning | Cat# 354230 |
| Bleomycin Sulfate | Sigma | Cat# B5507 |
| 4-OHT | Sigma | Cat# H7904 |
| Critical Commercial Assays | ||
| Click-iT™ EdU Imaging Kit | ThermoFisher | Cat# C10086 |
| RNAScope Multiplex Fluorescent Reagent Kit v2 | ACDBio | Cat# 323100 |
| RNA ISH Probe against AW112010 | ACDBio | Cat# 487351 |
| Deposited Data | ||
| Raw and analyzed data | This Paper | GSE129937; GSE130077 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Scgb1a1-CreERT | (Rawlins et al., 2009) | B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J; RRID:IMSR_JAX:016225 |
| Mouse: SPC-CreERT2 | (Chapman et al., 2011) | |
| Mouse: Sox2-CreERT2 | (Arnold et al., 2011) | B6;129S-Sox2tm1(cre/ERT2)Hoch/J; RRID:IMSR_JAX:017593 |
| Mouse: Ai14-tdTomato | (Madisen et al., 2010) | B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J; RRID:IMSR_JAX:007914 |
| Mouse: mTmG | (Muzumdar et al., 2007) | B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J; RRID:IMSR_JAX:007676 |
| Mouse: C57BL/6-Tg(UBC-GFP)30Scha/J | (Schaefer et al., 2001) | RRID:IMSR_JAX:004353 |
| Mouse: C57/Bl6 | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Software and Algorithms | ||
| R 3.4.1 | R Project | https://r-project.org/ |
| Seurat v2.0 | (Butler et al., 2018) | https://satijalab.org/seurat/ |
| Seurat v3.1 | (Stuart et al., 2019) | https://satijalab.org/seurat/ |
| Velocyto | (La Manno et al., 2018) | velocyto.org |
| Ingenuity Pathway Analysis | Qiagen | www.qiagen.com/ingenuity |
| GraphPad Prism 8 | GraphPad | https://graphpad.com |
Highlights.
Supervised scRNA-seq uncovers unexpected heterogeneity within club-like cells
Specialized epithelial progenitors with unique features hide among mature cells
Different stem/progenitors are activated in region and injury-dependent manner
Transplantation of expanded progenitors rescues lung function of injured mice
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01-HL128484 (H.A.C.), U01-HL134766 (H.A.C.), NHLBI_1F32HL143931-01A1 (J.J.K.), and China Scholarship Council (X.D.T.), and DRC Center Grant NIH P30 DK063720 (UCSF Flow Cytometry Core). We thank E. Wan of the Institute for Human Genetics core facility, UCSF, for assistance with scRNA-sequencing.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, and Hochedlinger K (2011). Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BL (2013). Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley C, and Matunis E (2004). Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331–1334. [DOI] [PubMed] [Google Scholar]
- Brockes JP, and Kumar A (2002). Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nat Rev Mol Cell Biol 3, 566–574. [DOI] [PubMed] [Google Scholar]
- Buchweitz JP, Harkema JR, and Kaminski NE (2007). Time-dependent airway epithelial and inflammatory cell responses induced by influenza virus A/PR/8/34 in C57BL/6 mice. Toxicol Pathol 35, 424–435. [DOI] [PubMed] [Google Scholar]
- Campisi J (2016). Cellular Senescence and Lung Function during Aging. Yin and Yang. Ann Am Thorac Soc 13 Suppl 5, S402–S406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, and Vu TH (2011). Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121, 2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J (2017). Origin and regulation of a lung repair kit. Nat Cell Biol 19, 885–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur P, Munjhal A, and Lal SK (2011). Influenza virus and cell signaling pathways. Med Sci Monit 17, RA148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsanathan S, Alder JK, Sellares J, Rojas M, Gurkar AU, and Mora AL (2019). Cellular Senescence: The Trojan Horse in Chronic Lung Diseases. Am J Respir Cell Mol Biol 61, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. (2014). Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T, and Spradling A (2004). Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature 428, 564–569. [DOI] [PubMed] [Google Scholar]
- Kester L, and van Oudenaarden A (2018). Single-Cell Transcriptomics Meets Lineage Tracing. Cell Stem Cell 23, 166–179. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, and Jacks T (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, and Tanaka EM (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. [DOI] [PubMed] [Google Scholar]
- Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, et al. (2011). Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. (2017). Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163 e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, Qin Z, Li Y, Yang R, Pu W, et al. (2019). Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat Genet 51, 728–738. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell AM, Yao C, Yeckes AR, Wang Y, Selvaggio AS, Tang J, Kirsch DG, and Stripp BR (2016). p53 Regulates Progenitor Cell Quiescence and Differentiation in the Airway. Cell Rep 17, 2173–2182. [DOI] [PubMed] [Google Scholar]
- McQualter JL (2019). Endogenous lung stem cells for lung regeneration. Expert Opin Biol Ther, 1–8. [DOI] [PubMed] [Google Scholar]
- McQualter JL, Yuen K, Williams B, and Bertoncello I (2010). Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci U S A 107, 1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, and Desai TJ (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichane M, Javed A, Sivakamasundari V, Ganesan M, Ang LT, Kraus P, Lufkin T, Loh KM, and Lim B (2017). Isolation and 3D expansion of multipotent Sox9(+) mouse lung progenitors. Nat Methods 14, 1205–1212. [DOI] [PubMed] [Google Scholar]
- Ostrin EJ, Little DR, Gerner-Mauro KN, Sumner EA, Rios-Corzo R, Ambrosio E, Holt SE, Forcioli-Conti N, Akiyama H, Hanash SM, et al. (2018). beta-Catenin maintains lung epithelial progenitors after lung specification. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, Morty RE, Vadasz I, Mayer K, Gattenloehner S, et al. (2016). Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. PLoS Pathog 12, e1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, and Hogan BL (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, and Zammit PS (2012). Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development 139, 2845–2856. [DOI] [PubMed] [Google Scholar]
- Salwig I, Spitznagel B, Vazquez-Armendariz AI, Khalooghi K, Guenther S, Herold S, Szibor M, and Braun T (2019). Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer BC, Schaefer ML, Kappler JW, Marrack P, and Kedl RM (2001). Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell Immunol 214, 110–122. [DOI] [PubMed] [Google Scholar]
- Strunz M, Simon LM, Ansari M, Mattner LF, Angelidis I, Mayr CH, Kathiriya J, Yee M, Ogar P, Sengupta A, et al. (2019). Longitudinal single cell transcriptomics reveals Krt8+ alveolar epithelial progenitors in lung regeneration. bioRxiv.
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. (2013). Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AI, J.S., Zhao G, Quansah KK, Farshchian JN, Neupauer KM, Littauer EQ, Paris AJ, Liberti DC, Worthen GS, Morrisey EE, Vaughan AE (2019). Mesenchyme-Free Expansion and Transplantation of Adult Alveolar Progenitor Cells: Steps Toward Cell-Based Regenerative Therapies. npj Reg Med In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, Jackson JR, Xu J, Lee DK, Gotts JE, et al. (2017). Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol 19, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Riccio P, Schotsaert M, Mori M, Lu J, Lee DK, Garcia-Sastre A, Xu J, and Cardoso WV (2018). Spatial-Temporal Lineage Restrictions of Embryonic p63(+) Progenitors Establish Distinct Stem Cell Pools in Adult Airways. Dev Cell 44, 752–761 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, and Brody SL (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 283, L1315–1321. [DOI] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Limmon GV, Yin L, Leung NH, Yu H, Chow VT, and Chen J (2012). Regeneration of alveolar type I and II cells from Scgb1a1-expressing cells following severe pulmonary damage induced by bleomycin and influenza. PLoS One 7, e48451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Soh BS, Yin L, Hu G, Chen Q, Choi H, Han J, Chow VT, and Chen J (2017). Differentiation of Club Cells to Alveolar Epithelial Cells In Vitro. Sci Rep 7, 41661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M et al. (2015). p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.