Abstract
Despite rapid advances in the human healthcare, the infection caused by certain viruses results in high morbidity and mortality accentuate the importance for development of new antivirals. The existing antiviral drugs are limited, due to their inadequate response, increased rate of resistance and several adverse side effects. Therefore, one of the newly emerging field “peptide-based therapeutics” against viruses is being explored and seems promising. Over the last few years, a lot of scientific effort has been made for the identification of novel and potential peptide-based therapeutics using various advanced technologies. Consequently, there are more than 60 approved peptide drugs available for sale in the market of United States, Europe, Japan, and some Asian countries. Moreover, the number of peptide drugs undergoing the clinical trials is rising gradually year by year. The peptide-based antiviral therapeutics have been approved for the Human immunodeficiency virus (HIV), Influenza virus and Hepatitis virus (B and C). This review enlightens the various peptide sources and the different approaches that have contributed to the search of potential antiviral peptides. These include computational approaches, natural and biological sources (library based high throughput screening) for the identification of lead peptide molecules against their target. Further the applications of few advanced techniques based on combinatorial chemistry and molecular biology have been illustrated to measure the binding parameters such as affinity and kinetics of the screened interacting partners. The employment of these advanced techniques can contribute to investigate antiviral peptide therapeutics for emerging infections.
Keywords: Binding evaluation techniques, High throughput screening methods, In silico approaches, Peptide based therapeutics
Introduction
The infections caused by viral pathogens including clinical viruses or naturally emerging viruses pose a serious threat worldwide. Unfortunately, only few therapeutics are available for limited viruses like Human immunodeficiency virus (HIV), Hepatitis virus, Herpes simplex virus (HSV) and Influenza virus (Rider et al. 2011). Researchers are currently working to extend the range of specific and novel antivirals to other families of pathogens. Since, viruses depend on host cell organism for replication, the selection of target for the designing of effective and safe antiviral drugs without harming the host cell, is an extremely difficult process. Besides this, owing to the evolution, mutations occur in the viral genome, which contribute to the development of resistance to drugs and thus rendered many drugs ineffective (Lee et al. 2019). The peptides can block infection by targeting either virus or its host. The virus specific antiviral peptides are known as virucidal, as they directly target the viral proteins. Most of the antivirals have been reported to inhibit the development of viruses by targeting its specific regions or components. Various steps of viral life cycle have been targeted for the discovery of novel antiviral drugs, such as viral entry, viral synthesis, or assembly. Due to the extracellular site of action and blockage of viral infection, the viral entry inhibition is marked as an attractive strategy (Chew et al. 2017). Protein–protein interactions (PPIs) are the foundation of important cellular processes and are considered as primary targets for the drug discovery over the last decade (Lee et al. 2019; Teissier et al. 2011). The knowledge of crucial interactors involved in PPIs and their mechanism is necessary to pave way for the selection of suitable target for drug discovery. New approaches in therapeutics include the use of small cyclic molecules, proteins/peptides, nucleic acids such as small interfering RNA (siRNA) and small hairpin RNA (shRNA) molecules (Teissier et al. 2011). Among these advanced approaches, peptides as therapeutics is a promising field in the drug discovery (Lau and Dunn 2018). Peptides are the biologically active molecules composed of amino acids residues that disrupt the PPIs. They are small (less than 100 amino acids), and they can be easily synthesized. They are also highly specific and effective even in nanomolar range. The main benefit of using peptide as therapeutics is its hydrolysis by peptidases present in the body, which prevents its accumulation in specific organs and minimizes the toxic side effects (Ali et al. 2013). Previously, the pharmaceutical industries have shown poor interest for the expansion of peptide-based therapeutics because of their extremely poor ADME (absorption, distribution, metabolism and elimination) properties. However, the advanced research enables modifications of the peptides such as synthesis of amino acid enantiomers, addition of chemical compounds and their nanoparticle formulation to overcome the pharmacodynamic flaws of peptides (Gentilucci et al. 2010; Zeng et al. 2018). The advantages offered by the modified peptides have sparked the interest amongst the researchers and companies. Now-a-days peptide as therapeutics has come to forte with nearly 20 new peptides added in clinical trials annually. In fact, the global market of peptides as drugs has reached to billion dollars with currently more than 60 peptides approved by US Food and Drug Administration (FDA) and over 400 peptides being under clinical phase trials (Lau and Dunn 2018). Peptide as therapeutics are approved or are being considered for the treatment of diseases such as cancer, diabetes, cardiovascular diseases and even infection caused by few viruses such as HIV, Herpes, Hepatitis and Influenza virus. Thus, over the years peptide-based therapeutics has added a new dimension as the potential antiviral candidate. This review focuses on the types of peptides approaches that can be used for the identification of the lead peptides against the target protein and the selected advanced techniques reported for the validation of the peptide binding affinity to their targets.
Antiviral Peptides as Therapeutics
The peptides possessing potential to inhibit the virus are considered as antiviral peptides (AVPs). Usually, the AVPs exhibit antiviral effects by inhibiting the virus directly, but their inhibition sites and the mechanism of action vary within the viral replication cycle (Rider et al. 2011). The AVPs can be obtained through different approaches: (1) Computational approach (2) Natural sources and (3) Biological source such as High-throughput screening (Fig. 1). There are many online databases available which contain information regarding experimentally tested antiviral peptides such as Antiviral peptide database (AVPdp) (Qureshi et al. 2013) where 2683 entries of peptides including 624 modified AVPs are compiled till December 2019, while many others are unreported. Since the field of peptides as antiviral is not entirely explored, therefore, many research studies are being undertaken to elucidate the role of peptides in blocking viral infections. The first peptide drug approved for clinical indication, Enfuvirtide (Enf), a 36-amino acid residue peptide, against HIV corresponds to the heptad-repeat (HR2) domain of gp41 (HIV envelope protein). Enf prevents the fusion of HR1 domain to HR2 during HIV formation and blocks HIV infection (Teissier et al. 2011). Similarly, Boceprevir and Telaprevir, both synthetic peptides against Hepatitis C virus (HCV), got approval by FDA in 2011. These peptides act on NS3/4, a protease inhibitor, and interfere with viral replication (Divyashree et al. 2020). Other peptide candidates such as Myrcludex for Hepatitis B and D viruses (HBV and HDV) (Bogomolov et al. 2016), Flufirvitide for Influenza virus (Skalickova et al. 2015), Sifuvirtide for HIV-1 (Yu et al. 2018), IM862 and SCV-07 for HCV and Thymosin α-1 for HBV as well as HCV (Jenssen 2009), are under various phase trials of pre-clinical and clinical studies. We have discussed the different approaches used for the identification of AVPs. Moreover, the few selected methodologies used for the validation of identified peptides as potential AVPs have also been described here. The techniques used in identification and validation of peptides are compared in Table 1.
Fig. 1.
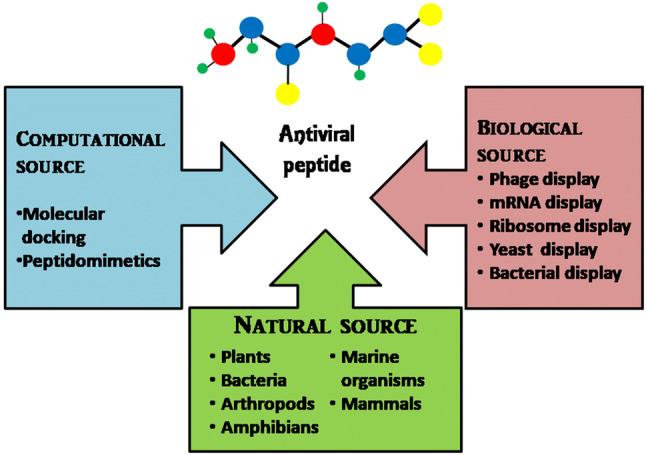
Different sources of antiviral peptides
Table 1.
Comparative analysis of selected techniques applied to identify or validate viral peptides
| S. No | Approach | Technique | Characteristics | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|
| 1 | Computational | Docking | Virtual screening of target based hits |
Expedite the process of drug discovery Size, shape, charge distribution, polarity, hydrogen bonding, and hydrophobic interactions of ligand-receptor complexes can be identified Identification of target sites of the ligand and the receptor molecule Structure–activity based studies |
Requires multiple runs to obtain reliable results Limits the flexibility of receptor Less accurate Docking calculations are complex |
Nevola and Giralt (2015) |
| Peptidomimetics |
Mimics of the natural peptides Prevent the protein–protein/protein-peptide interaction by competitive binding |
Design the mimic of peptide with enhanced bioavailability Overcome the proteolytic instability of natural peptides Improved receptor selectivity |
Offers restricted conformational structures Termini are exchanged with the inversion of their sequence Requires entire understanding of target interaction |
Sen et al. (2019) | ||
| 2 | Biological | Phage display |
Utilizes phages to display foreign protein/peptide Most adopted system |
Rapid identification of target specific phages System suitable for delivering small peptides (< 20 aa) High throughput screening approach Selection of disease specific antigen mimics Selection of organ specific peptides Used in B-cell and T-cell epitope mapping |
Library size is limited by phage transformation efficiency (~ 109) Allows only natural amino acids Complicated affinity maturation process because of large diversity of proteins/peptides displayed on surface Limits the rapidity of library generation Restricts the size of expressed proteins Limits the intractability of some targets |
Matsubara (2012) and Fukunaga and Taki (2012) |
| m-RNA display |
Uses the transcription and translation machinery extracted from prokaryotic/eukaryotic cells Utilizes covalent mRNA-polypeptide complexes linked through puromycin |
Library size as large as (~ 1012–15) Protein expression free from cellular constraints Allows the incorporation of unnatural amino acids Greater diversity as transformation is not required Increased probability of higher affinity hits |
Solid surface based biopanning often results in non specific binding Likely to interfere with other molecules due to single stranded form of m-RNA Display larger proteins (> 300 aa) with lower efficiency Not suitable for displaying membrane bound proteins Ribonuclease free environment is required |
Wang and Liu (2011) | ||
| Ribosome display |
Living cell free technology Utilizes non-covalent ribosome-mRNA-polypeptide complexes for display system |
Library size as large as (~ 1012–15) Greater diversity due to not being dependent on transformation efficiency Increased probability of higher affinity hits Suitable for generating toxic, proteolytically sensitive and unstable proteins Random mutations can be introduced Allows the incorporation of unnatural amino acids |
Selection stringency is limited System is very sensitive to RNase activity Ribonuclease free environment is required |
Dreier and Pluckthun 2011 | ||
| Yeast display |
Proteins/peptides are usually fused to Aga2 protein subunit of yeast Can be displayed as N- or C-terminal fusion |
Displays fully nativemonomeric as well as oligomeric proteins on their surface due to eukaryotic machinery More diverse library as compared to phage system |
Library size is smaller (~ 106–7) than other display systems Allows only natural amino acids Allows the expression of extracellular proteins Complicated affinity maturation process because of large diversity of proteins/peptides displayed on surface |
Linciano et al. (2019) | ||
| 3 | Advanced Techniques | Surface Plasmon resonance (SPR) |
Label-free binding technique Quantitatively analyzes the real time binding kinetics of two bimolecular interactions |
Label-free detection Real-time data monitoring Sensitive and accurate Small sample quantity Ability to Handle Complex Samples Ability to Replicate Measurements |
Expensive instrument and sensors Expensive maintenance Low throughput |
Patching (2014) |
| Biolayer interferometry |
Optical analytical technique Quantify real time binding kinetics of bimolecular interactions |
Label-free detection Real-time data monitoring No reference channel required Crude sample compatibility System requires less maintenance High throughput Low vibrational/mechanical noise Simple, fast and more accurate results |
Requires immobilization of ligand to surface of tip No temperature control Low sensitivity as compared to SPR Poor reproducibility Relatively high sample consumption Results should be cross-validated with SPR |
Shah and Duncan (2014) | ||
| Fluorescence Resonance Energy Transfer (FRET) |
Radiation less transmission of energy from donor to acceptor Distance-dependant |
Simple, sensitive and easily automated Non-radioactive High time resolution High throughput Low reagent consumption |
Requires fluorophore labeled molecules Distance of donor–acceptor pair is limited (< 10 nm) |
Rogers et al. (2012) |
Antiviral Peptides Designed from Computational Approach
Computer assisted drug designing process is based on the understanding of structural and functional aspects of the viral machinery. The rational knowledge of the viral proteins and the interactors/cellular partners assists in the selection of target protein. This approach has expedited the process of drug designing. Peptides can be identified computationally; via in silico screening using molecular docking. A docking program predicts the target site which is usually known as pocket or protrusion with hydrogen bond donors and acceptors, hydrophobic characteristics and different molecular shapes. Subsequently, library of peptides docked with these pockets results in the highest binding peptide (Nevola and Giralt 2015). For example, peptides have been designed computationally using virtual docking against the surface protein of Zika virus (ZIKV) for its detection. Tetra, penta, hexa and heptapeptide libraries were docked using Open Eye Scientific Software against envelope protein of ZIKV; subsequently, eight peptides were selected. They were further tested by Direct ELISA and out of them, three were delineated with best performance for Zika detection (Mascini et al. 2019). Another method is based on peptidomimetics which mimics the designed targets, prevents the interaction of proteins by competitive binding. Four putative peptide inhibitors were designed against Nipah virus (NiV) proteins using the approach of peptidomimetics and the stability of peptide-protein complexes were analysed using MD simulation (Sen et al. 2019). The in silico methods (Docking and peptidomimetics) can predict the peptide sequence but further validation using in vitro/in vivo approaches is required to establish its biological activity.
Antiviral Peptides Derived from Natural Source
The peptides that exhibit immunomodulatory and inhibitory properties against infection caused by bacteria, fungi, viruses or protozoa, are expressed as naturally occurring antimicrobial peptides (AMPs). The AMPs have been extensively used as substitutes of antibiotics for bacterial infections, but recently their use has been expanded to antiviral therapeutics also (Ahmed et al. 2019a). The naturally occurring AVPs are amphipathic and cationic in nature with net positive charge (Bulet et al. 2004). Moreover, it has been proven that the hydrophobicity of the peptides is an essential property for targeting enveloped viruses (Badani et al. 2014; Wang et al. 2017). These AVPs can be derived from different sources such as plants, bacteria, arthropods, amphibians, marine organisms and mammals with their varied mechanism of action (Zhang and Gallo 2016). A peptide family called cyclotides derived from different plant sources has been proven successfully for their antiviral activity against HIV, Influenza virus and Dengue virus (Ireland et al. 2008; Sencanski et al. 2015; Gao et al. 2010). The small size, cationic and amphipathic nature of the cyclotides facilitates its effective binding to the target and rupture the membrane. This allows the leakage of cell components which further leads to the cell death (Weidmann and Craik 2016). In a study, kalata B1, a cyclotide isolated from the leaves of Oldenlandia affins plant, showed destruction of viral particle at entry step along with inhibition of viral-host membrane fusion, thus exhibiting anti-HIV activity (Henriques et al. 2011). Similarly, a peptide derived from arthropod, Hyalophora ceecropia, known as cecropin A showed the inhibitory activity against HIV, Junin virus (JUNV) and HSV by suppression of their gene expression (Wachinger et al. 1998; Albiol Matanic and Castilla 2004). Moreover, in recent studies bovine lactoferrin (bLF) has showed the antiviral activity against three Aedes mosquito transmitted viruses: Dengue (DENV), Chikungunya (CHIKV) and Zika virus apart from anti-HBV activity established in a previous study (Li et al. 2009; Carvalho et al. 2017; Chen et al. 2017). The bLF blocks the viral binding to its target site and thus prevents its spread to host cells. Many other AVPs originated from natural source are summarized in Table 2 with their varied mode of action. However, despite promising efficacy, the utility of these peptides is constrained due to weak binding, low stability, other side effects and virus resistance. The shortcomings of AVPs from natural resource need to be addressed so that they can be considered as mainstay antiviral therapeutics.
Table 2.
Selected AVPs derived from natural sources and their mode of action against virus
| S. No | Peptide | Source | Characteristics | Targeted virus | Mode of action/ activity | Reference |
|---|---|---|---|---|---|---|
| I | Plant | |||||
| 1 | 2 kDa peptide | Seeds of Sorghum bicolor | 2 kDa, cationic and amphipathic peptide | HSV-1 | Inhibition of cell entry by masking essential viral envelope proteins | Camargo et al. (2008) |
| 2 | Cyclotide: vhl-1 | Leaves of Viola hederaceae | 31 amino acid cyclic cystine knot like peptide | HIV | Inhibition of viral fusion by disrupting the lipid envelope | Wang et al. (2008) |
| 3 | Cycloviolacin VY1 | Viola yedoensis | 31 amino acid peptide, three conserved disulphide bonds, bridge like 3-D structure | Influenza H1N1 | Inhibition of virus at cell entry | Liu et al. (2014) |
| 4 | Kalata B1 | Leaves of Oldenlandia affinis | Cyclic backbone, knot-like rigid structure, three conserved disulphide bonds | HIV | Inhibition of viral-host membrane fusion | Henriques et al. (2011) |
| 5 | Kalata B1-inspired peptide | Derivative of Kalata B1 peptide | Amino acid modifications in kalata B1peptide | DENV | Inhibition of viral replication | Gao et al. (2010) |
| 6 | Peptides 2 and 4 | Acacia catechu | 15 amino acids, cationic and amphipathic peptide | DENV | Inhibition at early steps of viral entry | Panya et al. (2019) |
| 7 | Sesquin |
Seeds of Vigna sesquipedalis |
7 kDa, cationic, defensin like peptide | HIV | Inhibition of viral replication by hindering reverse transcriptase activity | Jack and Tzi (2005) |
| II | Bacteria | |||||
| 8 | Locillomycin | Bacillus subtilis | Cyclic lipononapeptide | PEDV | unknown | Luo et al. (2015) |
| 9 | Surfactin | Bacillus subtilis | Cyclic lipopeptide, amphiphilic | HIV, HSV-1, HSV-2, VSV, SIV, NDV, PEDV | Disintegration of lipid envelope and, capsid through ion channel formations | Yuan et al. (2019) |
| III | Arthropod | |||||
| 10 | Alloferon 1 and 2 | Hemolymph of blowfly: Calliphora vicina | Linear, nonglycosylated oligopeptide of 13 and 12 amino acid residues, respectively | Influenza virus | Inhibition by activation of natural killer cells and release of interferon | Chernysh et al. (2002) |
| HSV-1 | Inhibition of viral replication | |||||
| 11 | Alloferon 1-derived peptide | Hemolymph of blowfly: Calliphora vicina | Modifications in their N-terminal portions of Alloferon1 peptide | CBV-2 | Inhibition of viral replication | Kuczer et al. (2010) |
| 12 | Bmkn2-7 | Venom of scorpion: Mesobuthus martensii | 13-amino acid residues, basic, alpha-helical peptide | HIV-1 | Inhibition by direct interaction with viral particle | Chen et al. (2012) |
| 13 | Cecropin A | Moth Hyalophora cecropia | 37-amino acid linear, cationic peptide | HIV; HSV-1 and 2; JUNV | Suppression of viral gene expression | Wachinger et al. (1998) and Hultmark et al. (2005) |
| 14 | Ctry2459 | Chaerilus tryznai | 13-amino-acid residue, helical and amphipathic peptide | HCV | Inhibition by inactivating the viral particles, virucidal activities and suppressed the established infection at cellular level | Hong et al. (2013) |
| 15 | Ctry2459-H2 | Histidine-rich Ctry2459 peptide | ||||
| 16 | Ctry2469-H3 | Histidine-rich Ctry2459 peptide | ||||
| 17 | Eva1418 | Venom of scorpion: Euscorpiops validus | Helical and amphipathic peptide | HSV-1 | Inhibition by disruption of initial steps of infection | Zeng et al. (2018) |
| 18 | Eva1418-FH5 | Histidine rich derivative of Eva1418 peptide | Enhanced inhibition activity with lowest cytotoxicity | |||
| 19 | Hp1090 | Venom of Scorpion: Heterometrus petersii | Amphipathic α-helical peptide | HCV | Inhibition of viral replication | Yan et al. (2011) |
| 20 | Hp1239 | Venom of Scorpion: Heterometrus petersii | Amphipathic α-helical peptide | HSV-1 | Inhibition of cell entry by blocking viral-host membrane fusion | Hong et al. (2014) |
| 21 | Hp1036 | |||||
| 22 | Lactarcin 1 | Venom of spider: Lachesana tarabaeve | Amphipathic α-helical peptide | DENV | Inhibition of viral replication by binding to viral protease | Rothan et al. (2014) |
| 23 | Mastoparan | Venom of wasp: Vespula lewisii | 14 amino acid residues, cationic, amphipathic α-helical peptide | VSV, HSV-1, flaviviruses | Inhibition of cell entry by disruption of envelope | Moreno and Giralt (2015) |
| 24 | Mastoparan 7 | Venom of wasp: Vespula lewisii | Derivative of Mastoparan peptide | VSV | Inhibition of cell entry by disruption of envelope | Sample et al. (2013) |
| 25 | Melittin | Venom of bee: Apis mellifera | 26 amino acid linear cytolytic peptide with no disulfide bridge | HIV-1; HSV-1 and 2; JUNV | Inhibition of cell entry by disruption of envelope | Albiol Matanic and Castilla (2004), Galdiero et al. (2013), and Hood et al. (2013) |
| 26 | Mucroporin-M1 | Scorpion: Lychas mucronatus | Cationic host defense peptide | MeV, Influenza-H5N1; SARS-CoV; HIV-1 | Virucidal activity | Li et al. (2011) |
| HBV | Inhibition of viral replication by decreasing expression of important HBV replication factors | Zhao et al. (2012) | ||||
| IV | Amphibians | |||||
| 27 |
Dermaseptins S3, S4 |
Frogs of Phyllomedusa genus | cationic, amphipathic and α-helical peptide | HSV-1 and 2, HIV | Inhibition at cell entry step by targeting viral envelopes | Lorin et al. (2005) |
| 28 | Dermaseptin derived peptide | Frogs of Phyllomedusa genus | cationic, amphipathic and α-helical peptide | Rabies virus |
Inhibition at cell entry step Virucidal mechanism of action |
Bergaoui et al. (2013) |
| 29 | HS-1 | Skin of Anuran: Hypsiboas semilineatus | cationic, amphipathic and α-helical peptide | DENV 2 and 3 | Inhibition at cell entry step by targeting viral envelopes | Monteiro et al. (2018) |
| 30 | Magainin I and II | Frog: Xenopus laevis | 23 amino acid residues, lysine-rich, cationic, amphipathic and α-helical peptide | HSV-1 and 2 | Inhibition at cell entry step by disrupting the structure of viral envelope proteins; virucidal activity | AlbiolMatanic and Castilla (2004) |
| 31 | Magainin-II derived peptide | Frog: Xenopus laevis | alanine-substituted magainin-2 amide along with three other peptides | VV | Virucidal activity by attacking its envelope | |
| 32 | Temporin B | Frog: Rana temporaria | 10–50 amino acids, cationic, amphipathic and α-helical peptide | HSV-1 | Inhibition at entry step by interfering in cell-to-cell spread of the virus | |
| 33 | Urumin | Indian frog: Hydrophylax bahuvistara | 27-residues, cationic, amphipathic and α-helical peptide | Influenza-H1N1 and H1N2 | Inhibition at cell entry step by targeting cell receptors | Holthausen et al. (2017) |
| V | Marine organisms | |||||
| 34 | Callipeltin A | Callipeltin sp. | Cyclic depsidecapeptide | HIV-1 | Inhibition of virion entry | Zampella et al. (1996) |
| 35 | Celebeside A-C | Siliquariaspongia mirabilis | Cyclic depsipeptides | HIV-1 | Inhibition of virion entry | Plaza et al. (2009) |
| 36 | Clavanin A | Tunicate: Styelaclava | 23 residues alpha-helical peptide with amidated C-terminal |
Rotavirus Denovirus |
Inhibition by interfering with viral membranes | Carriel-Gomes et al. (2007) |
| 37 | Homophymine A | Homophymia sp. | Cyclic depsipeptide | HIV-1 | Inhibition of virion entry | Zampella et al. (2008) |
| 38 | Koshikamides F | Theonella swinhoei | 17-residue cyclic depsipeptides | HIV-1 | Inhibition by blocking HIV entry into T cells | Plaza et al. (2010) |
| 39 | Koshikamides H | Theonella cupola | 17-residue cyclic depsipeptides | HIV-1 | Inhibition by blocking HIV entry into T cells | |
| 40 | LvHcL48 | Hemocyanin of shrimp: Litopenaeus vannamei | 79 amino acid fragment | WSSV | Inhibition of the transcription and proliferation possibly by binding to the viral envelope protein | Zhan et al. (2018) |
| 41 | Microspinosamide | Sidonops microspinosa | cyclic depsipeptide incorporating 13 amino acid residues | HIV | Inhibition of cytopathic effects of the infection | Rashid et al. (2001) |
| 42 | Mirabamide A, C and D | Siliquaria spongia mirabilis | Cyclic depsipeptides | HIV-1 | Inhibition at the early stages of virus entry | Plaza et al. (2007) |
| 43 |
Mirabamides E, F, G, and H |
Sponge: Stelletta clavosa | Cyclic depsipeptides | HIV-1 | Inhibition at entry step by disruption of viral membrane fusion | Lu et al. (2011) |
| 44 | Mollamides B | Tunicate: Didemmummolle | Cyclic hexapeptide | HIV | unknown | Donia et al. (2008) |
| 45 | Mutremdamide A | Theonella swinhoei | Sulfated cyclic depsipeptide | HIV-1 | Inhibition by blocking HIV entry into T cells | Plaza et al. (2010 |
| 46 | Neamphamide A | Neamphius huxleyi | Cyclic depsipeptide | HIV-1 | Inhibition of virion entry | Oku et al. (2004) |
| 47 | Nkl71-100 | Turbot: Scophthalmus maximus | five-helix bundled structure stabilized by three intra chain disulphide bonds | SVC | Inhibition by not binding of viral particles to host cells and fusion of virus and cell membranes | Falco et al. (2019) |
| 48 | Papuamide A | Tunicate: Didemmum molle | Cyclic depsipeptides | HIV | virucidal mechanism | Andjelic et al. (2008) |
| 49 | Piscidin 1 | Mast cells of hybrid Striped bass (fish) | 22 amino acid, α–helical and amphipathic peptide | PRV | Inhibition by direct interaction with virus | Hu et al. (2019) |
| 50 | Pa-MAP 1 | Polar fish: Pleunorectus americanus | an alanine-rich α-helix peptide composed of eleven amino acid residues with three imperfect motif repetitions | HSV-1 and 2 | Virucidal mechanism of action, Inhibition at entry step by interacting viral surface glycoprotein | Migliolo et al. (2012) |
| 51 | P34 | Intestinal contents of Leporinus sp. (fish) | Anionic, thermostable, hydrophobic, lipidic peptide | EAV, FHV-1 | Virucidal activity | Castro et al. (2014) |
| BoHV-1 | Inhibition of the viral penetration | Castro et al. (2017) | ||||
| 52 | Stellettapeptin A and B | Stelletta clavosa | Cyclic and nonribosomal depsipeptides | HIV-1 | Inhibition of cytopathic effects of the infection | Shin et al. (2015) |
| 53 | TheopapuamideA | Theonella swinhoei | Cyclic depsipeptides | HIV-1 | Inhibition of virion entry | Andjelic et al. (2008) |
| 54 | Theopapuamide B-D | Siliquariaspongia mirabilis | undecapeptides with an N-terminal fatty acid moiety | HIV-1 | Inhibition by disruption of viral membrane | Plaza et al. (2009) |
| VI | Mammals | |||||
| 55 | α-Defensin HNPs 1, 2 and 4 | Human neutrophil |
18 to 45 amino acid residues cationic charge, amphipathic properties and predominance of β sheets stabilized by three disulfide bonds |
HIV-1 | Inhibition at cell entry step | Wu et al. (2005) |
| 56 | α-Defensin HNPs 1 | Human neutrophil | Influenza A | Inhibition of viral replication | Salvatore et al. (2007) | |
| 57 | β-defensins hDB-2 | Epithelial cells | cysteine-rich, cationic peptides | HPV; VZV; HIV | Inhibition at cell entry as well as viral replication by late reverse transcripts and nuclear import | Meyer-Hoffert et al. (2008) and Crack et al. (2012) |
| 58 | β-defensins hBD-3 | Epithelial cells | HPV; VV; VZV; HIV | Inhibition of viral replication | Quinones-Mateu et al. (2003), Howell et al. (2007), Gwyer Findlay et al. (2013) | |
| 59 | Cathelicidin LL-37 | Human neutrophil granules | 12 to 88 amino acid residues, cationic, α-helical and amphipathic peptide | VZV; VV; HSV-1; HIV; RSV; Influenza A; HCV; DENV; ZIKV; VEEV | Inhibition of cell entry by disruption of envelope | Sørensen et al. (2001), Barlow et al. (2011), Tripathi et al. (2015), Matsumura et al. (2016), Alagarasu et al. (2017) |
| Adenovirus; Aichi virus; Rhinovirus | Inhibition of cell entry | Gordon et al. (2005), Sousa et al. (2017), Ahmed et al. (2019b) | ||||
| 60 | CYVIP | Human hemofiltrate | 71-amino-acid, cationic peptide |
HCMV HSV-1 |
Inhibition of cell entry by interacting the host cell receptors | Borst et al. (2013) |
| 61 | Indolicidin | Bovine neutrophils | Tridecapeptide amide | HIV | Inhibition by membrane-disruption | Robinson et al. (1998) |
| 62 | Lactoferrin | Mammals’ milk | Hydrophobic, cationic, and helical peptide | CMV; HSV-1and 2; Adenovirus; Rotavirus; Poliovirus; RSV; HIV; Influenza; HCV; HBV DENV; CHIKV; ZIKV | Inhibition at cell entry as well as viral replication | Van der Strate et al. (2001), Li et al. (2009), Carvalho et al. (2017), Chen et al. (2017) |
| 63 | Lactoferricin | Derivative of lactoferrin | Amphipathic, cationic peptide corresponds to lactoferrin fragment 17–41 | CMV, HIV-1, HPV | Inhibition at cell entry step | Andersen et al. (2001), Mistry et al. (2007), Li et al. (2009), Wang et al. (2016) |
| HSV-1and 2 | Inhibition of viral replication by interfering the host cell microtubules | Marr et al. (2009) | ||||
| 64 | Protegrin-1 | White blood cells of swine | 18 amino acid residues, cyclical, β -sheets and cationic | DENV | Inhibition of viral replication by binding to viral protease | Rothan et al. (2012) |
Human immunodeficiency virus: HIV; Dengue virus: DENV; Herpes simplex virus 1 and 2: HSV-1 and HSV-2; Porcine epidemic diarrhea virus: PEDV; Vesicular stomatitis virus: VSV; Simian immunodeficiency virus: SIV; Newcastle disease virus: NDV; Coxsackie virus B2: CBV-2; Junin virus: JUNV; Hepatitis C virus: HCV; Measles morbillivirus: MeV; Severe acute respiratory syndrome coronavirus: SARS-CoV; Hepatitis B virus: HBV; Vaccinia virus: VV; White spot syndrome virus: WSSV; Carp sprivivirus: SVC; Pseudorabies virus: PRV; Equine arteritis virus: EAV; Feline herpes virus type-1: FHV-1; Bovine herpesvirus1: BoHV-1; Human papillomavirus: HPV; Varicella zoster virus: VZV; Respiratory syncytial virus: RSV; Zika virus: ZIKV; Venezuelan equine encephalitis virus: VEEV; Human cytomegalovirus: HCMV; Cytomegalovirus: CMV; Chikungunya virus: CHIKV
Antiviral Peptides Identified Through Biological Approach
Methodologies based on in vitro display approach, usually offer genetically encoded peptides with superior quality and high affinity to their targets. Among these methodologies, phage display, mRNA display, ribosome display, yeast display and bacteria display are the most common technologies to generate peptides. The phage display technology is widely used and considered as the most appropriate for the screening of high efficiency peptides. While these technologies have already been thoroughly illustrated in other reviews (Nevola and Giralt 2015), we have focused on the use of selected methodologies for the identification of antiviral peptides and compared them in Table 1.
Phage Display
Phage display technology is an efficient in vitro screening method for the selection of high affinity and target specific peptide binder from a randomly displayed peptide library. The technology involves the fusion of exogenous peptide sequence into the genome of phage, its expression on the surface as fusion product to phage surface protein. The phage displayed libraries thus constructed have 109–10 variants at a time. In this method, biopanning is performed in which the target molecule is immobilized on surface and incubated with phage library. The unbound or excess of phage particles are removed by washing and potentially bound phages are eluted by acidic/basic buffer or with appropriate ligand. These recovered phages are amplified by infecting bacterial cells Escherichia coli and are used for subsequent rounds of biopanning to obtain target specific phages using affinity selection. The sequencing of DNA isolated from binding phage, validated by ELISA, helps to identify peptide sequence (Fukunaga and Taki 2012; Matsubara 2012). A peptide named P3 against Japanese encephalitis virus (JEV) host fusion has been identified as the potential AVP using phage display library. The screened peptide has shown the highest affinity to domain III of JEV envelope glycoprotein assessed by Biolayer interferometry and IC90 of ~ 100 µM and IC50 of ~ 1 µM in JEV infected BHK-21 cells (Wei et al. 2019). Similarly, an analogous study conducted by de la Guardia et al. (2017) identified three peptides against the domain III of DENV envelope protein to block the DENV infection. Further these peptides were non-toxic to the target cells. Moreover, the same approach has also been used to identify peptides targeting non-structural viral protein: RNA-dependent RNA polymerase (NS5B) of HCV, by screening a library composed of disulfide-constrained heptapeptides (Amin et al. 2003). In another study, a novel heptapeptide was identified using random peptide phage library which inhibited the integration of HIV genome into the host (Desjobert et al. 2004). The most important advantage of this technology over others is its high rate of mutability with affinity selection, which widely employs the screening of phage displayed peptides for identification of potential AVPs. There are many other AVPs derived from the utilization of phage display technology which are summarised in Table 3.
Table 3.
Characteristics of the AVPs derived from Phage display technology
| S. No | Peptide sequence | Library used | Targeted virus | Targeted protein | References |
|---|---|---|---|---|---|
| 1 | GSHHRHVHSPFV | 12-mer peptide library: New England Biolabs (NEB) | Avian infectious bronchitis virus | Purified whole virus | Peng et al. (2006) |
| 2 | HAWDPIPARDPF | 12-mer peptide library (NEB) | Avian influenza A virus-subtype H5N1 | H5N1 viruses | Wu et al. (2011) |
| 3 | AAWHLIVALAPN | ||||
| 4 | ATSHLHVRLPSK | ||||
| 5 | NDFRSKT | 7-mer disulfide constrained peptide library (NEB) | Avian influenza virus H9N2 | AIV sub-type H9N2 virus particles | Rajik et al. (2009) |
| 6 | HSIRYDF | 7-mer peptide Library (NEB) | Bovine ephemeral fever virus | Neutralization site 1 of glycoprotein: G1 | Hou et al. (2018) |
| 7 | YSLRSDY | ||||
| 8 | DRATSSNA | Octapeptides peptide library | Classical swine fever virus | Envelope protein: E2 | Yin et al. (2014) |
| 9 | SYQSHYY | 7-mer peptide Library (NEB) | Dengue virus | Recombinant dengue envelope protein and its domain III | de la Guardia et al. (2017) |
| 10 | STSFWIT | ||||
| 11 | ELLASPW | ||||
| 12 | CWSFFSNIC | 7-mer disulfide constrained peptide library (NEB) | Hepatitis B virus | Full-length HBcAg | Ho et al. (2003) |
| 13 | KHMHWHPPALNT | 12-mer peptide library (NEB) | Hepatitis B virus | PreS1 region of L-protein | Wang et al. (2011) |
| 14 | WTDMFTAWWSTP | M13-based 12-mer peptide library | Hepatitis B virus | Thio-PreS | Deng et al. (2007) |
| 15 | FPWGNTW | 7-mer disulfide constrained peptide library (NEB) | Hepatitis C virus | NS5B (del 21-His) protein | Amin et al. (2003) |
| 16 | ATWVCGPCT | Phage-displayed nonapeptide library (PVIII9aa) | Hepatitis C virus | mAb JS-81 against CD81 | Cao et al. (2007) |
| 17 | WPWHNHR | heptapeptide M13 phage-display library | Hepatitis C virus | Truncated envelope protein E2 | Lu et al. (2014) |
| 18 | RINNIPWSEAMM | libraries of random 12-mers, 7-mers, and cyclic 9-mers | Human immunodeficiency virus | Envelope glycoprotein gp120 | Ferrer and Harrison (1999) |
| 19 | VSWPELYKWTWS | 7-mer disulfide constrained peptide library; 12-mer peptide library (NEB) | Human immunodeficiency virus | mAb VRC01 | Chikaev et al. (2015) |
| 20 | FHNHGKQ | 7-mer peptide library (NEB) | Human immunodeficiency virus | HIV-1 Integrase | Desjobert et al. (2004) |
| 21 | GWWYKGRARPVSAVA | Pentadecapeptides peptide library | Influenza virus A | Monolayer of the ganglioside:GM3 | Matsubara et al. (2009) |
| 22 | RAVWRHSVATPSHSV | ||||
| 23 | SENRKVPFYSHS | 12-mer peptide library (NEB) | Japanese encephalitis virus | Domain III of the virus envelope glycoprotein | Zu et al. (2014) |
| 24 | TPDCTRWWCPLT | 12-mer peptide library (NEB | Japanese encephalitis virus | E protein | Wei et al. (2019) |
| 25 | RLNNRARIILRA | 12-mer peptide library (NEB) | Mink enteritis virus | Purified whole virus | Zhang et al. (2012) |
| 26 | LAHKSRLYERHM | ||||
| 27 | CTLTTKLYC | 7-mer disulfide constrained peptide library (NEB) | Newcastle disease virus | Inactivated whole virus | Ramanujam et al. (2002) |
| 28 | EVSHPKVG | Heptapeptide library-pSKAN8-HyA library | Newcastle disease virus | Inactivated whole virus | Ozawa et al. (2005) |
| 29 | SGGSNRSP | ||||
| 30 | WVTTSNQW | ||||
| 31 | IQTAFNQGA | 7-mer disulfide constrained peptide library (NEB) | Porcine reproductive and respiratory syndrome virus | mAb N3H2 against nucleocapsid protein | Liu et al. (2012) |
| 32 | HRILMRIR | 12-mer peptide library (NEB) | Porcine reproductive and respiratory syndrome virus | ORF1b | An et al. (2005) |
| 33 | CHWMFSPWC |
Random heptapeptide library flanked by cysteines |
Puumala orthohantavirus | Inactivated whole virus | Heiskanen et al. (1997) |
| 34 | TATTEK | 12-mer peptide library (NEB) | West Nile virus | Non-structural protein 1 | Sun et al. (2011) |
| 35 | VVDGPETKEC | ||||
| 36 | P9 peptide | Peptide library (Spring Bioscience) | West Nile virus | Recombinant E protein | Bai et al. (2007) |
mRNA Display
mRNA display technology utilizes the covalently bonded mRNA-polypeptide complexes formed during in vitro translation, which are linked through puromycin (an analogue of the 3′tyrosyl-tRNA along with mimics of adenosine and tyrosine) via A- site of ribosome. The complexes with desired functions are allowed to bind to the immobilized target protein, reverse transcribed to cDNA and amplified via Polymerase chain reaction (PCR). This enables the reinforcement of DNA template library for next round of screening (Cotten et al. 2012; Newton et al. 2019). The most successful use of mRNA displayed peptide library was described by Litovchick and Szostal (2008), in which they have screened potential AVPs using cyclic peptide-mRNA fusion library targeting Internal ribosomal entry site (IRES) of HCV for inhibition of virions. Another use of this technology was reported for the reverse engineering of peptide vaccines for HCV. High affinity peptides to neutralizing monoclonal antibodies (mAbs) of HCV were selected in this study and used for peptide-based vaccine development (Guo et al. 2015).
Ribosome Display
Ribosome display is an entirely in vitro and cell-free system which makes it efficient in comparison to other display systems (Nevola and Giralt 2015). In this system, the coupling of genotype and phenotype is essential for the selection of high affinity peptides from their pool. During in vitro translation, the association between the mRNA, ribosome and the nascent polypeptide leads to a stabilized protein-ribosome-mRNA complex. This ternary complex is feasible due to the presence of spacer sequence, without stop codon, inserted into the DNA library coding for proteins/ peptides. The spacer ensures that the peptide folds properly and stays attached to the mRNA and ribosomes. These specific ribosomal complexes that display folded peptides are then allowed to bind to the immobilized target and the non-specific ones are washed off. The mRNA complexes having bound polypeptide chains are recovered and their sequences are obtained (Zahnd et al. 2007). A large library that contains 1013–1014 clones can be screened as it is not dependent on the living cell system and is free of any bias. This technology has numerous advantages above others as the diversity of library depends on the number of available ribosomes and mRNA in the system rather restricted by the bacterial transformation efficiency (Dreier and Pluckthun 2011). Moreover, such system allows insertion of random mutations at any round of selection since library has not been transformed after any diversification step. The ribosomal display technology has opened a new insight for using peptide inhibitors for early diagnostic as well as therapeutic agent. For instance, the peptide inhibitor against envelope protein E2 of HCV was identified using ribosomal display library. After extensive selection of 13 rounds, 12-mer peptides were generated. This peptide named PE2D has not only being verified to bind E2 protein but also blocks the virus entry inside hepatocyte cells (Chen et al. 2010).
Yeast Display
The main advantage of the yeast display system over the others is the complete exposure of the peptides/protein for fusion and its compatibility with the fluorescence-activated cell sorting (FACS), which enables the high-throughput screening and characterization of protein/peptide combinatorial libraries (Linciano et al. 2019). Moreover, it also allows the expression of proteins with post translational modifications which has encountered the problem of misfolding in the field of antibody engineering (Mei et al. 2017). Saccharomyces cerevisiae strain based on the Aga1–Aga2 proteins is the most widely used display system. In this system, the protein/peptide is displayed either as N- or C-terminal fusion to the Aga2 protein of yeast cell, which is linked to the Aga1 via disulphide bonds. Every yeast cell exhibits ~ 104–105 copies of the Aga2 fusion protein/peptide on its surface though the expression of individuals may vary. The construct of yeast cell also contains two epitope tags at the N and C terminus of Aga2 fusion protein, which facilitates the real time quantification of their expression using flow cytometry. Moreover, the tags also enable to estimate and quantify the binding of the target via different labelling approaches (Linciano et al. 2019). Though yeast display system is a valuable platform for screening purpose, however, the library size is restricted due to limited transformation efficiency of yeast. Another major limitation of yeast display is complicated affinity maturation process in comparison to other systems. Besides, these drawbacks, this technology has provided a wide application of high throughput screening in peptide engineering and a platform to study protein–protein/peptide interactions in vivo. In a recent study, this technology was used to screen the hits from a grafted C-peptide library of HIV gp41 against N-peptide trimer of HIVgp41. As a result, four hits suppressed the HIV entry better than others (Tennyson et al. 2018).
Application of Advanced Techniques to Validate Identified Peptides
Another challenging task in the intervention of antiviral peptides is the corroboration of the binding of selected peptides to the target protein, and their antiviral efficacy. Various techniques have been developed to evaluate the PPIs in vitro/in vivo such as Surface Plasmon Resonance (SPR), Optical based Biolayer interferometry, Fluorescence Resonance Energy Transfer (FRET), Nuclear Magnetic Resonance (NMR), Isothermal Calorimetry (ITC), yeast two hybrid display, microscopic visualization and many more. Some of them can be used for the binding evaluation of the peptides to their target proteins. Since various reviews and reports are available on PPIs detection methods in detail (Nevola and Giralt 2015), this review focuses only on the recent techniques used to determine/ validate the peptide binding efficiency (Table 1).
Surface Plasmon Resonance (SPR)
SPR is an optical based detection and label-free technique which utilizes the protein in small amount for the real time quantification and evaluation of the binding affinity as well as kinetics between peptide and target protein (Patching 2014). The binding affinity between interacting partners is measured via small variation in the refractive index at sensor surface. This response change is calculated as the change in the angle of resonance of refracted light when flowing analyte binds to the immobilized ligand. The change in the angle of resonance is measured in the form of resonance unit (RU), where 1RU is equivalent to the 10–4 deg/10–12 gmm−2 angle shift. It has become the gold standard in research, typically characterizes the interaction between two molecules in which one is in mobile state and the other is fixed on a gold film. This technique can be used to screen the library of molecules for their binding affinity against a single soluble protein which is immobilized on the sensor surface (Tang et al. 2010). Thus, SPR has emerged as a powerful technique in therapeutic intervention. It can also be adapted to study the interactions involving complicated proteins in situ, such as, membrane-bound proteins, ion channels and other growth, immune and cellular receptors, which are considered as potential targets for drug discovery (Patching 2014). Bai et al. (2007) have investigated the affinity interaction of screened peptides to the Envelope protein of West Nile virus (WNV) using SPR, in which they found peptide P9 to have the highest affinity to the target. Besides this, in another study, the binding of Helix-A peptide to the neuronal microtubules (MTs): β-tubulin was determined by SPR. Helix-A peptide prevents the binding of the gp120 protein of HIV to the β-tubulin, a neuronal MT and possesses neuroprotective activity (Avdoshina et al. 2019). Moreover, the SPR has been used as a ligand screening strategy for Influenza virus and HSV-1, in which the technique enables the continuous screening of inhibitors that inhibit the viral entry. The major advantage of this technique is the use of minimal amount of immobilized viral surface proteins or receptors as compared to other techniques (Kumar 2017).
Biolayer Interferometry
Another optical based and label-free technique is the Biolayer interferometry (BLI) which validates the interaction between two molecules by quantifying the change in an interference pattern. The target molecule is immobilized on the tip of fiber optic biosensor that moves toward the wells containing the binding partner present in solution. The association and dissociation of the binding partner with the immobilized molecule is monitored by BLI, leading to the generation of optical thickness at the tip of biosensor that produces an optical interference pattern. This pattern can be quantified and used to determine real time kinetic rates of binding and dissociation (Shah and Duncan 2014). Thus, it has become a valuable tool for monitoring interactions between small molecules in the field of drug discovery. This technique is advantageous over others as nonspecific and non ideal interactions can be differentiated in initial steps by examining their binding response and moreover, it has low false positive rate. Besides, the varied flow rate, available unbound molecules and the refractive index of adjacent medium do not affect the obtained interference pattern, which is the unique property of this technique (Wartchow et al. 2011). In this context Zu et al. (2014) have analysed the real time binding affinity of chemically synthesized screened peptides to the Domain III of JEV envelope protein and reported peptide P3 possessed the highest affinity.
Fluorescence Resonance Energy Transfer (FRET)
FRET is a sensitive method to investigate the interaction of proteins with large diverse set of peptides/proteins libraries for high throughput screening efficiently. This technique is reliable on the distance-dependent transfer of energy between dye-labelled molecules, where the excited donor fluorophore transfers its energy to an acceptor chromophore (Rogers et al. 2012). This energy transfer determines the ratio metric signal generated by the reduction in fluorescence of donor molecule and the increment in fluorescence of acceptor molecule. The technique of FRET can be used as both screening as well as validation method. In view of this, various FRET based studies have been reported for the identification of potent inhibitors against several viral proteases such as SARS coronavirus 3CLpro protease, DENV NS2B-NS3 protease, WNV Serine Protease, HCV NS3/4A protease and HIV protease. Similarly, a FRET based proteolytic assay was used to screen the compounds against CHIKV capsid protein (Aggarwal et al. 2015).
Challenges to the Peptide as Therapeutic Use
Several limitations that obstruct the way of peptide to be a successful therapeutic drug, are its instability, short half-life, lower potency, inability to cross membrane barriers and poor bioavailability due to protease degradation (Ali et al. 2013). The main challenge is to overcome these limitations and to achieve the desired efficacy for the required time span. Various modifications have been employed to enhance the stability and physiochemical properties of the peptides (Gentilucci et al. 2010). For instance, the conjugation of peptide to polymers such as polyethylene glycol (PEG) has enhanced the stability of peptides by increasing their molecular weight (Chew et al. 2017). Likewise, the bioavailability was improved by balancing the aqueous solubility via replacement of redundant hydrophobic amino acids to charged/polar residues (Mant et al. 2009; Wu et al. 2010). Moreover, there are two computational softwares based on support-vector machine (SVM) to predict the solubility of the peptides, thus assisting in the designing and optimising the peptide bioavailability (Lee et al. 2019). In addition to the strategies involved in the improvement of peptide properties, the delivery of peptides has also been improved by linking of peptides to the cell penetrating peptides (CPPs) to enhance their cell permeability (Chew et al. 2017). CPPs are general peptides (< 30 amino acids) derived from natural/unnatural sources or chimeric sequences, considered as promising carrier for successful delivery of therapeutic molecules varying from small chemical molecules, liposomes, proteins, peptides and nucleic acids for in vitro as well as in vivo applications (Heitz et al. 2009). Alternatively, peptides can be encapsulated in nanoparticles for efficient delivery, or administered through primary parental or transdermal routes with variations such as prefilled syringes, auto injectors and biodegradable micro needles (Lee et al. 2019). These modifications help to address the challenges of poor ADME properties of non-modified peptides (Lau and Dunn 2018).
Conclusion
In summary, the discovery of the peptide-based therapies have made a significant impact in the research. Many peptide therapies are available in the clinical and pre-clinical trials which are expected to yield positive results. The various approaches including computational, natural and biological sources provide a wide repository for identification of peptides involved in viral therapeutics. These promising therapeutics/inhibitors are advantageous because of their high specificity, selectivity against target and can be easily developed without the prior structural knowledge of target (except docking). Novel technologies like SPR, BLI and FRET validate the binding of identified peptides to their target which further aid in the selection of high affinity potential AVPs. However, there is a dearth of knowledge in the field of identification and characterization of antiviral therapeutics; further advancement and commercialization of peptide-based therapeutics is warranted.
Acknowledgements
The authors acknowledge Jaypee Institute of Information Technology (JIIT), Noida for providing infrastructure facilities.
Author Contributions
GA reviewed the literature and wrote the manuscript. RG conceived the idea and finalized the manuscript.
Funding
GA is funded by the Department of Science and Technology, Government of India: DST-INSPIRE (IF 150104).
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Ethical Approval
This is a review article which does not contain any type of studies related to human or animal participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aggarwal M, Sharma R, Kumar P, Parida M, Tomar S. Kinetic characterization of trans-proteolytic activity of Chikungunya virus capsid protease and development of a FRET-based HTS assay. Sci Rep. 2015;5:14753. doi: 10.1038/srep14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Siman-Tov G, Hall G, Bhalla N, Narayanan A. Human antimicrobial peptides as therapeutics for viral infections. Viruses. 2019;11:704. doi: 10.3390/v11080704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Siman-Tov G, Keck F, Kortchak S, Bakovic A, Risner K, Lu TK, Bhalla N, de la Fuente-Nunez C, Narayanan A. Human cathelicidin peptide LL-37 as a therapeutic antiviral targeting Venezuelan equine encephalitis virus infections. Antiviral Res. 2019;164:61–69. doi: 10.1016/j.antiviral.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Alagarasu K, Patil PS, Shil P, Seervi M, Kakade MB, Tillu H, Salunke A. In-vitro effect of human cathelicidin antimicrobial peptide LL-37 on dengue virus type 2. Peptides. 2017;92:23–30. doi: 10.1016/j.peptides.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Albiol Matanic VC, Castilla V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int J Antimicrob Agents. 2004;23:382–389. doi: 10.1016/j.ijantimicag.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Ali R, Rani R, Kumar S. New peptide based therapeutic approaches. In: Ashraf GMd, Sheikh IA., editors. Advances in protein chemistry. Jeddah: OMICS Group eBooks; 2013. [Google Scholar]
- Amin A, Zaccardi J, Mullen S, Olland S, Orlowski M, Feld B, Labonte P, Mak P. Identification of constrained peptides that bind to and preferentially inhibit the activity of the hepatitis C viral RNA-dependent RNA polymerase. Virology. 2003;313:158–169. doi: 10.1016/S0042-6822(03)00313-1. [DOI] [PubMed] [Google Scholar]
- An TQ, Zhou YJ, Qiu HJ, Tong GZ, Wang YF, Liu JX, Yang JY. Identification of a Novel B cell epitope on the nucleocapsid protein of porcine reproductive and respiratory syndrome virus by phage display. Virus Genes. 2005;31:81–87. doi: 10.1007/s11262-005-2203-1. [DOI] [PubMed] [Google Scholar]
- Andersen JH, Osbakk SA, Vorland LH, et al. Lactoferrin and cyclic lactoferricin inhibit the entry of human cytomegalovirus into human fibroblasts. Antiviral Res. 2001;51:141–149. doi: 10.1016/s0166-3542(01)00146-2. [DOI] [PubMed] [Google Scholar]
- Andjelic CD, Planelles V, Barrows LR. Characterizing the anti-HIV activity of Papuamide A. Mar Drugs. 2008;6:528–549. doi: 10.3390/md20080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdoshina V, Taraballi F, Tasciotti E, Uren A, Mocchetti I. Helix-A peptide prevents gp120-mediated neuronal loss. Mol Brain. 2019;12:61. doi: 10.1186/s13041-019-0482-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badani H, Garry RF, Wimley WC. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim Biophys Acta Biomembr. 2014;1838:2180–2197. doi: 10.1016/j.bbamem.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Town T, Pradhan D, Cox J, Ledizet M, Anderson JF, Flavell RA, Krueger JK, Koski RA, Fikrig E. Antiviral peptides targeting the West Nile virus envelope protein. J Virol. 2007;81:2047–2055. doi: 10.1128/JVI.01840-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PG, Svoboda P, Mackellar A, Nash AA, York IA, Pohl J, Davidson DJ, Donis RO. Antiviral activity and increased host defense against influenza infection elicited by the human cathelicidin LL-37. PLoS ONE. 2011;6:1–9. doi: 10.1371/journal.pone.0025333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergaoui I, Zairi A, Tangy F, Aouni M, Selmi B, Hani K. In vitro antiviral activity of dermaseptin S4 and derivatives from amphibian skin against herpes simplex virus type 2. J Med Virol. 2013;85:272–281. doi: 10.1002/jmv.23450. [DOI] [PubMed] [Google Scholar]
- Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, Lehr T, Lempp FA, Wedemeyer H, Haag M, Schwab M. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: First results of a phase Ib/IIa study. J Hepatol. 2016;65:490–498. doi: 10.1016/j.jhep.2016.04.016. [DOI] [PubMed] [Google Scholar]
- Borst EM, Ständker L, Wagner K, et al. A peptide inhibitor of cytomegalovirus infection from human hemofiltrate. Antimicrob Agents Chemother. 2013;57:4751–4760. doi: 10.1128/aac.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulet P, Stöcklin R, Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- Camargo Filho I, Cortez DAG, Ueda-Nakamura T, Nakamura CV, Dias Filho BP. Antiviral activity and mode of action of a peptide isolated from Sorghum bicolor. Phytomedicine. 2008;15:202–208. doi: 10.1016/j.phymed.2007.07.059. [DOI] [PubMed] [Google Scholar]
- Cao J, Liao XL, Wu SM, Zhao P, Zhao LJ, Wu WB, Qi ZT. Selection of a phage-displayed peptide recognized by monoclonal antibody directed blocking the site of hepatitis C virus E2 for human CD81. J Microbiol Methods. 2007;68:601–604. doi: 10.1016/j.mimet.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Carriel-Gomes MC, Kratz JM, Barracco MA, Bachére E, Barardi CR, Simões CM. In vitro antiviral activity of antimicrobial peptides against herpes simplex virus 1, adenovirus, and rotavirus. Mem Inst Oswaldo Cruz. 2007;102:469–472. doi: 10.1590/s0074-02762007005000028. [DOI] [PubMed] [Google Scholar]
- Carvalho CAM, Casseb SMM, Gonçalves RB, Silva EV, Gomes AM, Vasconcelos PF. Bovine lactoferrin activity against Chikungunya and Zika viruses. J Gen Virol. 2017;98:1749–1754. doi: 10.1099/jgv.0.000849. [DOI] [PubMed] [Google Scholar]
- Castro CC, Sant'anna V, Vargas GD, Lima MD, Fischer G, Brandelli A, Motta AD, Hübner SD. Antiviral activity of a Bacillus sp: P34 peptide against pathogenic viruses of domestic animals. Braz J Microbiol. 2014;45:1089–1094. doi: 10.1590/s1517-83822014000300043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro CC, Silva DS, Costa GA, Fischer G, Vargas GD, Brandelli A, Lima MD, Motta AD, Hubner SD. Activity of the antimicrobial peptide P34 against bovine alpha herpes virus type 1. Ciência Rural. 2017;47:e20160668. doi: 10.1590/0103-8478cr20160668. [DOI] [Google Scholar]
- Chen F, Zhao Y, Liu M, Li D, Wu H, Chen H, Zhu Y, Luo F, Zhong J, Zhou Y, Qi Z. Functional selection of hepatitis C virus envelope E2-binding Peptide ligands by using ribosome display. Antimicrob Agents Chemother. 2010;54:3355–3364. doi: 10.1128/aac.01357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao L, Zhong M, Zhang Y, Han C, Li Q, Yang J, Zhou D, Shi W, He B, Liu F. Anti-HIV-1 activity of a new scorpion venom peptide derivative Kn2-7. PLoS ONE. 2012;7:1–9. doi: 10.1371/journal.pone.0034947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Fan YC, Lin JW, Chen YY, Hsu WL, Chiou SS. Bovine lactoferrin inhibits dengue virus infectivity by interacting with heparan sulfate, low-density lipoprotein receptor, and DC-SIGN. Int J Mol Sci. 2017;18:1–13. doi: 10.3390/ijms18091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernysh S, Kim SI, Bekker G, Pleskach VA, Filatova NA, Anikin VB, Platonov VG, Bulet P. Antiviral and antitumor peptides from insects. Proc Natl Acad Sci. 2002;99:12628–12632. doi: 10.1073/pnas.192301899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew MF, Poh KS, Poh CL. Peptides as therapeutic agents for dengue virus. Int J Med Sci. 2017;14:1342–1359. doi: 10.7150/ijms.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikaev AN, Bakulina AY, Burdick RC, Karpenko LI, Pathak VK, Ilyichev AA. Selection of Peptide Mimics of HIV-1 Epitope Recognized by Neutralizing Antibody VRC01. PLoS ONE. 2015;10:e0120847. doi: 10.1371/journal.pone.0120847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten SW, Zou J, Wang R, Huang BC, Liu R. mRNA display-based selections using synthetic peptide and natural protein libraries. Methods Mol Biol. 2012;805:287–297. doi: 10.1007/978-1-61779-379-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crack LR, Jones L, Malavige GN, Patel V, Ogg GS. Human antimicrobial peptides LL-37 and human β-defensin-2 reduce viral replication in keratinocytes infected with varicella zoster virus. Clin Exp Dermatol. 2012;37:534–543. doi: 10.1111/j.1365-2230.2012.04305.x. [DOI] [PubMed] [Google Scholar]
- de la Guardia C, Quijada M, Lleonart R. Phage-displayed peptides selected to bind envelope glycoprotein show antiviral activity against dengue virus serotype 2. Adv Virol. 2017;2017:1827341. doi: 10.1155/2017/1827341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Zhai JW, Michel ML, Zhang J, Qin J, Kong YY, Zhang XX, Budkowska A, Tiollais P, Wang Y, Xie YH. Identification and characterization of peptides that interact with hepatitis B virus via the putative receptor binding site. J Virol. 2007;81:4244–4254. doi: 10.1128/jvi.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjobert C, de Soultrait VR, Faure A, Parissi V, Litvak S, Tarrago-Litvak L, Fournier M. Identification by phage display selection of a short peptide able to inhibit only the strand transfer reaction catalyzed by human immunodeficiency virus type 1 integrase. Biochemistry. 2004;43:13097–13105. doi: 10.1021/bi049385e. [DOI] [PubMed] [Google Scholar]
- Divyashree M, Mani MK, Reddy D, Kumavath R, Ghosh P, Azevedo V, Barh D. Clinical applications of antimicrobial peptides (AMPs): where do we stand now? Protein Pept Lett. 2020;27:120–134. doi: 10.2174/0929866526666190925152957. [DOI] [PubMed] [Google Scholar]
- Donia MS, Wang B, Dunbar DC, Desai PV, Patny A, Avery M, Hamann MT. Mollamides B and C, Cyclic Hexapeptides from the Indonesian Tunicate Didemnum molle. J Nat Prod. 2008;71:941–945. doi: 10.1021/np700718p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier B, Pluckthun A. Ribosome display: a technology for selecting and evolving proteins from large libraries. Methods Mol Biol. 2011;687:283–306. doi: 10.1007/978-1-60761-944-4_21. [DOI] [PubMed] [Google Scholar]
- Falco A, Medina-Gali R, Poveda J, Bello-Perez M, Novoa B, Encinar JA. Antiviral activity of a Turbot (Scophthalmus maximus) NK-lysin peptide by inhibition of low-pH virus-induced membrane fusion. Mar Drugs. 2019;17:87. doi: 10.3390/md17020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Harrison SC. Peptide ligands to human immunodeficiency virus type 1 gp120 identified from phage display libraries. J Virol. 1999;73:5795–5802. doi: 10.1128/JVI.73.7.5795-5802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Taki M. Practical tips for construction of custom peptide libraries and affinity selection by using commercially available phage display cloning systems. J Nucleic Acids. 2012;2012:295719. doi: 10.1155/2012/295719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero S, Falanga A, Tarallo R, Russo L, Galdiero E, Cantisani M, Morelli G, Galdiero M. Peptide inhibitors against herpes simplex virus infections. J Pept Sci. 2013;19:148–158. doi: 10.1002/psc.2489. [DOI] [PubMed] [Google Scholar]
- Gao Y, Cui T, Lam Y. Synthesis and disulfide bond connectivity-activity studies of a kalata B1-inspired cyclopeptide against dengue NS2B-NS3 protease. Bioorganic Med Chem. 2010;18:1331–1336. doi: 10.1016/j.bmc.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Gentilucci L, De Marco R, Cerisoli L. Chemical modifications designed to improve peptide stability: incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr Pharm Des. 2010;16:3185–3203. doi: 10.2174/138161210793292555. [DOI] [PubMed] [Google Scholar]
- Gordon YJ, Huang LC, Romanowski EG, et al. Human cathelicidin (LL-37), a multifunctional peptide, is expressed by ocular surface epithelia and has potent antibacterial and antiviral activity. Curr Eye Res. 2005;30:385–394. doi: 10.1080/02713680590934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Duan H, Kachko A, Krause BW, Major ME, Krause PR. Reverse engineering of vaccine antigens using high throughput sequencing enhanced mRNA display. EBioMedicine. 2015;2:859–867. doi: 10.1016/j.ebiom.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwyer Findlay E, Currie SM, Davidson DJ. Cationic host defence peptides: potential as antiviral therapeutics. BioDrugs. 2013;27:479–493. doi: 10.1007/s40259-013-0039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen T, Lundkvist A, Vaheri A, Lankinen H. Phage-displayed peptide targeting on the Puumala hantavirus neutralization site. J Virol. 1997;71(5):3879–3885. doi: 10.1128/JVI.71.5.3879-3885.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques ST, Huang YH, Rosengren KJ, Franquelim HG, Carvalho FA, Johnson A, Sonza S, Tachedjian G, Castanho MA, Daly NL, Craik DJ. Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization on hemolytic and anti-HIV activities. J Biol Chem. 2011;286:1–24. doi: 10.1074/jbc.m111.253393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KL, Yusoff K, Seow HF, Tan WS. Selection of high affinity ligands to hepatitis B core antigen from a phage-displayed cyclic peptide library. J Med Virol. 2003;69:27–32. doi: 10.1002/jmv.10266. [DOI] [PubMed] [Google Scholar]
- Holthausen DJ, Lee SH, Kumar VT, Bouvier NM, Krammer F, Ellebedy AH, Wrammert J, Lowen AC, George S, Pillai MR, Jacob J. An amphibian host defense peptide is virucidal for human H1 hemagglutinin bearing influenza viruses. Immunity. 2017;46:587–595. doi: 10.1016/j.immuni.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Hong W, Zhang R, Di Z, He Y, Zhao Z, Hu J, Wu Y, Li W, Cao Z. Design of histidine-rich peptides with enhanced bioavailability and inhibitory activity against hepatitis C virus. Biomaterials. 2013;34:3511–3522. doi: 10.1016/j.biomaterials.2013.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Li T, Song Y, Zhang R, Zeng Z, Han S, Zhang X, Wu Y, Li W, Cao Z. Inhibitory activity and mechanism of two scorpion venom peptides against herpes simplex virus type 1. Antiviral Res. 2014;102:1–10. doi: 10.1016/j.antiviral.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, Jallouk AP, Campbell N, Ratner L, Wickline SA. Cytolytic nanoparticles attenuate HIV-1 infectivity. Antivir Ther. 2013;18:95–103. doi: 10.3851/imp2346. [DOI] [PubMed] [Google Scholar]
- Hou P, Zhao G, He C, Wang H, He H. Biopanning of polypeptides binding to bovine ephemeral fever virus G 1 protein from phage display peptide library. BMC Vet Res. 2018;14:3. doi: 10.1186/s12917-017-1315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Streib JE, Leung DYM. Antiviral activity of human beta-defensin 3 against vaccinia virus. J Allergy Clin Immunol. 2007;119:1022–1025. doi: 10.1016/j.jaci.2007.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Guo N, Chen S, Guo X, Liu X, Ye S, Chai Q, Wang Y, Liu B, He Q. Antiviral activity of Piscidin 1 against pseudorabies virus both in vitro and in vivo. Virol J. 2019;16:95. doi: 10.1186/s12985-019-1199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. 2005;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- Ireland DC, Wang CKL, Wilson JA, Gustafson KR, Craik DJ. Cyclotides as natural anti-HIV agents. Biopolym - Pept Sci Sect. 2008;90:51–60. doi: 10.1002/bip.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack HW, Tzi BN. Sesquin, a potent defensin-like antimicrobial peptide from ground beans with inhibitory activities toward tumor cells and HIV-1 reverse transcriptase. Peptides. 2005;26:1120–1126. doi: 10.1016/j.peptides.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Jenssen H. Therapeutic approaches using host defence peptides to tackle herpes virus infections. Viruses. 2009;1:939–964. doi: 10.3390/v1030939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczer M, Dziubasik K, Midak-Siewirska A, Zahorska R, Luczak M, Konopinska D. Studies of insect peptides alloferon, Any-GS and their analogues. Synthesis and antiherpes activity. J Pept Sci. 2010;16:186–189. doi: 10.1002/psc.1219. [DOI] [PubMed] [Google Scholar]
- Kumar PKR. Systematic screening of viral entry inhibitors using surface plasmon resonance. Rev Med Virol. 2017;27:e1952. doi: 10.1002/rmv.1952. [DOI] [PubMed] [Google Scholar]
- Lau JL, Dunn MK. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg Med Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Lee AC, Harris JL, Khanna KK, Hong JH. A comprehensive review on current advances in peptide drug development and design. Int J Mol Sci. 2019;20:2383. doi: 10.3390/ijms20102383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhou H, Huang G, Liu N. Inhibition of HBV infection by bovine lactoferrin and iron, zinc-saturated lactoferrin. Med Microbiol Immunol. 2009;198:19–25. doi: 10.1007/s00430-008-0100-7. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhao Z, Zhou D, Chen Y, Hong W, Cao L, Yang J, Zhang Y, Shi W, Cao Z, Wu Y. Virucidal activity of a scorpion venom peptide variant mucroporin-M1 against measles, SARS CoV and influenza H5N1 viruses. Peptides. 2011;32:1518–1525. doi: 10.1016/j.peptides.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linciano S, Pluda S, Bacchin A, Angelini A. Molecular evolution of peptides by yeast surface display technology. Med Chem Comm. 2019;10:1569–1580. doi: 10.1039/c9md00252a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick A, Szostak JW. Selection of cyclic peptide aptamers to HCV IRES RNA using mRNA display. Proc Natl Acad Sci USA. 2008;105:15293–15298. doi: 10.1073/pnas.0805837105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu K, Feng X, Ma Z, Luo C, Zhou B, Cao R, Huang L, Miao D, Pang R, He D, Lian X, Chen P. Antiviral activity of phage display selected peptides against Porcine reproductive and respiratory syndrome virus in vitro. Virology. 2012;432:73–80. doi: 10.1016/j.virol.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Liu MZ, Yang Y, Zhang SX, Tang L, Wang HM, Chen CJ, Shen ZF, Cheng KD, Kong JQ, Wang W. A cyclotide against influenza A H1N1 virus from Viola yedoensis. Yao xue xue bao. Acta Pharm Sin. 2014;49:905–912. [PubMed] [Google Scholar]
- Lorin C, Saidi H, Belaid A, et al. The antimicrobial peptide Dermaseptin S4 inhibits HIV-1 infectivity in vitro. Virology. 2005;334:264–275. doi: 10.1016/j.virol.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lu Z, Van Wagoner RM, Harper MK, Baker HL, Hooper JNA, Bewley CA, Ireland CM. Mirabamides E-H, HIV-Inhibitory Depsipeptides from the Sponge Stelletta clavosa. J Nat. 2011;74:185–193. doi: 10.1021/np100613p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Yao M, Zhang JM, Yang J, Lei YF, Huang XJ, Jia ZS, Ma L, Lan HY, Xu ZK, Yin W. Identification of peptides that bind hepatitis C virus envelope protein E2 and inhibit viral cellular entry from a phage-display peptide library. Int J Mol Med. 2014;33:1312–1318. doi: 10.3892/ijmm.2014.1670. [DOI] [PubMed] [Google Scholar]
- Luo C, Liu X, Zhou X, Guo J, Truong J, Wang X, Zhou H, Li X, Chen Z. Unusual biosynthesis and structure of locillomycins from Bacillus subtilis 916. Appl Environ Microbiol. 2015;81:6601–6609. doi: 10.1128/aem.01639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mant CT, Kovacs JM, Kim HM, Pollock DD, Hodges RS. Intrinsic amino acid side-chain hydrophilicity/hydrophobicity coefficients determined by reversed-phase high-performance liquid chromatography of model peptides: comparison with other hydrophilicity/hydrophobicity scales. Biopolymers. 2009;92:573–595. doi: 10.1002/bip.21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr AK, Jenssen H, Moniri MR, Hancock RE, Panté N. Bovine lactoferrin and lactoferricin interfere with intracellular trafficking of Herpes simplex virus-1. Biochimie. 2009;91:160–164. doi: 10.1016/j.biochi.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Mascini M, Dikici E, Robles Mañueco M, Perez-Erviti JA, Deo SK, Compagnone D, Wang J, Pingarrón JM, Daunert S. Computationally designed peptides for Zika virus detection: an incremental construction approach. Biomolecules. 2019;9:498. doi: 10.3390/biom9090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T. Potential of peptides as inhibitors and mimotopes: selection of carbohydrate-mimetic peptides from phage display libraries. J Nucleic Acids. 2012 doi: 10.1155/2012/740982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T, Sumi M, Kubota H, Taki T, Okahata Y, Sato T. Inhibition of influenza virus infections by sialylgalactose-binding peptides selected from a phage library. J Med Chem. 2009;52:4247–4256. doi: 10.1021/jm801570y. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Sugiyama N, Murayama A, Yamada N, Shiina M, Asabe S, Wakita T, Imawari M, Kato T. Antimicrobial peptide LL-37 attenuates infection of hepatitis C virus. Hepatol Res. 2016;46:924–932. doi: 10.1111/hepr.12627. [DOI] [PubMed] [Google Scholar]
- Mei M, Zhou Y, Peng W, Yu C, Ma L, Zhang G, Yi L. Application of modified yeast surface display technologies for non-Antibody protein engineering. Microbiol Res. 2017;196:118–128. doi: 10.1016/j.micres.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U, Schwarz T, Schroder J-M, Gläser R. Expression of human beta-defensin-2 and -3 in verrucae vulgares and condylomata acuminata. J Eur Acad Dermatol Venereol. 2008;22:1050–1054. doi: 10.1111/j.1468-3083.2008.02675.x. [DOI] [PubMed] [Google Scholar]
- Migliolo L, Silva ON, Silva PA, Costa MP, Costa CR, Nolasco DO, Barbosa JA, Silva MR, Bemquerer MP, Lima LM, Romanos MT. Structural and functional characterization of a multifunctional alanine-rich peptide analogue from pleuronectes americanus. PLoS ONE. 2012;7:e47047. doi: 10.1371/journal.pone.0047047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry N, Drobni P, Näslund J, Sunkari VG, Jenssen H, Evander M. The anti-papillomavirus activity of human and bovine lactoferricin. Antiviral Res. 2007;75:258–265. doi: 10.1016/j.antiviral.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Monteiro JMC, Oliveira MD, Dias RS, Nacif-Marçal L, Feio RN, Ferreira SO, Oliveira LL, Silva CC, Paula SO. The antimicrobial peptide HS-1 inhibits dengue virus infection. Virology. 2018;514:79–87. doi: 10.1016/j.virol.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Moreno M, Giralt E. Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: melittin, apamin and mastoparan. Toxins. 2015;7:1126–1150. doi: 10.3390/toxins7041126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevola L, Giralt E. Modulating protein-protein interactions: the potential of peptides. Chem Commun (Camb) 2015;51:3302–3315. doi: 10.1039/c4cc08565e. [DOI] [PubMed] [Google Scholar]
- Newton M, Cabezas-Perusse Y, Tong CL, Seelig B. In vitro selection of peptides and proteins–advantages of mRNA display. ACS Synth Biol. 2019;9:181–190. doi: 10.1021/acssynbio.9b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku N, Gustafson KR, Cartner LK, Wilson JA, Shigematsu N, Hess S, Pannell LK, Boyd MR, McMahon JB. Neamphamide A, a new HIV-inhibitory depsipeptide from the papua new guinea marine sponge Neamphius huxleyi. J Nat. 2004;67:1407–1411. doi: 10.1021/np040003f. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Ohashi K, Onuma M. Identification and characterization of peptides binding to newcastle disease virus by phage display. J Vet Med Sci. 2005;67:1237–1241. doi: 10.1292/jvms.67.1237. [DOI] [PubMed] [Google Scholar]
- Panya A, Yongpitakwattana P, Budchart P, Sawasdee N, Krobthong S, Paemanee A, Roytrakul S, Rattanabunyong S, Choowongkomon K, Yenchitsomanus P. Novel bioactive peptides demonstrating anti-dengue virus activity isolated from the Asian medicinal plant Acacia Catechu. Chem Biol Drug Des. 2019;93:100–109. doi: 10.1111/cbdd.13400. [DOI] [PubMed] [Google Scholar]
- Patching SG. Surface plasmon resonance spectroscopy for characterization of membrane protein–ligand interactions and its potential for drug discovery. Biochim Biophys Acta. 2014;1838:43–55. doi: 10.1016/j.bbamem.2013.04.028. [DOI] [PubMed] [Google Scholar]
- Peng B, Chen H, Tan Y, Jin M, Chen H, Guo A. Identification of one peptide which inhibited infectivity of avian infectious bronchitis virus in vitro. Sci China C. 2006;49:8. doi: 10.1007/s11427-006-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza A, Gustchina E, Baker HL, Kelly M, Bewley CA. Mirabamides A-D, Depsipeptides from the Sponge Siliquariaspongia mirabilis that inhibit HIV-1 Fusion. J Nat Prod. 2007;70:1753–1760. doi: 10.1021/np070306k. [DOI] [PubMed] [Google Scholar]
- Plaza A, Bifulco G, Keffer JL, Lloyd JR, Baker HL, Bewley CA. Celebesides A-C and theopapuamides B-D, depsipeptides from an Indonesian sponge that inhibit HIV-1 entry. J Org Chem. 2009;74:504–512. doi: 10.1021/jo802232u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza A, Bifulco G, Masullo M, Lloyd JR, Keffer JL, Colin PL, Hooper JN, Bell LJ, Bewley CA. Mutremdamide A and koshikamides C-H, peptide inhibitors of HIV-1 entry from different Theonella species. J Org Chem. 2010;75:4344–4355. doi: 10.1021/jo100076g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, Marotta ML, Mirza M, Jiang B, Kiser P, Medvik K. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS. 2003;17:F39–F48. doi: 10.1097/01.aids.0000096878.73209.4f. [DOI] [PubMed] [Google Scholar]
- Qureshi A, Thakur N, Himani T, Kumar M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucl Acids Res. 2013;42:D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajik M, Jahanshiri F, Omar AR, Ideris A, Hassan SS, Yusoff K. Identification and characterisation of a novel anti-viral peptide against avian influenza virus H9N2. Virol J. 2009;6:74. doi: 10.1186/1743-422X-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanujam P, Tan WS, Nathan S, Yusoff K. Novel peptides that inhibit the propagation of Newcastle disease virus. Arch Virol. 2002;147:981–993. doi: 10.1007/s00705-001-0778-y. [DOI] [PubMed] [Google Scholar]
- Rashid MA, Gustafson KR, Cartner LK, Shigematsu N, Pannell LK, Boyd MR. Microspinosamide, a new HIV-inhibitory cyclic depsipeptide from the marine sponge Sidonops microspinosa. J Nat Prod. 2001;64:117–121. doi: 10.1021/np0002379. [DOI] [PubMed] [Google Scholar]
- Rider TH, Zook CE, Boettcher TL, Wick ST, Pancoast JS, Zusman BD. Broad-spectrum antiviral therapeutics. PLoS ONE. 2011;6:e22572. doi: 10.1371/journal.pone.0022572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WE, McDougall B, Tran D, Selsted ME. Anti-HIV-1 activity of indolicidin, an antimicrobial peptide from neutrophils. J Leukoc Biol. 1998;63:94–100. doi: 10.1002/jlb.63.1.94. [DOI] [PubMed] [Google Scholar]
- Rogers MS, Cryan LM, Habeshian KA, Bazinet L, Caldwell TP, Ackroyd PC, Christensen KA. A FRET-based high throughput screening assay to identify inhibitors of anthrax protective antigen binding to capillary morphogenesis gene 2 protein. PLoS ONE. 2012;7:e39911. doi: 10.1371/journal.pone.0039911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, Abdulrahman AY, Sasikumer PG, Othman S, Abd Rahman N, Yusof R. Protegrin-1 inhibits dengue NS2B-NS3 serine protease and viral replication in MK2 cells. J Biomed Biotechnol. 2012;2012:1–6. doi: 10.1155/2012/251482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan HA, Bahrani H, Rahman NA, Yusof R. Identification of natural antimicrobial agents to treat dengue infection: in vitro analysis of latarcin peptide activity against dengue virus. BMC Microbiol. 2014;14:1–10. doi: 10.1186/1471-2180-14-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore M, García-Sastre A, Ruchala P, Lehrer RI, Chang T, Klotman ME. α-defensin inhibits influenza virus replication by cell-mediated mechanism(s) J Infect Dis. 2007;196:835–843. doi: 10.1086/521027. [DOI] [PubMed] [Google Scholar]
- Sample CJ, Hudak KE, Barefoot BE, Koci MD, Wanyonyi MS, Abraham S, Staats HF, Ramsburg EA. A mastoparan-derived peptide has broad-spectrum antiviral activity against enveloped viruses. Peptides. 2013;48:96–105. doi: 10.1016/j.peptides.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Kanitkar TR, Roy AA, Soni N, Amritkar K, Supekar S, Nair S, Singh G, Madhusudhan M. Predicting and Designing therapeutics against the Nipah virus. PLoS Negl Trop Dis. 2019;13:e0007419. doi: 10.1371/journal.pntd.0007419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sencanski M, Radosevic D, Perovic V, Gemovic B, Stanojevic M, Veljkovic N, Glisic S. Natural products as promising therapeutics for treatment of influenza disease. Curr Pharm Des. 2015;21:5573–5588. doi: 10.2174/1381612821666151002113426. [DOI] [PubMed] [Google Scholar]
- Shah NB, Duncan TM. Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J Vis Exp. 2014;18:e51383. doi: 10.3791/51383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HJ, Rashid MA, Cartner LK, Bokesch HR, Wilson JA, McMahon JB, Gustafson KR. Stellettapeptins A and B, HIV-inhibitory cyclic depsipeptides from the marine sponge Stelletta sp. Tetrahedron Lett. 2015;56:4215–4219. doi: 10.1016/j.tetlet.2015.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalickova S, Heger Z, Krejcova L, Pekarik V, Bastl K, Janda J, Kostolansky F, Vareckova E, Zitka O, Adam V, Kizek R. Perspective of use of antiviral peptides against influenza virus. Viruses. 2015;7:5428–5442. doi: 10.3390/v7102883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen OE, Follin P, Johnsen AH, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- Sousa FH, Casanova V, Findlay F, Stevens C, Svoboda P, Pohl J, Proudfoot L, Barlow PG. Cathelicidins display conserved direct antiviral activity towards rhinovirus. Peptides. 2017;95:76–83. doi: 10.1016/j.peptides.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun EC, Ma JN, Liu NH, Yang T, Zhao J, Geng HW, Wang LF, Qin YL, Bu ZG, Yang YH, Lunt RA, Wang LF, Wu DL. Identification of two linear B-cell epitopes from West Nile virus NS1 by screening a phage-displayed random peptide library. BMC Microbiol. 2011;11:160. doi: 10.1186/1471-2180-11-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zeng X, Liang J. Surface plasmon resonance: an introduction to a surface spectroscopy technique. J Chem Educ. 2010;87:742–746. doi: 10.1021/ed100186y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissier E, Penin F, Pécheur EI. Targeting cell entry of enveloped viruses as an antiviral strategy. Molecules. 2011;16:221–250. doi: 10.3390/molecules16010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson RL, Walker SN, Ikeda T, Harris RS, McNaughton BR. Evaluation of sequence variability in HIV-1 gp41 C-peptide helix-grafted proteins. Bioorg Med Chem. 2018;26:1220–1224. doi: 10.1016/j.bmc.2017.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Wang G, White M, Qi L, Taubenberger J, Hartshorn KL. Antiviral activity of the human cathelicidin, LL-37, and derived peptides on seasonal and pandemic influenza A viruses. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0124706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Strate BWA, Beljaars L, Molema G, Harmsen MC, Meijer DKF. Antiviral activities of lactoferrin. Antiviral Res. 2001;52:225–239. doi: 10.1016/s0166-3542(01)00195-4. [DOI] [PubMed] [Google Scholar]
- Wachinger M, Kleinschmidt A, Winder D, von Pechmann N, Ludvigsen A, Neumann M, Holle R, Salmons B, Erfle V, Brack-Werner R. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J Gen Virol. 1998;79:731–740. doi: 10.1099/0022-1317-79-4-731. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu R. Advantages of mRNA display selections over other selection techniques for investigaiu Rtion of protein–protein interactions. Expert Rev Proteomic. 2011;8:335–346. doi: 10.1586/epr.11.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CKL, Colgrave ML, Gustafson KR, Ireland DC, Goransson U, Craik DJ. Anti-HIV cyclotides from the Chinese medicinal herb Viola yedoensis. J Nat Prod. 2008;71:47–52. doi: 10.1021/np070393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Liu Y, Zu X, Jin R, Xiao G. Blocking peptides against HBV: PreS1 protein selected from a phage display library. Biochem Biophys Res Commun. 2011;412:633–637. doi: 10.1016/j.bbrc.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Wang WY, Wong JH, Ip DT, Wan DC, Cheung RC, Ng TB. Bovine lactoferrampin, human lactoferricin, and lactoferrin 1–11 inhibit nuclear translocation of HIV integrase. Appl Biochem Biotechnol. 2016;179:1202–1212. doi: 10.1007/s12010-016-2059-y. [DOI] [PubMed] [Google Scholar]
- Wang CK, Shih LY, Chang K. Large-scale analysis of antimicrobial activities in relation to amphipathicity and charge reveals novel characterization of antimicrobial peptides. Molecules. 2017;22:2037. doi: 10.3390/molecules22112037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartchow CA, Podlaski F, Li S, Rowan K, Zhang X, Mark D, Huang KS. Biosensor-based small molecule fragment screening with biolayer interferometry. J Comput Aided Mol Des. 2011;25:669–676. doi: 10.1007/s10822-011-9439-8. [DOI] [PubMed] [Google Scholar]
- Wei J, Hameed M, Wang X, Zhang J, Guo S, Anwar MN, Pang L, Liu K, Li B, Shao D, Qiu Y. Antiviral activity of phage display-selected peptides against Japanese encephalitis virus infection in vitro and in vivo. Antiviral Res. 2019;174:104673. doi: 10.1016/j.antiviral.2019.104673. [DOI] [PubMed] [Google Scholar]
- Weidmann J, Craik DJ. Discovery, structure, function, and applications of cyclotides: circular proteins from plants. J Exp Bot. 2016;67:4801–4812. doi: 10.1093/jxb/erw210. [DOI] [PubMed] [Google Scholar]
- Wu Z, Cocchi F, Gentles D, Ericksen B, Lubkowski J, DeVico A, Lehrer RI, Lu W. Human neutrophil α-defensin 4 inhibits HIV-1 infection in vitro. FEBS Lett. 2005;579:162–166. doi: 10.1016/j.febslet.2004.11.062. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Luo J, O'Neil KT, Kang J, Lacy ER, Canziani G, Baker A, Huang M, Tang QM, Raju TS, Jacobs SA. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng Des Sel. 2010;23:643–651. doi: 10.1093/protein/gzq037. [DOI] [PubMed] [Google Scholar]
- Wu D, Li G, Qin C, Ren X. Phage displayed peptides to avian H5N1 virus distinguished the virus from other viruses. PLoS ONE. 2011;6:e23058. doi: 10.1371/journal.pone.0023058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhao Z, He Y, Wu L, Cai D, Hong W, Wu Y, Cao Z, Zheng C, Li W. A new natural α-helical peptide from the venom of the scorpion Heterometrus petersii kills HCV. Peptides. 2011;32:11–19. doi: 10.1016/j.peptides.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Yin L, Luo Y, Liang B, Wang F, Du M, Petrenko VA, Qiu HJ, Liu A. Specific ligands for classical swine fever virus screened from landscape phage display library. Antivir Res. 2014;109:68–71. doi: 10.1016/j.antiviral.2014.06.012. [DOI] [PubMed] [Google Scholar]
- Yu D, Ding X, Liu Z, Wu X, Zhu Y, Wei H, Chong H, Cui S, He Y. Molecular mechanism of HIV-1 resistance to sifuvirtide, a clinical trial–approved membrane fusion inhibitor. J Biol Chem. 2018;293:12703–12718. doi: 10.1074/jbc.ra118.003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Zhang S, Peng J, Li Y, Yang Q. Synthetic surfactin analogues have improved anti-PEDV properties. PLoS ONE. 2019;14:e0215227. doi: 10.1371/journal.pone.0215227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnd C, Amstutz P, Plückthun A. Ribosome display: selecting and evolving proteins in vitro that specifically binds to a target. Nat Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- Zampella A, D’Auria MV, Paloma LG, Casapullo A, Minale L, Debitus C, Henin Y. Callipeltin A, an anti-hiv cyclic depsipeptide from the new caledonian lithistida sponge Callipelta sp. J Am Chem Soc. 1996;118:6202–6209. doi: 10.1021/ja954287p. [DOI] [Google Scholar]
- Zampella A, Sepe V, Luciano P, Bellotta F, Monti MC, D’Auria MV, Jepsen T, Petek S, Adeline MT, Laprévôte O, Aubertin AM. Homophymine A, an anti-HIV cyclodepsipeptide from the sponge Homophymia sp. J Org Chem. 2008;73(14):5319–5327. doi: 10.1021/jo800583b. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Zhang R, Hong W, Cheng Y, Wang H, Lang Y, Ji Z, Wu Y, Li W, Xie Y, Cao Z. Histidine-rich modification of a scorpion-derived peptide improves bioavailability and inhibitory activity against HSV-1. Theranostics. 2018;8:199–211. doi: 10.7150/thno.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Aweya JJ, Wang F, Yao D, Zhong M, Chen J, Li S, Zhang Y. Litopenaeus vannamei attenuates white spot syndrome virus replication by specific antiviral peptides generated from hemocyanin. Dev Comp Immunol. 2018;91:50–61. doi: 10.1016/j.dci.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Zhang LJ, Gallo RL. Antimicrobial peptides. Curr Biol. 2016;26:R14–R19. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang Y, Ji Q, Gu J, Liu S, Feng X, Sun C, Li Y, Lei L. Selection of antiviral peptides against mink enteritis virus using a phage display peptide library. Curr Microbiol. 2012;66:379–384. doi: 10.1007/s00284-012-0284-3. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Hong W, Zeng Z, Wu Y, Hu K, Tian X, Li W, Cao Z. Mucroporin-M1 inhibits hepatitis B virus replication by activating the mitogen-activated protein kinase (MAPK) pathway and down-regulating HNF4α in vitro and in vivo. J Biol Chem. 2012;287:30181–30190. doi: 10.1074/jbc.m112.370312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu X, Liu Y, Wang S, Jin R, Zhou Z, Liu H, Gong R, Xiao G, Wang W. Peptide inhibitor of Japanese encephalitis virus infection targeting envelope protein domain III. Antiviral Res. 2014;104:7–14. doi: 10.1016/j.antiviral.2014.01.011. [DOI] [PubMed] [Google Scholar]


