Abstract
Background
The present study demonstrates the potential of flavanoid narcissoside against the novel corona virus (COVID-19) complications using molecular docking studies.
Methods
The computation molecular docking screening was performed using Molegro Virtual Docker software (MVD) with grid resolution of 30 Å. Protein of COVID 19 virus was taken from protein data bank.
Results
The standard inhibitor X77 (N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide) identified from the protein inhibitor complex 6W63 from protein data bank was docked with COVID 19 protein 6W63 which showed MolDock score of −156.913, rerank Sore −121.296 and H Bond −5.7369, while the flavanoid narcissoside had showed MolDock score −180.739, Rerank Sore −137.092 and H Bond −18.6771. The narcissoside showed potent inhibitory effect which is greater than standard X77. The result showed that narcissoside have high affinity towards 6W63 as it showed thirteen hydrogen bonds with nine amino acids (Arg 188, Glu 166, His 164, Cys 145 (2 bonds), Asn 14 (2 bonds), Cys 44 (2 bonds), His 41 (2 bonds), Gln 192, Thr 190) while X777 showed four hydrogen bonds with amino acids (Gly 143, Cys 145, Glu 166, Ser 144).
Conclusion
From computation approach it was concluded that narcissoside is a potent inhibitor of viral COVID 19 protein 6W63. The narcissoside have high affinity and inhibition potential than standard inhibitor X77 (N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide). The narcissoside predicted as more potent inhibitor which can be further optimize, pharmacologically and clinically evaluated for the treatment of novel coronavirus COVID-19.
Keywords: Molecular docking, COVID-19, Flavonoid, MVD, X77, 6W63
At a glance of commentary
Scientific background on the subject
The need of an hour to develop the potent inhibitor of novel coronavirus (COVID-19) causing pandemic respiratory syndrome has brought the dry laboratories to work out on existing and novel medicine for their potential against the deadly virus.
What this study adds to the field
The narcissoside, a glycosyloxyflavone had showed potent inhibitory potential against coronavirus protein 6W63 which observed to more than the reported inhibitor X77. The potent molecule can be further be evaluated for preclinical and clinical studies and can be formulated in suitable dosage form for maximum bioavailability.
An ongoing outbreak of respiratory syndrome caused by a novel coronavirus called as COVID-19 which was reported in Wuhan, China in December 2019 [[1], [2], [3]].In the following months the infection was spread across the world and is becoming an important causative agent for the fatal disease [[4], [5], [6], [7]].
It is a great global public health concern and extensive measures are taken to reduce transmission of COVID-19 to control the current outbreak. With the emergence of new virus strains, there is a need for the development of the novel and effective antiviral for the treatment of COVID-19. In the present work, an attempt was made using computational approach to identify the potential flavones inhibitors narcissoside that binds to coronavirus for the treatment of COVID-19. Despite of various system of medicine available for the treatment, the recent curiosity is particularly in medicinal plants which have been important sources of new drugs, leads, and new chemical entities [[8], [9], [10], [11]].
Molecular modeling and structure-based drug design has been a promising technique to discover novel inhibitors and has become a famous practice in modern drug discovery. Until the computational drug discovery was adopted, the drugs were identified by chance in a manner of trial and error. Computer-aided drug design helps to facilitate and speed up the drug designing process, which involves a variety of methods to identify novel compounds.
Flavonoids are the natural substance widely distributed in grains, fruits, vegetables, leaves, barks, stems, roots and flowers. With more than 4000 variable phenolic structures, flavonoids have been discovered from plant species. Flavonoids are more investigated and are interesting compounds and almost many of them are of pharmacological importance. It has been demonstrated through numerous scientific studies that the flavonoids have antibacterial, anti-angiogenesis, anti-inflammatory, antiviral, antiatherosclerotic, antitumor, anti-thrombogenic, and antifungal activity [12,13].
The narcissoside, also known as narcissin is an Isorhamnetin-3-O-rutinoside flavonoid that belongs to monomethoxyflavone derivative and has been isolated from number of medicinal plants some are wild plant Peucedanum aucheri Boiss [14], leaves of Manihot escylenta Crantz (Cassava leaves) [15], Shoots of Caragana spinosa [16], flowers of Flos Sophorae Immaturus [17], plants of Nitaria Genus [18], leaves of Gynura divaricata [19], and aerial parts of Atriplex halimus L [20].
Material and methods
Molecular docking
Preparation of ligand
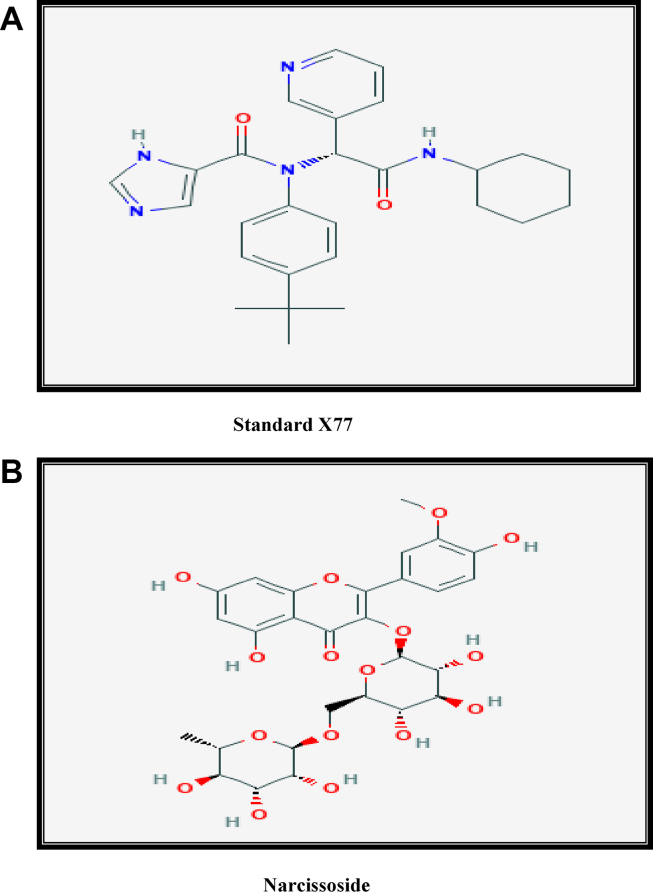
The COVID-19 inhibitor N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide reported as inhibitor in binding state with the protein 6w63 in protein data bank and 3-O-Methylquercetin 3-rutinoside (Fig. 1) were identified from PubChem chemical database. The structures were drawn using Chem3D and energy minimization using the MM2 force field and saved in.mol format. The missing bond orders, charges, bonds, and hybridization states of the structures were assigned using MVD software [21].
Fig. 1.
Chemical structure of standard X77(A) and narcissoside (B).
Preparation of protein
The 3D structure of the viral protein was retrieved from the Protein Data Bank (PDB ID: 6W63) and the proteins were prepared by removal of all water molecules, ligand, cofactors and assigning bonds, bond order, hybridization, and charges using MVD software [21].
Docking search algorithms and scoring functions
Piecewise Linear Potential (PLP) algorithm is used for scoring function in the computation screening. MolDock simplex evolution search algorithm with grid resolution 30 Å was used to carried out docking process [22]. The cavity were predicted and restricted to three using cavity prediction wizard, the cavity with the large volume was selected as the origin for the binding site. The ligands were docked with viral protein and best-generated poses were selected based on the docking scores [23].
Parameters for scoring functions
The energies are calculated in two scoring function Moldock Score and Rerank Score.
MolDock score
E score which is a docking scoring function, defined by the following energy terms:
| E score = E inter + E intra |
Where E inter is the sum of ligand–protein interaction energy, ligand–water interaction energy and ligand–cofactor interaction energy while E intra is the internal energy of the ligand [24]. The E intra energy includes sum of interactions energies of electronic, H bond, clash, sp2-sp2 hybridization, steric, torsal, torsal (ligand atoms), vanderwaal, soft constraint penalty and E-solvation.
Re rank score
In MVD, the rerank score provides an estimation of the interaction strength.This is not measured in chemical units and does not require complex contributions [24].
Results
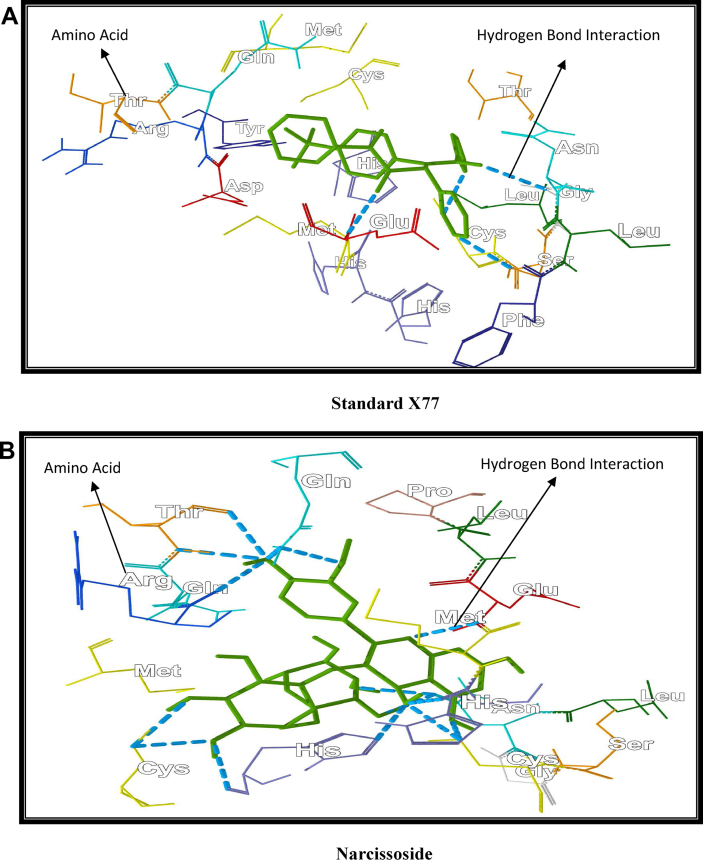
The results reported in Table 1, Table 2 and Fig. 2 clearly indicated that Narcissoside fits perfectly at the active site of Protein 6W63, a novel corona virus COVID-19.
Table 1.
The Docking Score of Standard X77 and Narcissoside with corona virus protein 6W63 (COVID -19).
| S.No. | Ligand | Protein | Interaction Score |
||
|---|---|---|---|---|---|
| MolDock | Rerank | H Bond | |||
| 1 | Standard X77 | 6W63 | −156.913 | −121.296 | −5.7369 |
| 2 | Narcissoside | 6W63 | −180.739 | −137.092 | −18.6771 |
Table 2.
The Total Interaction energy of Standard X77 and Narcissoside with corona virus protein 6W63 COVID -19.
| S.No. | Target Atoms: Molecule | Residue | ID | Total Energy of Interaction |
|
|---|---|---|---|---|---|
| Standard X77 | Narcissoside | ||||
| 1 | 6W63 [A] | Arg | 188 | −7.66744 | −10.9985 |
| 2 | 6W63 [A] | Asn | 142 | −15.5609 | −19.9549 |
| 3 | 6W63 [A] | Asp | 187 | −5.02839 | −2.15432 |
| 4 | 6W63 [A] | Cys | 44 | −0.895733 | −1.73084 |
| 5 | 6W63 [A] | Cys | 145 | −10.3877 | −5.53185 |
| 6 | 6W63 [A] | Gln | 189 | −9.77781 | −12.3962 |
| 7 | 6W63 [A] | Gln | 192 | −0.400777 | −7.60682 |
| 8 | 6W63 [A] | Glu | 166 | −23.2155 | −21.4249 |
| 9 | 6W63 [A] | Gly | 143 | −7.30909 | −2.03337 |
| 10 | 6W63 [A] | His | 41 | −23.5486 | −25.5306 |
| 11 | 6W63 [A] | His | 163 | −2.50682 | −0.714996 |
| 12 | 6W63 [A] | His | 164 | −4.38194 | −6.59603 |
| 13 | 6W63 [A] | His | 172 | −1.35267 | NA |
| 14 | 6W63 [A] | Leu | 27 | −3.6484 | NA |
| 15 | 6W63 [A] | Leu | 141 | −7.1357 | −3.79481 |
| 16 | 6W63 [A] | Leu | 167 | NA | −5.21798 |
| 17 | 6W63 [A] | Met | 49 | −4.62123 | −14.7328 |
| 18 | 6W63 [A] | Met | 165 | −20.9047 | −23.4037 |
| 19 | 6W63 [A] | Phe | 140 | −5.14266 | −0.782012 |
| 20 | 6W63 [A] | Pro | 52 | NA | −0.357936 |
| 21 | 6W63 [A] | Pro | 168 | NA | −2.51004 |
| 22 | 6W63 [A] | Ser | 144 | −6.14224 | −1.10625 |
| 23 | 6W63 [A] | Thr | 25 | −0.777035 | −0.430475 |
| 24 | 6W63 [A] | Thr | 26 | −1.97101 | NA |
| 25 | 6W63 [A] | Thr | 190 | −0.547215 | −6.94543 |
| 26 | 6W63 [A] | Tyr | 54 | −1.20212 | −0.778733 |
| 27 | 6W63 [A] | Val | 186 | −0.313283 | −0.87103 |
Fig. 2.
Interaction of Standard X77(A) and Narcissoside (B) with active cavity of corona virus protein 6W63 COVID -19.
It involves complex interaction with 24 active interaction sites of protein CWC6 which were Arg 188, Asn 142, Asp 187, Cys 44, Cys 145, Gln 189, Gln 192, Glu 166, Gly 143, His 41, His 163,His 164, Leu 141, Leu 167, Met49, Met 185, Phe 140, Pro 52, Pro 168, Ser 144, Tr 25, Thr 190, Tyr 54, Val 186.
The Narcissoside showed same complex interaction as showed by the standard inhibitor N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide(X77) in binding state with the protein 6w63 which were Arg 188, Asn 142, Asp 187, Cys 44, Cys 145, Gln 189, Gln 192, Glu 166, Gly 143, His 41, His 163,His 164, His 172, Leu 27, Leu 141, Met49, Met 185, Phe 140, Ser 144, Thr 25, Thr 26, Thr 190, Tyr 54, Val 186. However three amino acid interactions were changed in both the inhibitors. It was observed that Narcissoside interacted with Leu 167, Pro 52 and Pro 168 while X77 interacted with His 172, Leu 27 and Thr 26.
The Docking score indicated that the standard X77 showed MolDock score of −156.913, rerank Sore −121.296 and H Bond −5.7369, while the flavanoid narcissoside had showed MolDock score −180.739, Rerank Sore −137.092 and H Bond −18.6771. The narcissoside showed potent inhibitory effect which is greater than standard X77.
Discussion
In Jan 2020, 25 fatalcases of 2019-nCoV infections were reported in China. The number of deaths in the world is rising quickly. Reports of exported cases have been released in several parts in China and other countries.transmission dynamics and the full spectrum of clinical illness are not yet fully understood. Severe pneumonia, respiratory failure, acute respiratory distress syndrome (ARDS), and cardiac injury, including fatal outcomes are the complications of COVID 19. Many of the patients were infected by exposure to the environment and transmission from person to person. Our present study is aimed to find out the potent inhibitor from natural origin against this corona virus.
In our study both the inhibitors has bound the active cavity of protein and showed significant binding with amino acids. The binding energies of Narcissoside with amino acid (Asn 142 (−19.955), Glu 166 (−21.425), His 41 (−25.531) & Met 165 (−23.04)) were compared with the binding energy of standard X77 with amino acids (Asn 142 (−15.561), Glu 166 (−23.216), His 41 (−23.549) & Met 165 (−20.905)).The result showed that the Narcissoside required least energy to bind with the protein.
The interaction showed that Narcissoside showed high affinity then the standard X77 as Narcissoside showed thirteen hydrogen bonds with nine amino acids (Arg 188, Glu 166, His 164, Cys 145 (2 bonds), Asn 14 (2 bonds), Cys 44 (2 bonds), His 41 (2 bonds), Gln 192, Thr 190) while X77 showed four hydrogen bonds with amino acids (Gly 143, Cys 145, Glu 166, Ser 144) showed in Fig. 2. It was also observed that the andrews affinity i.e. function group contribution to drug receptor interaction, for the narcissoside is 24.6248 which is 2.5 times more than standard X77 (i.e. 9.161).
Conclusion
The outbreak of the pandemic respiratory syndrome caused by a novel coronavirus (COVID-19) requires the attention of global researcher to discover the possible safe and effective medicine to cure the same. In the present work the standard inhibitor X77 and Narcissoside were docked with the protein of novel corona virus COVID-19 6W63.
It was observed that the affinity of the Narcissoside was more as compare to standard X77. Narcissoside get fits perfectly at the active site of the 6W63 and showed thirteen hydrogen bonds with nine amino acids in the cavity, while the standard X77 showed only four hydrogen bonds with amino acids. Further research can be performed to identify the safety and efficacy parameters at both preclinical and clinical stages to evaluate the effectiveness of Narcissoside for the treatment of novel coronavirus COVID-19.
Conflicts of interest
The authors declare that they have no competing interests.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J Med Virol. 2020;92:518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rates S.M.K. Plants as source of drugs. Toxicon. 2001;39:603–613. doi: 10.1016/s0041-0101(00)00154-9. [DOI] [PubMed] [Google Scholar]
- 9.Cordell G.A., Colvard M.D. Some thoughts on the future of ethnopharmacology. J Ethnopharmacol. 2005;100:5–14. doi: 10.1016/j.jep.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 10.Hogan J.C. Jr. Combinatorial chemistry in drug discovery. Nat Biotechnol. 1997;15:328–330. doi: 10.1038/nbt0497-328. [DOI] [PubMed] [Google Scholar]
- 11.Dahanukar S.A., Kulkarni R.A., Rege N.N. Pharmacology of medicinal plants and natural products. Indian J Pharmacol. 2000;32:S81–S118. [Google Scholar]
- 12.Astuya A. Pharmacological activities of flavonoids : a review. Aquacult Res. 2017;48:9006–9014. [Google Scholar]
- 13.Tungmunnithum D., Thongboonyou A., Pholboon A., Yangsabai A. Flavonoids and other phenolic compounds from medicinal plants for pharmaceutical and medical aspects: an overview. Medicines. 2018;5:93. doi: 10.3390/medicines5030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehaghani Zahra Ahmadian, Asghari Gholamreza, Sadeghi Dinani Masoud. Isolation and identification of nicotiflorin and narcissin from the aerial parts of Peucedanum aucheri Boiss. J Agric Sci Technol. 2017;7:45–51. [Google Scholar]
- 15.Tao H., Cui B., Zhang H., Bekhit A.E.D., Lu F. Identification and characterization of flavonoids compounds in cassava leaves (Manihot esculenta Crantz) by HPLC/FTICR-MS. Int J Food Prop. 2019;22:1134–1145. [Google Scholar]
- 16.Olennikov D.N., Partilkhaev V.V. Isolation and densitometric HPTLC analysis of rutin, narcissin, nicotiflorin, and isoquercitrin in Caragana spinosa shoots. J Planar Chromatogr - Mod TLC. 2012;25:30–35. [Google Scholar]
- 17.Xie Z., Sun Y., Lam S., Zhao M., Liang Z., Yu X. Extraction and isolation of flavonoid glycosides from Flos Sophorae Immaturus using ultrasonic-assisted extraction followed by high-speed countercurrent chromatography. J Separ Sci. 2014;37:957–965. doi: 10.1002/jssc.201301340. [DOI] [PubMed] [Google Scholar]
- 18.Tulyaganov T.S., Nazarov O.M., Makhmudov O.E., Vdovin A.D., Abdullaev N.D. N-allylisonitrarine and narcissin from plants of the Nitraria genus. Chem Nat Compd. 2001;37:470–473. [Google Scholar]
- 19.Isolated G., Atriplex F., Herb L., El-aasr M., Kabbash A., El-seoud K.A.A. Antimicrob Immunomodul Activ Flavonol. 2016;8:1159–1168. [Google Scholar]
- 20.Wan C., Yu Y., Zhou S., Tian S., Cao S. Isolation and identification of phenolic compounds from Gynura divaricata leaves. Phcog Mag. 2011;7:101–108. doi: 10.4103/0973-1296.80666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubey Kushagra, Dubey Raghvendra, Gupta Revathi G.A. In-silico Reverse Docking Studies for the identification of potential of Betanin on some enzymes involved in diabetes and its complications. J Drug Deliv Therapeut. 2019;9:72–74. [Google Scholar]
- 22.Madhuri M., Prasad C., Rao Avupati V. In silico protein-ligand docking studies on thiazolidinediones as potential anticancer agents. Int J Comput Appl. 2014;95:13–16. [Google Scholar]
- 23.Heble N.K., Mavillapalli R.C., Selvaraj R., Jeyabalan S. Molecular docking studies of phytoconstituents identified in Crocus sativus, Curcuma longa, Cassia occidentalis and Moringa oleifera on thymidylate synthase - an enzyme target for anti-cancer activity. J Appl Pharmaceut Sci. 2016;6:131–135. [Google Scholar]
- 24.Mavillapalli R.C., Jayabalan S., Muthusamy S. Molecular docking studies of phytoconstituents identified in cinnamomum verum and coriandrum sativum on HMG COA reducatse – an enzyme target for antihyperlipidemic activity. Int J Pharma Sci Res. 2017;8:4172–4179. [Google Scholar]