Abstract
Viruses are the most abundant organisms on our planet, affecting all living beings: some of them are responsible for massive epidemics that concern health, national economies and the overall welfare of societies. Although advances in antiviral research have led to successful therapies against several human viruses, still some of them cannot be eradicated from the host and most of them do not have any treatment available. Consequently, innovative antiviral therapies are urgently needed.
In the past few years, research on G-quadruplexes (G4s) in viruses has boomed, providing powerful evidence for the regulatory role of G4s in key viral steps. Comprehensive bioinformatics analyses have traced putative G4-forming sequences in the genome of almost all human viruses, showing that their distribution is statistically significant and their presence highly conserved. Since the genomes of viruses are remarkably variable, high conservation rates strongly suggest a crucial role of G4s in the viral replication cycle and evolution, emphasizing the possibility of targeting viral G4s as a new pharmacological approach in antiviral therapy. Recent studies have demonstrated the formation and function of G4s in pathogens responsible for serious diseases, such as HIV-1, Hepatitis B and C, Ebola viruses, to cite a few. In this chapter, we present the state of the art on the structural and functional characterization of viral G4s in RNA viruses, DNA viruses and retroviruses. We also present the G4 ligands that provide further details on the viral G4 role and which, showing promising antiviral activity, which could be exploited for the development of innovative antiviral agents.
Keywords: DNA virus, RNA virus, Retrovirus, G-quadruplex, Antiviral therapy, G-quadruplex-ligand, Virus life cycle, Latency, Chronic infection, Non-canonical nucleic acid structure
1. Introduction
Viruses, the most abundant organisms on our planet, affect all living beings, including animals, plants, parasites, fungi and bacteria. Their subtle ability to develop strategies to evade the host immune system, along with the high mutation and replication rates, has made viruses plague terrestrial ecosystems since their appearance. Indeed, virus-borne infectious diseases tend to spread abruptly over wide geographical areas, leading to outbreaks that can dramatically affect health, national economies, and the overall well-being of societies.1 In the past few decades, pharmaceutical and biotechnological advance has succeeded in developing new therapeutics for the management of different viral diseases: for example, anti-retroviral therapy against the human immunodeficiency virus (HIV), or the pan-genotypic direct-acting antiviral drugs used for hepatitis C (HCV) management. Nevertheless, the majority of current therapies are unable to accomplish virus eradication or sustained virological response, therefore the battle against viral pathogens still represents a primary focus of medical research. In addition, the constant onset of new viral pathogens, generally correlated with massive outbreaks, imperils researchers to determine the pathogenic mechanisms of the new viruses, with the ultimate goal to provide new therapeutic approaches in the antiviral field.2, 3
Viruses are obligate intracellular parasites characterized by great diversity in terms of morphology, size, genome nature, and protein composition. A virus particle, also called a virion, consists of the viral genome, which can be either DNA or RNA in a single- or double-stranded state, surrounded by a protein coat, the capsid. Some virus families additionally present an external lipid bilayer called envelope, derived from cellular membranes (Fig. 1 ).
Fig. 1.
Cartoon representation of a generic virus structure. Viruses can be either “naked,” i.e., formed by the genome and capsid only, or “enveloped,” i.e., with the additional presence of the outer lipid membrane (envelope) and surface proteins, as shown.
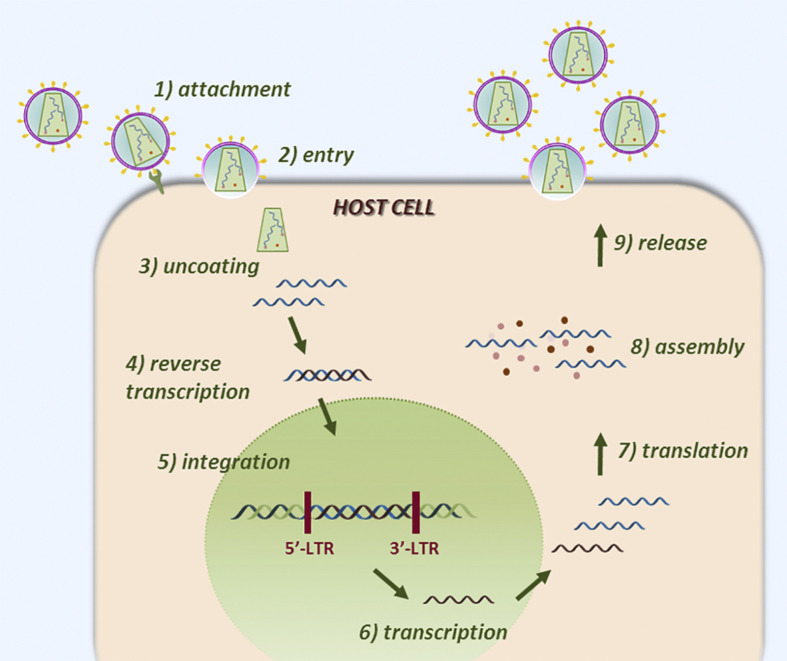
In order to replicate, viruses induce the host cell to synthesize all the essential components necessary to form new virus particles. As a consequence, new virions are assembled in the cell and egress from it to infect other cells, completing their replicative life cycle (Fig. 2 ). Some viruses do not always actively replicate while within the host but remain dormant in defined cell types by integrating their genome into the host DNA, or through formation of circular viral genome molecules (episomes). This process is termed viral latency: through it the virus can evade the host immune system, usually establishing a lifelong infection.4 It is worth noting that each viral class has its own peculiar viral cycle: their details will be described later in this chapter.
Fig. 2.
Cartoon representation of a simplified virus life cycle. The virus attaches to the host cell through surface receptors or proteins (step 1) and enters the cell, according to different mechanisms (step 2). After penetration, the viral genome is uncoated (step 3) and released into the cytoplasm. At this stage, the DNA or RNA genome nature defines the location and the strategy adopted for genome replication (step 4): the majority of DNA viruses replicate in the cell nucleus. Most RNA viruses replicate in the cytoplasm of infected cells. For retroviruses, retrotranscription occurs in the cytoplasm, while integration takes place in the cell nucleus. After viral mRNA synthesis, viral proteins are expressed in the cytoplasm (steps 5–6). The newly synthesized viral elements (genomes and proteins) are assembled into new virions (step 7) and released outside the cell (step 8).
Adapted from Ruggiero, E.; Richter, S. N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res.2018,46, 3270–3283.
Viruses can be classified according to different parameters, such as genome nature, capsid structure, presence of the envelope, type of infection. The most commonly used system is the Baltimore classification, which allocates viruses into seven different classes, according to the nucleic acid type and the resulting mRNA synthesis mechanism (Table 1 ).5
Table 1.
Baltimore classification of viruses.
| Class | Genomea | mRNA synthesis | Virus families |
|---|---|---|---|
| I | dsDNA | mRNA transcribed directly from DNA template |
Herpesviridae Adenoviridae Papillomaviridae Polyomaviridae Poxviridae |
| II | ssDNA | ssDNA converted to dsDNA before transcription |
Anelloviridae Parvoviridae |
| III | dsRNA | mRNA transcribed from RNA genome | Reoviridae |
| IV | ssRNA (+) | RNA genome acts as mRNA |
Coronaviridae Astroviridae Caliciviridae Faviviridae Picornaviridae Togaviridae |
| V | ssRNA (−) | mRNA transcribed from RNA genome |
Rhabdoviridae Filoviridae Paramyxoviridae Arenaviridae Bunyaviridae Orthomyxoviridae |
| VI | ssRNA – RT | Genome is converted to dsDNA by RT, it is integrated into the host's genome and then transcribed to mRNA | Retroviridae |
| VII | dsDNA – RT | DNA is transcribed through an RNA intermediate which acts as mRNA or as template for mRNA synthesis | Hepadnaviridae |
ds = double-stranded; ss = single stranded; RT = reverse transcriptase; + and − indicate strand orientation.
G-quadruplexes (G4s) have been identified in viruses belonging to almost all groups of the Baltimore classification. Indeed, in the past few years, research on the biological role of G4s in virology has boomed, providing strong evidence of the relationship between G4s and the regulation of key viral steps. A recent comprehensive bioinformatics analysis traced putative G4-forming sequences (PQSs) in all human viruses, showing that their distribution is statistically relevant, making them a distinctive feature of the different virus families. In addition, such PQSs are highly conserved, despite the high recombination rates that characterize viruses, suggesting a crucial role for G4s in viral evolution and replication, and corroborating their targeting as a valid pharmacological approach for antiviral therapy.6
2. DNA viruses
DNA viruses include a great variety of viruses in terms of host types, genome length and structure, replication site, replication machinery source and associated diseases. Based on their unique characteristics, they can adopt different replication strategies.7 Most of these viruses share the ability to cause latent infections, where the virus remains quiescent in a specific cellular reservoir, in a non-replicative state until reactivation.8 DsDNA viruses have been found to be particularly enriched in PQSs.6 Indeed PQS-related functional studies have been reported for most herpesviruses, papillomaviruses and also the hepatitis B virus (Fig. 3 ).
Fig. 3.
Summary of G4s reported in DNA virus genomes. For each virus the following information is provided: first column, virion structure and dimension; second column, genome size and organization; third and fourth columns, schematic representation of the G4s (red dots) location in the genome or in the mRNA; fifth column, G4 ligands tested so far on viral G4s and corresponding references.
Adapted from Ruggiero, E.; Richter, S. N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res.2018,46, 3270–3283.
2.1. Herpesviruses
The Herpesviridae family includes a large number of viruses with long linear dsDNA genomes: it comprises more than 100 pathogens classified into three subfamilies, alpha-, beta- and gammaherpesvirinae, which mainly differ in the host target cells. Among all human herpesviruses (HHVs), nine species routinely infect humans, are extremely widespread and responsible for the onset of several different diseases, such as orolabial and genital herpes, chickenpox and shingles, mononucleosis and different types of cancer (Table 2 ). Interestingly, more than 90% of adults are infected with at least one of these.9
Table 2.
Human herpesviruses classification.
| Subfamily | Virus | Acronym | Major related diseases |
|---|---|---|---|
| Alpha | Herpes simplex virus type 1 | HSV-1 | Orolabial herpes, encephalitis |
| Herpes simplex virus type 2 | HSV-2 | Genital herpes | |
| Varicella zoster virus | VZV | Chickenpox, shingles | |
| Beta | Cytomegalovirus | CMV | CMV-mononucleosis, congenital cytomegalic inclusion disease |
| Human herpesvirus 6A/6B | HHV-6A/B | Roseola infantum | |
| Human herpesvirus 7 | HHV-7 | Roseola infantum | |
| Gamma | Epstein-Barr virus | EBV | Mononucleosis, lymphomas, leukemia, lymphoproliferative syndromes |
| Kaposi's sarcoma-associated herpesvirus | KSHV | Kaposi's sarcoma, Castleman disease |
HHVs differ significantly in terms of DNA base composition and sequence arrangement; nonetheless, they share many biological properties, including the ability to sustain, after a primary infection, a latent state through maintenance of their genome in the infected organism as an extrachromosomal nuclear episome.10 During their life cycle, HHVs trigger an intricate chain of events to achieve replication, which involves both cellular and viral structures. Briefly, after attachment to the host's cell surface, the virus penetrates into the cell cytoplasm and migrates to the nucleus, where it releases the viral genome. Here, different complex steps lead to the synthesis of concatemer copies of viral DNA, which, along with the newly translated viral proteins, are assembled into new virions and finally released.11
Pharmacological research in the antiherpetic field has been quite successful, since to date, several efficient drugs are available on the market. One of the most efficient antiviral agents is acyclovir, along with its derivatives. These compounds are classified as nucleosides analogues: they block viral replication by being incorporated into the nascent viral DNA. They have been widely employed for the treatment of alpha-HHVs and are still considered the first line drugs of choice in the management of these viruses. However, the excessive use of anti-HHVs agents over large periods, especially in immunocompromised patients, has led to the onset of resistant viral strains. Moreover, considering that HHVs undergo latency, and that, to date, there is no way to eradicate the virus from the host cells, the development of new compounds with innovative targets and mechanisms of action is urgently needed.11
Genome-wide bioinformatics analysis has revealed remarkable PQS density in all HHV species, mostly in genome regulatory and repeated regions, which are crucial to sustain the episomal form. The enrichment, distribution and conservation rate of these sequences strongly corroborate G4s as functional elements in the regulation of the viral life cycle.12, 13 In addition, a recent study demonstrated an interesting correlation between G4 sequence composition in HHVs and their hosts, suggesting a possible long-term virus-host co-evolution process.14 Consequently, G4s may represent efficient pharmacological targets, alternative to those aimed at by the current anti-herpetic therapies.
2.1.1. Alpha-herpesviruses
The subfamily of alpha-HHVs includes HSV type 1 and 2, and VZV: these are characterized by the establishment of a persistent infection in sensory neuronal cells. Most of the studies conducted on the role of G4s in HHVs regulation focused on the HSV-1 virus, as its genome is characterized by 68% GC content, with multiple and highly stable G4-forming sequences.15 HSV-1 G4s formation has been monitored in infected cells through a G4-specific antibody at different time points post-infection. G4s were found to form in a virus cycle-dependent fashion. During viral replication, viral G4s fold massively in the cell nucleus and localize in different compartments according to the viral genome movements in the cell during the viral cycle.16 Bioinformatics and the following experimental analysis traced conserved PQSs in: (i) the promoter region of immediate-early genes of all alpha-HHVs, genes that are expressed at the very early stage of viral infection and encode for regulatory proteins involved in all subsequent viral steps17 (ii) the terminal and internal repeated regions, which include packaging signals involved in concatemer cleavage during replication13 (iii) the proximity of recombination breakpoints.18 Most of the identified G4s proved to be exceptionally stable when tested in vitro through biochemical and biophysical assays.
Employment of known G4 ligands has provided interesting insights in the regulation of HSV-1 G4s, first and foremost the reduction in virus replication, obtained after treatment with either BRACO-19 (IC50 ~ 8 μM)15 or a core-extended naphthalene diimide derivative (IC50 ~ 18 nM).19 Both compounds led to a significant reduction in viral DNA synthesis and late transcripts production. Furthermore, in vitro assays revealed the ability of G4 ligands to improve further HSV-1 G4s stability and their ability to stall polymerase progression.
2.1.2. Beta-herpesviruses
Viruses belonging to the Betaherpesvirinae subfamily are the human CMV, HHV-6 and HHV-7, which mainly exploit leukocytes as target cells for latency. Recent work on this class of viruses has analyzed the regulatory role of G4s in the human CMV and HHV-6.
2.1.2.1. Human cytomegalovirus
The human CMV is estimated to affect more than 80% of the worldwide population. It has the largest dsDNA genome among HHVs (~ 235 kb), which was found to include a remarkably high number of PQS, distributed among the promoters of immediate-early, early and late genes. The majority of the sequences have been demonstrated to actually fold into G4 structures, and to be stabilized after treatment with two different porphyrin-based G4 ligands, TMPyP4 and NMM. Interestingly, in a luciferase reporter system that included different PQSs in viral promoters, NMM was shown to suppress the expression of several mRNAs and to reduce both intracellular and extracellular viral DNA levels.20 Recently, the anti-CMV activity of a polymerase inhibitor CX-5461, a known G4 ligand currently in phase I clinical trial, was also demonstrated. Interestingly, the compound induced a 2.0 log reduction in viral titer, together with a significant decrease in the amount of viral DNA and pUL44, the viral processivity factor: these data indicate that the effect of CX-5461 is exerted at the viral DNA replication stage of the viral life cycle. Considering that the compound is known to induce a DNA damage response (DDR), its antiviral activity may be related to a double G4-related mechanism of action, involving both the viral targets and the activation of cellular stress response pathways.21
2.1.2.2. Human herpesviruses 6A/6B
HHV-6 consists of two related viruses known as HHV-6A and 6B, which infect almost 100% of the human population and cause the febrile illness roseola infantum, also called the sixth childhood eruptive disease. The HHV-6 genome is characterized by the presence of variable-length telomere-like repeat regions at its termini, which can integrate by homologous recombination into the human chromosome at the telomeres; however, the mechanisms regulating this process remain poorly understood. Considering that telomeres are commonly known to fold into G4s, involvement of these non-canonical arrangements in the regulation of HHV-6 chromosomal integration is plausible. Indeed, treatment of HHV-6 infected telomerase-expressing cells with the G4 ligand BRACO-19 resulted in a significant reduction in chromosomal integration, likely due to the prevention of telomere end elongation.22
2.1.3. Gamma-herpesviruses
The human viruses EBV and KSHV, classified as gamma-HHVs, establish latency in lymphocytes. They are both associated with the development of lymphomas originating from B cells, especially in immunocompromised patients.
2.1.3.1. Epstein-Barr virus
EBV is one the most common human viruses in the world, affecting more than 90% of the world's population. Its primary infection tends to be generally asymptomatic. However it could lead to infectious mononucleosis and, in the worst case, to various types of cancer. Its genome comprises genes encoding for six EBV nuclear proteins that are expressed only in the latent infection and sustain the viral episome maintenance.23 Studies on the characterization and role of G4s in EBV showed that the EBV-encoded nuclear antigen-1 (EBNA1), the viral protein pivotal to immune evasion, promotes viral DNA replication by interacting with RNA G4s and recruiting the cellular origin replication complex. The EBNA1 mRNA itself is enriched in folded G4s, which behave as cis-acting modulators of viral mRNA translation, producing ribosome dissociation.24, 25 EBNA1 mRNA G4s have also been found to regulate the endogenous presentation of EBNA1-specific CD8 + T-cell epitopes, seen in persistent infections.26 The employment of G4 ligands has helped the investigation of the functional and biochemical characteristics of EBNA1. BRACO-19, for instance, stabilized the viral RNA G4 and, in infected lymphocytes, reduced EBV genome copy numbers. It also negatively modulated transcription levels of EBNA2 and EBNA3A, and inhibited EBNA1-dependent DNA replication.24 Another G4 ligand, pyridostatin (PDS), enhanced the stability of the EBV G4s, reducing EBNA1 synthesis level in a concentration-dependent way, both in vitro and in vivo. As a result, EBV-infected cells were found to be less efficiently recognized by virus-specific T cells, although such a mechanism still needs to be elucidated. These data show that G4-mediated inhibition affects processes that are crucial for viral DNA replication,25 reinforcing the importance of alternative DNA structures in the regulation of virus processes. The cellular protein nucleolin, which has been widely shown to bind G4s,27 recognizes and also binds viral EBNA1 mRNA G4s. As a consequence it downregulates EBNA1 expression and antigen presentation, counteracting EBV stealthiness. Such interactions can be targeted to modulate EBV progression: indeed the PhenDC3 G4 ligand was found to prevent nucleolin binding to EBNA1 mRNA G4s and to increase the endogenous EBNA1 levels in EBV-infected B cells and in cells derived from a nasopharyngeal carcinoma.28 Following on from PhenDC3, variously substituted bis(acylhydrazones) derivatives have also been investigated for their ability to interfere with the nucleolin/EBNA1 mRNA G4s interaction. Two compounds, PhenDH2 and PyDH2, have been found to be more active and less toxic with respect to the lead compound.29 However, it is worth noting that although many of the tested PhenDC3 analogues displayed strong in vitro binding and stabilization of RNA G4s, only a few of them were able to interfere with EBV immune evasion. This result suggested that the ability to hamper nucleolin/EBNA1 interaction might require additional activities, independent from G4 stabilization. Indeed, different G4 ligands can generate opposite effects: this is the case of PDS, which, in contrast to PhenDC3, further suppressed EBNA1 mRNA translation,25 as also demonstrated by quantitative proximity ligation assay.30 All these data support the ability of EBNA1 G4s to operate as nucleolin-binding platform. A very recent study reported that they also promote nuclear retention,31 but the relationship between the two processes needs further investigation. Notably, EBV nuclear antigens are involved in B-cells transformation, especially EBNA1, which is expressed in all EBV-related tumors.32 Therefore, it is tempting to speculate that the nucleolin/EBNA1 G4 complex could be involved in virus-related oncogenesis and it might be exploited as a pharmacological target to impair both virus and tumor progression.
G4s have also been observed in mRNAs of additional gamma-HHV maintenance proteins, known to regulate their self-synthesis, corroborating the structural regulatory role of G4s in the virus system.25
2.1.3.2. Kaposi's sarcoma-associated herpesvirus
KSHV is the causative agent of Kaposi's sarcoma and other lymphoproliferative disorders, such as primary effusion lymphoma and a plasmablastic form of multicentric Castleman's disease. It mostly concerns AIDS patients, as KSHV seroprevalence is directly proportional to HIV seropositivity rates. At the moment, there are no treatments for the lytic or latent infections.33 The KSHV genome is arranged in a ~ 140 kb long central region, encoding viral genes, flanked by repeated regions (terminal repeats, or TRs), which harbor the viral episome unique origin of replication. TRs are highly enriched in G/C bases and have been demonstrated to form stable G4s, located both in the forward and reverse strands, and involved in the regulation of episomal replication. Stabilization of viral G4s by means of different G4 ligands, PhenDC3 and TMPyP4, stalled the replication fork at the TR level, with consequent replicative stress and inhibition of KSHV DNA replication, which resulted in the dramatic reduction (60%) of viral episome copy numbers.34 In addition, the KSHV latency associated nuclear antigen 1 (LANA1) protein, which is the master regulatory protein of KSHV latency, forms G4 structures in its own mRNA, analogous to EBNA1. G4 stabilization induced by TMPyP4 treatment downregulated LANA1 translation, with consequent reduction of protein expression in KSHV infected cells, suggesting that LANA1 mRNA G4s are involved in the regulation of the translation machinery. Interestingly, reduced translation has been found to be strictly related to reduction of antigen presentation, one of the main mechanisms adopted by LANA1 to promote immune evasion.35
Altogether, these data confirm that G4s are key regulatory element in the regulation of host-pathogen interaction in the Herpesviridae family and support the feasible exploitation of these structures as innovative pharmacological targets in the antiherpetic therapy.
2.2. Other DNA viruses
Among DNA viruses, G4s have been also studied in the human papillomavirus (HPV) and the hepatitis B virus (HBV).
2.2.1. Human papillomavirus
HPVs include more than 100 characterized viruses, which infect and replicate within epithelial structures, inducing specific hyperplastic lesions. Among them, those defined as “high-risk” HPVs can efficiently induce malignant transformation of infected cells, leading to the onset of cervical, anogenital, and several oropharyngeal cancers. Notably, high-risk HPVs infections cause almost 99% of cervical cancers and approximately 5% of all cancers worldwide.36 Currently, despite the availability of anti-HPV vaccines, access to effective treatments is limited in many parts of the world; furthermore, no specific anti-HPV drugs are available. G4 forming sequences have been found in 8 out of the 120 identified HPV types, all belonging to the “high risk” HPV subgroup.37 Nevertheless, the majority of the identified sequences are characterized by high polymorphism,38 which allows the formation of different alternative structures, i.e., hairpins.39 Such dynamism makes HPV G4s difficult to study and, above all, to target; therefore, further in-depth studies will be necessary to disclose the involvement of HPV G4s in the viral life cycle.
2.2.2. Hepatitis B virus
HBV is a hepatotropic virus, characterized by a partially double-stranded, circular DNA genome, which replicates through an RNA intermediate, synthesized by a reverse transcriptase. Persistent HBV infections are related to serious liver damage, eventually leading to cirrhosis and hepatocellular carcinoma. The pronounced genetic variability associated with HBV determines the existence of 10 genotypes (A–J) which differ in terms of transmissibility, virus loads, response to antiviral therapy, and ability to cause liver disease. Hundreds of millions of people worldwide are chronically infected with HBV, and despite the availability of a prophylactic vaccine and antiviral drugs that keep HBV levels under control, full eradication is not yet achievable. Treatments are urgently needed, as almost 1 million deaths per year are estimated to be caused by HBV-related cancers and associated diseases.40
Very recently, a G4-forming sequence was identified as a genotype-specific regulator of HBV replication, located in the preS2/S promoter of HBV genotype B. The sequence was shown to fold into an intramolecular hybrid G4 structure, the stabilization of which surprisingly led to enhanced transcription of the preS2/S gene promoter, with consequent production of the HBV surface antigen (HBsAg). The positive modulatory effect was demonstrated through the employment of the G4 ligands BRACO-19 and PDS, revealing a yet unknown role for DNA secondary structures in the viral context.41
3. RNA viruses
RNA viruses constitute the major part of the global virome, occupying four out of seven of Baltimore's classes. They display the highest mutation rates, which often generate multiple complex mixtures of related virus variants, feature responsible for their adaptability to environment selection. The high number of mutations is generated by the RNA-dependent RNA polymerase that lacks proofreading activity.42 This enzyme synthesizes RNA from the RNA template and, therefore, it is essential for replication of most RNA viruses.
PQSs have been abundantly found in both positively- and negatively-stranded ssRNA viruses, while very few have been observed in dsRNA viruses.6 Most of the studies reported so far regard flavi-, filo- and coronaviruses (Fig. 4 ).
Fig. 4.
Summary of G4s reported in RNA virus genomes. For each virus the following information is provided: first column, virion structure and dimension; second column, genome size and organization; third and fourth columns, schematic representation of the G4s (red dots) location in the genome or in the mRNA; fifth column, G4 ligands tested so far on viral G4s and corresponding references.
Adapted from Ruggiero, E.; Richter, S. N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res.2018,46, 3270–3283.
3.1. Flaviviruses
The Flaviviridae family comprises a large number of (+)-ssRNA viruses, the genomes of which serve also as mRNA for viral protein translation. More specifically, the whole genome is translated into a unique polyprotein, which subsequently undergoes different cleavages by means of viral and host proteases, to finally provide three structural proteins, namely the pre-membrane/membrane (prM/M), the capsid (C) and the envelope (E) proteins, and seven non-structural (NS) proteins. At both ends of the viral genome, there are two untranslated regions (UTR) of different length, which are essential for viral replication and immune modulation and whose activity is strongly related to the secondary structures adopted, mainly stem-loops.43 Flavivirus infections represent a serious global threat because of the continuous outbreaks reported worldwide. Pathogenic flaviviruses are mainly transmitted by mosquitoes or ticks and subsequent infections can lead to the onset of severe neurological and non-neurological diseases. The family comprises over 100 virus species, including emerging and re-emerging arboviruses of global significance, such as Japanese encephalitis (JEV), West Nile (WNV), Zika (ZIKV), dengue (DENV), yellow fever (YFV), and tick-borne encephalitis (TBEV) virus, and also the hepatitis C virus (HCV), which belongs to the Hepacivirus genus. Albeit commercially available vaccines for some of these viruses have been developed, nowadays no effective specific treatments exist, besides the recent one established against HCV. Considering the global threat of flaviviral pandemics, specific treatments are craved for.44
3.1.1. ZIKA virus
ZIKV is transmitted to humans by mosquito bites: in adults, it usually causes mild to no symptoms, while it might be devastating if contracted during pregnancy, as it is related to microcephaly in the unborn child. Several G4 forming sequences have been discovered in the positive strand of the ZIKV genome, while the negative one is devoid of PQSs. Seven of the identified sequences were found to be surprisingly conserved within more than 50 analyzed flavivirus genomes. Given the great viral genome variability, this observation suggests a significant involvement of these PQS in the regulation of the life cycle of these viruses. An additional G4 in the unique 3′-UTR region is present in ZIKV: this region is pivotal to viral replication of the negative-sense strand, therefore it could be a target to impair viral progression.45
3.1.2. Hepatitis C virus
HCV affects more than 70 million people worldwide. It can cause both acute and chronic hepatitis, possibly leading to cirrhosis, liver failure and cancer. Indeed, HCV is responsible for 25% of liver cancers. The HCV genome, in common with that of all flaviviruses, is organized into a long open reading frame, which produces a unique polyprotein that is processed by proteases to form 10 structural and non-structural proteins. Notwithstanding the numerous virus variants emerging constantly in patients, the core protein is the least variable and therefore it represents an attractive antiviral target.46 In the last decade, the development of direct-acting antiviral agents (DAAs), a combination of drugs targeting specific nonstructural proteins, has dramatically changed the management of HCV infections, improving sustained virological response by 90%.47 Nevertheless, because of the possible emergence of resistant strains (https://www.hcvguidelines.org/evaluate/resistance, accessed Jan 23, 2020), identification of new targets is always highly valued.
Bioinformatics and biophysical analysis of HCV G4s has demonstrated the existence of six conserved PQSs throughout the whole viral genome, mostly in the negative strand. The one located in the core gene was characterized as the most stable G4.48 Its stabilization with the G4 ligands TMPyP4 and a PDS derivative (PDP) promoted viral RNA-dependent RNA polymerase stalling, with consequent reduction in HCV core protein levels. In vivo analysis revealed that PDP led to G4-mediated antiviral activity in the low micromolar range, by inhibiting intracellular replication of different HCV genotypes.49 A deeper investigation also showed that the HCV core RNA G4 is recognized and bound by the cellular protein nucleolin. This interaction, which has been found to be structure-related, resulted in inhibition of HCV replication in cells. Such activity was counteracted by the employment of G4 ligands such as PDS and PDP, which competed for the same binding site. TMPyP4 did not produce the same effect, however, this last compound is in general less selective for G4s.50 Moreover, HCV infection promoted nucleolin overexpression, which in turn induced a drastic reduction of HCV total RNA levels. Conversely, nucleolin silencing was directly correlated with increased HCV RNA expression.48 These results strongly suggest the existence of a host anti-viral immunity mechanism involving G4 structures. Additional bioinformatic studies based on a different algorithm showed the presence of a very interesting G4-forming sequence in the stem-loop IIy of the HCV negative strand, precisely in the 3’-UTR, which is the initiation site for the (+)-strand replication. The G4 sequence, highly conserved in many different HCV strains, was strongly stabilized by the bisquinolinium derivative PhenDC3 and as a consequence, RNA synthesis was hampered by more than 70%. Moreover, in a luciferase system in which luciferase expression was directly related to viral replication, 60% reduction was observed upon treatment with the compound.51
HCV was also used as the reference RNA virus to develop a fluorescence light-up probe, a thioflavine T derivative (ThT-NE), for the direct visualization of the native HCV RNA genome in living cells. ThT-NE showed remarkable specificity for HCV RNA G4s with respect to other G4s in different cell models, probably because during infections viral G4s outnumber cellular G4s in order to maximize viral replication. Moreover, the HCV replication machinery promotes formation of vesicle-like viral replication complexes that contain most of the newly synthesized viral RNA, generating a localized G4 enrichment that improves ThT-NE recognition.52 If this strategy were applicable also to other pathogens, it would represent a striking turning point in pathological research and clinical diagnosis.
3.2. Filoviruses
Filoviruses comprises five genera of viruses, including deadly pathogens such as Ebola (EBOV) and Marburg (MARV) viruses, which cause fatal hemorrhagic fever in humans and primates, with no cure available thus far. These viruses are characterized by filamentous virions with a ~ 19 kb-long (−)ssRNA, which includes seven genes. Filoviruses RNA replication consists in the generation of a (+)ssRNA intermediate, namely the anti-genome, which acts as template for the synthesis of new genomes. Studies on the presence of G4s in filovirus genomes has revealed a highly conserved G-rich sequence in the EBOV L gene, which encodes for the viral RNA-dependent RNA polymerase and modulates viral replication and transcription. In vitro analysis has demonstrated that this sequence folds into a dynamic parallel G4, stabilized by treatment with the TMPyP4 ligand. Interestingly, this compound was able to reduce L gene transcription and to impair replication of the EBOV mini-genome, a cell-based method involving firefly luciferase as reporter protein.53 Moreover, induced circular dichroism experiments performed in the presence of the fluorescent probe thiazole orange, revealed the folding into G4s of several sequences derived from original viral isolates of both EBOV and MARV.54 Additional investigations will be necessary to assess the biological role of the identified sequences in the viral life cycle.
3.3. SARS coronavirus
The severe acute respiratory syndrome coronavirus (SARS-CoV) was identified after a massive outbreak in 2003, which led to the death of almost 10% of the affected people. The virus is one of the most pathogenic viruses in humans and it is highly contagious. Furthermore, no effective drugs are available against it. SARS-CoV belongs to the Coronaviridae family and has a (+)-ssRNA genome of 29.7 kb in length, encoding for 13 viral genes. The nonstructural protein 3, also known as SARS unique domain (SUD), plays a crucial role in viral replication and transcription. It was demonstrated to preferentially bind G4-forming oligonucleotides that form in the 3′-UTR of mRNAs coding for host-cell proteins implicated in the regulation of several processes, such as apoptosis and signal transduction. For this reason, the SUD/G4 interaction has been speculated to be involved in the modulation of genome replication and host cell response to viral infection.55, 56
In December 2019, a novel coronavirus named SARS-CoV2 was identified in Wuhan, China; it causes the coronavirus disease (COVID-19), which in a minority of people progresses to severe atypical pneumonia that requires hospitalization and has extremely high mortality among the elderly and immunocompromised people.57 SARS-CoV-2, being highly contagious, caused a pandemic (COVID-19) in the early months of 2020. SARS-CoV and SARS-CoV-2 share 86% genome homology: they both infect humans by entering the lower respiratory tract through the angiotensin-converting enzyme 2 (ACE2).58 However, an amino acid composition difference of about 25% was found between the SUD of the two viruses,59 therefore further characterization of the SARS-CoV-2 SUD will be necessary to assess its function and possible interaction with G4 nucleic acids.
4. Retroviruses
Retroviruses are the most ancient discovered viruses: they are able to infect almost all vertebrates worldwide, causing severe diseases, such as immunodeficiencies, neurological disorders, and cancer. Retroviruses are classified into two subfamilies: Orthoretrovirinae, including alpha-, beta-, gamma-, delta-, epsilon-retroviruses and lentiviruses; Spumavirinae, represented by the genus spumavirus. They possess two copies of (+)-ssRNA genome, formed by four coding genes, gag, pro, pol and env, several genes encoding for accessory proteins and two flanking repeat segments followed by untranslated regions. The pol gene encodes for the reverse transcriptase (RT) enzyme, the RNA-dependent DNA polymerase that allows synthesis of viral DNA from the genomic RNA. The newly formed dsDNA, which presents two identical long terminal repeats (LTRs) at both ends, once integrated into the host's chromosomal DNA forms the provirus. The provirus is transcribed and translated to form new virions, or it may remain dormant, i.e., latent: in both cases, it can be no longer eliminated from the host. The LTR located at the 5′-end represents the control center for viral gene expression. It is organized in three sections, U3, R, and U5. The U3 region includes binding sites for transcription factors and acts as the unique promoter for retroviral transcription.60
Interestingly, highly conserved PQSs have been found in the majority of retrovirus subfamilies, mainly located in the LTRs. Biophysics assays have demonstrated the actual ability of these sequences to fold into G4s, which were strongly stabilized by G4 ligands, such as BRACO-19 and a core-extended NDI, supporting the possibility of targeting the LTR G4s as innovative antiretroviral therapy.61, 62
4.1. Lentiviruses
Viruses belonging to the Lentiviridae family infect a broad range of mammals: they are responsible for the onset of several neurological and immunological deficiencies, the most known of which is the human acquired immunodeficiency syndrome (AIDS) caused by HIV-1. Related diseases, associated to different lentiviruses, can be observed also in species like simians, felines, equines and cattle.
Analysis of PQSs in these viruses has mainly focused on the promoter region, the 5′-LTR, revealing that almost all members of the family share the possibility to fold into G4s. The most intriguing result was the abundance of PQSs in relation to species: all lentiviruses affecting primates showed PQSs in their 5′-LTR. A lower percentage was observed in the bovine lentiviruses, while none, or very low, in the phylogenetically more distant species. These data suggest that the G4 structures are beneficial to the overall virus biology and have consequently been selected for throughout evolution. Furthermore, a strong correlation between G4s and the binding site for the transcription factors Sp1 and NF-κB was observed in almost all primate lentiviruses, corroborating the key role played by G4s in the regulation of viral transcription.61
4.1.1. Human immunodeficiency virus type 1
The HIV-1 virus, lead representative of the lentivirus subfamily, is still one of the most serious public health challenges. Since its discovery in the early 1980s, successful and significant efforts have been reached in terms of preventions and treatment: to date, almost 38 million people worldwide are affected by the virus, more than half of which are receiving antiretroviral therapy (ART), a combination of three or more antiretroviral drugs that reduces virus production and correlated disease progression (https://www.who.int/gho/hiv/en/, accessed Jan 23, 2020). Nevertheless, there is still no cure to eradicate the virus from the host, and HIV-related deaths continue to impact society and economy: consequently, identification of alternative and effective antiviral targets is urged.
G4s perfectly fit into this picture, as they could represent key elements in HIV-1 targeting, both in the lytic and latent state. Research on G4s in this virus has been quite productive, on both the viral genomic RNA and the integrated provirus (Fig. 4).
The HIV-1 genome, as mentioned above, consists in two copies of positive, single-stranded RNA that, once synthesized, dimerize in order to be encapsulated into the new virion: this step is critical for viral recombination (Fig. 5 ). Several G-rich sequences have been found in proximity of the dimerization site63 and in the central portion of the genome, adjacent to a central polypurine tract.64 Both RNA regions are associated with dimerization and primer-strand transfer during reverse transcription, events that promote recombination. Collected data infer that the G4s in this region might be involved in the regulation of the aforementioned processes, making RNA G4s fundamental for the virus.
Fig. 5.
Cartoon representation of the HIV-1 life cycle. The virus attaches to the host cell through surface receptors (step 1) to enter the cell (step 2). After penetration, the viral genome is uncoated (step 3) and released into the cytoplasm. At this stage, the viral RNA is retro-transcribed into a double-stranded intermediate (step 4) which enters the nucleus, where it can be integrated into the host genome (step 5). After transcription (step 6), mRNA acts as template for the synthesis of new viral genome copies and for the expression of viral proteins in the cytoplasm (steps 6). The newly synthesized viral elements (genomes and proteins) are assembled into new virions (step 7) and released outside the cell (step 8).
Adapted from Ruggiero, E.; Richter, S. N. G-Quadruplexes and G-Quadruplex Ligands: Targets and Tools in Antiviral Therapy. Nucleic Acids Res.2018,46, 3270–3283.
At the 5′-UTR, specifically in the U3 segment, a highly G-rich sequence, overlapping with three Sp1 binding sites, has been found to fold into multiple G4s. These structures provide a contact point for dimerization between the two RNA strands, contributing to viral recombination.65 In detail, three G4s have been identified, U3-II, U3-III and U3-IV, which showed remarkable stability and were well recognized by the G4 ligand BRACO-19, which was able to stall RT progression in vitro, therefore impairing viral DNA synthesis and thus viral production.66 The nucleocapsid protein (NCp7), a viral protein that assists RT by unwinding HIV-1 RNA genome secondary structures in order to promote polymerase progression, could counteract such mechanism. NCp7 is known to preferentially bind single-stranded nucleic acids compared to duplex states, and works as a chaperone, as it promotes folding of nucleic acids in their most thermodynamically stable conformations.67 It was recently demonstrated that NCp7 contributes to RT processivity by recognition and subsequent unfolding of HIV-1 RNA G4s.68 NCp7 has also been reported to interact in vitro with a G4 sequence located in the HIV-1 central DNA flap overlapping positive-strand, which forms a tetramolecular G4.69 The G4s in this region might be involved in the protection of the pre-integrated genome from nuclease degradation. However, the biological outcome of the G4-NCp7 interaction still needs to be elucidated in vivo.
Regarding the provirus, the LTR U3 promoter sequence is highly enriched in guanines, especially at the Sp1 and NF-κB binding sites.70 In this region, three overlapping G4 structures can form: LTR-II and LTR-III that are constitutively formed, and LTR-IV, the folding of which is induced by proteins or ligands binding.71 LTR-III and LTR-IV structures, which have been characterized through NMR spectroscopy, showed very peculiar features. LTR-III G4 folds into a hybrid, three-layered quadruplex, with a conserved duplex stem-loop in the central loop.72 LTR-IV G4 has a parallel topology with a bulged G-tract at its 3′-end.73 Such distinctive structures could be exploited to design specific ligands able to selectively target HIV-1 LTR G4s, overcoming the low selectivity that characterizes currently available G4 ligands. The two LTR G4s are mutually exclusive and induce opposite effects on the LTR promoter activity. LTR-III G4 stabilization inhibits viral transcription,71 while LTR-IV G4 hampers this activity.73 The folding/unfolding of this region is controlled by at least two cellular proteins: nucleolin, which greatly stabilizes the LTR G4s, with a consequent overall silencing effect,74 and the human ribonucleoprotein A2B1, which in turn unwinds the G4s to promote transcription.75 These data infer that the fine balance between G4s might be one of the regulatory mechanisms in the regulation of HIV-1 promoter activity. Moreover, it has been very recently shown that the LTR transcription machinery also involves another alternative structure, the i-motif,76 which can form in the provirus negative strand, further supporting the regulatory role of non-canonical nucleic acids structures in virus biology.77 Several G4 ligands have been tested to assess the effect on HIV-1 G4s on the viral promoter and promising stabilizing activity was observed, leading to an overall antiviral effect. BRACO-19 significantly stabilized the naturally occurring G4s, LTR-II and LTR-III, and induced folding of LTR-IV. Upon treatment with increasing compound concentrations, LTR promoter activity was reduced by almost 70% when compared to the untreated condition: a mutated non-G4 sequence used as control showed no variation in promoter response and thus confirmed the effect to be G4-related.71 BRACO-19 was then tested in various cell lines, using different HIV-1 strains. It exerted promising G4-mediated antiviral activity, in the micromolar range. Considering that the G4 ligand also stabilizes HIV-1 RNA G4s, the antiviral effect could be ascribed to a dual mechanism of action, both at the pre- and the post-integration level.66 The development of a new series of core-extended NDI derivatives has significantly improved the anti-HIV-1 activity of the G4 ligands. In fact, these molecules, characterized by exceptional solubility properties, displayed outstanding in vitro stabilizing activity on HIV-1 LTR G4s, with preference for the viral over the human telomeric G4s, used as a cellular reference. Most importantly, the series exhibited strong antiviral activity in the low nanomolar range (IC50 < 25 nM). Time-related antiviral assays combined with reporter assays, have confirmed the G4-mediated mechanism of action.78
Further investigation on G4s in the HIV-1 integrated genome have identified three PQSs in the nef gene. This encodes for the viral accessory protein Nef, a factor involved in the maintenance of a persistent HIV infection in vivo. The nef gene is located at the 3′-end of the viral genome, with its coding region that partially overlaps with the 3′-LTR. The observed PQSs were highly conserved and able to fold into two-tetrads G4s. Nef G4s were stabilized in vitro by different G4 ligands, such as TMPyP4, BRACO-19 and PIPER. Interestingly, employment of TMPyP4 in the TZM-bl reporter cell line, which supports Nef-dependent HIV-1 replication, resulted in a dose-dependent inhibition of viral infectivity.79 Altogether, these data support the possibility of exploiting viral G4s as novel anti-HIV-1 targets, possibly in both the lytic and latent infection.
Indeed, one of the most appealing and interesting aspects about G4s in HIV-1 is their possible involvement in the regulation of viral latency. The virus establishes a state of latent infection in T cells: at this stage, it is not susceptible to antiretroviral drugs. The molecular mechanisms underlying this process are complex and not completely understood; nonetheless some forms of latency entail transcriptional interference, thus the 5′-LTR and its G4s might be involved.80 G4 stabilizing agents have been tested in cells infected with latent HIV-1, where they showed strong antiviral activity, effectively linking G4s to HIV-1 latency. BRACO-19 in latently infected cells reduced viral titer to undetectable levels, an effect that was still observed after long-term treatment. TMPyP4 administration also blocked viral replication in two lymphocyte T cell lines with established HIV-1 latency. The antiviral activity was found to be coupled with higher apoptosis/death rates, with respect to untreated cells and the effect was enhanced upon association with DNA damage repair inhibitors.81
5. Discussion and final remarks
The involvement of G4s in the regulation of cellular processes, such as transcription, replication, translation and telomere maintenance, has been widely accepted by the scientific community. The development of specific G4 ligands with promising anticancer effects has encouraged identification of new ways to harness G4 modulation in other human diseases, including viral infections. Altogether, the data collected in the past few years on G4s in viruses, their interaction with specific ligands and the consequent effect on the modulation of the viral machinery validate the possibility of developing G4-based antiviral drugs. The first aspect to acknowledge is that all the characterized viral G4s are located in highly-conserved genomic regions. In virology, base-conservation analysis is a critical issue, since viruses are characterized by high mutation rates.82 The limited availability of deposited sequences for many viruses, especially the emerging ones, limits a comprehensive conservation analysis; however, data accumulated so far clearly show that PQSs are conserved elements within viruses, thus representing efficient targets in antiviral research.
Taking into account that both hosts and viruses have genomic sequences able to organize into G4s, one of the most demanding tasks in the design of antiviral G4 ligands is to accomplish selectivity toward viral versus cellular G4s. As a matter of fact, one of the major limitations in the development of these compounds is in respect to the common chemical features that characterize the currently employed G4 ligands: the large flat aromatic core that stacks on the G quartet, increasing discrimination among different G4s; high-molecular weights and positive-charged side chains, which guarantee loops and grooves interaction to the detriment of cellular uptake.83 Indeed, low selectivity and poor drug-like properties are major factors in the lack of progression of current G4 ligands beyond phase II clinical trials.
In order to achieve selectivity, deep knowledge about the structural characteristics of each identified G4 is necessary. In fact, loop and groove regions are distinctive marks of the G4 structure and are amenable to selective recognition. Structural data on cellular G4/G4 ligand complexes has demonstrated the involvement of the grooves in the stabilization of such interactions, contributing to formation of a compact arrangement that would likely efficiently impair the transcription machinery.84 It is therefore possible to speculate that enhanced selectivity could be mediated by interaction with loop/groove moieties. To this end, NMR and crystallographic determinations of viral G4s and their complexes with ligands are necessary, in order to allow ad hoc design of new antiviral molecules. To date, only two viral G4 structures have been solved through NMR spectroscopy: the HIV-1 LTR-III72 and LTR-IV73 G4s. Molecular docking analysis, coupled to cleavage experiments, has enabled the identification of groove interactions between LTR-III G4 and the NDI-Cu-DETA derivative, a highly selective G4-cleaving agent.85 Structure resolution is thus a valid approach for the development of specific compounds.
Notwithstanding their poor selectivity, most of the G4 ligands tested in viruses have demonstrated promising antiviral activity and, in the majority of cases, the activity was shown to be G4-dependent. One possible explanation for this outcome is the amount of viral G4s, which, during infection, largely exceeds that of cellular G4s, especially during replication, as reported for HSV-1.16 Considering that generally G4s are located in regulatory regions crucial for viral life cycle progression, a combination of abundance of viral G4s and impairment of key viral functions might explain the observed antiviral effect. Exploring the effective folding of G4s during different steps could help not only help understand the role of G4s in different viruses, but it would heighten the possibility of targeting them in the most useful and effective way. Ultimately, extending this kind of investigation to other viruses would likely lead to a more rapid and successful application of G4 ligands in antiviral research.
However, it must be realized that each virus has complex and unique characteristics, therefore investigation of the effective target and mechanism of action of compounds at the molecular level may not be straightforward. The case of the EBV virus is a clear example of this entangled picture: PDS impairs EBNA1 synthesis, while PhenDC3 exerts the exact opposite effect.25, 28 Probably, the overall outcome could be ascribed to multiple G4-mediated mechanisms.
Finally, one of the most exciting applications of G4 ligands in antiviral therapy is the possibility to fight viruses that undergo latency. Current available therapies cannot eradicate the latent virus from its host, and consequently infective agents such as HIV and HHVs still cause non-eradicable diseases. In this scenario, a G4-mediated pharmacological approach would affect both the replicating and the latent virus, possibly leading to virus eradication. To this end, the virus-induced manipulation of host chromatin needs to be considered. Recently, several studies reported virus-host chromatin interactions and chromatin machinery modulation by viral proteins.86 Epigenetic modifications of viral chromatin and viral transcriptome might be crucial in conveying the virus toward either lytic or latent infection.87, 88 Since G4 formation has been directly linked to the suppressive role of heterochromatin and it seems to occur only in highly transcribed nucleosome-depleted chromatin regions,89 it would be fascinating to understand how viral G4s affect and could be affected by such a complex mechanism.
In conclusion, all the data reported so far on G4s in viruses clearly show that G4 structures are pivotal elements in the regulation of the virus life cycle, both at the lytic and latent levels. G4 ligands can be envisaged as efficient and promising antiviral agents; therefore research on the discovery of new G4-ligand small molecules should be encouraged. The availability of selective molecules will also help to clarify and further understand the role of G4s in the viral life cycle and ultimately in viral pathogenesis.
G4-mediated antiviral drugs could represent a striking turning point in the management of viral infectious diseases, especially in people who cannot access immunization, such as immunocompromised and elderly people. Finally, the G4-mediated antiviral effects observed in latent systems may lead to cutting-edge therapeutic approaches in the treatment of human deadly malignancies, such as AIDS, herpes- and HPV-related cancers.
References
- 1.Krupovic M., Dolja V.V., Koonin E.V. Origin of Viruses: Primordial Replicators Recruiting Capsids from Hosts. Nat. Rev. Microbiol. 2019;17:449–458. doi: 10.1038/s41579-019-0205-6. [DOI] [PubMed] [Google Scholar]
- 2.Blair W., Cox C. Current Landscape of Antiviral Drug Discovery. F1000Res. 2016;5:1–7. doi: 10.12688/f1000research.7665.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E.A. Novel Coronavirus Emerging in China—Key Questions for Impact Assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 4.Modrow S., Falche D., Truyen U., Schätzl H. Molecular Virology. Springer; Berlin, Heidelberg: 2013. Viruses: Definition, Structure, Classification; pp. 17–30. [Google Scholar]
- 5.Ryu W.-S. Molecular Virology of Human Pathogenic Viruses. Elsevier; 2017. Discovery and Classification; pp. 3–20. [DOI] [Google Scholar]
- 6.Lavezzo E., Berselli M., Frasson I., Perrone R., Palù G., Brazzale A.R., Richter S.N., Toppo S. G-Quadruplex Forming Sequences in the Genome of all Known Human Viruses: A Comprehensive Guide. PLoS Comput. Biol. 2018;14(12) doi: 10.1371/journal.pcbi.1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne S. Viruses. Elsevier; 2017. Introduction to DNA Viruses; pp. 231–236. [DOI] [Google Scholar]
- 8.Lieberman P.M. Epigenetics and Genetics of Viral Latency. Cell Host Microbe. 2016;19(5):619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan K., Luo M.H. Herpesviruses: Epidemiology, Pathogenesis, and Interventions. Virol. Sin. 2017;32(5):347–348. doi: 10.1007/s12250-017-4108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grinde B. Herpesviruses: Latency and Reactivation—Viral Strategies and Host Response. J. Oral Microbiol. 2013;5(1):22766. doi: 10.3402/jom.v5i0.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole C.L., James S.H. Antiviral Therapies for Herpesviruses: Current Agents and New Directions. Clin. Ther. 2018;40(8):1282–1298. doi: 10.1016/j.clinthera.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas B., Kandpal M., Jauhari U.K., Vivekanandan P. Genome-Wide Analysis of G-Quadruplexes in Herpesvirus Genomes. BMC Genomics. 2016;17(1):949. doi: 10.1186/s12864-016-3282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas B., Kumari P., Vivekanandan P. Pac1 Signals of Human Herpesviruses Contain a Highly Conserved G-Quadruplex Motif. ACS Infect. Dis. 2018;4(5):744–751. doi: 10.1021/acsinfecdis.7b00279. [DOI] [PubMed] [Google Scholar]
- 14.Puig Lombardi E., Londoño-Vallejo A., Nicolas A. Relationship between G-Quadruplex Sequence Composition in Viruses and Their Hosts. Molecules. 2019;24(10):1942. doi: 10.3390/molecules24101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artusi S., Nadai M., Perrone R., Biasolo M.A., Palu G., Flamand L., Calistri A., Richter S.N. The Herpes Simplex Virus-1 Genome Contains Multiple Clusters of Repeated G-Quadruplex: Implications for the Antiviral Activity of a G-Quadruplex Ligand. Antiviral Res. 2015;118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artusi S., Perrone R., Lago S., Raffa P., Di Iorio E., Palu G., Richter S.N. Visualization of DNA G-Quadruplexes in Herpes Simplex Virus 1-Infected Cells. Nucleic Acids Res. 2016;44(21):10343–10353. doi: 10.1093/nar/gkw968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frasson I., Nadai M., Richter S.N. Conserved G-Quadruplexes Regulate the Immediate Early Promoters of Human Alphaherpesviruses. Molecules. 2019;24(13):2375. doi: 10.3390/molecules24132375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saranathan N., Biswas B., Patra A., Vivekanandan P. G-Quadruplexes May Determine the Landscape of Recombination in HSV-1. BMC Genomics. 2019;20(1):382. doi: 10.1186/s12864-019-5731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callegaro S., Perrone R., Scalabrin M., Doria F., Palu G., Richter S.N. A Core Extended Naphtalene Diimide G-Quadruplex Ligand Potently Inhibits Herpes Simplex Virus 1 Replication. Sci. Rep. 2017;7(1):2341. doi: 10.1038/s41598-017-02667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravichandran S., Kim Y.-E., Bansal V., Ghosh A., Hur J., Subramani V.K., Pradhan S., Lee M.K., Kim K.K., Ahn J.-H. Genome-Wide Analysis of Regulatory G-Quadruplexes Affecting Gene Expression in Human Cytomegalovirus. PLoS Pathog. 2018;14(9) doi: 10.1371/journal.ppat.1007334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westdorp K.N., Terhune S.S. Impact of RNA Polymerase I Inhibitor CX-5461 on Viral Kinase-Dependent and -Independent Cytomegalovirus Replication. Antiviral Res. 2018;153:33–38. doi: 10.1016/j.antiviral.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert-Girard S., Gravel A., Artusi S., Richter S.N., Wallaschek N., Kaufer B.B., Flamand L. Stabilization of Telomere G-Quadruplexes Interferes with Human Herpesvirus 6A Chromosomal Integration. J. Virol. 2017;91(14) doi: 10.1128/jvi.00402-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smatti M.K., Al-Sadeq D.W., Ali N.H., Pintus G., Abou-Saleh H., Nasrallah G.K. Epstein-Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene among Healthy Population: An Update. Front. Oncol. 2018;8:211. doi: 10.3389/fonc.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norseen J., Johnson F.B., Lieberman P.M. Role for G-Quadruplex RNA Binding by Epstein-Barr Virus Nuclear Antigen 1 in DNA Replication and Metaphase Chromosome Attachment. J. Virol. 2009;83(20):10336–10346. doi: 10.1128/jvi.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murat P., Zhong J., Lekieffre L., Cowieson N.P., Clancy J.L., Preiss T., Balasubramanian S., Khanna R., Tellam J. G-Quadruplexes Regulate Epstein-Barr Virus-Encoded Nuclear Antigen 1 MRNA Translation. Nat. Chem. Biol. 2014;10(5):358–364. doi: 10.1038/nchembio.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tellam J.T., Zhong J., Lekieffre L., Bhat P., Martinez M., Croft N.P., Kaplan W., Tellam R.L., Khanna R. MRNA Structural Constraints on EBNA1 Synthesis Impact on in Vivo Antigen Presentation and Early Priming of CD8 + T Cells. PLoS Pathog. 2014;10(10) doi: 10.1371/journal.ppat.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brázda V., Hároníková L., Liao J., Fojta M. DNA and RNA Quadruplex-Binding Proteins. Int. J. Mol. Sci. 2014;15(10):17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lista M.J., Martins R.P., Billant O., Contesse M.A., Findakly S., Pochard P., Daskalogianni C., Beauvineau C., Guetta C., Jamin C. Nucleolin Directly Mediates Epstein-Barr Virus Immune Evasion through Binding to G-Quadruplexes of EBNA1 MRNA. Nat. Commun. 2017;8:16043. doi: 10.1038/ncomms16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reznichenko O., Quillévéré A., Martins R.P., Loaëc N., Kang H., Lista M.J., Beauvineau C., González-García J., Guillot R., Voisset C. Novel Cationic Bis(Acylhydrazones) as Modulators of Epstein–Barr Virus Immune Evasion Acting Through Disruption of Interaction Between Nucleolin and G-Quadruplexes of EBNA1 MRNA. Eur. J. Med. Chem. 2019;178:13–29. doi: 10.1016/j.ejmech.2019.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Prado Martins R., Findakly S., Daskalogianni C., Teulade-Fichou M.-P., Blondel M., Fåhraeus R. In Cellulo Protein-MRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules. Molecules. 2018;23(12):3124. doi: 10.3390/molecules23123124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prado Martins R., Malbert-Colas L., Lista M.J., Daskalogianni C., Apcher S., Pla M., Findakly S., Blondel M., Fåhraeus R. Nuclear Processing of Nascent Transcripts Determines Synthesis of Full-Length Proteins and Antigenic Peptides. Nucleic Acids Res. 2019;47(6):3086–3100. doi: 10.1093/nar/gky1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Sharkawy A., Al Zaidan L., Malki A. Epstein-Barr Virus-Associated Malignancies: Roles of Viral Oncoproteins in Carcinogenesis. Front. Oncol. 2018;8:265. doi: 10.3389/fonc.2018.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goncalves P.H., Ziegelbauer J., Uldrick T.S., Yarchoan R. Kaposi Sarcoma Herpesvirus-Associated Cancers and Related Diseases. Curr. Opin. HIV AIDS. 2017;12(1):47–56. doi: 10.1097/coh.0000000000000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madireddy A., Purushothaman P., Loosbroock C.P., Robertson E.S., Schildkraut C.L., Verma S.C. G-Quadruplex-Interacting Compounds Alter Latent DNA Replication and Episomal Persistence of KSHV. Nucleic Acids Res. 2016;44(8):3675–3694. doi: 10.1093/nar/gkw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dabral P., Babu J., Zareie A., Verma S.C. LANA and HnRNP A1 Regulate the Translation of LANA MRNA Through G-Quadruplexes. J. Virol. 2020;94(3):01508–01519. doi: 10.1128/JVI.01508-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doorbar J., Egawa N., Griffin H., Kranjec C., Murakami I. Human Papillomavirus Molecular Biology and Disease Association. Rev. Med. Virol. 2015;25:2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tluckova K., Marusic M., Tothova P., Bauer L., Sket P., Plavec J., Viglasky V. Human Papillomavirus G-Quadruplexes. Biochemistry. 2013;52(41):7207–7216. doi: 10.1021/bi400897g. [DOI] [PubMed] [Google Scholar]
- 38.Marusic M., Hosnjak L., Krafcikova P., Poljak M., Viglasky V., Plavec J. The Effect of Single Nucleotide Polymorphisms in G-Rich Regions of High-Risk Human Papillomaviruses on Structural Diversity of DNA. Biochim. Biophys. Acta. 2017;1861(5 Pt. B):1229–1236. doi: 10.1016/j.bbagen.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Marušič M., Plavec J. Towards Understanding of Polymorphism of the G-Rich Region of Human Papillomavirus Type 52. Molecules. 2019;24(7):1294. doi: 10.3390/molecules24071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoulim F., Lebossé F., Levrero M. Current Treatments for Chronic Hepatitis B Virus Infections. Curr. Opin. Virol. 2016;18:109–116. doi: 10.1016/j.coviro.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Biswas B., Kandpal M., Vivekanandan P. A G-Quadruplex Motif in an Envelope Gene Promoter Regulates Transcription and Virion Secretion in HBV Genotype B. Nucleic Acids Res. 2017;45(19):11268–11280. doi: 10.1093/nar/gkx823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf Y.I., Kazlauskas D., Iranzo J., Lucía-Sanz A., Kuhn J.H., Krupovic M., Dolja V.V., Koonin E.V. Origins and Evolution of the Global RNA Virome. MBio. 2018;9(6) doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chong H.Y., Leow C.Y., Abdul Majeed A.B., Leow C.H. Flavivirus Infection—A Review of Immunopathogenesis, Immunological Response, and Immunodiagnosis. Virus Res. 2019;274:197770. doi: 10.1016/j.virusres.2019.197770. [DOI] [PubMed] [Google Scholar]
- 44.Baharuddin A., Hassan A., Sheng G., Nasir S., Othman S., Yusof R., Othman R., Rahman N. Current Approaches in Antiviral Drug Discovery Against the Flaviviridae Family. Curr. Pharm. Des. 2014;20(21):3428–3444. doi: 10.2174/13816128113199990635. [DOI] [PubMed] [Google Scholar]
- 45.Fleming A.M., Ding Y., Alenko A., Burrows C.J. Zika Virus Genomic RNA Possesses Conserved G-Quadruplexes Characteristic of the Flaviviridae Family. ACS Infect. Dis. 2016;2(10):674–681. doi: 10.1021/acsinfecdis.6b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gawlik K., Gallay P.A. HCV Core Protein and Virus Assembly: What We Know Without Structures. Immunol. Res. 2014;60(1):1–10. doi: 10.1007/s12026-014-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourlière M., Pietri O. Hepatitis C Virus Therapy: No One Will Be Left behind. Int. J. Antimicrob. Agents. 2019;53(6):755–760. doi: 10.1016/j.ijantimicag.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Bian W.-X., Xie Y., Wang X.-N., Xu G.-H., Fu B.-S., Li S., Long G., Zhou X., Zhang X.-L. Binding of Cellular Nucleolin With the Viral Core RNA G-Quadruplex Structure Suppresses HCV Replication. Nucleic Acids Res. 2019;47(1):56–68. doi: 10.1093/nar/gky1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S.R., Min Y.Q., Wang J.Q., Liu C.X., Fu B.S., Wu F., Wu L.Y., Qiao Z.X., Song Y.Y., Xu G.H. A Highly Conserved G-Rich Consensus Sequence in Hepatitis C Virus Core Gene Represents a New Anti-Hepatitis C Target. Sci. Adv. 2016;2(4) doi: 10.1126/sciadv.1501535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J., Chaires J.B. Sequence and Structural Selectivity of Nucleic Acid Binding Ligands. Biochemistry. 1999;38(49):16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 51.Jaubert C., Bedrat A., Bartolucci L., Di Primo C., Ventura M., Mergny J.L., Amrane S., Andreola M.L. RNA Synthesis Is Modulated by G-Quadruplex Formation in Hepatitis C Virus Negative RNA Strand. Sci. Rep. 2018;8(1):8120. doi: 10.1038/s41598-018-26582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo X., Xue B., Feng G., Zhang J., Lin B., Zeng P., Li H., Yi H., Zhang X.-L., Zhu H. Lighting up the Native Viral RNA Genome With a Fluorogenic Probe for the Live-Cell Visualization of Virus Infection. J. Am. Chem. Soc. 2019;141(13):5182–5191. doi: 10.1021/jacs.8b10265. [DOI] [PubMed] [Google Scholar]
- 53.Wang S.R., Zhang Q.Y., Wang J.Q., Ge X.Y., Song Y.Y., Wang Y.F., Li X.D., Fu B.S., Xu G.H., Shu B. Chemical Targeting of a G-Quadruplex RNA in the Ebola Virus L Gene. Cell Chem. Biol. 2016;23(9):1113–1122. doi: 10.1016/j.chembiol.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 54.Krafcikova P., Demkovicova E., Viglasky V. Ebola Virus Derived G-Quadruplexes: Thiazole Orange Interaction. Biochim. Biophys. Acta. 2017;1861(5 Pt. B):1321–1328. doi: 10.1016/j.bbagen.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 55.Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-Unique Domain (SUD) of SARS Coronavirus Contains Two Macrodomains That Bind G-Quadruplexes. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R. A G-Quadruplex-Binding Macrodomain Within the “SARS-Unique Domain” Is Essential for the Activity of the SARS-Coronavirus Replication-Transcription Complex. Virology. 2015;484:313–322. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss P., Murdoch D.R. Clinical Course and Mortality Risk of Severe COVID-19. Lancet. 2020;395:1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilder-Smith A., Chiew C.J., Lee V.J. Can We Contain the COVID-19 Outbreak With the Same Measures as for SARS? Lancet. Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fung S.-Y., Yuen K.-S., Ye Z.-W., Chan C.-P., Jin D.-Y. A Tug-of-War Between Severe Acute Respiratory Syndrome Coronavirus 2 and Host Antiviral Defence: Lessons From Other Pathogenic Viruses. Emerg. Microbes Infect. 2020;9(1):558–570. doi: 10.1080/22221751.2020.1736644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jern P., Coffin J.M. Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 2008;42(1):709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 61.Perrone R., Lavezzo E., Palu G., Richter S.N., Palù G., Richter S.N. Conserved Presence of G-Quadruplex Forming Sequences in the Long Terminal Repeat Promoter of Lentiviruses. Sci. Rep. 2017;7(1):2018. doi: 10.1038/s41598-017-02291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruggiero E., Tassinari M., Perrone R., Nadai M., Richter S.N. Stable and Conserved G-Quadruplexes in the Long Terminal Repeat Promoter of Retroviruses. ACS Infect. Dis. 2019;5(7):1150–1159. doi: 10.1021/acsinfecdis.9b00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sundquist W.I., Heaphy S. Evidence for Interstrand Quadruplex Formation in the Dimerization of Human Immunodeficiency Virus 1 Genomic RNA. Proc. Natl. Acad. Sci. U. S. A. 1993;90(8):3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piekna-Przybylska D., Sharma G., Bambara R.A. Mechanism of HIV-1 RNA Dimerization in the Central Region of the Genome and Significance for Viral Evolution. J. Biol. Chem. 2013;288(33):24140–24150. doi: 10.1074/jbc.M113.477265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piekna-Przybylska D., Sullivan M.A., Sharma G., Bambara R.A. U3 Region in the HIV-1 Genome Adopts a G-Quadruplex Structure in its RNA and DNA Sequence. Biochemistry. 2014;53(16):2581–2593. doi: 10.1021/bi4016692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perrone R., Butovskaya E., Daelemans D., Palu G., Pannecouque C., Richter S.N. Anti-HIV-1 Activity of the G-Quadruplex Ligand BRACO-19. J. Antimicrob. Chemother. 2014;69(12):3248–3258. doi: 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- 67.Rajendran A., Endo M., Hidaka K., Tran P.L.T., Mergny J.-L., Gorelick R.J., Sugiyama H. HIV-1 Nucleocapsid Proteins as Molecular Chaperones for Tetramolecular Antiparallel G-Quadruplex Formation. J. Am. Chem. Soc. 2013;135(49):18575–18585. doi: 10.1021/ja409085j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butovskaya E., Soldà P., Scalabrin M., Nadai M., Richter S.N. HIV-1 Nucleocapsid Protein Unfolds Stable RNA G-Quadruplexes in the Viral Genome and Is Inhibited by G-Quadruplex Ligands. ACS Infect. Dis. 2019;5(12):2127–2135. doi: 10.1021/acsinfecdis.9b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyonnais S., Gorelick R.J., Mergny J.-L., Le Cam E., Mirambeau G. G-Quartets Direct Assembly of HIV-1 Nucleocapsid Protein along Single-Stranded DNA. Nucleic Acids Res. 2003;31(19):5754–5763. doi: 10.1093/nar/gkg716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amrane S., Kerkour A., Bedrat A., Vialet B., Andreola M.L., Mergny J.L. Topology of a DNA G-Quadruplex Structure Formed in the HIV-1 Promoter: A Potential Target for Anti-HIV Drug Development. J. Am. Chem. Soc. 2014;136(14):5249–5252. doi: 10.1021/ja501500c. [DOI] [PubMed] [Google Scholar]
- 71.Perrone R., Nadai M., Frasson I., Poe J.A., Butovskaya E., Smithgall T.E., Palumbo M., Palu G., Richter S.N. A Dynamic G-Quadruplex Region Regulates the HIV-1 Long Terminal Repeat Promoter. J. Med. Chem. 2013;56(16):6521–6530. doi: 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Butovskaya E., Heddi B., Bakalar B., Richter S.N., Phan A.T. Major G-Quadruplex Form of HIV-1 LTR Reveals a (3 + 1) Folding Topology Containing a Stem-Loop. J. Am. Chem. Soc. 2018;140(42):13654–13662. doi: 10.1021/jacs.8b05332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Nicola B., Lech C.J., Heddi B., Regmi S., Frasson I., Perrone R., Richter S.N., Phan A.T. Structure and Possible Function of a G-Quadruplex in the Long Terminal Repeat of the Proviral HIV-1 Genome. Nucleic Acids Res. 2016;44(13):6442–6451. doi: 10.1093/nar/gkw432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tosoni E., Frasson I., Scalabrin M., Perrone R., Butovskaya E., Nadai M., Palu G., Fabris D., Richter S.N. Nucleolin Stabilizes G-Quadruplex Structures Folded by the LTR Promoter and Silences HIV-1 Viral Transcription. Nucleic Acids Res. 2015;43(18):8884–8897. doi: 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scalabrin M., Frasson I., Ruggiero E., Perrone R., Tosoni E., Lago S., Tassinari M., Palù G., Richter S.N. The Cellular Protein HnRNP A2/B1 Enhances HIV-1 Transcription by Unfolding LTR Promoter G-Quadruplexes. Sci. Rep. 2017;7:45244. doi: 10.1038/srep45244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Day H.A., Pavlou P., Waller Z.A.E. I-Motif DNA: Structure, Stability and Targeting with Ligands. Bioorg. Med. Chem. 2014;22(16):4407–4418. doi: 10.1016/j.bmc.2014.05.047. [DOI] [PubMed] [Google Scholar]
- 77.Ruggiero E., Lago S., Šket P., Nadai M., Frasson I., Plavec J., Richter S.N. A Dynamic I-Motif With a Duplex Stem-Loop in the Long Terminal Repeat Promoter of the HIV-1 Proviral Genome Modulates Viral Transcription. Nucleic Acids Res. 2019;47(21):11057–11068. doi: 10.1093/nar/gkz937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perrone R., Doria F., Butovskaya E., Frasson I., Botti S., Scalabrin M., Lago S., Grande V., Nadai M., Freccero M. Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 2015;58(24):9639–9652. doi: 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perrone R., Nadai M., Poe J.A., Frasson I., Palumbo M., Palu G., Smithgall T.E., Richter S.N. Formation of a Unique Cluster of G-Quadruplex Structures in the HIV-1 Nef Coding Region: Implications for Antiviral Activity. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0073121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siliciano R.F., Greene W.C. HIV Latency. Cold Spring Harb. Perspect. Med. 2011;1(1):a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Piekna-Przybylska D., Sharma G., Maggirwar S.B., Bambara R.A. Deficiency in DNA Damage Response, a New Characteristic of Cells Infected With Latent HIV-1. Cell Cycle. 2017;16(10):968–978. doi: 10.1080/15384101.2017.1312225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peck K.M., Lauring A.S. Complexities of Viral Mutation Rates. J. Virol. 2018;92(14) doi: 10.1128/jvi.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spiegel J., Adhikari S., Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2019;2:123–136. doi: 10.1016/j.trechm.2019.07.002. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell N.H., Parkinson G.N., Reszka A.P., Neidle S. Structural Basis of DNA Quadruplex Recognition by an Acridine Drug. J. Am. Chem. Soc. 2008;130(21):6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 85.Nadai M., Doria F., Scalabrin M., Pirota V., Grande V., Bergamaschi G., Amendola V., Winnerdy F.R., Phan A.T., Richter S.N. A Catalytic and Selective Scissoring Molecular Tool for Quadruplex Nucleic Acids. J. Am. Chem. Soc. 2018;140(44):14528–14532. doi: 10.1021/jacs.8b05337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Knipe D.M., Lieberman P.M., Jung J.U., McBride A.A., Morris K.V., Ott M., Margolis D., Nieto A., Nevels M., Parks R.J. Snapshots: Chromatin Control of Viral Infection. Virology. 2013;435(1):141–156. doi: 10.1016/j.virol.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knipe D.M., Cliffe A. Chromatin Control of Herpes Simplex Virus Lytic and Latent Infection. Nat. Rev. Microbiol. 2008;6(3):211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 88.Fleming A.M., Nguyen N.L.B., Burrows C.J. Colocalization of m 6 A and G-Quadruplex-Forming Sequences in Viral RNA (HIV, Zika, Hepatitis B, and SV40) Suggests Topological Control of Adenosine N 6-Methylation. ACS Cent. Sci. 2019;5(2):218–228. doi: 10.1021/acscentsci.8b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. G-Quadruplex Structures Mark Human Regulatory Chromatin. Nat. Genet. 2016;48(10):1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]