Abstract
From December 2019, a novel coronavirus, SARS-CoV-2, caused an outbreak of pneumonia in Wuhan city and rapidly spread throughout China and globally. However, the clinical characteristics and co-infection with other respiratory pathogens of patients with COVID-19 and the factors associated with severity of COVID-19 are still limited. In this retrospective cohort study, we included 354 inpatients with COVID-19 admitted to Renmin Hospital of Wuhan University from February 4, 2020 to February 28, 2020. We found levels of interleukin-6, interleukin-10, C-reactive protein, D-dimer, white blood cell count and neutrophil count were clearly elevated in males and critical cases compared with females and severe and mild cases, respectively. However, lymphopenia was more severe in males than females and levels of tumor necrosis factor alpha were reduced significantly in critical cases than severe and mild cases. 23.5% of severe cases and 24.4% of critical cases were co-infected with other respiratory pathogens. Additionally, stepwise multivariable regression analysis suggested that co-infection, lymphocyte count and levels of D-dimer were associated with severity of COVID-19.These findings provide crucial clues for further identification of the mechanisms, characteristics and treatments of patients with COVID-19.
Keywords: COVID-19, Laboratory factors, Gender, Co-infection
Since December 2019, an outbreak of unexplained pneumonia, now known as coronavirus disease 2019 (COVID-19), occurred in Wuhan city and rapidly spread throughout China and globally [[1], [2], [3], [4], [5]]. A novel coronavirus named as the 2019-nCoV previously and renamed as SARS-CoV-2 by International Committee on Taxonomy of Viruses [6] was isolated from these patients in Wuhan by Chinese scientists On Jan 10, 2020. Full-genome sequencing and phylogenic analysis suggested that SARS-CoV-2 originated via natural selection [7] differs from Middle East respiratory syndrome-CoV and severe acute respiratory syndrome-CoV [[8], [9], [10]]. It was found that the SARS-CoV-2 infection could cause not only clusters of severe respiratory illness similar to SARS-CoV-1, but also mild upper respiratory diseases and asymptomatic infection [[11], [12], [13]].
Although previous studies have demonstrated certain clinical characteristics of patients with COVID-19 [8,11,12], their detail clinical characteristics are still limited and the sex differences in clinical characteristics of COVID-19 patients have not been well studied. Moreover, details of the laboratory assessments such as complete blood count, coagulation profile, serum biochemical tests and inflammatory factors associated with severity of COVID-19 have not yet been well described. In addition, previous studies have shown that patients with COVID-19 can co-infected with other respiratory virus [14] and will also have secondary infections with bacteria and fungi [13,15], however, the information of co-infection with other respiratory pathogens especially atypical pathogens is still scared. Therefore, we present details of 354 inpatients with COVID-19 admitted to Renmin Hospital of Wuhan University to further explore the clinical characteristics and co-infections with other respiratory pathogens of patients with COVID-19 as well as the sex differences in clinical characteristics and the factors associated with severity of COVID-19.
2. Materials and methods
2.1. Study design and participants
This retrospective cohort study included 354 inpatients admitted to Renmin Hospital of Wuhan University and diagnosed with COVID-19 according to World Health Organization interim guidance from February 4, 2020 to February 28, 2020. This study was approved by the Ethics Committee of the Renmin Hospital of Wuhan University (WDRY2020-K066). Data were collected from routine clinical practice, and informed consent was not required.
2.2. Data collection
The clinical characteristics of patients were analyzed by the research team of the Department of Clinical Laboratory, Renmin Hospital of Wuhan University. The epidemiological, clinical and laboratory assessments were obtained with data collection forms from electronic medical records. Laboratory assessments consisted of complete blood count, coagulation profile, serum biochemical tests, C-reactive protein (CRP), procalcitonin (PCT), interleukin-6 (IL-6), interleukin-2 (IL-2), interleukin-4 (IL-4), interleukin-10 (IL-10), tumor necrosis factor alpha (TNF-α), interferon-gamma (IFN-γ) and D-dimer, which is a significant prognostic factor in patients with suspected infection and sepsis [16].
2.3. Laboratory procedures
Throat-swab specimens from the upper respiratory tract that were obtained from all patients at admission were maintained in viral–transport medium. All patients included were verified as positive for SARS-CoV-2 infection in throat swabs analyzed by real-time RT-PCR using the same protocol described previously [11].
Sputum were obtained for identification of 13 respiratory pathogens, including adenovirus, boca virus, influenza A virus, H1N1, H3N2, influenza B viruses, coronavirus (OC43, HKU1, NL63 and 229E), metapneumovirus, parainfluenza virus, respiratory syncytial virus, rhinovirus, Mycoplasma pneumoniae, and Chlamydia pneumoniae. The detection of 13 respiratory pathogens was performed of Pathogenic Nucleic Acid Detection Kit (Health, Ningbo, China) following the manufacturer’s instructions based on electrophoresis fragment analysis with PCR.
Bronchoalveolar lavage fluids or blood were obtained for identification of possible causative bacteria or fungi. Routine bacteria culture was performed independently in accordance with following respiratory pathogenic microorganisms operating standards: the samples were seeded on bacteriological media such as blood agar plate, chocolate agar plates and blue agar plates using sterile wire loops and incubated at 35 °C for 72 h in a thermostatic incubator. Routine fungus culture was performed independently in accordance with following respiratory pathogenic microorganisms operating standards: 2–5 ml of the lavage fluids was treated with equal volumes of sterile DTT (0.3 mg/ml) for 15 min at room temperature in order to dissolve viscous mucus. Then, 0.5 ml of the samples were inoculated on two Sabouraud/glucose (4%) agar plates (Becton–Dickinson) each containing chloramphenicol (0.4 g/L) and gentamicin (0.04 g/L).The plates were incubated at 37 and 30 °C, respectively. Subsequently the dominant colonies were picked for bacterial and fungus detection using the VITEK MS system (bioMérieux, Marcy l’Etoile, France).
Routine blood examinations were complete blood count, coagulation profile, serum biochemical tests (including renal and liver function, creatine kinase, lactate dehydrogenase, and electrolytes), myocardial enzymes, interleukin-6 (IL-6), serum ferritin, and procalcitonin.
2.4. Statistical analysis
SPSS (Statistical Package for the Social Sciences) version 20.0 software was used for statistical analysis. Categorical variables were expressed as proportions and compared between groups using the X 2 test. Continuous data were expressed as mean±SD for normally distributed variables or median (inter quartile range) for others. The paired t-test and Mann–Whitney U test were used to compare continuous variables of normal distribution and non-normal distribution, respectively. Comparison of the groups by ANOVA was followed by SNK-q test to determine differences between individual groups. The patients were grouped by genders, clinical classification and co-infection or not. Finally, factors associated with severity of COVID-19 were analyzed with stepwise regression analysis with adjusted for age and sex. A 2-sided α of less than 0.05 was considered statistically significant.
3. Results
3.1. Presenting characteristics
This study population enrolled 354 hospitalized patients with COVID-19.175 (49.44%) were men and the median age was 62 years (range, 23–90 years) (Table 1 ). Comorbidities were present in nearly one third of patients, with hypertension being the most common comorbidity, followed by diabetes and coronary heart disease (Table 1). Based on clinical characteristics the study group was further divided into subjects presenting mild symptoms (n = 115; 50.43% males), subjects presenting severe symptoms (n = 155; 49.68% males) and subjects presenting critical symptoms (n = 84; 47.62% males). The median ages of subjects in the mild, severe, and critical groups were 61 years (range, 23–79 years), 62 years (range, 25–89 years), and 65.5 years (range, 35–90 years), respectively. No significant differences were observed in ages among these three groups. In total, 11 of the 354 patients (3.11%) with confirmed COVID-19 died following progression. 343 patients survived to hospital discharge, giving a survival to hospital discharge rate of 96.89%. And 8.3% (7/84) died in the 84 patients of critical group and 2.59% (4/155) died in the 155 patients of severe group. Of 175 male patients, 4.57% (8/175) died and 1.68% (3/179) died in the 179 female patients.
Table 1.
Baseline characteristics of 354 patients infected with SARS-CoV-2.
| Characteristics | No.(%) |
|||
|---|---|---|---|---|
| Total (n = 354) | Mild (n = 115) | Severe (n = 155) | Critical (n = 84) | |
| Sex | ||||
| Female | 179 (50.56) | 57 (49.57) | 78 (50.32) | 44 (52.38) |
| Male | 175 (49.44) | 58 (50.43) | 77 (49.68) | 40 (47.62) |
| Median age (range) y | 62 (23–90) | 61 (23–79) | 62 (25–89) | 65.5 (35–90) |
| 15-44 y | 49 (13.84) | 21 (18.26) | 19 (12.26) | 9 (10.71) |
| 45-64 y | 149 (42.09) | 48 (41.74) | 69 (44.52) | 32 (38.10) |
| ≥65 y | 156 (44.07) | 46 (40.00) | 67 (43.23) | 43 (51.19) |
| Comorbidity | 114 (32.20) | 39 (33.91) | 47 (30.32) | 28 (33.33) |
| Hypertension | 74 (20.90) | 23 (20.00) | 33 (21.29) | 18 (21.43) |
| Diabetes | 35 (9.89) | 9 (7.83) | 18 (11.61) | 8 (9.52) |
| Coronary heart disease | 18 (5.08) | 5 (4.35) | 7 (4.52) | 6 (7.14) |
| Chronic obstructive lung disease | 6 (1.69) | 2 (1.74) | 3 (1.94) | 1 (1.19) |
| Carcinoma | 2 (0.56) | 2 (1.74) | 0 | 0 |
| Death | 11 (3.11, male 8) | 0 | 4 (2.59, male 3) | 7 (8.30, male 5) |
3.2. Laboratory parameters among mild, severe and critical groups
A number of laboratory parameters showed significant differences among patients presented as mild, severe and critical symptoms, including white blood cell and neutrophil counts, lymphocyte counts as well as levels of D-dimer and CRP (Table 2 ). The levels of IL-6 and IL-10 were increased substantially in patients of severe and critical groups. Differently, the levels of TNF-αwere decreased substantially in patients of severe and critical groups. However, there were no significant differences in levels of PCT among these three groups.
Table 2.
Laboratory findings of 354 patients infected with SARS-CoV-2.
| Characteristics | Normal Range | Mean (Standard Deviation) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n = 354) | Male (n = 175) | Female (n = 179) | P Value | Mild (n = 115) | Severe (n = 155) | Critical (n = 84) | P Value | Co-infection (n = 23) | Non-co-infection (n = 93) | P Value | ||
| White blood cell count, × 109/L | 3.5–9.5 | 6.59 (3.02) | 6.99 (3.48) | 6.19 (2.44) | 0.012 | 6.04 (2.68) | 6.67 (3.00) | 7.17 (3.40) | 0.030 | 8.00 (3.25) | 6.45 (2.97) | 0.006 |
| Neutrophil count, × 109/L | 1.8–6.3 | 4.90 (3.02) | 5.39 (3.51) | 4.42 (2.36) | 0.002 | 4.15 (2.67) | 5.00 (2.91) | 5.73 (3.42) | 0.001 | 6.60 (3.34) | 4.73 (2.94) | 0.001 |
| Lymphocyte count, × 109/L | 1.1–3.2 | 1.13 (0.54) | 1.02 (0.43) | 1.23 (0.61) | <0.001 | 1.31 (0.59) | 1.09 (0.50) | 0.93 (0.47) | <0.001 | 0.89 (0.38) | 1.14 (0.55) | 0.012 |
| D-dimer, mg/L | 0–0.55 | 5.97 (15.53) | 7.81 (17.51) | 4.17 (13.10) | 0.027 | 1.90 (3.78) | 5.74 (13.15) | 11.97 (25.00) | <0.001 | 16.22 (25.08) | 4.99 (13.95) | <0.001 |
| IL2, pg/mL | ≤11.4 | 4.56 (12.01) | 5.46 (17.07) | 3.68 (0.81) | 0.165 | 5.70 (20.65) | 3.92 (2.25) | 4.14 (2.74) | 0.456 | 4.46 (2.84) | 4.56 (12.46) | 0.965 |
| IL4, pg/mL | ≤12.9 | 3.87 (7.10) | 3.64 (2.02) | 4.10 (9.78) | 0.542 | 4.61 (12.11) | 3.41 (1.12) | 3.70 (2.54) | 0.380 | 4.64 (4.92) | 3.81 (7.25) | 0.566 |
| IL6, pg/mL | ≤20.0 | 38.96 (127.72) | 53.96 (165.00) | 24.30 (73.28) | 0.029 | 17.44 (38.29) | 29.82 (82.47) | 85.29 (227.55) | <0.001 | 178.68 (358.19) | 25.55 (62.21) | <0.001 |
| IL10, pg/mL | ≤5.9 | 7.67 (8.15) | 9.21 (10.73) | 6.18 (3.85) | <0.001 | 6.50 (4.84) | 6.78 (6.10) | 11.04 (13.03) | <0.001 | 14.02 (16.64) | 7.16 (6.84) | <0.001 |
| IFN-γ, pg/mL | ≤5.5 | 18.72 (103.92) | 28.3 (133.84) | 9.42 (61.42) | 0.09 | 15.75 (73.21) | 23.62 (131.91) | 13.70 (78.72) | 0.733 | 32.76 (138.31) | 17.59 (100.84) | 0.475 |
| TNF-α, pg/mL | ≤18 | 10.07 (16.90) | 10.57 (18.28) | 9.58 (15.48) | 0.582 | 13.63 (22.11) | 9.64 (15.37) | 5.81 (7.79) | 0.005 | 8.38 (11.95) | 10.20 (16.90) | 0.598 |
| PCT, ng/mL | <0.1 | 0.22 (0.74) | 0.30 (0.96) | 0.14 (0.41) | 0.039 | 0.11 (0.33) | 0.29 (1.04) | 0.22 (0.38) | 0.146 | 0.68 (1.62) | 0.17 (0.58) | <0.001 |
| CRP, mg/L | <10.0 | 47.23 (51.74) | 54.13 (54.38) | 40.49 (48.23) | 0.013 | 33.22 (45.08) | 47.87 (52.13) | 65.23 (54.33) | <0.001 | 82.99 (65.46) | 43.80 (49.00) | <0.001 |
| C3, g/L | 0.9–1.8 | 1.04 (0.22) | 1.07 (0.24) | 1.01 (0.20) | 0.015 | 1.04 (0.22) | 1.07 (0.21) | 0.99 (0.24) | 0.053 | 1.06 (0.25) | 1.04 (0.22) | 0.714 |
| C4, g/L | 0.1–0.4 | 0.25 (0.13) | 0.27 (0.15) | 0.24 (0.10) | 0.020 | 0.25 (0.10) | 0.26 (0.11) | 0.25 (0.18) | 0.723 | 0.25 (0.13) | 0.25 (0.13) | 0.972 |
| IgA, g/L | 0.7–4.0 | 2.62 (1.15) | 2.71 (1.12) | 2.53 (1.18) | 0.146 | 2.49 (1.25) | 2.67 (1.05) | 2.71 (1.18) | 0.317 | 2.84 (1.23) | 2.60 (1.15) | 0.264 |
| IgG, g/L | 7.0–16.0 | 13.00 (3.99) | 13.09 (3.95) | 12.91 (4.04) | 0.684 | 12.35 (3.53) | 13.04 (3.82) | 13.80 (4.73) | 0.042 | 12.66 (3.21) | 13.03 (4.06) | 0.625 |
| IgM, g/L | 0.4–2.3 | 0.99 (0.44) | 0.94 (0.41) | 1.04 (0.45) | 0.019 | 1.01 (0.46) | 0.97 (0.43) | 1.01 (0.43) | 0.643 | 1.07 (0.56) | 0.98 (0.42) | 0.317 |
Abbreviations: IL-2, interleukin-2; IL-4, interleukin-4; IL-6,interleukin-6; IL-10, interleukin-10; IFN-γ, Interferon-gamma; TNF-α, Tumor necrosis factor alpha; CRP, C-reactive protein; PCT, procalcitonin. P < 0.05 was considered statistically significant.
3.3. Laboratory parameters between genders
To investigate the differences between genders, we compared some laboratory parameters of 175 males and 179 females. Preliminary analysis indicated that higher white blood cell and neutrophil counts, as well as higher levels of D-dimer, IL-6, IL-10, CRP and PCT were found in male patients compared to those of females, which was similar to patients in critical and severe groups compared with those of mild groups (Table 2). Differently, lymphocytopenia was significantly more severe in males than females (Table 2). However, there were no significant differences in levels of INF-αbetween genders (Table 2).
3.4. Co-infection with other respiratory pathogens
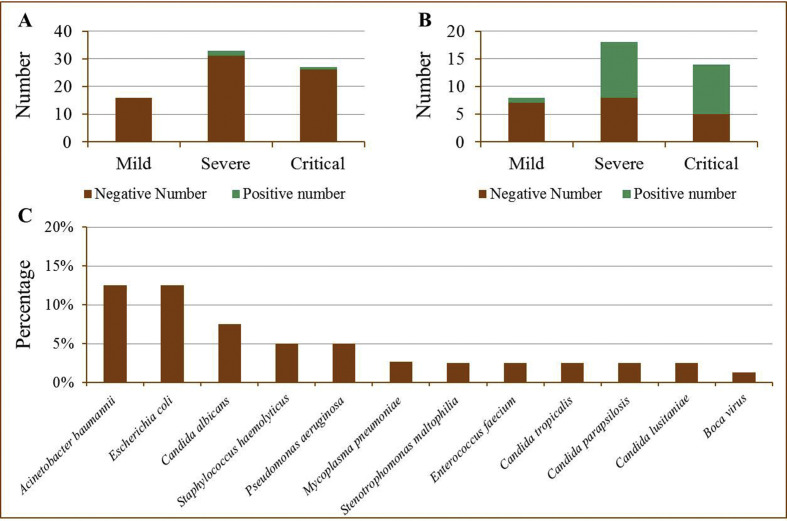
76 patients (mild n = 16, severe n = 33, critical n = 27) were suspected of co-infection with other respiratory viruses especially atypical pathogens during the course of hospitalization combined with the results of CT and infection markers, and their sputum were obtained for identification of 13 respiratory pathogens. In total, 3 (3.95%) of the 76 patients had a pathogen infection (Fig. 1 A). Among them, one severely ill patient and one critically ill patient were co-infected with Mycoplasma pneumonia, respectively.
Fig. 1.
The information of patients co-infected with other respiratory pathogens. A: Pathogen-positive numbers for all 13 respiratory pathogens tested in different groups according to clinical classification. B: Pathogen-positive numbers for lower respiratory pathogens such as bacterial and fungi tested for traditional culture in previously different groups. C: Positive rate of other respiratory pathogens in the test population.
Bronchoalveolar lavage fluids or blood of 40 patients (mild n = 8, severe n = 18, critical n = 14) suspected of co-infection with bacteria and fungi especially Acinetobacter baumannii and Candida albicans were collected for traditional culture detection. We found that 50% (20/40) of the 40 patients were co-infected with bacterial and fungi pathogens (Fig. 1B). And 64.3% (9/14), 55.6% (10/18) and 12.5% (1/8) of patients in critical, severe and mild groups were co-infected with bacterial and fungi pathogens. It was worth noting that there was one critical patient infected with two species of bacterial (A. baumannii and Staphylococcus haemolyticus) and one critical patient infected with bacterial and fungi (Escherichia coli and Candida tropicalis) simultaneously. Meanwhile, three cases of C. albicans and four cases of drug-resistant A. baumannii were detected only in critically ill patients. The other pathogens detected included Pseudomonas aeruginosa Stenotrophomonas maltophilia, Enterococcus faecium, Candida parapsilosis, Candida lusitaniae and boca virus (Fig. 1C).
Higher white blood cell and neutrophil counts, as well as higher levels of D-dimer, IL-6, IL-10, CRP and PCT were observed in patients co-infected with other respiratory pathogens than those of infected with SARS-CoV-2 homogeneously (Table 2).
3.5. Factors associated with severity of COVID-19
Stepwise multivariable regression models were used to find the association of severity of COVID-19 with each of the other factors with adjusted age and sex. Those factors white blood cell count, neutrophil count, lymphocyte count, levels of D-dimer, IL-2, IL-4, IL-6, IL-10, IFN-γ, TNF-α, CRP, PCT, C3, C4, IgA, IgE, IgG, IgM and co-infection with other respiratory pathogens were incorporated into the regression models. For stepwise multivariable regression analysis, we selected the variables that were allowed to enter the model in advance. Co-infection with other respiratory pathogens, lymphocyte count and levels of D-dimer, were associated with severity of COVID-19 (R = 0.375, P < 0.001) (Table 3 ).
Table 3.
Stepwise multivariable regression analysis of factors associated with severity of COVID-19.
| Factors | Severity of COVID-19 |
||
|---|---|---|---|
| F | R | P-value | |
| Co-infection | 10.507 | 0.257 | 0.014 |
| Co-infection + lymphocyte count | 9.722 | 0.341 | <0.001 |
| Co-infection + lymphocyte count + levels of D-dimer | 8.022 | 0.375 | <0.001 |
4. Discussion
In this retrospective cohort study, we used 354 samples to make a preliminary assessment of the clinical characteristics of patients with COVID-19 from the following and aspects such as gender, clinical classification and co-infection with other respiratory pathogens.
Previous studies suggested that increased age was associated with death in patients with SARS-CoV-1, MERS and COVID-19 [[17], [18], [19]]. However, no significant differences were found in ages among these three clinical classification groups in this study. Whereas, we found 63.64% (7/11) of the 11 non-survivors were co-infected with other respiratory pathogens, especially C. albicans and drug-resistant A. baumannii. Meanwhile, 75% patients co-infected with other bacterial and fungi were found to be concentrated on patients older than 50 years. In addition, we found co-infection with other respiratory pathogens, lymphocyte count and levels of D-dimer were associated with severity of COVID-19. And the patients grouped in critical cases presented the highest mortality rate and displayed the lowest lymphocyte count and the highest levels of D-dimer and the highest rate of co-infection with other bacterial and fungi significantly. Thus, untimely detection and treatment of co-infections with other respiratory pathogens especially fungi and drug-resistant bacterial may be an important cause to mortality in COVID-19. In particular, the highest levels of IL-6 and IL-10 and the lowest levels of TNF-αwere observed in subjects of critical groups. These results suggested that levels of IL-10 and TNF-αcould be used for clinical classification of COVID-19.
COVID-19 was more likely to affect men than women, and the symptoms seems to more severe in men [13]. In this study, sex bias in the fatality rate was observed. And lower lymphocyte count, higher white blood cell and neutrophil counts, as well as higher levels of D-dimer, IL-6, IL-10, CRP and PCT were observed in male patients, which was similar to patients in critical and severe groups compared with those of mild groups. However, the mechanisms underlying these differences are still not clear. The reduced susceptibility of females to SARS-CoV-2 infections could be depended on the protection of X chromosome and sex hormones, which played an important role in innate and adaptive immunity [20]. Previous studies have found higher percentages of SARS-CoV-1 infection in male mice than in female mice and provided mechanistic insights related to estrogen [21]. Additionally, 17 β-estradiol could down-regulate lung ACE2 mRNA, the putative receptor of SARS-CoV-2 [4], and protect females from influenza A virus pathogenesis [22].
Previous studies suggested that patients with COVID-19 can also co-infect with other respiratory pathogens such as viruses, bacteria and fungi [[13], [14], [15]]. In this study, bronchoalveolar lavage fluids or blood of 40 patients suspected of co-infection with bacteria and fungi during hospital admission were collected for traditional culture detection. Since the samples were collected 3–5 days after patients admitted to hospital, the bacteria and fungi detected were more likely to be secondary to COVID-19. And we found 45% cases (18/40) infected with single bacterial or fungi and 5% cases (2/40) were infected with two and more species of bacterial and fungi. The rate of bacterial/fungal co-infection was higher than that previously reported [13,15]. On the one hand, the number of samples in this study may be small, on the other hand, all the samples were collected and tested when patients suspected of co-infection with bacteria and fungi combined with the results of CT and infection markers. Furthermore, previous studies have shown that human metapneumovirus and other viruses can be detected from SARS-CoV-1 patients [23,24], and one case co-infected with SARS-CoV-2 and human metapneumovirus was reported [14]. However, only 1 case was infected with boca virus in 79 patients detected of 13 respiratory pathogens. These issues demonstrate that susceptibility to co-infection with other viruses may be reduced in SARS-CoV-2 patients. Additionally, for the samples were collected and tested 3–5 days after patients admitted to hospital, the viruses co-infected with SARS-CoV-2 may be cured.
Taken together, in this single-center case series of 354 hospitalized patients with COVID-19 in Wuhan, males had higher levels of interleukin-6, interleukin-10, C-reactive protein, D-dimer, white blood cell count and neutrophil count and more severe lymphopenia compared with females. Lower Levels of tumor necrosis factor alpha and higher levels of interleukin-10 were observed in critical cases than severe and mild cases. Stepwise multivariable regression analysis suggested that co-infection, lymphocyte count and levels of D-dimer were associated with severity of COVID-19.23.5% of severe cases and 24.4% of critical cases were co-infected with other respiratory pathogens. It is essential to identify pathogens, judge the patient’s condition, and avoid blind administration of drugs in treatment of patients with COVID-19.
Author contributions
Z-HL, S-HC, JL, J-TH, and L-NF had roles in the study design, data collection, data analysis, data interpretation, literature search, and writing of the manuscript. B-HZ and YL contributed to critical revision of the manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
No founding was received. We thank all clinicians and inspectors at the front line of COVID-19 epidemic.
Contributor Information
Binghong Zhang, Email: zbhong6288@163.com.
Yan Li, Email: yanlitf1120@163.com.
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. J Am Med Assoc. 2020 doi: 10.1001/jama.2020.1097. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Organization WH . 2020. Novel coronavirus(2019-nCoV): situation report-15.http://wwwwhoint/docs/default-source/coronaviruse/situationreports/20200204-sitrep-15-ncovpdf [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J Am Med Assoc. 2020 doi: 10.1001/jama.2020.1585. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Touzard-Romo F., Tape C., Lonks J.R. Co-infection with SARS-CoV-2 and human metapneumovirus. R I Med J. 2020;103:75–76. [PubMed] [Google Scholar]
- 15.Rawson T.M., Moore L.S.P., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa530. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joaquín R., Rodelo R., la Rosa G.D., Valencia M.L., Ospina S., Arango C.M. D-dimer is a significant prognostic factor in patients with suspected infection and sepsis. Am J Emerg Med. 2012;30:1991–1999. doi: 10.1016/j.ajem.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 17.Choi K.W., Chau T.N., Tsang O., Tso E., Chiu M.C., Tong W.L. Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann Intern Med. 2003;139:715–723. doi: 10.7326/0003-4819-139-9-200311040-00005. [DOI] [PubMed] [Google Scholar]
- 18.Hong K.H., Choi J.P., Hong S.H., Lee J., Kwon J.S., Kim S.M. Predictors of mortality in Middle East respiratory syndrome (MERS) Thorax. 2018;73:286–289. doi: 10.1136/thoraxjnl-2016-209313. [DOI] [PubMed] [Google Scholar]
- 19.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaillon S., Berthenet K., Garlanda C. Sexual dimorphism in innate immunity. Clin Rev Allergy Immunol. 2019;56:308–321. doi: 10.1007/s12016-017-8648-x. [DOI] [PubMed] [Google Scholar]
- 21.Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson D.P., Lorenzo M.E., Jian W., Klein S.L. Elevated 17beta-estradiol protects females from influenza A virus pathogenesis by suppressing inflammatory responses. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee N., Chan P.K., Yu I.T., Tsoi K.K., Lui G., Sung J.J. Co-circulation of human metapneumovirus and SARS-associated coronavirus during a major nosocomial SARS outbreak in Hong Kong. J Clin Virol. 2007;40:333–337. doi: 10.1016/j.jcv.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louie J.K., Hacker J.K., Mark J., Gavali S.S., Yagi S., Espinosa A. SARS and common viral infections. Emerg Infect Dis. 2004;10:1143–1146. doi: 10.3201/eid1006.030863. [DOI] [PMC free article] [PubMed] [Google Scholar]