Abstract
Osteosarcoma remains the most common primary bone tumor in dogs with half of affected dogs unable to survive one year beyond diagnosis. New therapeutic options are needed to improve outcomes for this disease. Recent investigations into potential therapeutic targets have focused on cell surface molecules with little clear therapeutic benefit. Transcription factors and protein interactions represent underdeveloped areas of therapeutic drug development. We have utilized allosteric inhibitors of the core binding factor transcriptional complex, comprised of core binding factor beta (CBFβ) and RUNX2, in four canine osteosarcoma cell lines Active inhibitor compounds demonstrate anti-tumor activities with concentrations demonstrated to be achievable in vivo while an inactive, structural analog has no activity. We show that CBFβ inhibitors are capable of inducing apoptosis, inhibiting clonogenic cell growth, altering cell cycle progression, and impeding migration and invasion in a cell-line dependent manner. These effects coincide with a reduced interaction between RUNX2 and CBFβ and alterations in expression of RUNX2 target genes. We also show that addition of CBFβ inhibitors to the commonly used cytotoxic chemotherapeutic drugs doxorubicin and carboplatin leads to additive and/or synergistic anti-proliferative effects in canine osteosarcoma cell lines. Taken together we have identified the interaction between components of the core binding factor transcriptional complex, RUNX2 and CBFβ, as a potential novel therapeutic target in canine osteosarcoma and provide justification for further investigations into the anti-tumor activities we describe here.
Keywords: canine sarcoma, osteosarcoma, novel therapeutic targets, transcription factor, RUNX2, core-binding factor beta (CBFβ)
Introduction
The treatment for osteosarcoma (OSA) has remained unchanged since the addition of adjuvant cytotoxic chemotherapy regimens (consisting of methotrexate, doxorubicin and cisplatin/carboplatin) to amputation or limb-sparing approaches in the late 1970s.1,2 Although these advances on OSA treatment represented an improvement of the survival rates in dogs and humans in the non-metastatic disease setting, 10-15% after 2 years and 65-70% after 5 years, respectively, little advancements have been achieved in patients with metastatic and/or relapsed disease despite great efforts to improve the prognosis in these patients through treatment intensification.3–5 Hence, there is an unmet need to find new therapeutic targets in order to increase the survival rates not only in patients with metastatic OSA but also with non-metastatic disease.
Among molecular targets investigated for therapeutic intervention in human OSA are cell surface receptor tyrosine kinases that are overexpressed, such as insulin-like growth factor 1 receptor (IGF1R), epidermal growth factor receptor 2 (HER-2/ERBB2), platelet-derived growth factor receptor (PDGFR) and hepatocyte growth factor receptor (HGFR/MET),5 yet there is still not strong evidence supporting their therapeutic effectiveness.6–10 In canine OSA, the multi-kinasetargeting drug toceranib phosphate has been evaluated in the adjuvant setting with no observable therapeutic benefit. A retrospective evaluation of dogs with macroscopic pulmonary metastases found no improvement to outcome in dogs treated with toceranib.11 Another study prospectively evaluated the addition of toceranib to metronomic cyclophosphamide/piroxicam therapy following amputation and adjuvant carboplatin; no improvement in disease free interval (DFI) or overall survival (OS) was noted with the addition of toceranib.12 These studies in human and canine patients exemplify the difficulty in developing targeted therapies against cell surface receptors in OSA and underscore the importance of identifying alternate therapeutic targets for this disease.
Transcription factors (TFs) are multi-protein complexes that serve as convergence points for intracellular signaling pathways and ultimately lead to modulation of gene expression. Historically, TFs as targets for cancer therapy has been a little-developed field but there is a growing interest in creating therapeutic strategies to alter the function of TFs as some of the most relevant oncogenes are in fact TFs, such as mutant p53 and MYC.13 RUNX2 is a transcription factor that is critical in normal skeleton development by regulating chondrocyte maturation and osteoblast differentiation.14 RUNX2 forms a heterodimer with Core-binding factor β (CBFβ), conferring stability to RUNX2 and enhancing Runx2-dependent transcriptional modulation; this interaction is essential for the inhibition of osteoblast differentiation by metastatic breast cancer cells and is associated with malignant phenotype of other tumor types.15,16 RUNX2 and CBFβ protein expression is deregulated in OSA and levels are increased in cell lines and tissues and RUNX2 overexpression is involved in the chemoresistance of human OSA cells.17,18 Canine OSA tumors also have increased copy number of the RUNX2 locus and highly express the RUNX2 protein.19,20 Furthermore, silencing of RUNX2 in human OSA cell lines sensitizes them to doxorubicin-induced apoptosis via c-Myc activity.21 Thus, the heterodimer RUNX2/CBFβ may be a promising candidate as a therapeutic target for this disease in both species.
Recently, Illendula et al have developed small molecules capable of allosterically inhibiting the binding of CBFβ to RUNX transcription factors.22 These compounds have demonstrated anti-tumor activity in leukemia and ovarian cancer cell lines, compromising cell viability, cell cycle and migration/invasiveness capabilities.22,23 With these novel compounds, we have examined the effects of inhibiting CBFβ/RUNX2 interaction in canine OSA cell lines and the mechanisms underlying these effects. We found that small molecule inhibitors of CBFβ/RUNX2 binding decrease cell viability and clonogenic growth, arrest cell cycle, impair metastatic capabilities and trigger apoptotic cell death in canine OSA cell lines. These effects are likely due to reduced CBFβ/RUNX2 interaction and subsequent decreased expression of RUNX2 target genes. Altogether, our findings support the CBFβ/RUNX2 interplay as a viable, druggable target in OSA and provide valuable information for potential future clinical development of these compounds for this disease.
Methods
Cell lines and culture conditions
The previously validated and characterized canine OS cell lines Abrams, Grade, HMPOS (p53 mutant) and D-17 (p53 wild-type) were generously supplied by Dr. Dawn Duval (Flint Animal Cancer Center, Colorado State University, Fort Collins, CO).24–27 Cells were routinely cultured in Corning® RPMI-1640 culture medium with L-Glutamine (Mediatech Inc., Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) and penicillin (50 U/mL)/streptomycin (50 μg/mL) (Mediatech Inc). Cell cultures were maintained at 37°C in a humidified environment with 5% CO2 and all assays were performed in this culture media.
Core binding factor (CBF) beta inhibitors
The small molecule inhibitors of CBFβ/RUNX binding AI-10-104 and AI-14-91, and control compound AI-4-88 employed in this work were synthetized by Dr. J.H. Bushweller’s laboratory as previously described.22 All compounds were dissolved in dimethyl sulfoxide (DMSO).
Cell viability
Cell proliferation and viability were evaluated by means of the fluorometric, bioreductive resazurin assay (Sigma-Aldrich, St. Louis, MO). Following a preliminary experiment to determine optimal cell numbers, cells were seeded in 96-well plates (Corning® Costar™, Corning, NY) at 4x103 (24 hour assay), 2.5x103 (48 hour assay) and 1.5x103 (72 hour assay) cells per well. We performed two experimental approaches: 1) cells were treated with vehicle control, CBFβ inhibitors, or negative control compound for 48 hours in a range of concentrations (0, 20, 30, 50, 60, 75 or 100 μM); and 2) and cells were treated with 20 μM CBFβ inhibitors (approximate IC50 from 48 hour proliferation assays), control compound, or vehicle for 24, 48 or 72 hours. All the treatments were performed in triplicate and repeated three times. Following the treatment, culture medium was replaced and cells were incubated with 10% resazurin in fresh culture media for 1-2 hours. Fluorescence was then measured at excitation/emission 560/590 nm on a microplate reader (Synergy HI, BioTek, Winooski, VT).
Clonogenic assay
Abrams, HMPOS and D-17 cells were treated with 20 μM CBFβ/RUNX inhibitors, control compound, or vehicle for 24 hours. Cells were subsequently detached by trypsinization and 300 single cells per well in 2 mL RPMI culture medium were seeded in triplicate in a 6-well plate (Corning® Costar™). Cells were incubated to form colonies for 7 days. Finally, colonies were gently washed with warm IX PBS and then fixed with 95% ethanol:acetic acid solution (3:1), stained with crystal violet (0.5% w/v) and manually counted using a microscope. Representative images of the assays were taken with a digital image analyzer (FluorChem E, ProteinSimple, San Jose, CA).
Caspase-3/-7 activity
Measurement of the enzymatic activities of caspase-3/-7 was carried out with a fluorometric SensoLyte® Homogeneous AMC Caspase-3/-7 Assay Kit (AnaSpec, Fremont, CA), according to manufacturer’s instructions. Cells were treated with 20 μM CBFβ inhibitors, control compound, or vehicle for 48 hours in a 96-well microplate (Corning® Costar™). Fluorescence intensity was measured using 354 nm excitation and 442 nm emission detection on a fluorescence microplate reader (Synergy HI, BioTek).
Bivariate Annexin V/PI analysis
Abrams and D-17 cells were collected after 48 hours treatment with 20 μM CBFβ inhibitors or control compound or vehicle, and subsequently washed twice with IX Binding Buffer and stained employing the eBioscience™ Annexin-V Apoptosis Detection Kit FITC (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Stained cells were processed on a CytoFLEX S flow cytometer (Beckman Coulter, Indianapolis, IN) and data were analyzed using FlowJo v4.0 software (FlowJo LLC, Ashland, OR). The HMPOS and Grade cell lines were not included in this analysis because they have a stably transfected nuclear red protein which was found to interfere with the compensation for AnnexinV/PI flow cytometric analysis.
Cell cycle analysis
Cell cycle was analyzed in D-17 and Abrams cells treated with 20 μM CBFβ inhibitors or control compound or vehicle for 24 hours. Following the treatment, cells were washed in PBS and fixed in cold 70% ethanol drop by drop while vortexing and then incubated at −20°C for 2 hours. Subsequently, cells were washed twice in stain buffer (PBS, 2% FBS) and stained using a PI/RNase Staining Buffer (BD Pharmingen™, BD Biosciences, San Diego, CA). Stained samples were evaluated on a CytoFLEX S flow cytometer (Beckman Coulter) and data were then analyzed using FlowJo v4.0 software (FlowJo LLC).
Cell migration and invasion assay
HMPOS (4.5 x 104) or D-17 (1.0 x 105) cells in FBS-free medium were added to the inserts of Corning® Matrigel® Invasion chambers (Coming, Bedford, MA) and culture medium containing 10% FBS was added to the lower chamber. Following the attachment of the cells to matrigel (around 1 hour after plating), cells were treated with 20 μM CBFβ inhibitors, control compound, or vehicle and incubated at 37°C. After 20 hours, non-invading cells were removed from the upper surface of the insert, those on the lower surface were fixed with 95% ethanol:acetic acid and then stained with crystal violet. Images (seven random 20x fields) were taken using a Leica DM2000 phase-contrast inverted microscope (Leica Mycrosystems). Cells were treated in duplicate and assayed separately. Cell migration was evaluated in parallel employing Corning® BioCoat™ Control Inserts with 8,0pm pores without matrigel.
Quantitative RT-PCR
All four canine OS cell lines were treated with CBFβ inhibitors (20 μM), control compound (20 μM), or vehicle. After 24 hours, total RNAs were isolated using TRIzol™ Reagent (Invitrogen), according to manufacturer’s instructions. Reverse transcription was performed employing a Quantitec® Reverse Transcription Kit (Qiagen Inc., Germantown, MD) on a SimpliAmp Thermal Cycler (Applied Biosystems®, ThermoFisher Scientific, Waltham, MA). Quantitative RT-PCR (qRT-PCR) assays were performed in duplicate employing a QuantiFast® SYBR® Green PCR Kit (Qiagen) on an AriaMx Realtime PCR System (Agilent Technologies, Santa Clara, CA). Sequences of canine gene primers are shown in Table 1. The geometric mean of housekeeping genes Rps5, Gapdh and Hnrnph1 was used to normalize the expression of genes of interest. The threshold cycle (CT) was determined and relative gene expression was calculated using the comparative Ct method 2 −Δ(ΔCT).28
Table 1.
Canine primer pairs used in qRT-PCR.
| Gene Symbol | Sequence | Sequence (5’→3’) |
|---|---|---|
| Bglap | Forward | TTACCAGCGCTTCTATGGC |
| Reverse | TATTCTGGAGCAGCTGTGATG | |
| Cdkn1a | Forward | TGCTACATGGGCTAGTTGTG |
| Reverse | CCAGGATGTTACAGGAGCTTAC | |
| Vegfa | Forward | CTGACGAGATCGAGTACATCTTC |
| Reverse | CACTCTAGGCCCTCATCATTAC | |
| Rps5 43 | Forward | TCACTGGTGAGAACCCCCT |
| Reverse | CCTGATTCACACGGCGTAG | |
| Hnrnph1 44 | Forward | CTCACTATGATCCACCACG |
| Reverse | TAGCCTCCATAACCTCCAC | |
| Gapdh | Forward | AACATCATCCCTGCTTCCAC |
| Reverse | AGACCACCTGGTCCTCAGTG | |
Immunoblotting for protein expression analysis
Whole-cell protein lysates from all four untreated cell lines were extracted, quantified and immunoblotted as previously described.29 Following protein transfer and blocking, PVDF membranes were incubated with primary antibodies: anti-RUNX2, anti-CBFβ, anti-H3 (Cell Signaling, Boston, MA) or anti-β-actin (ThermoFisher Scientific); and then with a horseradish peroxidase-conjugated secondary anti-rabbit or anti-mouse antibody (Invitrogen). Immunolabelling was detected employing Luminata™ Forte Western HRP substrate (Millipore, Billerica, MA) and was visualized with a digital chemiluminescent image analyzer (FluorChem E, ProteinSimple, San Jose, CA).
Co-immunoprecipitation of CBFβ/RUNX2
Protein lysates (500 μg) extracted from HMPOS (high basal RUNX2 expression) and D-17 (lower basal RUNX2 expression) treated with 20 μM CBFβ inhibitors for 6 hours, were incubated with anti-RUNX2 (2μg, Cell Signaling) overnight at 4°C under agitation, followed with a 4 hours incubation (4°C in a rotator mixer) with Protein A-Sepharose® CL-4B beads (GE Healthcare, San Ramon, CA). Next, immunonoprecipitates were washed three times with protein extraction buffer. Loading Buffer 2X was added to the samples and boiled at 95°C for 5 min in order to denature the proteins and separate them from the beads and antibody. Lastly, samples were analyzed by immunoblotting as explained above and PVDF membranes were incubated with anti-CBFβ (Cell Signaling). IgG protein (detected ≈55 kDa) was employed as loading control protein.
Combination of CBFβ inhibitor and chemotherapeutic drugs
The dose-effect of AI-14-91 (AI; active CBFβ inhibitor) and its combination with Doxorubicin (Dox) or Carboplatin (Carbo) on the proliferation of canine OSA cell lines was analyzed by Chou-Talalay method for drug combination which is based on the median-effect equation.30,31 Canine OSA cell lines plated in 96 well microplates were treated with AI (2.5-40 μM), Dox (12-500 ng/mL) or Carbo (12.5-500 μg/rnL) and combinations of AI with Dox or Carbo in a constant concentration ratio (Dox/Carbo:AI, 5:1). Following 48 hour incubation at 37°C, cell viability/proliferation was evaluated using Resazurin Assay as described previously. Combination index (CI), dose-reduction index (DRI), dose-effect levels of cell growth inhibition (ED50-ED95), and median-effect dose (Dm) were calculated using CompuSyn software (ComboSyn Inc, Paramus, NI). CI values <1, 1 and >1 suggest a synergistic, additive or antagonistic effect, respectively. DRI indicates the dose reduction of individual components allowed in a synergistic combination at a given effect level.31
Data and statistical analysis
All experiments were repeated independently at least twice and technical replicates in each independent experiment (at least in duplicate) were average and yielded a value for a biological replicate. Data (mean ± SEM) were statistically analyzed with GraphPad Prism® v7.04 (GraphPad Software, San Diego, CA). Comparisons of the effects of CBFβ inhibitors, negative control and vehicle control were compared by one- or two-ways ANOVA test followed by a Bonferroni multiple comparison test. For all comparisons, a P- value ≤0.05 was considered significant.
Results
RUNX2/CBFβ interaction is disrupted in canine OSA cells by CBFβ inhibitors, compromising cellular viability and clonogenic cell growth
Cellular viability/proliferation was analyzed in canine OSA cells (Abrams, HMPOS, D-17 and Grade) treated for 48 hours with CBFβ inhibitors (AI-10-104, AI-14-91) or the negative control compound (AI-4-88) employing a Resazurin Assay. Our results demonstrated that both AI-10-104 and AI-14-91 significantly decreased canine OSA cell viability/proliferation in a concentration-dependent fashion compared to AI-4-88 (P<0.001) with the HMPOS and Abrams cell lines. Cell proliferation was also significantly inhibited (P<0.001) with D-17 and Grade cell lines but this inhibition was already drastic with the lowest concentration used (20 μM) without enhanced effect with higher concentrations (Figure 1A). Given these results, we decided to employ 20 μM for all the compounds in the remaining experiments of this study. Cell proliferation was also analyzed in canine OSA cells treated with 20 μM CBFβ inhibitors or negative control compound at 24, 48 and 72 hours after treatment in order to evaluate the time-dependent effects. Our resazurin assays revealed a time-dependent anti-proliferative activity in cells treated with AI-10-104 and AI-14-91, with slight recovery in Grade cells exposed to AI-14-91 after 72 hours (Figure 1B). The negative control compound did not exert a negative effect in cellular viability in cells treated up to 72 hours. Moreover, we analyzed the long-term effect on cell proliferation by means of clonogenic assays (Figure 1C). Our findings showed that both surviving fraction and colony formation in canine OSA cells treated with CBFβ inhibitors were significantly suppressed when compared to AI-4-88 or vehicle control (P<0.01). To support that the anti-proliferative effects in canine OSA cell lines by CBFβ inhibitors were associated with a disruption of CBFβ/RUNX2 interaction, we performed an immunoprecipitation of RUNX2 and then examined CBFβ levels by immunoblotting. After confirming that all canine OSA cell lines express both proteins (Figure 2A), we selected HMPOS and D-17 to perform the co-immunoprecipitation due to their different p53 status (mutant and wild type, respectively). Our findings showed a decreased interaction of CBFβ and RUNX2 in HMPOS and D-17 after 6 hours of treatment, although this reduction was greater in D-17 cells (Figure 2B).
Figure 1. Cell proliferation and clonogenic growth is suppressed by small molecule inhibitors of CBFβ/RUNX2 binding in canine OSA cell lines.

Resazurin assay was used to assess cellular proliferation/viability in (A) cells treated with a range of concentrations (0-100 μM) of AI-10-104, AI-14-91 or AI-4-88 (negative control compound), or vehicle control for 48 hours, and (B) cells treated with 20 μM of AI-10-104, AI-14-91 or AI-4-88, or vehicle control after 24, 48, or 72 hours-treatment. (C) Representative images from clonogenic assays with respective surviving fractions. Data (mean±SEM) reported as percentage of control and analyzed by one-way or two-way ANOVA followed by a Bonferroni test (**p <0.01, ***p <0.001, ****p <0.0001).
Figure 2. Inhibitors of CBFβ reduce CBFβ/RUNX2 binding in canine OSA cell lines.

(A) Representative western blot images of CBFβ (22 kDa) and RUNX2 (55 kDa) in untreated Abrams, HMPOS, D-17 and Grade cells. (B) Representative images of the immunoblotting analysis and histograms expressing quantification of CBFβ after immunoprecipitation of RUNX2 in protein lysates from HMPOS and D-17 cells treated with AI-10-104, AI-14-91 or AI-4-88 at 20 μM, or vehicle control for 6 hours.
Small molecule inhibitors of CBFβ/RUNX binding trigger apoptotic cell death in canine OSA cells
In order to elucidate whether the inhibition of canine OSA cell proliferation by inhibitors of CBFβ/RUNX involves apoptosis pathways, we analyzed caspase-3/-7 activity employing a fluorometric assay in canine OSA cells (HMPOS, D-17 and Abrams) after a 48 hour treatment (Figure 3A). Both AI-10-104 and AI-14-91 (20 μM) led to a significant increase in caspase-3/-7 activity in all canine OSA cells as compared to AI-4-88 or vehicle control (P<0.01). We further evaluated the apoptotic profiles in Abrams and D-17 cells treated for 48 hours (20 μM) using a bivariate PI/Annexin-V assay analyzed by flow cytometry. Small molecule inhibitors significantly enhanced late apoptosis (PI+/Annexin-V+) and necrosis (PI+/Annexin-V−) features in Abrams (P<0.001) and D-17 cells (P<0.05), compared to AI-4-88 or vehicle control. Furthermore, AI-10-104 and AI-14-91 significantly triggered early apoptosis (PI7Annexin-V+) in D-17 cells (P<0.001). Unlike small molecule inhibitors of CBFβ/RUNX binding, AI-4-88 did not have any effect on either apoptosis profile or caspase-3/-7 activity in canine OSA cells (Figure 3B). Together, our results indicate that inhibitors of CBFβ/RUNX binding greatly trigger apoptotic cell death in canine OSA cells, partially explaining the decrease in cell viability and proliferation.
Figure 3. Necrotic and apoptotic cell death was triggered in canine OSA cell lines treated with small molecule inhibitors of CBFβ/RUNX2 binding.

Canine OSA cells treated with AI-10-104, AI-14-91 or AI-4-88 at 20 μM, or vehicle control for 48 hours were employed to evaluate the cell-death by means of (A) a caspase-3/-7 activity assay and (B) a bivariate PI/Annexin-V analysis. Data (mean±SEM) were calculated as percentage of control (untreated cells) and analyzed by two-way ANOVA multiple test followed by a Bonferroni test (*p <0.05, **p <0.01, ***p <0.001, ****p <0.0001).
Cell cycle progression is affected by inhibitors of CBFβ/RUNX binding in canine OSA cells
To evaluate whether cell cycle progression is compromised in canine OSA cells by small molecule inhibitors of CBFβ/RUNX binding, cell cycle was analyzed by flow cytometry in Abrams and D-17 cells treated for 24 hours, after staining DNA with a PI/RNase buffer. Cell cycle progression was slowed in a cell line dependent manner. Cell cycle was significantly arrested in G1 phase in Abrams cells treated with AI-10-104 or AI-14-91 as compared to AI-4-88 or vehicle control (P<0.001), accompanied with a significant decrease of cells in G2 phase (Figure 4A; P<0.05). However, cell cycle progression in D-17 was differently influenced by inhibitors of CBFβ/RUNX depending on the compound. AI-10-104 led to a significant accumulation of D-17 cells in G2 phase (P<0.05) while cells in G1 phase are significantly reduced (P<0.01) (Figure 4B).
Figure 4. CBFβ inhibitors disrupted the cell cycle in canine OSA cells.

Flow cytometry histograms (left) and quantitation of cell cycle distribution (right) of (A) Abrams and (B) D-17 cells stained with PI after treatment with vehicle, or 20 μM AI-10-104, AI-14-91 or AI-4-88 (negative control compound) for 24 hours. Data (mean±SEM) were calculated as percentage of control (untreated cells) and analyzed by two-way ANOVA multiple test followed by a Bonferroni test (*p <0.05, **p <0.01, ***p <0.001, ****p <0.0001).
Small molecules inhibitors of CBFβ/RUNX binding suppress in vitro metastatic ability of canine OSA cells
The impact of inhibitors of CBFβ/RUNX binding, specifically AI-10-104, on migratory behavior and invasion capability of canine OSA cells were further examined, because both processes are critical for tumor progression and metastasis. Migration and invasiveness of AI-10-104-treated HMPOS and D-17 cells were significantly inhibited (P<0.001) after 20 hours of treatment (Figures 5A and 5B). While neither vehicle control nor AI-4-88 affected any of these processes in D-17 cells, the latter significantly suppressed the invasion capability of HMPOS cells. However, the calculated invasion index, which represents the percentage of migrating cells that are also capable of invasion through the Matrigel, showed that only AI-10-104 had a critical effect in invasiveness of HMPOS and D-17 cells (Figure 5C).
Figure 5. Migration and invasive capabilities were significantly inhibited in canine OSA cells by CBFβ inhibitors.
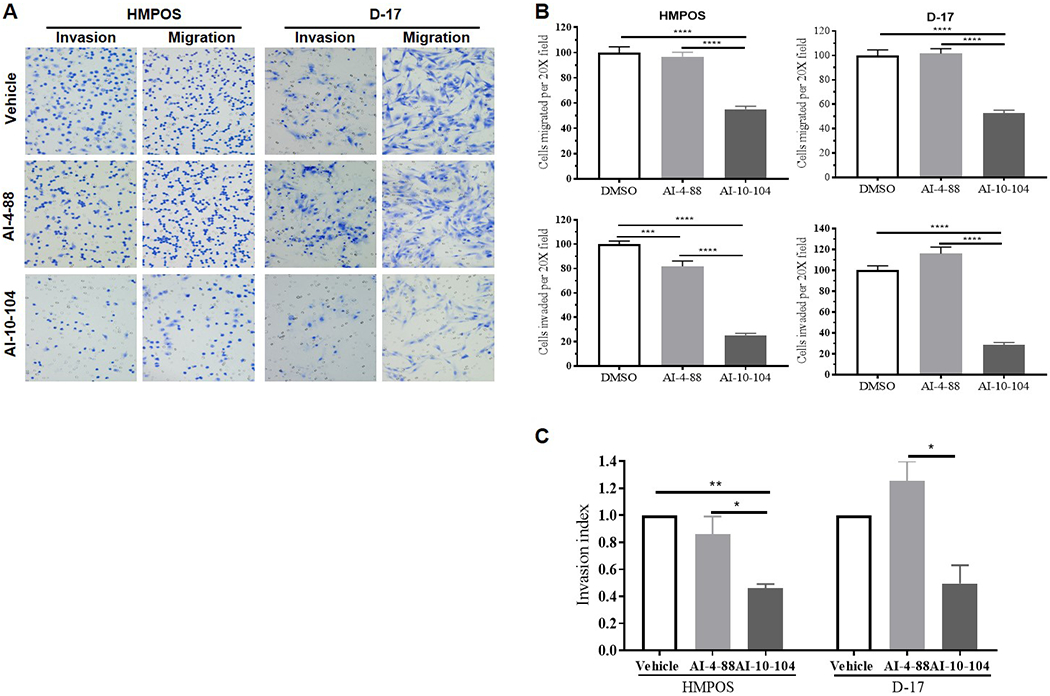
D-17 and HMPOS cells plated on upper matrigel-coated or control inserts were treated with 20 μM AI-10-104 or AI-4-88 (negative control compound), or vehicle and subjected to transwell invasion/migration assays for 20 hours. (A) Representative phase-contrast microscopy images (20X) of transwell invasion and migration assays. (B) Histograms expressing percentage of migrating and invading cells after 20 hours of incubation. (C) Invasion index was calculated by dividing percentage invasion of treated cells by the percentage invasion of vehicle cells. Data (mean±SEM) analyzed by one-way or two-way ANOVA followed by a Bonferroni test (*p <0.05, **p <0.01, ***p <0.001, ****p <0.0001).
Small molecules inhibitors of CBFβ/RUNX binding regulate the gene expression of RUNX2 targets
We next evaluated the gene expression of RUNX2 targets in order to elucidate whether anti-tumor effects in canine OSA cells by small molecules inhibitors of CBFβ/RUNX binding are associated with the disruption in the interaction of these proteins and subsequent alterations in RUNX2-mediated gene expression. We analyzed the mRNA expression levels by qRT-PCR for three targets genes of RUNX2, including Vegfa, Cdkn1a (p21) and Bglap (osteocalcin), in Abrams, D-17, HMPOS and Grade cells treated with 20 μM AI-10-104, AI-14-91 or negative compound (AI-4-88), or vehicle control (Figure 6). We found that all four canine OSA cell lines showed a decreased expression of Bglap mRNA following the AI-10-104 or AI-14-91 treatment, with significant reduction seen in HMPOS and Grade (P<0.05). The mRNA levels of Vegfa were greatly decreased in Abrams and Grade although not significantly, while D-17 showed a significant enhanced expression (P<0.01) and Vegfa levels were not altered in HMPOS. With regard to Cdkn1a expression, while AI-14-91 induced a reduction in its expression in D-17 and Grade, AI-10-104 triggered a significant increase in Abrams and HMPOS (P<0.01).
Figure 6. Small molecule inhibitors of CBFβ/RUNX2 binding modulated gene expression of RUNX2 targets in canine OSA cell lines.

Relative mRNA expression levels of VEGFA, Cdkn1a and Bglap were analyzed by qRT-PCR and normalized versus geometric mean of housekeeping genes GAPDH, Rps5, and Hnrnph1. Data (mean±SEM) were analyzed by two-way ANOVA multiple test followed by a Bonferroni test (*p <0.05, **p <0.01, ***p <0.001).
CBFβ inhibitors synergize with chemotherapeutics in a drug- and cell-dependent manner
The Chou-Talalay method was employed to evaluate the effect of combining a CBFβ inhibitor (AI-14-91 -AI-) with traditionally used chemotherapy drugs for canine OSA treatment, such as Doxorubicin (Dox) and Carboplatin (Carbo). Table 1 summarizes the combination effects of AI, Dox and Carbo in canine OSA cell lines. The AI/Dox combinations were synergistic at all dose-effect levels in Abrams and Grade cells, except for ED50 in the latter. This synergistic effect was also showed in D-17 and HMPOS cells, although the combinations were only additive at the highest dose levels (>ED90). Regarding AI/Carbo, combinations were synergistic in a dose-dependent manner in Grade and Abrams cells, with the exception of ED50 in Abrams cells. Table 2 shows that cytotoxic activity of both chemotherapeutic drugs is improved when canine OSA cells are co-treated with the inhibitor of CBFβ/RUNX binding, AI-14-91, except in the HMPOS cell line. The DRI values reflect that doses of chemotherapeutic drugs may be decreased due to their synergistic actions. For example, to inhibit cell proliferation by 95% (ED95), the concentration of Dox could be reduced 13.13-fold, 4.55-fold and 26.00-fold in Abrams, D-17 and Grade, respectively, when combined with AI. The concentration of Carbo can be reduced 6.04-fold, 1.72-fold and 5.55-fold to get the same inhibition of cell growth in Abrams, D-17 and Grade, respectively. However, the combination of AI with chemotherapeutic drugs did not exhibit a clear advantage in HMPOS cells.
Table 2.
Combination study of CBFbeta inhibitor with conventional chemotherapeutic drugs in canine OSA cell lines.
| CELL LINE | TREATMENT | PARAMETERS | CI VALUE | DRI VALUE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dm | m | r | ED50 | ED75 | ED90 | ED95 | ED50 | ED75 | ED90 | ED95 | ||
| Abrams | DOX | 33.84 | 0.91 | 0.82 | 2.10 | 4.16 | 8.25 | 13.13 | ||||
| Carbo | 10.02 | 0.68 | 1.00 | 0.82 | 1.73 | 3.64 | 6.04 | |||||
| AI | 12.75 | 1.76 | 0.93 | 5.23 | 4.07 | 3.16 | 2.67 | |||||
| AI:DOX | 19.31 | 2.11 | 0.87 | 0.81 | 0.49 | 0.31 | 0.23 | |||||
| AI:Carbo | 14.63 | 1.25 | 0.96 | 1.41 | 0.82 | 0.59 | 0.54 | |||||
| D-17 | DOX | 35.84 | 0.83 | 0.99 | 1.80 | 2.55 | 3.60 | 4.55 | ||||
| Carbo | 56.57 | 1.52 | 0.97 | 1.81 | 1.77 | 1.74 | 1.72 | |||||
| AI | 18.75 | 3.48 | 0.93 | 2.99 | 1.95 | 1.27 | 0.95 | |||||
| AI:DOX | 23.85 | 1.12 | 0.98 | 0.79 | 0.82 | 1.08 | 1.44 | |||||
| AI:Carbo | 37.56 | 1.48 | 0.98 | 0.89 | 1.08 | 1.36 | 1.63 | |||||
| Gracie | DOX | 8.61 | 0.57 | 0.93 | 0.56 | 2.35 | 9.82 | 26.00 | ||||
| Carbo | 19.28 | 1.15 | 0.99 | 1.21 | 2.14 | 3.77 | 5.55 | |||||
| AI | 13.29 | 1.35 | 0.92 | 4.33 | 5.91 | 8.08 | 9.98 | |||||
| AI:DOX | 18.43 | 2.18 | 0.91 | 2.02 | 0.60 | 0.23 | 0.14 | |||||
| AI:Carbo | 19.06 | 2.85 | 0.93 | 0.96 | 0.56 | 0.33 | 0.23 | |||||
| HMPOS | DOX | 1.88 | 0.77 | 0.80 | 30.98 | 2.57 | 0.21 | 0.04 | ||||
| Carbo | 6.85 | 2.07 | 0.94 | 0.75 | 0.84 | 0.94 | 1.02 | |||||
| AI | 15.67 | 2.59 | 0.98 | 4.26 | 4.30 | 4.34 | 4.36 | |||||
| AI:DOX | 0.08 | 0.28 | 0.64 | 0.03 | 0.45 | 6.15 | 38.40 | |||||
| AI:Carbo | 12.87 | 2.64 | 0.96 | 1.58 | 1.43 | 1.29 | 1.21 | |||||
AI: AI-14-91, Dox: Doxorubicin, Carbo: Carboplatin, Dm: median-effect dose, m: slope, r: linear correlation coefficient of the Dm plot, (conformity), CI: Combination index (<1 synergism, 1 additive, >1 antagonism), DRI: dose-reduction index, ED: dose-effect levels of cell growth inhibition.
Discussion
It is widely known that gene expression is profoundly altered in cancer, particularly those genes involved in regulating cellular survival, proliferation and self-renewal. In many cases, this altered gene expression in cancer is caused by a misregulation of transcription factors, due to a myriad of mechanisms. Therefore, transcription factors have emerged as promising targets for the development of new treatment approaches against cancer.32,33 Numerous strategies to target transcription factors have been considered, including direct targeting of transcription factors, modulating degradation or expression levels, preventing DNA binding, and blocking protein/protein interactions.34 However, transcription factors are challenging to target due to their typically flat and large interaction surfaces.13,32 Identification of molecules that work through allosteric inhibition is one strategy to overcome the difficulty in targeting the interaction surfaces of transcription factor protein complexes. In this regard, Illendula et al. have described the development of small molecules that bind to CBFβ at a site that is not part of the interface for RUNX protein binding, alter binding dynamics and reduce the affinity of CBFβ for RUNX.22 In their studies, the CBFβ inhibitors were shown to inhibit the growth of leukemia cells with AML-ETO and MLL-AF9 fusion proteins which have a requirement for RUNX1 activity. This growth inhibition was associated with reduced interactions between CBFβ and RUNX1, reduced RUNX1 occupancy on target genes and altered gene expression.35 Furthermore, Shin et al. recently demonstrated that knockdown of RUNX2 or CBFβ induces apoptosis in some human OS cell lines, and identified potential downstream effectors responsible for this activity, including MYC.36 Another recent study identified tumor suppressive activity of a microRNA through its targeting of RUNX2 in OS cells.37 Thus, in this study we aimed to identify whether inhibiting the interaction between RUNX2 and CBFβ, using allosteric inhibitors of CBFβ, would be sufficient to induce anti-tumor effects in canine OSA cell lines or if this activity may require knockdown of RUNX2 levels. We identified both concentration- and time-dependent anti-proliferative effects for two active CBFβ inhibitors while the negative control compound had no effect on cell proliferation. Clonogenic cell growth was also reduced by exposure to CBFβ inhibitors. The anti-proliferative effects were found to be due, in part, to both increased apoptosis as well as inhibition of cell cycle progression. The effect of CBFβ inhibition on cell cycle progression was cell-line dependent; a significant increase in G1 with a reduction in G2/M was seen in the Abrams cell line while the opposite was true for the D-17 cell line which also showed a significant increase in the sub-G1 population. This may be explained, in part, by differences in the relative expression of Cdkna (p21), an essential G1 checkpoint control protein, following exposure to inhibitors.38,39 We identified a significant increase in Abrams while the expression change in D-17 was less pronounced. Annexin V/PI staining showed differences between cell lines in the distribution of cells in early apoptosis, late apoptosis and necrosis, the overall effect in both cell lines was a significant increase in apoptosis.
We identified that all four of the cell lines utilized express RUNX2 and CBFβ and co-immunoprecipitation demonstrated that these proteins interact in canine OSA cells. We only recognized a slight reduction in the amount of CBFβ bound to RUNX2 in our study. This may have been due to sub-optimal sample timing, inhibitor concentration, or re-association of proteins during the early steps of the co-immunoprecipitation protocol. It is important to note that less than complete inhibition of the interaction was identified by Illendula et al. in their evaluation of CBFβ and RUNX1.35 We did not identify a reduction in the levels of RUNX2 or CBFβ protein following treatment with inhibitors; this is interesting because the CBFβ interaction has been suggested as a mechanism to stabilize RUNX2 and prevent its degradation through ubiquitination.40,41 It may be that the degree of inhibition was not complete enough to impact RUNX2 protein levels, yet was adequate to result in downstream effects such as reduced proliferation, increased apoptosis, reduced invasion and migration and altered gene expression. In this study, the alterations in expression of Vegfa following inhibitor treatment were cell-line dependent with reduced expression only seen in the Abrams and Grade cell lines. Relative expression of Cdkn1a (p21) was also cell-line dependent with regard to the degree of change, though all cell lines demonstrated some degree of increased expression. Furthermore, all four cell lines showed a reduction in relative expression of Bglap (osteocalcin) following CBFβ inhibition. Thus, we have demonstrated altered expression of RUNX2 target genes following exposure of OSA cells to inhibitors of the CBFβ-RUNX2 interaction.
We also evaluated the effect of combined CBFβ-RUNX2 inhibition and cytotoxic chemotherapy using drugs commonly employed in the treatment of OSA, given that RUNX2 has been implicated in the DNA damage response 42 In the Abrams and Grade cell lines, the addition of the CBFβ inhibitor to DOX or Carbo results in a synergistic anti-proliferative effect, particularly at the higher effect levels. Interestingly, in the D-17 and HMPOS cell lines these synergistic effects are not appreciated and may become antagonistic, particularly for the p53-mutant HMPOS cell line. However, given that these two cell lines had similar trends toward antagonism at higher effect levels, it is possible that p53 activity is not the determining factor in sensitivity to combinations of CBFβ inhibition and cytotoxic chemotherapy. Overall, these results suggest that the combination of cytotoxic chemotherapy and CBFβ inhibition may be beneficial in subsets of OSA, but may not provide therapeutic benefit in all cases. However, we have identified the interaction between RUNX2 and CBFβ as an important regulator of the malignant phenotype of canine OSA cells and provide evidence that inhibiting this interaction without the requirement for reducing RUNX2 or CBFβ levels results in anti-tumor effects, providing a framework for future investigations into the potential clinical utility of CBFβ-RUNX2 inhibitors as a therapeutic option for OSA.
Acknowledgements:
This work was supported by grants from the NIH (K01 OD026526-01) and the University of California, Davis, Center for Companion Animal Health (CCAH 2017-13-F) to L.A.W. and NIH R01 CA234478 to J.H.B.
Footnotes
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/vco.12526
Data Availability Statement
All data can be made available upon reasonable request.
Conflict of Interest
The authors declare no conflicts of interest.
References
- 1.Vos HI, Coenen MJ, Guchelaar HJ, et al. The role of pharmacogenetics in the treatment of osteosarcoma. Drug Discov Today 2016;21:1775–1786. [DOI] [PubMed] [Google Scholar]
- 2.Harrison DJ, Geller DS, Gill JD, et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther 2018;18:39–50. [DOI] [PubMed] [Google Scholar]
- 3.Maniscalco L Canine osteosarcoma: understanding its variability to improve treatment. Vet J 2015;203:135–136. [DOI] [PubMed] [Google Scholar]
- 4.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr 2016;28:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kansara M, Teng MW, Smyth MJ, et al. Translational biology of osteosarcoma. Nat Rev Cancer 2014;14:722–735. [DOI] [PubMed] [Google Scholar]
- 6.Weigel B, Malempati S, Reid JM, et al. Phase 2 trial of cixutumumab in children, adolescents, and young adults with refractory solid tumors: a report from the Children’s Oncology Group. Pediatr Blood Cancer 2014;61:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner LM, Fouladi M, Ahmed A, et al. Phase II study of cixutumumab in combination with temsirolimus in pediatric patients and young adults with recurrent or refractory sarcoma: a report from the Children’s Oncology Group. Pediatr Blood Cancer 2015;62:440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebb D, Meyers P, Grier H, et al. Phase II trial of trastuzumab in combination with cytotoxic chemotherapy for treatment of metastatic osteosarcoma with human epidermal growth factor receptor 2 overexpression: a report from the children’s oncology group. J Clin Oncol 2012;30:2545–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med 2012;367:1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grignani G, Palmerini E, Dileo P, et al. A phase II trial of sorafenib in relapsed and unresectable high-grade osteosarcoma after failure of standard multimodal therapy: an Italian Sarcoma Group study. Ann Oncol 2012;23:508–516. [DOI] [PubMed] [Google Scholar]
- 11.Kim C, Matsuyama A, Mutsaers AJ, et al. Retrospective evaluation of toceranib (Palladia) treatment for canine metastatic appendicular osteosarcoma. Can Vet J 2017;58:1059–1064. [PMC free article] [PubMed] [Google Scholar]
- 12.London CA, Gardner HL, Mathie T, et al. Impact of toceranib/piroxicam/cyclophosphamide maintenance therapy on outcome of dogs with appendicular osteosarcoma following amputation and carboplatin chemotherapy: A multi-institutional study. PLoS One 2015;10:e0124889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazo JS, Sharlow ER. Drugging undruggable molecular cancer targets. Annu Rev Pharmacol Toxicol 2016;56:23–40. [DOI] [PubMed] [Google Scholar]
- 14.Komori T Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res 2010;339:189–195. [DOI] [PubMed] [Google Scholar]
- 15.Mendoza-Villanueva D, Zeef L, Shore P. Metastatic breast cancer cells inhibit osteoblast differentiation through the Runx2/CBFbeta-dependent expression of the Wnt antagonist, sclerostin. Breast Cancer Res 2011;13:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis JN, Rogers D, Adams L, et al. Association of core-binding factor beta with the malignant phenotype of prostate and ovarian cancer cells. J Cell Physiol 2010;225:875–887. [DOI] [PubMed] [Google Scholar]
- 17.Sadikovic B, Thorner P, Chilton-Macneill S, et al. Expression analysis of genes associated with human osteosarcoma tumors shows correlation of RUNX2 overexpression with poor response to chemotherapy. BMC Cancer 2010;10:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.San Martin IA, Varela N, Gaete M, et al. Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfbeta in osteosarcoma cells. J Cell Physiol 2009;221:560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Khan AA, Gunn HJ, Day MJ, et al. Immunohistochemical validation of spontaneously arising canine osteosarcoma as a model for human osteosarcoma. J Comp Pathol 2017;157:256–265. [DOI] [PubMed] [Google Scholar]
- 20.Angstadt AY, Thayanithy V, Subramanian S, et al. A genome-wide approach to comparative oncology: high-resolution oligonucleotide aCGH of canine and human osteosarcoma pinpoints shared microaberrations. Cancer Genet 2012;205:572–587. [DOI] [PubMed] [Google Scholar]
- 21.Roos A, Satterfield L, Zhao S, et al. Loss of Runx2 sensitises osteosarcoma to chemotherapy-induced apoptosis. Br J Cancer 2015;113:1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Illendula A, Gilmour J, Grembecka J, et al. Small molecule inhibitor of CBFβ-RUNX binding for RUNX transcription factor driven cancers. EBioMedicine 2016;8:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlton AL, Illendula A, Gao Y, et al. Small molecule inhibition of the CBFβ/RUNX interaction decreases ovarian cancer growth and migration through alterations in genes related to epithelial-to-mesenchymal transition. Gynecologic Oncology 2018;149:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York D, Withers SS, Watson KD, et al. Enrofloxacin enhances the effects of chemotherapy in canine osteosarcoma cells with mutant and wild-type p53. Vet Comp Oncol 2017;15:1087–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marley K, Helfand SC, Edris WA, et al. The effects of taurolidine alone and in combination with doxorubicin or carboplatin in canine osteosarcoma in vitro. BMC Vet Res 2013;9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, Thamm DH, Misra V. The effect of Zhangfei/CREBZF on cell growth, differentiation, apoptosis, migration, and the unfolded protein response in several canine osteosarcoma cell lines. BMC Veterinary Research 2015;11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fowles JS, Dailey DD, Gustafson DL, et al. The Flint Animal Cancer Center (FACC) canine tumour cell line panel: a resource for veterinary drug discovery, comparative oncology and translational medicine. Vet Comp Oncol 2017;15:481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 29.Alegre F, Ormonde AR, Snider KM, et al. A genetically engineered microRNA-34a prodrug demonstrates anti-tumor activity in a canine model of osteosarcoma. PLoS One 2018;13:e0209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou T-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Research 2010;70:440–446. [DOI] [PubMed] [Google Scholar]
- 31.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006;58:621–681. [DOI] [PubMed] [Google Scholar]
- 32.Heppler LN, Frank DA. Targeting oncogenic transcription factors: Therapeutic implications of endogenous STAT inhibitors. Trends Cancer 2017;3:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aalders KC, Tryfonidis K, Senkus E, et al. Anti-angiogenic treatment in breast cancer: Facts, successes, failures and future perspectives. Cancer Treat Rev 2017;53:98–110. [DOI] [PubMed] [Google Scholar]
- 34.Lambert M, Jambon S, Depauw S, et al. Targeting transcription factors for cancer treatment. Molecules 2018;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Illendula A, Gilmour J, Grembecka J, et al. Small molecule inhibitor of CBFbeta-RUNX binding for RUNX transcription factor driven cancers. EBioMedicine 2016;8:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin MH, He Y, Marrogi E, et al. A RUNX2-mediated epigenetic regulation of the survival of p53 defective cancer cells. PLoS Genet 2016;12:e1005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Long F, Wan J, et al. MicroRNA-205 acts as a tumor suppressor in osteosarcoma via targeting RUNX2. Oncol Rep 2016;35:3275–3284. [DOI] [PubMed] [Google Scholar]
- 38.Xiong Y, Hannon GJ, Zhang H, et al. p21 is a universal inhibitor of cyclin kinases. Nature 1993;366:701–704. [DOI] [PubMed] [Google Scholar]
- 39.Deng C, Zhang P, Harper JW, et al. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995;82:675–684. [DOI] [PubMed] [Google Scholar]
- 40.Park NR, Lim KE, Han MS, et al. Core binding factor beta plays a critical role during chondrocyte differentiation. J Cell Physiol 2016;231:162–171. [DOI] [PubMed] [Google Scholar]
- 41.Zhao M, Qiao M, Oyajobi BO, et al. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem 2003;278:27939–27944. [DOI] [PubMed] [Google Scholar]
- 42.Wysokinski D, Pawlowska E, Blasiak J. RUNX2: A master bone growth regulator that may be involved in the DNA damage response. DNA Cell Biol 2015;34:305–315. [DOI] [PubMed] [Google Scholar]
- 43.Brinkhof B, Spee B, Rothuizen J, et al. Development and evaluation of canine reference genes for accurate quantification of gene expression. Anal Biochem 2006;356:36–43. [DOI] [PubMed] [Google Scholar]
- 44.Selvarajah GT, Bonestroo FAS, Timmermans Sprang EPM, et al. Reference gene validation for gene expression normalization in canine osteosarcoma: a geNorm algorithm approach. BMC Vet Res 2017;13:354. [DOI] [PMC free article] [PubMed] [Google Scholar]


