Abstract
Synovial fibroblasts are key cells orchestrating the inflammatory response in arthritis. Here we demonstrate that loss of miR-146a, a key epigenetic regulator of the innate immune response, leads to increased joint destruction in a TNF-driven model of arthritis by specifically regulating the behavior of synovial fibroblasts. Absence of miR-146a in synovial fibroblasts display a highly deregulated gene expression pattern and enhanced proliferation in vitro and in vivo. Deficiency of miR-146a induces deregulation of tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6) in synovial fibroblasts, leading to increased proliferation. In addition, loss of miR-146a shifts the metabolic state of fibroblasts towards glycolysis and augments the ability of synovial fibroblasts to support the generation of osteoclasts by controlling the balance of osteoclastogenic regulatory factors receptor activator of NF-κB ligand (RANKL) and osteoprotegerin (OPG). Bone marrow transplantation experiments confirmed the importance of miR-146a in the radioresistant mesenchymal compartment for the control of arthritis severity, in particular for inflammatory joint destruction. This study therefore identifies microRNA-146a as an important local epigenetic regulator of the inflammatory response in arthritis. It is a central element of an anti-inflammatory feedback loop in resident synovial fibroblasts, who are orchestrating the inflammatory response in chronic arthritis. MiR-146a restricts their activation, thereby preventing excessive tissue damage during arthritis.
1. Introduction
Inflammatory arthritides, such as rheumatoid arthritis (RA) or psoriatic arthritis (PsA), are systemic autoimmune diseases characterized by synovial infiltration of inflammatory cells, leading to clinical symptoms such as tender and swollen joints [1–5]. The local inflammatory milieu of hematopoietic and mesenchymal cells in the joint supports the development of bone damage by specifically equipped cells, namely osteoclasts, which are multinucleated cells derived from hematopoietic monocytic precursors and the only known cell type capable of bone resorption [1,6]. Although intensely studied, many mechanisms controlling joint inflammation and subsequent osteoclastogenesis are yet incompletely understood. The need for a better understanding of the pathogenesis of these diseases is highlighted by the fact that in spite of a steadily growing arsenal of therapies still up to 50% of patients with RA or PsA are not responding well to current treatment regimens [7,8].
Micro RNAs (miRs) are non-coding regulatory RNAs that have been shown to be important in the regulation of many aspects of biology, including cancer development and immunity [9–12]. MicroRNAs regulate gene expression at a post transcriptional level by translational repression or degradation of target gene mRNA [11,13]. Originally thought to be important for fine tuning of cellular responses, the generation of mice lacking specific microRNAs has demonstrated very specific functions of certain microRNAs [14–16]. Several miRs have been implicated in the pathogenesis of systemic autoimmune diseases including RA, although the in vivo relevance of individual microRNAs remains largely unknown [17–20].
MiR-146a is mainly expressed in immune cells of the innate and adaptive immune system and it is induced during their activation or maturation through NF-κB. Its main targets are interleukin-1 receptor-associated kinase 1 (IRAK1), TRAF6 and RelB, among others [16,21–23]. Functionally miR-146a regulates inflammatory responses in monocytes and macrophages as well as the resolution of Tcell responses and the maintenance of immunological tolerance [16,22,24,25].
MiR-146a expression has been demonstrated to be differentially regulated in patients with RA compared to healthy controls [26–30]. Furthermore, using agonistic miR-146a as a therapeutic agent in a commonly used disease model of arthritis, the collagen induced arthritis, revealed reduction of joint damage, by decreasing the capacity for osteoclastogenic differentiation of myeloid cells [31]. In addition, miR-146a has been shown to be an important regulator of the proinflammatory NF-κB induced cytokines IL-6 and IL-1β in Borrelia burgdorferi-induced arthritis [32]. In this study, we show that miR-146a, by regulating the expression of TRAF6, is a central regulator of the pathogenicity of synovial fibroblasts in inflammatory arthritis. Loss of miR-146a leads to increased proliferation and elevated metabolic activity in synovial fibroblasts and enhances their ability to support the generation of bone destructing osteoclasts during arthritis.
2. Materials and methods
2.1. Mice
Breeding pairs of miR-146a−/− and miR-146a+/+ littermate (B6.(FVB)-MIR146 TM1.1BAL/J) mice were provided by Mark Boldin/David Baltimore. These mice were crossed into Tg197 human TNF transgenic mice (hTNFtg; genetic background C57BL6; [33]) to obtain miR-146a−/−/hTNFtg mice. Mice used in all experiments were age- and sex-matched. All data were generated from littermates.
2.2. Antibodies and reagents
The following antibodies were used: CD11c (Clone: HL3) (Armenian hamster anti-mouse; BD Pharmingen); CD11b (Clone: M1/70) (rat anti-mouse; BD Pharmingen); CD11b (Clone: M1/70) (rat anti-mouse; eBioscience).
2.3. Arthritis disease model
For the human TNF alpha transgenic mouse model of arthritis [33] hTNFtg mice were crossed with miR-146a−/− mice.
2.4. Histological analysis
Mouse hind paws or tibiae were fixed in 4.5% formalin for 6 h, decalcified in 14% EDTA/ammonium hydroxide buffer (pH 7.2; Sigma-Aldrich) at 4 °C until the bones were pliable. Serial paraffin embedded sections from hind paws and tibiae (2 μm) were stained with hematoxylin and eosin (H&E) or for tartrate-resistant acid phosphatase (TRAP) activity. Staining for TRAP was done as previously described [34]. For quantification of inflammation and erosion areas, bone volume per tissue volume, trabecular number, trabecular thickness, trabecular separation, number of osteoclasts per bone perimeter and number of osteoblasts per bone perimeter, Zeiss Axioskop2 Mot microscope (Carl Zeiss Micro-Imaging) and Osteomeasure Analysis System (OsteoMetrics) were used. Osteoclasts were counted in TRAP stained sections of tarsal joints; to distinguish them from other TRAP+ cells such as macrophages and dendritic cells, we counted only TRAP+ cells displaying a minimum of 3 nuclei located at the eroded sites of bone surfaces in the metatarsal bone as osteoclasts. The sum of inflammation or erosion areas for each mouse was calculated from H&E-stained sections of tarsal joints from each mouse.
Microscopic analysis was performed on Zeiss Axioscop2 Mot microscope, at a magnification of 5× (numerical aperture NA: 0,16), 10× (NA: 0,45) and 20× (NA: 0,75) plan apochromat objective, at an illumination colour temperature of 3200 K, with air as imaging medium. Olympus DP73 was used as imaging camera. Cellsens Dimension was used as acquisition software. Images were processed by contrast optimization.
For immunohistochemistry rat monoclonal F4/80 (Serotec; diluted 1:200) and polyclonal rabbit anti Ki-67 (Abcam; diluted 1:100) were used. To quantify the percentage of F4/80 or Ki-67 positive cells, staining intensity of F4/80 or Ki-67 was analyzed by tissue cytometry using HistoQuest (Tissuegnostics).
Assessment of Ki-67 production in Ki-67 stained sections of tarsal joints, was done using a semi quantitative scoring system, ranging from 0 to 3 (0 = no colouring detectable, 1 = brown colouring on one or two spots, 2 = brown colouring on more than two spots, 3 = brown colouring of the whole section).
2.5. Cell culture
Osteoclast generation was performed as previously described [35].
For in vitro fibroblasts and coculture assays, fibroblasts were isolated from hind paws of wt or miR-146a−/− mice as previously described [40]. Fibroblasts were seeded at a density of 1 × 105cells/ml and proliferation was assessed by 3H-Thymidine incorporation. For coculture after 24 h of fibroblast adherence, macrophages were added to the culture and stimulated with 10 ng/ml IL-1 β [32].
2.6. Transfection
Transfection of fibroblasts with either TRAF6 or control siRNA (Dharmacon) was done as previously described [36].
2.7. Cell metabolism
Seahorse XF-24 cell culture plates (Agilent Technologies) were coated with 15 μg/cm2 rat collagen (Sigma), cells were seeded at a density of 4 × 104 cells/well. The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of cultured synovial fibroblasts were analyzed with an XF-24 Extracellular Flux Analyser (Seahorse Bioscience) according to the manufacturer’s protocols.
2.8. Micromass
Micromass cultures using mouse synovial fibroblasts were performed as previously described [37], followed by histological analysis of Ki-67 expression (see immunohistochemistry).
2.9. RNA sequencing
Total RNA was prepared from approximately 1 million cells by using mirVana miRNA Isolation Kit (AM1560, ABI). 200 ng of total RNA was subsequently used to prepare RNA-seq library by using TruSeq SR RNA sample prep kit (FC-122–1001, Illumina) by following manufacturer’s protocol. The libraries were sequenced for 50 cycles (single read) with a HiSeq 2000 (Illumina). Raw sequencing data were processed with CASAVA 1.8.2 to generate FastQ files. Sequence reads were mapped onto the mouse genome build mm9 using TopHat 2.0. Gene expression values (RPKM, reads per kilobase exon per million mapped reads) were calculated with Cufflinks 2.0. All downstream statistical analyses were performed with Partek Genomics Suite 6.6, R 3.0.1, and GeneSpring GX 12.1 (Agilent Technologies).
2.10. Generation of radiation chimeras
This was done as previously described [34].
2.11. MiRNA quantitative RT-PCR
Micro RNA was isolated using miRNeasy Mini Kit (Qiagen). MiR-146a expression was measured using TaqMan miRNA Assays hsa-miR-146a (Applied Biosystems) according to the manufacturer’s instruction using the Rotor-Gene Q PCR cycler (Qiagen). U6snRNA (miRNA assay U6 snRNA Applied Biosystems) was used as internal control. Relative expression of miR-146a was calculated by the 2ΔΔct method.
2.12. Messenger RNA (mRNA) quantitative RT-PCR
Total RNA was isolated using RNeasy mini kit (Qiagen). CDNA was prepared using Omniscript RT kit (Qiagen), followed by SYBR Green-based quantitative PCR (Roche Molecular Biochemicals) using the Light cycler 480 (Roche Molecular Biochemicals). The expression of genes of interest was normalized to GAPDH. The relative expression of the mRNA of the gene of interest was calculated by the 2ΔΔct method.
2.13. Flow cytometric analysis
Blood cells were washed with PBS supplemented with 1% FCS (GIBCO), stained with indicated antibodies and analyzed by flow cytometry (BD Biosciences Facs Canto II, with Facsdiva software).
2.14. Elisa
Supernatant concentration of RANKL (R&D Systems) as well as serum concentration of hTNF (R&D Systems) was assessed according to the manufacturer’s instructions.
2.15. Statistical analysis
Statistical significance of two different groups was calculated using the unpaired two tailed Student t-test. Area under the curve was used analysing the difference of cell proliferation, OCR and ECAR over the time followed by two tailed Student t-test. All analyses were performed using GraphPad Prism 5 software. Graphs present data as mean ± SEM. A p value less than or equal to 0.05 was considered significant (*p < 0.05, **p < 0.01, ***p < 0.001).
All animal studies were approved by the animal ethics committee of the Medical University Vienna and comply with institutional guidelines.
3. Results
3.1. Severity of arthritis is increased in hTNFtg mice lacking miR-146a
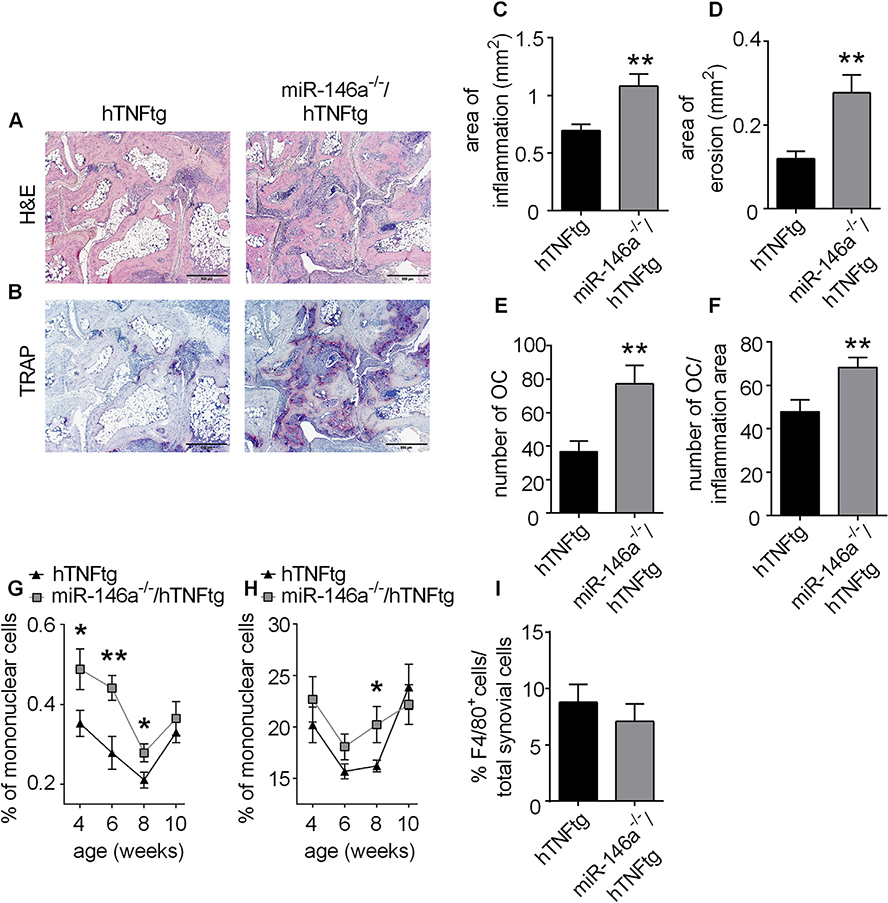
To test the effect of miR-146a deficiency in a model of chronic inflammatory arthritis mainly depending on the innate immune system displaying many features of human rheumatoid arthritis, we used the hTNFtg arthritis model [44, 45]. Therefore we crossed miR-146a−/− into hTNFtg mice to obtain miR-146a−/−/hTNFtg mice. While we were not able to detect differences in paw swelling and grip strength between hTNFtg and miR-146a−/−/hTNFtg animals (data not shown), when we analyzed the joints of these mice histologically, we observed a significant increase in the degree of synovial inflammation in miR-146a−/−/hTNFtg mice compared to hTNFtg mice. Moreover, local bone destruction was elevated more than twofold in miR-146a−/−/hTNFtg mice compared to their control mice. This increased bone destruction was accompanied by an about twofold increase in bone resorbing multinucleated synovial osteoclasts found locally within the eroded sites of bony surfaces of inflamed hind paws (Fig. 1A–E). Of note, in miR-146a−/−/hTNFtg mice, local osteoclastogenesis was elevated more profoundly than synovial inflammation, as the number of OCs in relation to the area of inflammation was significantly increased in these animals compared to hTNFtg mice, as was the area of erosion per area of inflammation, suggesting a small but significant dissociation of inflammation and erosion in miR-146a−/−/hTNFtg mice compared to hTNFtg mice (Fig. 1F, Fig. S 1A). We were not able to detect differences in the area of erosion per number of osteoclast (Fig. S 1B), suggesting that increased numbers of osteoclasts but not increased activity of individual osteoclasts account for elevated bone destruction in miR-146a−/−/hTNFtg mice compared to hTNFtg mice. When we analyzed the kinetics of inflammatory cells in blood during the course of arthritis, we noticed increased relative numbers of CD11b+ and CD11c+ myeloid cells in miR-146a−/−/hTNFtg mice compared to hTNFtg mice (Fig. 1G and H), suggesting enhanced mobilization of inflammatory myeloid cells from bone marrow in miR-146a−/−/hTNFtg mice compared to hTNFtg mice. Moreover measurement of the relative numbers of F4/80+ cells in histological sections of arthritic paws revealed no difference between hTNFtg and miR-146a−/−/hTNFtg animals suggesting that miR-146a does not impact the recruitment of myelomonocytic cells, which include potential osteoclast precursor cells. Nonetheless, OC precursors are not yet fully characterized and may also derive from other cell populations than just F4/80+ cells (Fig. 1I). Taken together, these data clearly demonstrate that miR-146a deficiency causes increased synovial inflammation and even more strikingly augmented local bone destruction. Furthermore, miR-146a regulates the egress of inflammatory myeloid cells from the bone marrow into the bloodstream during hTNF-driven arthritis but does not influence recruitment of specific inflammatory cell types into the inflamed synovial membrane.
Fig. 1. Human TNF transgenic mice lacking miR-146a (miR-146a−/−/hTNFtg) show more severe bone destruction compared to hTNFtg mice.
Histological sections of the hind paws of hTNFtg and miR-146a−/−/hTNFtg mice obtained at 10 weeks of age stained with A, Hematoxylin and eosin (H&E) B, Tartrate-resistant acid phosphatase (TRAP) (bars 500 μm, magnification x 5). Osteoclasts are displayed as purple-stained cells. C-F, Histomorphometric analysis of the extent of inflammation, bone erosion, number of osteoclasts and number of osteoclasts per inflammation area; in the tarsal area of the hind paws of hTNFtg (n = 16) and miR-146a−/−/hTNFtg (n = 15) mice. G and H, FACS analysis of CD11c+ (G) and CD11b+ (H) blood cells from hTNFtg (n = 19) and miR-146a−/−/hTNFtg (n = 20) mice at the indicated age, using anti-CD11c and -CD11b antibodies. I, Quantification of F4/80+ cells among total synovial cells, from immunohistochemically stained sections of hind paws from hTNFtg (n = 12) and miR-146a−/−/hTNFtg (n = 14) mice using anti-F4/80 antibody. *p < 0,05, **p < 0,01 Results are shown as mean ± SEM.
3.2. MiR-146a does not impact systemic bone loss
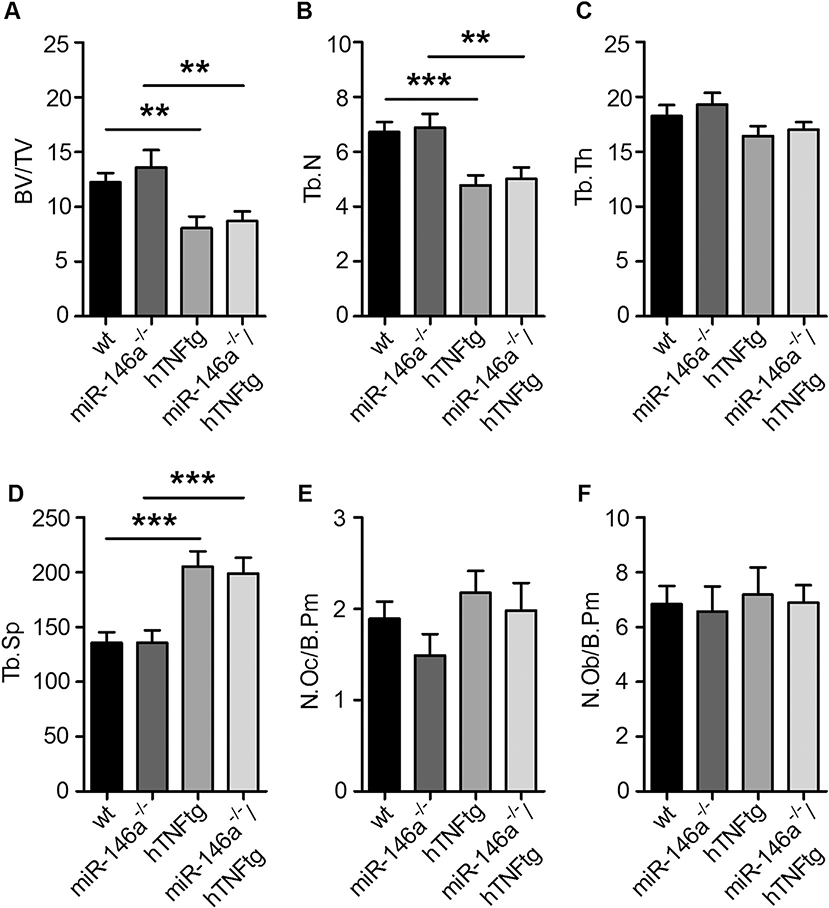
In hTNFtg mice, systemic overexpression of hTNF not only leads to arthritis and local bone destruction, but also to systemic bone loss. We therefore analyzed the influence of miR-146a deficiency on the development of osteoporosis induced by hTNF. We were not able to detect differences in wt and miR-146a−/− mice with respect to bone volume/tissue volume (bv/tv) and other variables of bone histomorphometry of tibial bones at 10 weeks of age (Fig. 2A–F). Consistent with previous reports, overexpression of hTNF decreased bone mass by 30% in hTNFtg animals compared to wt animals of the same age. Interestingly, however, miR-146a−/−/hTNFtg mice did not display more systemic bone loss compared to hTNFtg animals, in spite of their markedly higher local bone destruction (Fig. 2A–F). These data suggests, that miR-146a exerts its role locally in the inflamed joint. In line with this, we found miR-146a to be highly upregulated locally in the inflamed paws, but not in the spleen or in the bone marrow (S2 A-C), further demonstrating the important local role for miR-146a regulating joint pathology.
Fig. 2. Loss of miR-146a in hTNFtg mice does not have an impact on the development of osteoporosis.
A-F, Quantitative histological analysis of tartrate-resistant acid phosphatase (TRAP) stained sections of the tibiae from wt (n = 11), miR-146a−/− (n = 14), hTNFtg (n = 14) and miR-146a−/−/hTNFtg (n = 16) mice. A, Bone volume per tissue volume B, Trabecular number C, Trabecular thickness D, Trabecular separation E, Number of Osteoclasts per bone perimeter and F, Number of Osteoblasts per bone perimeter (n = 11).
3.3. MiR-146a controls activation and proliferation associated genes in synovial fibroblasts
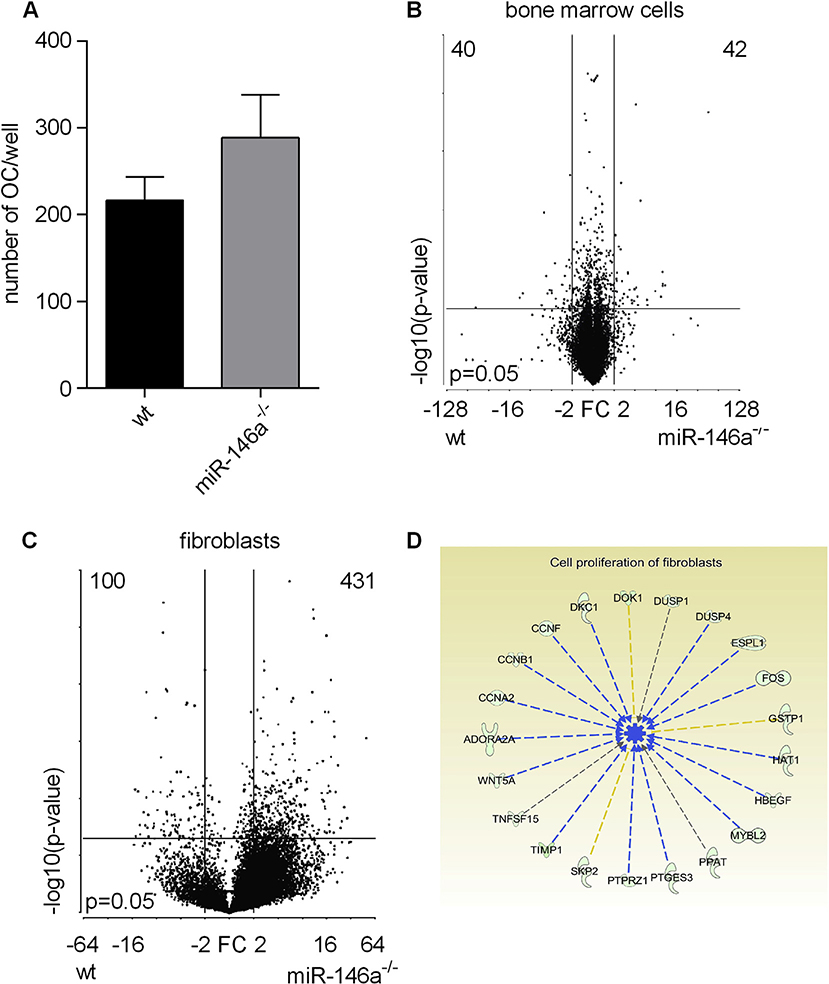
We first analyzed the potential of wt and miR-146a−/− bone marrow cells in response to macrophage colony-stimulating factor (MCSF) and RANKL. However, we did not detect differences in the numbers of osteoclasts between wt and miR-146a−/− bone marrow cells (Fig. 3A). We next analyzed the transcriptome of myeloid bone marrow cells and synovial fibroblasts, the two major cell types in the synovial membrane by RNA sequencing. This allowed us to analyze the impact of miR-146a deficiency specifically in these cells in more detail. While there were only marginal differences at the transcriptome levels in bone marrow cells from wt and miR-146a deficient animals (Fig. 3B), we found a high number of differently regulated genes in synovial fibroblasts between wt and miR-146a deficient cells (Fig. 3C). Pathway analysis of differently regulated genes revealed that proliferation of cells as well as de novo synthesis of pyrimidines might be deregulated in miR-146a deficient fibroblasts and we also observed high expression of activation associated genes in miR-146a−/− synovial fibroblasts (Fig. 3D and S3).
Fig. 3. Gene expression vary significantly in miR-146a deficient fibroblasts but not in bone marrow cells.
A, Bone marrow isolated from wt or miR-146a−/− mice was stimulated with MCSF (d0) and MCSF + RANKL (d3) to induce osteoclastogenesis. The number of OCs/well among TRAP positive stained multinucleated cells in wt and miR-146a−/− mice (d7) was analyzed (data shown are representative of 3 independent experiments). B and C, Volcano plots showing differentially expressed genes from wt versus miR-146a−/− bone marrow cells (n = 3) (B) and synovial fibroblasts (n = 2) (C). D, Pathways affected by loss of miR-146a in synovial fibroblasts as detected by ingenuity pathway analysis. Results are shown as mean ± SEM.
3.4. MiR-146a regulates synovial fibroblast proliferation by targeting TRAF6
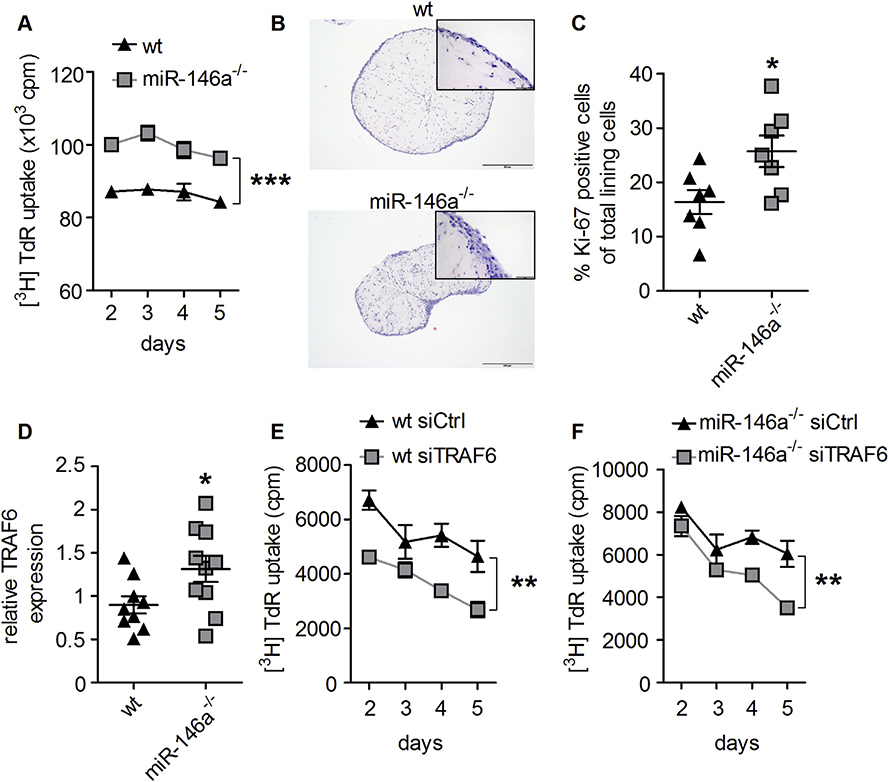
To test whether proliferation was indeed different in fibroblasts lacking miR-146a, we measured proliferation using 3H-Thymidine incorporation. As shown in Fig. 4A, proliferation of miR-146a−/− synovial fibroblasts was increased compared to wt synovial fibroblasts. We next analyzed the behavior of wt and miR-146a−/− synovial fibroblasts in an ex vivo three dimensional culture model [37]. MiR-146a knock out synovial fibroblasts were able to form a synovial lining layer similar to the wt controls (Fig. 4B). However, analysis of the proliferation marker Ki-67 in the lining layer of the 3D cultures revealed increased proliferation of miR-146a deficient compared to wt synovial fibroblasts (Fig. 4C). When we analyzed known target genes of miR-146a in synovial fibroblasts, we found only TRAF6 to be significantly upregulated compared to wt controls, whereas other miR-146a target genes, such as IRAK1 and RelB were not differently expressed (Fig. 4D and S4A and B). As TRAF6 has recently been implicated in regulating proliferation of lung fibroblasts [38], we tested, whether TRAF6 was involved in the increased proliferative capacity of miR-146a deficient synovial fibroblasts. Indeed, upon knock-down of TRAF6, the proliferation of synovial fibroblasts was significantly reduced, suggesting a key regulatory role of TRAF6 in controlling fibroblast proliferation (Fig. 4E and F).
Fig. 4. MiR-146a deficiency leads to increased proliferation of synovial fibroblasts.
A, In vitro proliferation of wt and miR-146a−/− synovial fibroblasts, measured by 3[H] thymidine incorporation (data shown are representative of 3 independent experiments). B, Hematoxylin and eosin staining from histological sections of wt and miR-146a−/− synovial fibroblasts cultured in micromasses (bars 500 μm, magnification x 5). C, Quantification of Ki-67+ synovial fibroblast cells among total synovial lining cells, from immunohistochemically stained sections of micromasses from wt (n = 7) and miR-146a−/− (n = 7) mice using anti Ki-67 antibody. D, Expression level of TRAF6 in synovial fibroblasts from wt (n = 9) and miR-146a−/− (n = 10) animals was analyzed using quantitative real time PCR. E and F, Synovial fibroblasts from wt or miR-146a deficient animals were transfected with either control (Ctrl) or TRAF6 siRNA, proliferation analysis was performed using 3[H]thymidine incorporation (data shown are mean values of 2 independent experiments). *p < 0,05, **p < 0,01, ***p < 0,001 Results are shown as mean ± SEM.
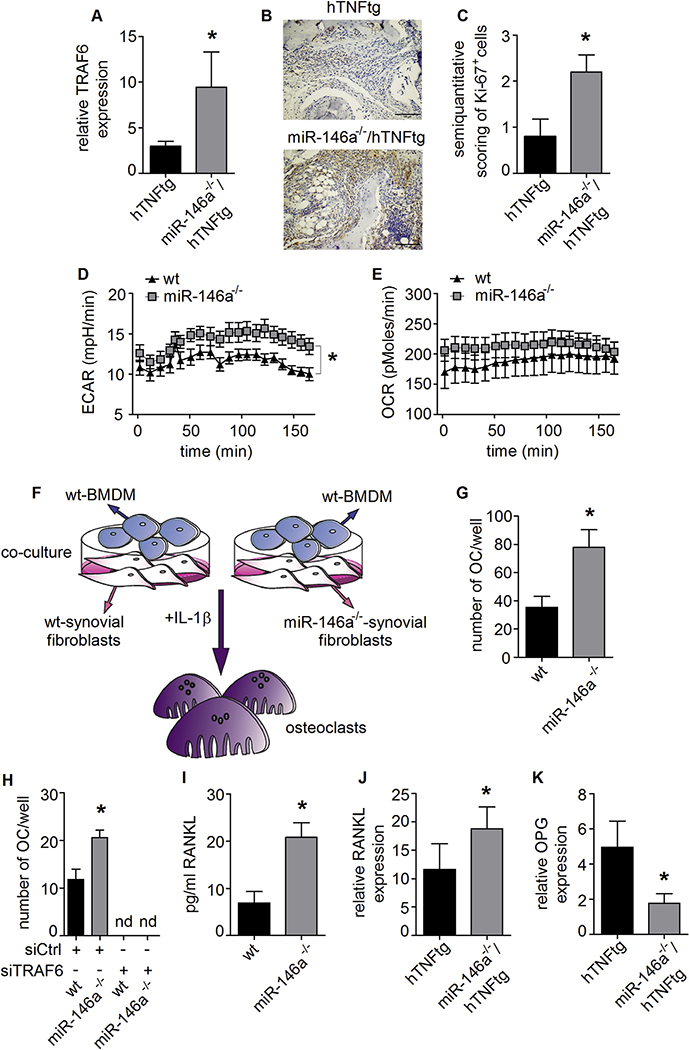
We also detected an increased expression of TRAF6 in the paws of miR-146a−/−/hTNFtg animals compared to hTNFtg animals in arthritis, demonstrating that TRAF6 was also upregulated in vivo. Again, other classical miR-146a regulated genes were not significantly altered (Fig. 5A and S5A and B). Furthermore, in line with our in vitro data, we could show significantly elevated amounts of Ki-67 expressed in the hind paws of miR-146a−/−/hTNFtg compared to hTNFtg mice (Fig. 5B and C).
Fig. 5. TRAF6 and consequential RANKL expression is controlled by miR-146a in synovial fibroblasts.
A, MRNA expression of TRAF6 in hind paws of hTNFtg (n = 10) and miR-146a−/−/hTNFtg (n = 6) animals, measured using Q-PCR. B, Synovial tissue of hTNFtg and miR-146a−/−/hTNFtg mice analyzed histologically for Ki-67 expression (bars 100 μm, magnification x 20). C, Ki-67 positive cells among total synovial cells in the inflamed tissue (n = 5). D and E, ECAR and OCR from wt and miR-146a deficient SF, analyzed using seahorse flux analyser (representative of 2 independent experiments). F, Schematic illustration of coculture assay from wt and miR-146a−/− SF and wt BMDM supplemented with IL-1 β. G, Number of osteoclasts from these cocultures was analyzed (representative of 3 independent experiments). H, Number of osteoclasts after transfection of wt or miR-146a−/− SF with control or TRAF6 siRNA following coculture with wt BMDM supplemented with IL-1 β (representative of 2 independent experiments). I, RANKL production of SF from wt and miR-146a deficient animals cultured with IL-1 β, analyzed by Elisa (mean values of 4 independent experiments). J and K, Expression level of RANKL and OPG mRNA in hind paws of hTNFtg (n = 4) and miR-146a−/−/hTNFtg (n = 6) animals measured using Q-PCR. *p < 0,05 Results are shown as mean ± SEM.
Recently, the analysis of the metabolic activity of immune cells has been of great interest, as it has become evident that changes in the metabolic activity of cells are often directly linked to their functional properties [39]. Increased levels of proliferation are associated with changes in metabolic activity of cells. Therefore, we examined the metabolic activity of synovial fibroblasts. The glycolytic activity (extracellular acidification rate; ECAR) was significantly augmented in synovial fibroblasts derived from miR-146a−/− compared to those derived from wt animals. There was a trend for an increased mitochondrial respiration (oxygen consumption rate; OCR), but it did not reach statistical significance (Fig. 5D and E). Therefore, these data identify miR-146a as a negative regulator of the metabolic activity of synovial fibroblasts.
3.5. MiR-146a controls the osteoclastogenic potential of synovial fibroblasts via TRAF6
Synovial fibroblasts have been shown to be essential for the induction of osteoclasts in inflammatory arthritis [40]. To test the ability of synovial fibroblasts to support the generation of osteoclasts from bone marrow precursors we set up a coculture system. Synovial fibroblasts of either wt or miR-146a deficient animals were cocultured with wt bone marrow cells (Fig. 5F). In this assay, we found that those fibroblasts lacking miR-146a were significantly superior to wt fibroblasts to support osteoclast differentiation (Fig. 5G). To analyze whether TRAF6 was involved also in this function of synovial fibroblasts, we knocked down TRAF6 in synovial fibroblasts prior to coculture. Upon knock down of TRAF6, the capacity of both wt and miR-146a deficient synovial fibroblasts to support the generation of osteoclasts was greatly reduced, suggesting that TRAF6-mediated signal transduction is essential for synovial fibroblast induced osteoclast generation (Fig. 5H). We next analyzed the production of RANKL, an important factor for osteoclastogenesis. In line with the increased formation of osteoclasts, we detected elevated amounts of RANKL in miR-146a deficient synovial fibroblasts (Fig. 5I). In addition, when we measured RANKL levels by qPCR in arthritic paws in vivo, we detected increased amounts of RANKL mRNA accompanied by reduced expression of OPG, the decoy receptor for RANKL, in miR-146a−/−/hTNFtg compared to hTNFtg mice (Fig. 5J and K). These data suggest, that loss of miR-146a causes deregulation of TRAF6, leading to fibroblasts with increased capacity to support the generation of bone resorbing osteoclasts via increased production of RANKL.
3.6. Mesenchymal miR-146a deficiency is responsible for increased local bone destruction
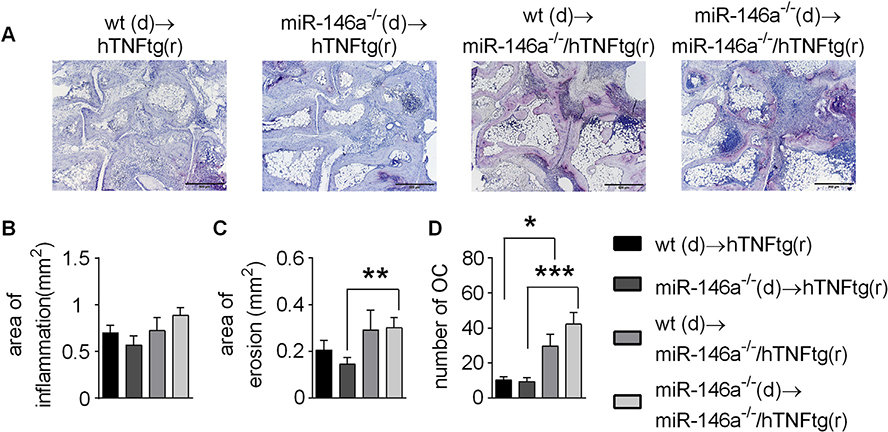
To dissect in vivo whether the radiosensitive hematopoietic compartment including monocytes, macrophages and osteoclasts or the radio resistant mesenchymal compartment (mainly synovial fibroblasts) are responsible for increased local bone destruction in miR-146a−/−/hTNFtg mice, we performed bone marrow transplants. We could not find significant differences in irradiated hTNFtg mice reconstituted with either wt or miR-146a−/− bone marrow (Fig. 6A–D). However, when we reconstituted miR-146a−/−/hTNFtg hosts with wt and miR-146a−/− bone marrow, we observed significantly increased erosions as well as local generation of osteoclasts compared to irradiated hTNFtg mice (Fig. 6A–D). These data demonstrate, that in chronic arthritis, miR-146a controls local joint destruction via controlling the pathogenicity of the radio resistant compartment, whose major component are synovial fibroblasts.
Fig. 6. Chimeric mice reveal an important regulatory function of miR-146a in mesenchymal cells, by controlling osteoclast development and bone destruction in arthritis.
A-D, Human TNFtg or miR-146a−/−/hTNFtg recipient mice were irradiated and challenged with either wt or miR-146a−/− bone marrow cells. A, TRAP stained sections of the hind paws of recipient mice obtained at 10 weeks of age (bars 500 μm, magnification x 5). B-D, Histomorphometric analysis of the extent of B, Inflammation C, Bone erosion and D, Number of Osteoclasts in the tarsal area of the hind paws of recipient mice (n = 5). *p < 0,05, **p < 0,01, ***p < 0,001 Results are shown as mean ± SEM.
4. Discussion
In recent years it has become evident that epigenetic regulation controls many aspects of health and disease, and especially microRNAs have been suggested as regulators of many pathological processes, including inflammatory joint diseases [41,42]. MiR-146a in particular was found to be strongly upregulated in synovial tissue of RA compared to OA patients as well as in RA synovial fibroblasts treated with proinflammatory stimuli especially with IL- 1β [28,29]. In the present study, we were able to show that miR-146a is an important anti-inflammatory microRNA that potently restricts the pathogenicity of synovial fibroblasts at least in part by controlling the expression of TRAF6. In our hTNFtg model of chronic inflammatory arthritis, lacking of miR-146a led to a significant increase in joint pathology, especially in erosive joint damage. Previous studies have focused primarily on the role of miR-146a in hematopoietic cells, and in these cells miR-146a has been shown to be upregulated upon stimulation with proinflammatory stimuli, especially in Ly6Chigh monocytes [21,23]. In another study, addition of exogenous agonistic miR-146a was found to regulate osteoclastogenesis, possibly by targeting TRAF6 in bone marrow cells [31]. However, in our experiments, using wt and miR-146a deficient cells, we saw only a slightly increased potential of miR-146a deficient bone marrow cells to differentiate into OCs. In contrast, we found several lines of evidence that, at least in our model, miR-146a most importantly controls synovial fibroblast behavior in arthritis. We find that miR-146a restricts the proliferation of cultured synovial fibroblasts, in both a conventional 2D as well as in a 3D culture system. Moreover we demonstrate that miR-146a inhibits the glycolytic activity of synovial fibroblasts, as evidenced by an increased extracellular acidification rate in miR-146a−/− cells. We could also detect increased numbers of proliferating cells in paws of arthritic miR-146a−/− mice.
Importantly, we show that miR-146a diminishes the ability of synovial fibroblasts to induce osteoclastogenesis of myeloid osteoclast precursors in vitro. We could confirm this increased ability of miR-146a−/− mesenchymal cells to induce osteoclasts in vivo, as we could clearly demonstrate in bone marrow transfer experiments that the radio resistant mesenchymal compartment is responsible for the increased erosive bone damage.
Of note, in contrast to the marked increase in local bone destruction, we did not detect differences in systemic bone loss between hTNFtg mice deficient or sufficient for miR-146a. In addition, the osteoclastogenic potential of bone marrow cells in vitro was not different between wt and miR-146a deficient cells. We therefore suggest, that the increased joint destruction in arthritis in miR146a deficient mice is predominantly caused by the local inflammatory environment shaped primarily by synovial fibroblasts.
Moreover, in our model miR-146a was not upregulated in tissues such as the bone marrow or the spleen, in contrast to the marked upregulation of miR-146a in the joint, also suggesting an important local role of this microRNA in arthritis.
Synovial fibroblasts have recently been demonstrated to be the essential source of RANKL for osteoclast generation in arthritis [40]. In line, we detected increased amounts of RANKL in cultured synovial fibroblasts as well as overexpression of RANKL accompanied by decreased amounts of OPG, the decoy receptor for RANKL, in the joints of miR-146a−/−/hTNFtg mice.
Mechanistically, and maybe a bit unexpectedly, we found TRAF6 to be involved in many aspects of synovial fibroblast biology. First of all, TRAF6 was upregulated locally in the arthritic paws as well as in cultured synovial fibroblasts of miR-146a deficient mice. TRAF6 is well recognized to be essential for osteoclastogenesis in myeloid cells, but has also been shown to regulate proinflammatory signaling in fibroblasts, as well as the proliferation of lung fibroblasts [38,41–43]. In our experiments, we found that TRAF6 positively regulates the proliferation of synovial fibroblasts, both in wt and miR-146a deficient cells. We also could demonstrate a role for TRAF6 in fibroblast-derived osteoclastogenic differentiation signals. The work of Wei et al. shows that IL-1 induces RANKL expression in stromal cells, demonstrating their key role in osteoclastogenesis under inflammatory conditions. Furthermore it is known that TRAF6 participates in IL-1 receptor signaling [44][46]. These data, in conjunction with those obtained in our study, identify TRAF6 as a potentially important molecule in the biology of synovial fibroblasts and suggest that miR-146a may regulate the pathological behavior of synovial fibroblasts by controlling the expression of TRAF6. However, the use of only the hTNFtg arthritis disease model may be a limitation of this study and therefore these findings might not be extrapolated to other disease models. Previous studies have shown that miR-146a expression was increased not only in synovial tissue, but also in PBMCs of patients with RA compared to healthy controls. However, although expression of miR-146a was increased in PBMCs of RA patients, expression of TRAF6 was not reduced [27–29].
It will be interesting to test the hypothesis proposed in our study using agonistic miR-146a to show its effects on FLS proliferations and RANKL expression.
In summary, we propose that enhanced proliferative and metabolic activity, in combination with increased production of RANKL and decreased OPG are the drivers of the increased bone destruction in hTNF-mediated arthritis in miR-146a deficient animals (See S6 for a graphical summary). The fact that increased expression of miR-146a has been demonstrated in RA is reminiscent of the observation of increased levels of the anti-inflammatory cytokine IL-10 in the synovia of arthritis patients [47, 48].
We therefore propose that miR-146a constitutes an important part of a local negative feedback loop preventing excessive inflammation, especially inflammatory tissue damage and joint destruction by restricting fibroblast pathogenicity in chronic inflammatory arthritis.
Supplementary Material
Acknowledgements
The authors thank Roman Ferdinand Stemberger, Tetyana Shvets, Karolina von Dalwigk and Carl-Walter Steiner for expert technical assistance. We thank Dr. George Kollias for providing the hTNFtg mice.
This research has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n°1151412 (BTCure), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme and EFPIA companies’ in kind contribution, Campus France, PHC Amadeus 33696WM and by a grant from the Austrian Science Fund (FWF), number P23730.
Abbrevations
- OPG
osteoprotegerin
- RANKL
receptor activator of NF-κB ligand
- miR
micro RNA
- FLS
fibroblast like synoviocytes
- PsA
psoriatic arthritis
- hTNFtg
human tumor necrosis factor transgene
- TRAF6
TNF receptor associated factor 6
- IRAK-1
interleukin-1 receptor-associated kinase 1
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jaut.2017.05.006.
References
- [1].Bluml S, Redlich K, Smolen JS, Mechanisms of tissue damage in arthritis, Semin. Immunopathol 36 (5) (2014. September) 531–540. [DOI] [PubMed] [Google Scholar]
- [2].McInnes IB, Schett G, Cytokines in the pathogenesis of rheumatoid arthritis, Nat. Rev. Immunol. 7 (6) (2007. June) 429–442. [DOI] [PubMed] [Google Scholar]
- [3].Scott DL, Smith C, Kingsley G, Joint damage and disability in rheumatoid arthritis: an updated systematic review, Clin. Exp. Rheumatol. 21 (5 Suppl 31) (2003. Sep-Oct) S20–S27. [PubMed] [Google Scholar]
- [4].Tak PP, Breedveld FC, Current perspectives on synovitis, Arthritis Res 1 (1) (1999) 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].McInnes IB, Schett G, The pathogenesis of rheumatoid arthritis, N. Engl. J. Med. 365 (23) (2011. December 8) 2205–2219. [DOI] [PubMed] [Google Scholar]
- [6].Redlich K, Smolen JS, Inflammatory bone loss: pathogenesis and therapeutic intervention, Nat. Rev. Drug Discov. 11 (3) (2012. March) 234–250. [DOI] [PubMed] [Google Scholar]
- [7].Scheinecker C, Redlich K, Smolen JS, Cytokines as therapeutic targets: advances and limitations, Immunity 28 (4) (2008. April) 440–444. [DOI] [PubMed] [Google Scholar]
- [8].Redlich K, Schett G, Steiner G, Hayer S, Wagner EF, Smolen JS, Rheumatoid arthritis therapy after tumor necrosis factor and interleukin-1 blockade, Arthritis Rheum. 48 (12) (2003. December) 3308–3319. [DOI] [PubMed] [Google Scholar]
- [9].Bartel DP, Chen CZ, Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs, Nat. Rev. Genet. 5 (5) (2004. May) 396–400. [DOI] [PubMed] [Google Scholar]
- [10].Lodish HF, Zhou B, Liu G, Chen CZ, Micromanagement of the immune system by microRNAs, Nat. Rev. Immunol. 8 (2) (2008. February) 120–130. [DOI] [PubMed] [Google Scholar]
- [11].Carthew RW, Sontheimer EJ, Origins and Mechanisms of miRNAs and siRNAs, Cell. 136 (4) (2009. February 20) 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang Y, Blelloch R, Cell cycle regulation by MicroRNAs in embryonic stem cells, Cancer Res. 69 (10) (2009. May 15) 4093–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Winter J, Jung S, Keller S, Gregory RI, Diederichs S, Many roads to maturity: microRNA biogenesis pathways and their regulation, Nat. Cell Biol. 11 (3) (2009. March) 228–234. [DOI] [PubMed] [Google Scholar]
- [14].Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. , Requirement of bic/microRNA-155 for normal immune function, Science 316 (5824) (2007. April 27) 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. , Regulation of the germinal center response by microRNA-155, Science 316 (5824) (2007. April 27) 604–608. [DOI] [PubMed] [Google Scholar]
- [16].Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. , miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice, J. Exp. Med. 208 (6) (2011. June 6) 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. , MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development, Immunity 33 (4) (2010. October 29) 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bluml S, Bonelli M, Niederreiter B, Puchner A, Mayr G, Hayer S, et al. , Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice, Arthritis Rheum. 63 (5) (2011. May) 1281–1288. [DOI] [PubMed] [Google Scholar]
- [19].Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, et al. , MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis, Proc. Natl. Acad. Sci. U. S. A. 108 (27) (2011. July 5) 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA, miRNAs and related polymorphisms in rheumatoid arthritis susceptibility, Autoimmun. Rev 11 (9) (2012. July) 636–641. [DOI] [PubMed] [Google Scholar]
- [21].Taganov KD, Boldin MP, Chang KJ, Baltimore D, NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses, Proc. Natl. Acad. Sci. U. S. A. 103 (33) (2006. August 15) 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. , Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses, Cell. 142 (6) (2010. September 17) 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Etzrodt M, Cortez-Retamozo V, Newton A, Zhao J, Ng A, Wildgruber M, et al. , Regulation of monocyte functional heterogeneity by miR-146a and Relb, Cell Rep. 1 (4) (2012. April 19) 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, et al. , miR-146a controls the resolution of T cell responses in mice, J. Exp. Med. 209 (9) (2012. August 27) 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhao JL, Rao DS, O’Connell RM, Garcia-Flores Y, Baltimore D, MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice, eLife 2 (2013) e00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Abou-Zeid A, Saad M, Soliman E, MicroRNA 146a expression in rheumatoid arthritis: association with tumor necrosis factor-alpha and disease activity, Genet. Test. Mol. Biomark. 15 (11) (2011. November) 807–812. [DOI] [PubMed] [Google Scholar]
- [27].Filkova M, Aradi B, Senolt L, Ospelt C, Vettori S, Mann H, et al. , Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis, Ann. Rheum. Dis. 73 (10) (2014. October) 1898–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. , Expression of microRNA-146 in rheumatoid arthritis synovial tissue, Arthritis Rheum. 58 (5) (2008. May) 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK, Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients, Arthritis Res. Ther 10 (4) (2008) R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. , Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis, Arthritis Rheum. 58 (4) (2008. April) 1001–1009. [DOI] [PubMed] [Google Scholar]
- [31].Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M, The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis, Arthritis Rheum. 63 (6) (2011. June) 1582–1590. [DOI] [PubMed] [Google Scholar]
- [32].Lochhead RB, Ma Y, Zachary JF, Baltimore D, Zhao JL, Weis JH, et al. , MicroRNA-146a provides feedback regulation of lyme arthritis but not carditis during infection with Borrelia burgdorferi, PLoS Pathog. 10 (6) (2014. June) e1004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Keffer J, Probert L, Cazlaris H, Georgopoulos S, Kaslaris E, Kioussis D, et al. , Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis, EMBO J. 10 (13) (1991. December) 4025–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bluml S, Binder NB, Niederreiter B, Polzer K, Hayer S, Tauber S, et al. , Antiinflammatory effects of tumor necrosis factor on hematopoietic cells in a murine model of erosive arthritis, Arthritis Rheum. 62 (6) (2010. June) 1608–1619. [DOI] [PubMed] [Google Scholar]
- [35].Bluml S, Friedrich M, Lohmeyer T, Sahin E, Saferding V, Brunner J, et al. , Loss of phosphatase and tensin homolog (PTEN) in myeloid cells controls inflammatory bone destruction by regulating the osteoclastogenic potential of myeloid cells, Ann. Rheum. Dis. 74 (1) (2015. January) 227–233. [DOI] [PubMed] [Google Scholar]
- [36].Rosner M, Siegel N, Fuchs C, Slabina N, Dolznig H, Hengstschlager M, Efficient siRNA-mediated prolonged gene silencing in human amniotic fluid stem cells, Nat. Protoc 5 (6) (2010. June) 1081–1095. [DOI] [PubMed] [Google Scholar]
- [37].Kiener HP, Watts GF, Cui Y, Wright J, Thornhill TS, Skold M, et al. , Synovial fibroblasts self-direct multicellular lining architecture and synthetic function in three-dimensional organ culture, Arthritis Rheum. 62 (3) (2010. March) 742–752. [DOI] [PubMed] [Google Scholar]
- [38].Starczynowski DT, Lockwood WW, Delehouzee S, Chari R, Wegrzyn J, Fuller M, et al. , TRAF6 is an amplified oncogene bridging the RAS and NFkappaB pathways in human lung cancer, J. Clin. Investig 121 (10) (2011. October) 4095–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Assmann N, Finlay DK, Metabolic regulation of immune responses: therapeutic opportunities, J. Clin. Investig 126 (6) (2016. June 1) 2031–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Danks L, Komatsu N, Guerrini MM, Sawa S, Armaka M, Kollias G, et al. , RANKL expressed on synovial fibroblasts is primarily responsible for bone erosions during joint inflammation, Ann. Rheum. Dis. 75 (6) (2016. June) 1187–1195. [DOI] [PubMed] [Google Scholar]
- [41].Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y, TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL, PLoS One 3 (12) (2008) e4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yamada T, Fujieda S, Yanagi S, Yamamura H, Inatome R, Yamamoto H, et al. , IL-1 induced chemokine production through the association of Syk with TNF receptor-associated factor-6 in nasal fibroblast lines, J. Immunol. 167 (1) (2001. July 1) 283–288. [DOI] [PubMed] [Google Scholar]
- [43].Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, et al. , TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling, Genes & Dev. 13 (8) (1999. April 15) 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wei S, Kitaura H, Zhou P, Ross FP, Teitelbaum SL , IL-1 mediates TNF-induced osteoclastogenesis, J. Clin. Investig 115 (2) (2005. February) 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.