Abstract
γδ T cells are major providers of proinflammatory cytokines. They are preprogrammed in the mouse thymus into distinct subsets producing either interleukin-17 (IL-17) or interferon-γ (IFN-γ), which segregate with CD27 expression. In the periphery, CD27− γδ (γδ27−) T cells can be induced under inflammatory conditions to coexpress IL-17 and IFN-γ; the molecular basis of this functional plasticity remains to be determined. On the basis of differential microRNA (miRNA) expression analysis and modulation in γδ T cell subsets, we identified miR-146a as a thymically imprinted post-transcriptional brake to limit IFN-γ expression in γδ27− T cells in vitro and in vivo. On the basis of biochemical purification of Argonaute 2–bound miR-146a targets, we identified Nod1 to be a relevant mRNA target that regulates γδ T cell plasticity. In line with this, Nod1-deficient mice lacked multifunctional IL-17+ IFN-γ+ γδ27− cells and were more susceptible to Listeria monocytogenes infection. Our studies establish the miR-146a/NOD1 axis as a key determinant of γδ T cell effector functions and plasticity.
INTRODUCTION
The proinflammatory cytokines interferon-γ (IFN-γ) and interleukin-17A (IL-17) are critical mediators of T cell responses against intracellular bacteria and viruses, or extracellular bacteria and fungi, as illustrated by the high susceptibility of patients with mutations in the IFN-γ or IL-17 pathways to Mycobacterium tuberculosis or Candida albicans infections, respectively (1). In addition to their protective roles in host defense, it has been shown that deregulated IFN-γ or/ and IL-17 production promotes chronic inflammation and autoimmunity. Thus, exacerbated IL-17 responses underlie inflammatory bowel disease, psoriasis, ankylosing spondylitis, and multiple sclerosis (1); and excessive IFN-γ is linked to type 1 diabetes and Crohn’s disease (2).
Although IFN-γ and IL-17 can derive from multiple cellular sources, there is often a major contribution from γδ T cells at early stages of immune responses (3). Namely, IFN-γ–producing γδ T cells play protective roles in viral, parasitic, and intracellular bacterial infections, such as Listeria monocytogenes (4, 5). On the other hand, IL-17–producing γδ T cells promote neutrophil mobilization and host defense against Staphylococcus aureus, Escherichia coli, and C. albicans, among other infections (3). IL-17–producing γδ T cells have gathered much recent attention because of their impact on animal models of inflammatory and autoimmune disorders such as psoriasis, colitis, arthritis, or multiple sclerosis (3).
The prompt and abundant production of IFN-γ and IL-17 by γδ T cells is associated with their “developmental preprogramming” in the thymus (3, 6–9). Thus, we and others have shown that mouse γδ thymocytes can acquire the capacity to produce IFN-γ or IL-17, which associates with the expression of CD27 and CCR6, respectively (7, 10). Our study on chromatin-based and transcriptional mechanisms governing γδ T cell differentiation revealed that CD27+ γδ (γδ27+) T cells are committed to the expression of IFN-γ (but not of IL-17), whereas CD27− γδ (γδ27−) T cells exhibit a plastic behavior and can differentiate into cells that produce both IL-17 and IFN-γ (11). Ifnγ and its transcriptional regulator Tbx21 displayed abundant active histone H3 marks (methylation of lysine K4 and acetylation) and were transcribed in both γδ T cell subsets. The coexpression of IFN-γ and IL-17 proteins by γδ27− T cells was observed under strong inflammatory conditions seemingly driven by IL-1β and IL-23. The double- producing IL-17+ IFN-γ+ γδ27− T cells have been documented in animal models of cancer (11), autoimmunity (12), and infection (5) and also in HIV-1–infected patients (13).
However, γδ27− T cell plasticity seems limited when compared with that of CD4+ T helper 17 (TH17) cells, which rapidly acquire the capacity to make IFN-γ when exposed to IL-12 or IL-23 in vitro (14–16) and have been found in colitis, Crohn’s disease, arthritis, diabetes, multiple sclerosis, and experimental autoimmune encephalomyelitis (EAE) (14, 15). Furthermore, analysis of an IL-17 fate-mapping reporter mouse showed that, whereas CD4+ T cells that had expressed IL-17 promptly converted into IFN-γ producers in the spinal cord during EAE development, IL-17+ γδ T cells were much more resistant to the acquisition of IFN-γ production (14). Because the Ifnγ locus displays active histone H3 marks and active transcription in γδ27− T cells (11), we hypothesized that microRNAs (miRNAs) might control IFN-γ expression at the post-transcriptional level in γδ27− T cells.
miRNAs drive their mRNA targets into the RNA-inducing silencing complex (RISC), in which Argonaute (AGO) proteins play a central role in preventing protein expression. Several miRNAs have been shown to affect the differentiation of cytokine-producing CD4+ T cell subsets [reviewed in 16)]. To date, only two miRNAs have been characterized in γδ T cells, miR-133b and miR-206, which are co-regulated with IL-17 but have no impact on cytokine production (17). Here, we have identified miR-146a as highly expressed in γδ27− T cells compared with their γδ27+ T cell counterparts, and through combined gain-of-function and loss-of-function approaches, we demonstrate that miR-146a inhibits IFN-γ production in γδ T cells and thus restricts the functional plasticity of γδ27− T cells in vitro and in vivo.
RESULTS
miR-146a is highly expressed in γδ27− T cells
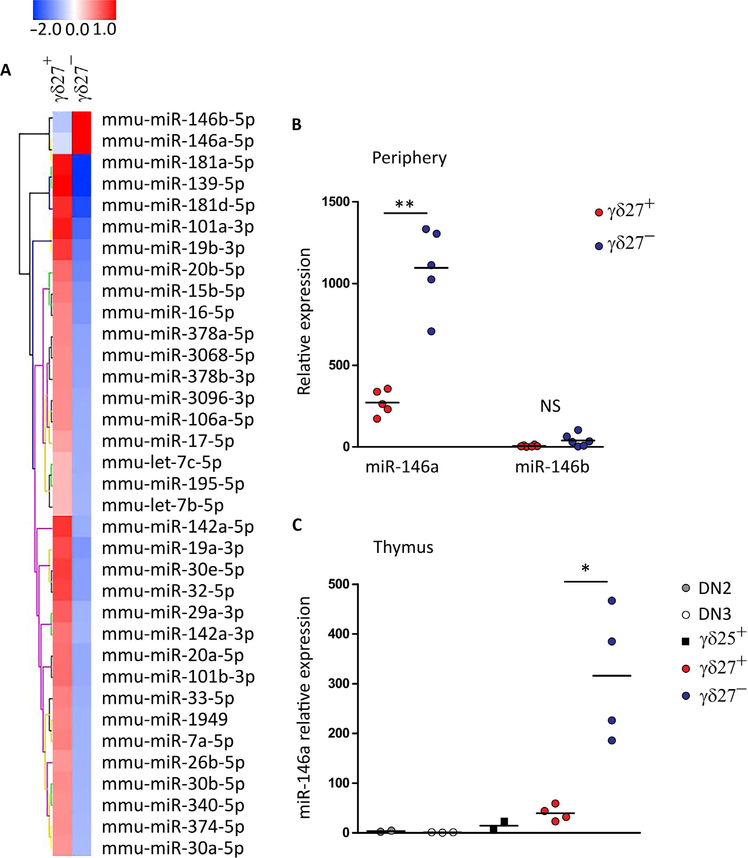
To address the role of miRNAs in the differentiation of proinflammatory γδ T cell subsets, we compared the ex vivo miRNA expression profile of highly purified CD27+ (γδ27+) and CD27− CCR6+ (γδ27−) γδ T cells isolated from pooled lymph nodes and spleen of naïve C57BL/6J mice. We identified 35 miRNAs differentially expressed (more than twofold) between γδ27+ and γδ27−CCR6+ cells (Fig. 1A). Only two related miRNAs, miR-146a and miR-146b, were expressed at higher levels in γδ27−CCR6+ T cells when compared with γδ27+ cells. The differential expression of miR-146a (about fourfold) was fully validated by quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis in peripheral γδ27+ and γδ27− T cell samples (Fig. 1B). In contrast to miR-146a, qPCR for miR-146b revealed very low levels of expression in all T cell subsets analyzed, including γδ27− T cells (Fig. 1B and fig. S1A).
Fig. 1. miR-146a is highly expressed selectively on γδ27− T cells.
(A) Microarray heat map of differentially expressed miRNAs in duplicate samples of γδ27+ (n = 4 mice per sample) and γδ27−CCR6+ T cells (n = 8 mice per sample) isolated from pooled lymph nodes and spleen of C57BL/6 mice (more than twofold enrichment). (B) RT-qPCR analysis of miR-146a and miR-146b expression in sorted γδ27+ and γδ27− T cells from pooled peripheral organs (lymph node and spleen) of C57BL/6 mice. NS, not significant. (C) RT-qPCR analysis of miR-146a expression in sorted DN2 (CD4−CD8−CD44+CD25+), DN3 (CD4−CD8−CD44−CD25+), γδ25+ (CD25+CD27+), γδ27+, and γδ27− thymocytes of C57BL/6 mice. Results are presented relative to miR-423–3p or RNU5G (reference small RNA) expression. Each symbol in (B) and (C) represents an individual mouse. *P < 0.05 and **P < 0.01 (Mann-Whitney two-tailed test).
The analysis of thymocyte subsets allowed us to establish that miR-146a is already differentially expressed during γδ T cell development, with a sharp increase as precursors mature into γδ27− (but not γδ27+) thymocytes (Fig. 1C). Furthermore, miR-146a expression was not modulated by exogenous T cell receptor (TCR) stimulation or inflammatory cytokines (fig. S1B). These data demonstrate that miR-146a expression is tightly regulated during γδ T cell differentiation, being confined to the subset that is preprogrammed in the thymus to produce IL-17 but no IFN-γ (7–9).
miR-146a inhibits IFN-γ production in γδ T cells
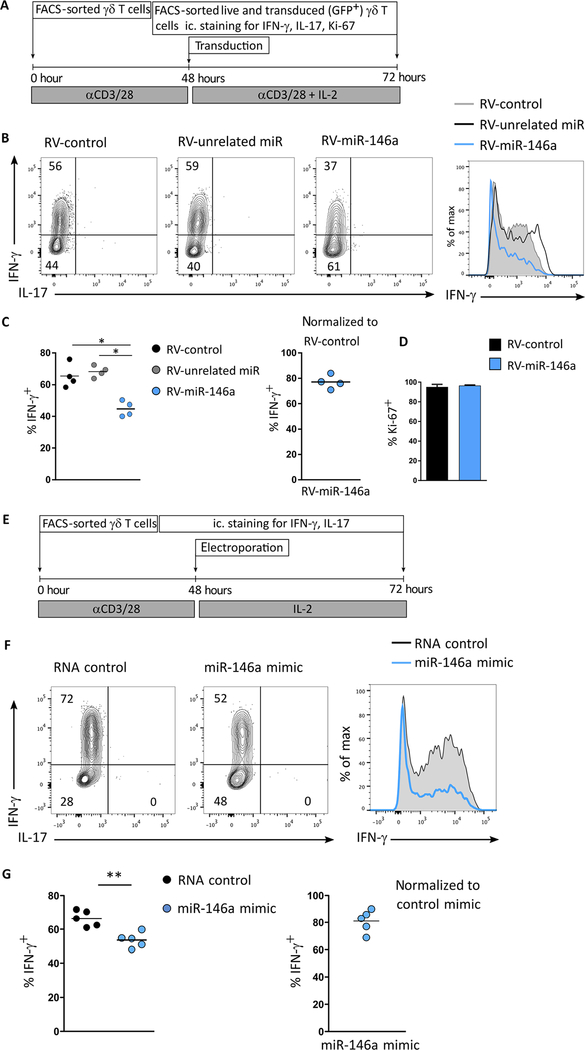
To gain insight into the function of miR-146a in effector γδ T cell differentiation, we performed gain-of-function studies either by retroviral overexpression of a miR-146a construct or by electroporation of miR-146a mimics in γδ T cells. Both strategies require T cell activation and therefore act on fully differentiated γδ T cells obtained from spleen and lymph nodes. Moreover, TCR/IL-2 stimulation is known to enhance IFN-γ production in γδ T cells (18). Retroviral transduction of the native stem loop of miR-146a in γδ T cells (Fig. 2A) led to marked overexpression of mature miR-146a (fig. S1C), and this resulted in a significant reduction of the frequency of IFN-γ+ γδ T cells compared with either an unrelated miR or a control vector alone (Fig. 2, B and C), without affecting γδ T cell proliferation, as evaluated by Ki-67 staining (Fig. 2D). These data were validated by an alternative gain-of-function approach based on the electroporation of synthetic miR-146a oligonucleotides (miR-146a mimics) in γδ T cells (Fig. 2E), which significantly decreased the frequency of IFN-γ+ γδ T cells when compared with control oligonucleotides (Fig. 2, F and G). Overexpression of miR-146a did not affect TH1 differentiation of CD4+ T cells (fig. S1, D and E). These in vitro data suggest that miR-146a inhibits IFN-γ production in γδ T cells, leading us to hypothesize that the high miR-146a levels in γδ27− T cells in vivo (Fig. 1, B and C) restrict their capacity to express IFN-γ.
Fig. 2. miR-146a inhibits IFN-γ production in γδ T cells.
Workflow and results of (A to D) retroviral (RV) overexpression of miR-146a and (E to G) electroporation of miR-146a mimics in sorted peripheral γδ T cells. (B) Flow cytometry analysis of intracellular IFN-γ and IL-17 expression; and frequency of (C) IFN-γ+ and (D) Ki-67+ cells in sorted GFP+ retrovirally transduced γδ T cells expressing either an unrelated miR (RV-unrelated miR), a control vector (RV-control), or miR-146a (RV-146a). (F) Flow cytometry analysis of intracellular IFN-γ and IL-17 expression and (G) frequency of IFN-γ+ in electroporated γδ T cells with either RNA control or miR-146a mimic. Numbers in quadrants of flow cytometry plots indicate percentages of cells. Data are representative of five independent experiments. *P < 0.05 and **P < 0.01 (Mann-Whitney two-tailed test).
miR-146a limits IFN-γ production and functional plasticity of γδ27− T cells
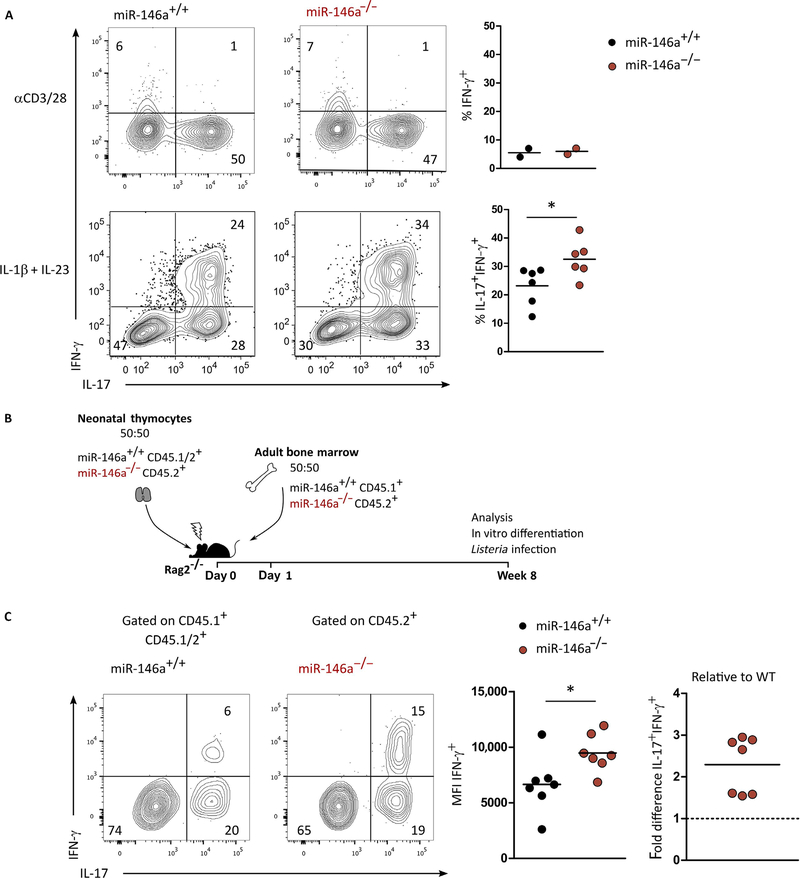
To explore the role of miR-146a in γδ T cell differentiation, we studied γδ T cells in miR-146a–deficient mice (19). In vitro, γδ T cell proliferation upon TCR/CD28 stimulation was not affected by the absence of miR-146a (fig. S2, A and B). The numbers of steady-state γδ27+ and γδ27− T cells, as well as their production of IFN-γ or IL-17, respectively, were similar between miR-146a–deficient and wild-type (WT) littermate control mice (fig. S2, C and D). We assessed the functional plasticity of γδ27− T cells, namely, their capacity to differentiate into IL-17+ IFN-γ+ cells upon IL-1β + IL-23 stimulation (fig. S3A). We observed a significantly increased frequency of double producers among miR-146a–deficient (miR-146a−/−) γδ27− T cells when compared with WT (miR-146a+/+) controls (Fig. 3A). As expected (11), TCR/CD28 activation (without IL-1β or IL-23) failed to generate double producers (Fig. 3A). Neither TH1 differentiation of CD4+ T cells (fig. S3, B and C) nor IFN-γ production by γδ27+ T cells (fig. S3, D and E) was affected by the loss of miR-146a. This is consistent with the very low abundance of miR-146a in γδ27+ T cells and thus ascribes a miR-146a function that is specific to the high expressing γδ27− T cell subset.
Fig. 3. miR-146a limits IFN-γ production and the functional plasticity of γδ27− T cells.
(A) Flow cytometry analysis of intracellular IFN-γ and IL-17 and frequency of IFN-γ+ and IL-17+ IFN-γ+ in γδ27− T cells isolated from peripheral organs and stimulated in vitro overnight with αCD3/28 or for 72 hours in the presence of IL-1β plus IL-23. (B) Schematic for 1:1 mixed miR-146a+/+ and miR-146a−/− adult BM and neonatal thymocyte chimeras in Rag2−/− hosts. (C) Flow cytometry analysis of intracellular IFN-γ and IL-17, mean fluorescence intensity (MFI) of IFN-γ+, and fold difference of IFN-γ+ IL-17+ in γδ27− T cells isolated from peripheral organs of above-introduced chimeras after in vitro stimulation for 72 hours with IL-1β plus IL-23. Numbers in quadrants of flow cytometry plots indicate percentages of cells. Each symbol in (A) and (C) represents an individual experiment. *P < 0.05 (Mann-Whitney two-tailed test).
To test whether miR-146a regulates IFN-γ production in γδ27− T cells in a cell-intrinsic manner, we generated (20) mixed (1:1) bone marrow (BM) and mixed (1:1) neonatal thymus chimeras by transferring of BM and neonatal thymocytes from miR-146a−/− mice (CD45.2+), together with BM (CD45.1+) and neonatal thymocytes (CD45.1/.2+) from WT mice into recombination activation gene 2 (RAG2)–deficient hosts (Fig. 3B and fig. S4A). The neonatal thymocytes are a prerequisite to generate IL-17–producing γδ T cells, because they are exclusively generated during fetal and perinatal life (21). This system allows the direct comparison of the progeny of miR-146a−/− versus miR-146a+/+ hematopoietic cells developing in the same host (miR-146a+/+ in the nonhematopoietic compartment). γδ27− T cells were isolated and activated with IL-1β plus IL-23 to drive the differentiation of IL-17+ IFN-γ+ double producers. We found that γδ27− T cells derived from miR-146a−/− were more prone to differentiate into double producers and expressed higher levels of IFN-γ than their WT counterparts (Fig. 3C). We also confirmed that the absence of miR-146a did not affect IFN-γ production by γδ27+ T cells (fig. S4B). Together, these results indicate that miR-146a acts as a cell-intrinsic brake to IFN-γ production specifically in γδ27− T cells, thereby limiting their functional plasticity.
miR-146a restricts functional plasticity of γδ27− T cells in Listeria infection
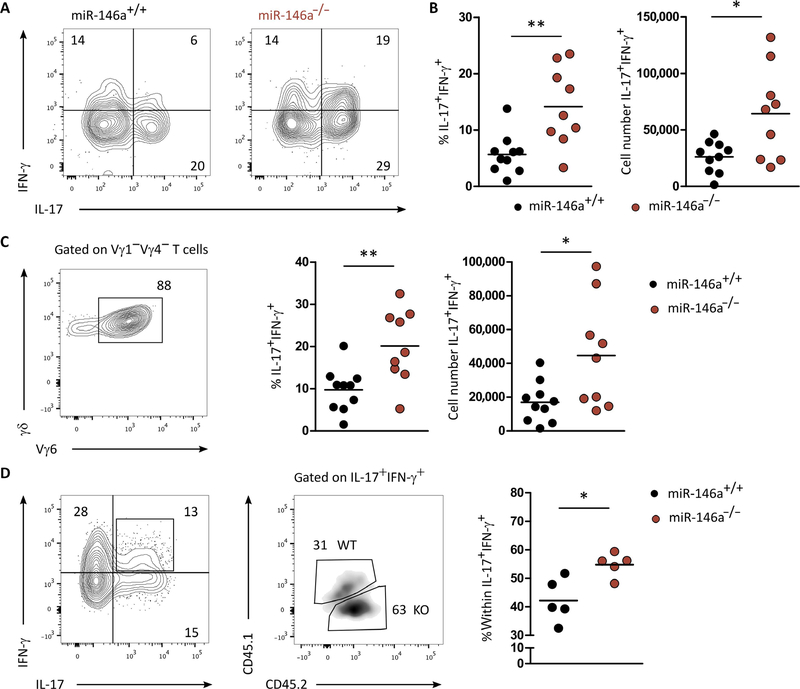
To validate miR-146a function in vivo, we analyzed miR-146a−/− mice upon oral infection with L. monocytogenes, where a sizeable population of IL-17+ IFN-γ+ γδ27− T cells had been previously reported (5). We infected miR-146a−/− and miR-146a+/+ (WT) littermate controls with L. monocytogenes and analyzed the γδ T cell response at day 7 post infection. γδ27− T cells increased their numbers by about five-fold in both WT and miR-146a−/− mice (fig. S5A). However, the frequency and absolute number of IL-17+ IFN-γ+ γδ27− T cells were significantly higher in miR-146a−/− when compared with WT mice (Fig. 4, A and B). This phenotype was largely accounted by Vγ6+ T cells (Fig. 4C), which are known to be the main γδ T cell population responding to L. monocytogenes infection (5). The frequency of single IFN-γ– or single IL-17–producing γδ T cells was the same in miR-146a−/− when compared to WT mice (fig. S5B). By using this infection model in the mixed BM and neonatal thymocyte chimeras introduced above (Fig. 3B), we found that the IL-17+ IFN-γ+ γδ27− T cells present in mesenteric lymph nodes were mostly from miR-146a−/− origin (Fig. 4D). As before, this phenotype was not because of a proliferative advantage of miR-146a−/− compared with WT-derived γδ T cells (fig. S5, C and D). These data demonstrate that γδ T cell plasticity under pathophysiological conditions in vivo is regulated by miR-146a.
Fig. 4. miR-146a restricts functional plasticity of γδ27− T cells in Listeria infection.
(A) Flow cytometry analysis of intracellular IFN-γ and IL-17 expression in γδ T cells isolated from spleens of miR-146a+/+ or miR-146a−/− mice, 7 days after infection with L. monocytogenes. (B) Frequency and total cell numbers of IL-17+ IFN-γ+ in γδ T cells isolated from spleens of miR-146a+/+ and miR-146a−/− mice, 7 days after infection with L. monocytogenes. (C) Representative flow cytometry analysis of Vγ6 chain usage (left), frequency of IFN-γ+ IL-17+ (middle), and total cell numbers of IFN-γ+ IL-17+ (right) of Vγ1Vγ4− γδ T cells isolated from spleen of miR-146a+/+ or miR-146a−/− mice, 7 days after infection with L. monocytogenes. (D) Representative flow cytometry analysis of IL-17+ IFN-γ+ γδ T cells isolated from the chimeras established in Fig. 3B. (Right) frequency within IFN-γ+ IL-17+ γδ T cells of either miR-146a+/+ or miR-146a−/− origin. Numbers in quadrants of flow cytometry plots indicate percentages of cells. Each symbol in (B) to (D) represents an individual mouse. *P < 0.05 and **P < 0.01 (Mann-Whitney two-tailed test). WT, wild-type; KO, knockout.
miR-146a targets Nod1 mRNA that is depleted in γδ27− T cells
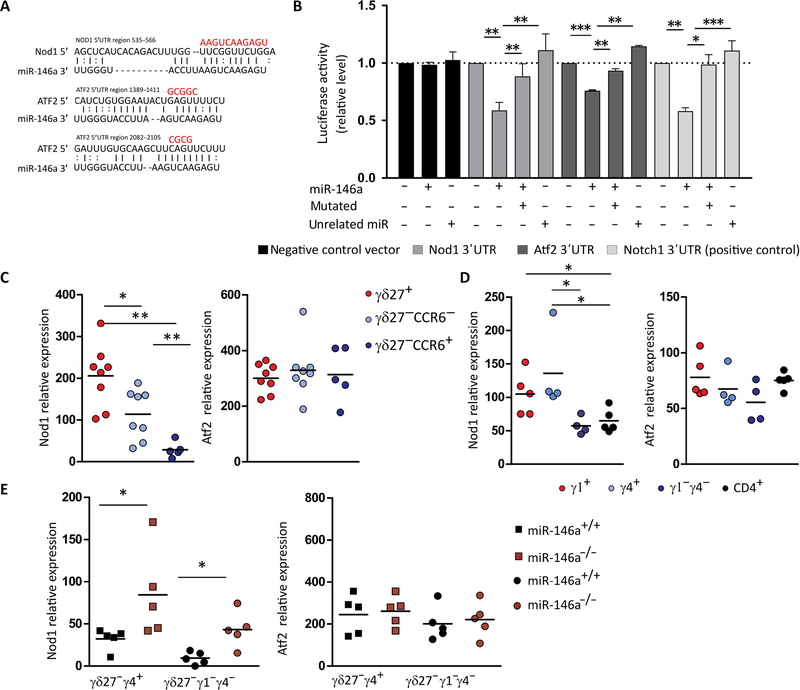
To unravel the molecular mechanism(s) underlying miR-146a function, namely, the relevant mRNA target(s), we undertook an unbiased approach based on differential Argonaute 2 (Ago2) RNA immunoprecipitation followed by deep sequencing (Ago2 RIP-seq). Ago2 is a crucial protein that engages with both miRNA and its mRNA target(s) in the RISC. To overcome the problem of lack of commercially available high affinity antibodies against Ago2, we took advantage of a genetically modified mouse in which endogenous Ago2 is replaced with a N-terminal–tagged version of the protein (3×FLAG-6×HIS), thus allowing stringent purification (i.e., with low unspecific background) of the RNA molecules bound to Ago2 [upon ultraviolet (UV) cross-linking)], followed by RNA sequencing (RNA-seq) (fig. S6, A and C). To enrich for miR-146a targets, we transduced purified CD3+ T cells (we could not use γδ T cells because of insufficient cell numbers for this approach) with a miR-146a–expressing retrovirus (RV) (~12-fold overexpression of miR-146a, as shown in fig. S1C). We thereby identified 225 putative mRNAs after differential Ago2 RIP-seq [data submitted to Gene Expression Omnibus (GEO)], from which 96 (42.7%) had a predicted miR-146a binding site in their 3′ untranslated region (3′UTR) (fig. S6B). This included two previously validated miR-146a targets, Zranb2 and Gtf2e2 [included in DIANA- TarBase v7.0 (22)]. Many of the other putative target transcripts had not been described in γδ T cells (table S1), and these data thus constitute a useful resource for future studies. For this study, to narrow down the potential mRNA targets of miR-146a involved in the regulation of IFN-γ production in γδ T cells, we focused on genes with transcription factor or receptor activity (Table 1) that had been associated with the IFN-γ pathway in other immune cells and, in particular, on two candidate genes, Atf2 (2.5-fold enriched in the RV-miR-146a sample), a transcription factor that binds to the IFN-γ promoter (23), and Nod1, an intracellular pattern recognition receptor recently shown to enhance IFN-γ production in αβ T cells (24, 25), which showed the highest fold difference (4.2-fold) among genes with receptor activity. Both putative targets have predicted miR-146a binding sites with canonical and noncanonical Watson-Crick base pairing (Fig. 5A). To examine direct miR-146a targeting of Atf2 and Nod1, we designed reporter constructs in a pmirGLO Dual-luciferase miRNA target expression vector for the 3′UTRs of Atf2 and Nod1. We additionally included a negative control vector containing a 3′UTR without miR-146a binding sites and a positive control containing the 3′UTR of Notch1, a previously validated miR-146a target (26, 27). We transiently transfected these constructs into human embryonic kidney (HEK) 293 T cells together with an expression plasmid for either miR-146a, an unrelated miRNA, or green fluorescent protein (GFP) control. Cotransfection of miR-146a (but not an unrelated miRNA) showed a significant repression of luciferase activity compared with GFP control for both mRNA targets (Fig. 5B). Mutations in the miR-146a binding sites of both Atf2 and Nod1 led to significant recoveries of luciferase levels (Fig. 5, A and B). We undertook various complementary mutational strategies to validate these findings, from mutating 3 to 11 individual base pairs (bp) to deleting the full binding region, always with consistent results (Fig. 5, A and B). Together, these data demonstrate that miR-146a can target Atf2 and Nod1.
Table 1.
Putative miR-146a targets based on differential Ago2-RNA immunoprecipitation followed by deep sequencing in T cells.
| Gene symbol | Fold change | No. of binding |
|---|---|---|
| Receptor activity | ||
| Nod1 | 4.21 | 1 |
| Tcp11 | 3.44 | 1 |
| Ghr | 3.07 | 1 |
| Paqr7 | 2.72 | 1 |
| Unc5a | 2.59 | 1 |
| Transcription factor activity | ||
| Ccnh | 6.48 | 1 |
| Tef | 4.49 | 2 |
| Myf6 | 3.80 | 1 |
| Zfp788 | 3.68 | 1 |
| Tfap2b | 3.67 | 2 |
| Grh12 | 3.34 | 1 |
| Foxc1 | 3.21 | 1 |
| Sp1 | 2.90 | 1 |
| Milt10 | 2.83 | 1 |
| Gtf2e2 | 2.83 | 3 |
| Atf2 | 2.53 | 2 |
| Yap1 | 2.50 | 2 |
| Phf21a | 2.32 | 3 |
Fig. 5. miR-146a targets Nod1 mRNA that is depleted in γδ27− T cells.
(A) Putative binding sites of miR-146a-5p in the 3′UTR region of Nod1 and Atf2. The line represents canonical Watson-Crick base pairing, whereas the dot represents noncanonical base pairing between miR-146a and Nod1 or Atf2 mRNAs. The red bases above illustrated the mutated sites in the region of Nod1 and Atf2 3′UTRs. (B) Dual luciferase reporter assay was performed to verify binding between miR-146a and Nod1 or Atf2 mRNAs. HEK293 T cells were cotransfected with a pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) containing either the WT or mutated 3′UTR target sites plus miR-146a, an unrelated miRNA, or GFP control expression vector. A negative construct (without miR-146a binding sites) and a positive construct (3′UTR of Notch1) were included. Data are from three to four independent experiments with technical replicates. *P < 0.05, **P < 0.005, ***P < 0.001, Student’s t test. RT-qPCR analysis of Nod1 and Atf2 and expression in (C) sorted γδ27+, γδ27−CCR6−, and γδ27−CCR6+ T cells from pooled peripheral organs (lymph node and spleen) of C57BL/6 mice; (D) sorted Vγ1+, Vγ4+, and Vγ1−γ4− γδ T cells and CD4+ T cells from pooled peripheral organs (lymph node and spleen) of C57BL/6 mice; and (E) sorted Vγ4+CD27− and Vγ1−γ4− CD27-γδ T cells from pooled lymph nodes of either miR-146a+/+ or miR-146a−/− mice. Results are presented relative to β-actin and HPRT expression. Each symbol in (C) to (E) represents an individual mouse. *P < 0.05 and **P < 0.01 (Mann-Whitney two-tailed test).
To investigate the potential impact of miR-146a on those mRNAs in γδ T cells, we analyzed their expression levels in γδ27+ and γδ27− T cell subsets. Whereas Atf2 was similarly expressed in γδ27+, γδ27−CCR6−, and γδ27−CCR6+ T cells (Fig. 5C), Nod1 expression (Fig. 5C) showed a notable negative association with miR-146a levels (Fig. 1, A and B). Namely, Nod1 mRNA was present in γδ27+ but essentially absent in γδ27−CCR6+ T cells (Fig. 5C), thus suggesting that Nod1 is targeted by miR-146a specifically in this γδ T cell subset. In addition, Nod1 showed the lowest expression levels in Vγ1−Vγ4− (mostly Vγ6+) γδ T cells (Fig. 5D). We further extended our expression analysis to ex vivo isolated Vγ4+ and Vγ1−γ4− γδ27− T cells and observed notable increases in Nod1 (but not Atf2) levels in both miR-146a−/− γδ27− T cell subsets when compared with miR-146a+/+ counterparts (Fig. 5E). In sum, these results strongly indicate that Nod1 is a bona fide target of miR-146a in γδ27− T cells. Although we were unable to perform luciferase assays in primary murine γδ27− T cells, we successfully validated Nod1 targeting by miR-146a in a human γδ T cell line (fig. S7) that we previously showed to be a suitable model to study human γδ T cell development (28).
Nod1 is required for IFN-γ production and functional plasticity of γδ T cells
A key parameter in miRNA research is the demonstration of inverse phenotypes between an miRNA and its mRNA target. We therefore analyzed the production of IFN-γ (and IL-17) in γδ T cells from Nod1−/− mice (as well as Atf2−/− mice as expected “negative controls”). Critically, Nod1−/− (but not Atf2−/−) γδ27− T cells showed a notable inability to differentiate into IL-17+ IFN-γ+ cells when compared with littermate controls (Fig. 6A). Again, this was not due to proliferative defects (Fig. 6A). The loss of Nod1 did not impair IL-17 production by γδ27− T cells (Fig. 6A).
Fig. 6. Nod1 is required for IFN-γ production and functional plasticity in γδ T cells.
(A) Flow cytometry analysis of intracellular IFN-γ and IL-17 expression and frequency of IFN-γ+ IL-17+ (left), IL-17+ (middle), and Ki-67+ (right) in γδ27− T cells isolated from peripheral organs (spleen and lymph nodes) of Nod1+/+ and Nod−/− littermates as well as Atf2+/+ and Atf2−/− littermates after overnight stimulation with IL-1β plus IL-23. (B) Flow cytometry analysis of intracellular IFN-γ and IL-17 expression and frequency of IFN-γ+ IL-17+ (left), IL-17+ (up) and Ki-67+ (bottom) in γδ27− T cells isolated from peripheral organs (spleen and lymph nodes) of miR-146a+/+ Nod1+/+, miR-146a−/−Nod1+/+, and miR-146a−/−Nod1+/− littermates stimulated for 72 hours in the presence of IL-1β plus IL-23. (C) Bacterial CFUs of L. monocytogenes bacterial burden was enumerated in the spleen 4 days after infection (left), weight loss (middle), and survival (right) of Nod1+/+, Nod+/−, Nod−/−, and TCRd−/− mice infected with Listeria (n = 7 to 18 in three independent experiments). (D) Flow cytometry analysis of intracellular IFN-γ and IL-17 expression in γδ T cells isolated from spleen of Nod1+/+, Nod+/−, and Nod−/− mice, 7 days after infection with Listeria. Numbers in quadrants of flow cytometry plots indicate percentages of cells. (E) Frequency of IFN-γ+ IL-17+ within γδ T cells (left) and Vγ1Vγ4− γδ T cells (right) isolated from spleen of Nod1+/+, Nod+/−, and Nod−/−, 7 days after infection with Listeria (n = 7 to 18 in three independent experiments). Each symbol in (A) and (D) represents an individual mouse. *P < 0.05 and **P < 0.01 (Mann-Whitney two-tailed test).
To functionally test the relevance of Nod1 as the target of miR-146a, we performed a genetic rescue experiment by crossing mR-146a−/− with Nod1−/− mice and analyzing littermate controls with various genotypes. We found that reducing Nod1 (miR-146a−/− Nod1+/− mice) prevented the accumulation of double producers among γδ27− T cells observed in miR-146−/− Nod1+/+ mice (relative to miR-146+/+ Nod1+/+ controls) (Fig. 6B), while not affecting γδ27+ T cell differentiation (fig. S8). These data strongly suggest that Nod1 is a dominant target of miR-146a in regulating γδ27− T cell plasticity.
Last, to address the physiological relevance of Nod1 in vivo, we performed L. monocytogenes infection. Nod1−/− mice were highly susceptible to Listeria infection, as illustrated by increased bacterial burden, severe weight loss, and death of Nod1−/− (compared with both Nod1+/+ and Nod1+/−) mice (Fig. 6C). This was associated with strikingly reduced IL-17+ IFN-γ+ γδ27− T cells in Nod1−/− γδ T cells (Fig. 6, D and E). TCRd−/− mice were extremely susceptible to Listeria infection even more than Nod1−/− mice (Fig. 6C). Together, these data establish a critical function of Nod1 in promoting IFN-γ production and functional plasticity in γδ27− T cells, which is inverse to the negative role of miR-146a in this process, and thus support the miR-146a/ Nod1 axis as a critical regulator of γδ T cell differentiation in vivo.
DISCUSSION
miRNAs are endowed with the capacity to regulate various molecular pathways of differentiation and functions in innate and adaptive immunity (29, 30). In T cells, miRNA repertoires are known to change dramatically upon activation and across distinct functional T cell subsets (31, 32). Building on our previous definition of proinflammatory γδ T cell subsets (7, 11), we show here that miR-146a is highly expressed selectively on γδ27− T cells, where it targets Nod1 and thus limits IFN-γ production by this IL-17–biased subset. The expression pattern of miR-146a was established during thymic γδ T cell development and was unaffected by exogenous TCR stimulation or inflammatory cytokine signals, further illustrating the importance of developmental preprogramming in γδ T cell biology (8, 9). In the periphery, inflammatory signals are major drivers of functional plasticity of γδ27− T cells, which manifests itself in the acquisition of IFN-γ coproduction by previous IL-17 single producers. We found that the genetic loss of miR-146a led to the accumulation of multifunctional IL-17+ IFN-γ+ double producers in vitro and in vivo, upon Listeria infection, where these cells had been associated with a protective memory response against the bacteria (5).
miR-146a has been previously associated with regulation of αβ T cell responses via dampening of nuclear factor κB (NF-κB) activation (33, 34). In the absence of miR-146a, enhanced TCR-induced NF-κB activation was linked to higher proliferation and increased production of effector cytokines including not only IFN-γ but also IL-17A and IL-2 by CD4+ and CD8+ T cells—that is, a general activation phenotype (33). However, more recently, miR-146a was shown to inhibit the production of autocrine IL-6 and IL-21 in autoreactive transgenic CD4+ T cells, which rather reduced their TH17 differentiation (35). By contrast, our results show normal proliferation and IL-17 production by miR-146a−/− γδ T cells, thus highlighting the selective impact on IFN-γ coproduction by γδ27− T cells, which may be linked to the high levels of expression of miR-146a in this subset. As a caveat of our study, we did not have the technical tools to specifically ablate miR-146a in γδ T cells, so we had to rely on full miR-146a “knockout” mice. No miRNA has yet been shown to play a role in TH17/TH1 plasticity. Looking globally at CD4+ T cell plasticity, miR-10a is thus far an isolated example of an miRNA implicated in limiting “lineage conversion”; being highly expressed in regulatory T cells, miR-10a prevents the acquisition of TH17 and follicular helper T cell features (36).
In contrast with the conventional targets (Traf6, Irak1, and Stat1, among others) of miR-146a in CD4+ T cells (33, 34), the key mRNA target we identified for miR-146a in γδ T cells was Nod1. This target was identified by an unbiased Ago2-RIP approach in Ago2-tagged T cells after miR-146a overexpression. Although overexpression or deletion of a given miRNA gene regulates distinct sets of target genes (37), we favored an overexpression approach because, as each mRNA is targeted by multiple miRNAs, critical mRNA targets of miR-146a could be unaffected in RISCs of miR-146a−/− T cells. Instead, by providing an excess of miR-146a, we increased the possibility of dragging its mRNA targets into RISCs. Moreover, because miR-146a is a low expressed miRNA, selectively enriched in a minor (γδ27−) T cell subset, using an overexpression system increases the signals attributable to this low abundant miRNA. The genome-wide Ago2 RIP-seq analysis allowed us to identify many potential previously unknown participants in the differentiation and activation of T cells. Here, we focused our analysis on Nod1, and our data suggest that miR-146a targets Nod1 not by canonical seed sequence–based pairing but by an alternative pathway already shown for other miRNAs [reviewed in (38)], such as miR-155 that targets more than 40% of its mRNAs by noncanonical interactions (39). This type of binding is very difficult to predict using standard strategies as those used by softwares like Targetscan (40). Thus, in this analysis, we modified the standard parameters used by miRWalk (41) by allowing a minimum seed binding of five nucleotides and further refined the results by calculating the Gibb’s function of the miRNA:mRNA hybrid. Because of 3′ compensatory binding, the results obtained for the selected miR-146a-5p target at the 3′UTR of Nod1 (ΔG= −22.5 kcal/mol) are consistent with a stable miR-146:Nod1 hybrid.
Nod1 is a crucial pattern recognition receptor initially thought to be restricted to innate myeloid cells. However, Nod1 has been shown to be expressed by CD8+ T cells, at comparable levels to macrophages, and to functionally coactivate T cell proliferation and IFN-γ production (24). We show here that γδ T cells, particularly the γδ27+ subset that makes IFN-γ, express Nod1. This is consistent with recent data showing that Nod1 ligation increases IFN-γ production by isolated CD8+ and CD4+ T cells in vitro, and Nod1-deficient T cells exhibited impaired IFN-γ production upon dextran sulfate sodium–induced acute inflammation and tumorigenesis in vivo (25). The other prototypic Nod-like receptor, Nod2, was also suggested to play a T cell–intrinsic role in IFN-γ production, which affected Toxoplasma gondii clearance (42).
As confirmed by our data, Nod1 is a major determinant of resistance to L. monocytogenes infection (43). The same applies to γδ T cells, but we could not directly link the two phenotypes because of lack of genetic tools to conditionally delete Nod1 specifically in γδ T cells. On the other hand, although in the infection setting Nod1 may be recognizing bacterial ligands such as peptidoglycan (44), the inability of Nod1–deficient γδ27− T cells to differentiate into IFN-γ producers in vitro suggests an additional mechanism of Nod1 activation. As another limitation of our study, we have dissected neither this upstream mechanism nor the downstream regulators of cytokine production. Nod1 (and Nod2) are important mediators of endoplasmic reticulum (ER) stress-induced production of proinflammatory cytokines (45). Future research should therefore address whether ER stress and the underlying unfolded protein response, known to be important drivers of inflammation and also to be induced upon T cell activation (46), play a role in Nod1-dependent γδ T cell differentiation.
In sum, our data demonstrate that high levels of miR-146a in γδ27− T cells suppress Nod1 expression, which is rescued upon ablation of miR-146a, thus promoting IFN-γ expression in thymically precommitted IL-17 producers. Multifunctional IL-17+ IFN-γ+ γδ T cells have been associated with pathogenic γδ T cell responses in experimental models of (ovarian) cancer (11) and autoimmunity (EAE) (12), and thus, the manipulation of miR-146a levels may have therapeutic potential.
MATERIALS AND METHODS
Mice
All mice used were adults 6 to 12 weeks of age. C57BL/6J and CD45.1 mice were from the Jackson Laboratory, and Rag2−/− mice were from Taconic Biosciences. Il17a-GFP knock-in mice were from Biocytogen, LLC. miR-146a−/− mice have been described previously (19). Nod1−/− and Atf2−/− mice were obtained from T. Mak (University Health Network, Toronto) and W. Breitwieser (Cancer Research UK Manchester Institute), respectively. The Ago2-tagged knock-in mice (EIF2c2-tag-KI) were generated by A.H.B.’s laboratory in conjunction with Biocytogen. A targeting vector was designed to insert a 147-bp N-terminal tag into the first exon of the Eif2c2 gene downstream of the ATG start site. The tag consists of 3×FLAG tag followed by a PreScission cleavage site and 6×HIS tag (sequence information available by request). A neomycin-selectable cassette was flanked by FLPe sites and inserted into the nonconserved region of intron 1 and targeted embryonic stem (ES) cells generated by homologous recombination. Selected ES cells were microinjected into C57BL/6 blastocysts and embryo transfer to pseudopregnant females. Chimeras were identified using standard procedures. Germline transmission was confirmed by Southern blotting and breeding program set up to establish the line. Both heterozygous and homozygous mice carrying the FLAG-HIS–tagged Ago2 are viable and fertile.
Mice were bred and maintained in the specific pathogen–free animal facilities of Instituto de Medicina Molecular (Lisbon, Portugal). All experiments involving animals were done in compliance with the relevant laws and institutional guidelines and were approved by the ethics committee of Instituto de Medicina Molecular.
Monoclonal antibodies
The following anti-mouse fluorescently labeled monoclonal antibodies (mAbs) were used (antigens and clones): TCRδ (GL3), Vγ1 (2.11), Vγ4 (UC3–10A6), CD3 (145.2C11), CD4 (GK1.5), CD8 (53–6.7), CD25 (PC61), CD27 (LG.7F9), CD44 (IM7), CD45.1 (A20), CD45.2 (104), Ki-67 (16A8), IFN-γ (XMG1.2), and IL-17A (TC11.18H10.1). Antibodies were purchased from BD Biosciences, eBiosciences, or BioLegend.
Cell preparation, flow cytometry, and cell sorting
Cell suspensions were obtained from spleens, lymph nodes, or thymus. Erythrocytes were osmotically lysed in red blood cell lysis buffer (BioLegend). Cells were filtered through 70-μm cell strainers (BD Biosciences). For cell surface staining, single-cell suspensions were incubated for 30 min with saturating concentrations of mAbs (see above). For intracellular cytokine staining, cells were stimulated with phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (1 μg/ml) in the presence of brefeldin A (10 μg/ml) (all from Sigma- Aldrich) for 3 hours at 37°C. Cells were stained for the above identified cell surface markers, fixed 30 min at 4°C, permeabilized with the Foxp3/Transcription Factor Staining Buffer set (eBioscience) in the presence of anti-CD16/CD32 (eBioscience) for 15 min at 4°C, and lastly incubated for 1 hour at 4°C with the above identified antibodies in permeabilization buffer. Cells were sorted on FACSAria (BD Biosciences), and samples were acquired using FACSFortessa (BD Biosciences). Data were analyzed using FlowJo software (Tree Star).
miRNA array
All experiments were conducted at Exiqon Services. The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. Total RNA (250 ng) from sample and reference was labeled with Hy3 and Hy5 fluorescent label, respectively, using the miRCURY LNA miRNA Hi-Power Labelling Kit, Hy3/Hy5 (Exiqon, Denmark), following the procedure described by the manufacturer. The Hy3- labeled samples and a Hy5- labeled reference RNA sample were mixed pairwise and hybridized to the miRCURY LNA miRNA Array (sixth generation: hsa, mmu, and rno) (Exiqon, Denmark), which contains capture probes targeting all miRNAs for human, mouse, or rat registered in the miRBASE 16.0. The hybridization was performed according to the miRCURY LNA miRNA Array instruction manual using a Tecan HS 4800 hybridization station (Tecan, Austria). After hybridization, the microarray slides were scanned and stored in an ozone-free environment (ozone level below 2.0 parts per billion) to prevent potential bleaching of the fluorescent dyes. The miRCURY LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies Inc.), and the image analysis was carried out using the ImaGene 9.0 software (BioDiscovery Inc.). The quantified signals were background-corrected (Normexp with an offset value of 10) and normalized using the Quantile normalization algorithm.
RNA isolation, complementary DNA production, and real-time PCR
RNA was isolated from fluorescence-activated cell sorting (FACS)–sorted cell populations using a High Pure RNA Isolation kit (Roche) for total RNA and an miRNeasy Mini kit (Qiagen) or a High Pure miRNA Isolation kit (Roche) for small RNA. For total RNA, reverse transcription was performed with random oligonucleotides (Invitrogen) using Moloney murine leukemia virus reverse transcriptase (Promega). For miRNA, reverse transcription was performed with a Universal cDNA Synthesis kit II (Exiqon). For total RNA samples, relative quantification of specific complementary DNA (cDNA) species to endogenous references HPRT or Actinb was carried out using SYBR on a ViiA7 cycler (Applied Biosystems). Primers were either designed manually or by the Universal ProbeLibrary Assay Design Center (Roche), and their sequences were as follows: HPRT, GACCTCTCGAAGTGTTGGAT (forward) and GCTTTGTATTTGGCTTTTCC (reverse); Actb, CGTGAAAAGATGACCCAGATCA (forward) and TGGTACGACCAGAGGCATACAG (reverse); Nod1, CCTTCGTCCTGCATCACTTC (forward) and CTGGTTGACGCTGAGTCTGA (reverse); and Atf2, GAAGAGTCTCGCCCACAGTC (forward) and TGGGTCTGAGGAGTTGTGTG (reverse). For miRNA samples, relative quantification of specific cDNA species to reference small RNA RNU5G or miR-423–3p was carried out using SYBR on a ViiA7 cycler (Applied Biosystems). The respective miRNA LNA primers were used (Exiqon).
Retroviral overexpression of miR-146a
The retroviral constructs encoding either mmu-miR-146a or an unrelated miRNA (mmu-miR-132) were generated by inserting the respective native pre-miRNA sequences flanked by about 200 bp into a modified pMig.IRES-GFP retroviral vector (Addgene #9044). miR-146a and miR-132 sequences were obtained by PCR from genomic DNA from C57BL/6J mice. The internal ribosomal entry site (IRES)–GFP sequence was removed and repIaced by PGK-GFP-WPRE sequence. The resulting vector encodes GFP under the control of the phosphoglycerate kinase (PGK) promoter and miR-146a or miR-132 under control of the retroviral long terminal repeat promoter. A Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element (WPRE) was inserted for increased viral titers. For retroviral transduction, γδ T cells and CD4+ T cells were sorted from lymph node and spleen of C57BL/6J mice and stimulated either with plate-bound anti-CD3ɛ (145.2C11) plus anti-CD28 mAb (37.51) (both 2.5 μg/ml) or plate-bound anti-CD3ɛ (145.2C11) (5 μg/ml) in the presence of soluble anti-CD28 (37.51) (2 μg/ml) with IL-12 (5 ng/ml) and anti–IL-4 (11B11) (10 μg/ml), respectively. The transduction was performed with polybrene (4 μg/ml; Sigma-Aldrich), and medium was supplemented with IL-2 (5 ng/ml). Forty-eight hours after transduction, live cells using a LIVE/DEAD stain kit (Life Technologies), and GFP+ cells were sorted and activated for intracellular staining.
γδ T cell electroporation
γδ T cells were sorted from lymph node and spleen of C57BL/6J mice and stimulated with plate-bound anti-CD3 and anti-CD28 (both 2.5 μg/ml). γδ T cells (2.5 × 104) were resuspended in T buffer and used per transfection with Neon electroporation transfection system (Invitrogen) with three pulses of 10 ms and 1550 V. miRIDIAN mimic of mouse mmu-miR-146a-5p (Dharmacon) and miRIDIAN mimic negative control #1 (Dharmacon) with an RNA concentration of 500 nM per electroporation were used. After electroporation, cells were cultured in media supplemented with IL-2 (5 ng/ml) and 48 hours later activated for intracellular cytokine staining.
In vitro γδ T cell stimulation and CD4+ TH cell polarization
γδ27+ and γδ27− T cells were FACS-sorted and subjected to various stimulation conditions overnight or for 24 to 72 hours. Cells were incubated on plate-bound anti-CD3ɛ (145.2C11) plus anti-CD28 mAb (37.51) (both at 2.5 μg/ml) supplemented with IL-12 (10 ng/ml) or, alternatively, in the presence of murine IL-1β plus IL-23 (both 50 ng/ml) or IL-12 (10 ng/ml). For TH1 cell culture conditions, FACS-sorted CD4+ T cells were incubated with plate-bound anti- CD3ɛ (145.2C11) and soluble anti-CD28 mAb (37.51) (both at 2 μg/ml) in the presence of IL-12 (5 ng/ml) and neutralizing anti–IL-4 mAb (11B11) (5 μg/ml) for 5 days. All cytokines were from PeproTech, except TGF-b and IL-23, which were from R&D systems.
L. monocytogenes infection and bacterial quantification
We used L. monocytogenes strain EGDe, carrying a recombinant InlA with a mutation in S192N and Y369S, as previously described (5). Briefly, all mice were deprived of food overnight before infection, housed individually, and given a ~0.5-cm3 piece of mashed food inoculated with 2 × 109 colony-forming units (CFUs) of L. monocytogenes in phosphate-buffered saline. CFUs were calculated from single-cell suspensions of spleens plated in serial dilutions on Brain Heart Infusion agar plates. Individual colonies were counted after 30 hours of incubation at 37°C.
Neonatal thymocytes and BM chimeras
Chimeras were generated as previously described (20). Briefly, Rag2 −/− mice were lethally irradiated (9.5 gray) and injected 6 hours after with a 1:1 mix of neonatal thymocytes (5 × 106 to 10 × 106 thymocytes per host) isolated from WT (CD45.1/CD45.2) and miR-146a−/− (CD45.2) donors within 48 hours after birth. After 24 hours, the host received a 1:1 mix of 5 × 106 to 10 × 106 BM cells isolated from WT (CD45.1/CD45.2) and miR-146a−/− (CD45.2) donors. Chimeras were kept on antibiotic-containing water (2% Bactrim; Roche) for the first 4 weeks after irradiation. The hematopoietic compartment was allowed to reconstitute for 8 weeks before the animals were used for experiments.
Differential Argonaute 2 RNA immunoprecipitation followed by deep sequencing
CD3+ T cells were FACS-sorted from pooled spleens of Ago2-tagged (3×FLAG-6×HIS tag) mice and stimulated with plate-bound anti- CD3ɛ (145.2C11) plus anti-CD28 mAb (37.51) (both 2.5 μg/ml) and transduced either with RV-miR-146a vector or RV-control vector. Twenty-four hours after transduction, GFP+ live cells [using a live/dead stain kit (Life Technologies)] were FACS-sorted. Cells (7 × 105 to 2 × 106) were UV–cross-linked in Stratalinker 2400 (400 mJ cm−2) and lysed. AGO-interacting RNA was immunoprecipitated using anti- Flag (M2) magnetic beads (Sigma-Aldrich). Following, Ago2-bound RNA was partially digested using RNase IT (Agilent Technologies) and, in a second step, purified using the HIS-tag and Ni–nitrilotriacetic acid beads (Qiagen) under high stringent denaturing wash conditions. After proteinase K treatment, RNA was isolated using phenol-chloroform isoamyl extraction (detailed protocol available upon request). The quality of the RNA was verified by an Agilent 2100 Bioanalyzer profile, and libraries were prepared using a Clontech SMARTer Stranded RNA-Seq Kit, starting with 8 μl of the immunoprecipitated RNA. The manufacturer’s solid phase reversible immobilization (SPRI) protocol was followed, using 4 min for RNA fragmentation, double bead cleanup after reverse transcription, and 20 PCR cycles for amplification. The resulting libraries were sequenced using Illumina HiSeq 2000 instrument with 50-bp single-read sequencing mode.
Ago2 RIP-seq data analysis
To assess overall RNA-seq data quality, sequence length distribution and the base calling accuracy as indicated by the Phred quality score (Q score) were calculated with high-throughput sequence data quality control software FastQC (version 0.10.1; Babraham Bioinformatics, UK). Adaptor sequences were trimmed using FASTX-toolkit. Sequence reads were mapped to the mouse reference genome (GRCm38) using Bowtie2 software (47). Aligned reads were thereafter sorted and indexed by SAMtools (48), Cuffdiff software, a part of the Cufflinks suite (49), was used to detect significant changes in mRNA enrichment between the experimental groups. Genes with a P value, for statistical significance, and q value, to detect the false discovery rate, of <0.05 were considered differentially enriched in Ago2-RIP experiments. Predicted miRNA targets for mmu-miR-146a were determined by miRWalk2 algorithm (41).
Dual luciferase reporter assay
The 3′UTR of Nod1, Atf2, Notch1 (positive control), and PTPBP2 (negative control) were cloned into pmirGLO vector (Promega) from genomic DNA. Primers are available upon request. Mutations in the predicted target sequences of the 3′UTRs were introduced by gene synthesis (GeneCust Europe). The luciferase reporter vectors carrying either the WT or mutant 3′UTR-sequence were transfected with the miR-146a expression vector, an unrelated miRNA (miR-132), or a control vector into HEK293 T cells (ATCC CRL-3216) using Lipofectamine 2000 (Thermo Fisher Scientific) or electroporated into PEER T-ALL cells [DSMZ (German Collection of Microorganisms and Cell Cultures), Braunschweig, Germany], following the manufacturer’s protocol. After 48 hours, firefly and Renilla luciferase activity were measured by using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity served as the internal control, and results were expressed as relative luciferase activity.
Carboxyfluorescein diacetate succinimidyl ester staining
For analysis of proliferation, FACS-sorted CD4+ and γδ T cells were stained by incubation for 5 min at 37°C with 5 mM CFSE (carboxyfluorescein diacetate succinimidyl ester; Molecular Probes). Reactions were quenched by washing with ice-cold RPMI medium supplemented with 10% (v/v) fetal calf serum. CFSE-labeled cells were stimulated with plate-bound anti-CD3ɛ (145.2C11) plus anti-CD28 mAb (37.51) (both 2.5 μg/ml). After 4 days, the proliferation was assessed by flow cytometry.
Bromo-deoxyuridine incorporation
Mice were given intraperitoneal injections of 1.5 mg of bromo-deoxyuridine (BrdU) on the day of L. monocytogenes infection, and BrdU at a concentration of 0.8 mg/ml was supplemented in the drinking water during the course of infection. Incorporation of BrdU was determined by a BrdU Flow kit (BD Pharmingen) following manufacturer’s protocol.
Statistical analysis
The statistical significance of differences between populations was assessed using a two-tailed nonparametric Mann-Whitney test or Student’s t test. P < 0.05 was considered significant and is indicated on the figures.
Supplementary Material
Fig. S1. miR-146a expression analysis.
Fig. S2. Loss of miR-146a does not affect steady-state γδ T cell subsets.
Fig. S3. Loss of miR-146a does not affect IFN-γ production by γδ27+ T cells or CD4+ TH1 cells upon in vitro polarization.
Fig. S4. Mixed BM and neonatal thymocyte chimeras.
Fig. S5. γδ T cell responses to L. monocytogenes infection.
Fig. S6. Differential Argonaute 2 immunoprecipitation for identification of mRNA targets of miR-146a.
Fig. S7. Nod1 is targeted by miR-146a in γδ T cells.
Fig. S8. Nod1 restriction does not affect IFN-γ production by γδ27+ T cells.
Table S1. Top 25 miscellaneous mRNAs differentially enriched upon Ago2-RNA immunoprecipitation in T cells overexpressing miR-146a.
Table S2. Raw data sets.
Acknowledgments
We thank F. Caiado, J. Ribot, S. Mensurado, N. Gonçalves-Sousa [Instituto de Medicina Molecular (iMM), Lisboa, Portugal], A. Helwak (Wellcome Centre for Cell Biology, Edinburgh, UK), and K. M. Ansel (University of California, San Francisco, USA) for discussions and advice; V. Benes and D. Pavlinic (European Molecular Biology Laboratory GeneCore facility) for assistance with library preparation for RNA-seq analyses; D. Denner for help in graphical output; and the staff of the Flow Cytometry and Rodent facilities of iMM Lisboa for valuable technical assistance. Ago2-tagged (3×FLAG-6×HIS tag) mice are available from A.H.B. under a material transfer agreement with the University of Edinburgh.
Funding: This work was funded by the European Research Council (CoG_646701) to B.S.-S., Fundação para a Ciência e Tecnologia (FCT; PTDC/BEX-BCM/3592/2014 to A.Q.G. and SFRH/BPD/108821/2015 to N.S.), Wellcome Trust Research Career Development Fellowships (WT097394A1A) to A.H.B., and NIH/National Institute of Allergy and Infectious Diseases and STOP Cancer Foundation to M.B. This publication was sponsored by LISBOA-01-0145-FEDER-007391, with the project cofunded by Fundo europeu de desenvolvimento regional (FEDER), through POR Lisboa 2020 (Programa Operacional Regional de Lisboa, PORTUGAL 2020) and FCT.
Footnotes
REFERENCES AND NOTES
- 1.Korn T, Bettelli E, Oukka M, Kuchroo VK, IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR, Bhan A, Autschbach F, Sullivan BM, Szabo SJ, Glimcher LH, Blumberg RS, The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J. Exp. Med. 195, 1129–1143 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papotto PH, Ribot JC, Silva-Santos B, IL-17+ γδ T cells as kick-starters of inflammation. Nat. Immunol. 18, 604–611 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hiromatsu K, Yoshikai Y, Matsuzaki G, Ohga S, Muramori K, Matsumoto K, Bluestone JA, Nomoto K, A protective role of gamma/delta T cells in primary infection with Listeria monocytogenes in mice. J. Exp. Med. 175, 49–56 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheridan BS, Romagnoli PA, Pham Q-M, Fu H-H, Alonzo F III, Schubert W-D, Freitag NE, Lefrançois L, γδ T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity 39, 184–195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen KDC, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, Baumgarth N, Chien Y.-h., Thymic selection determines γδ T cell effector fate: Antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon γ. Immunity 29, 90–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, Silva-Santos B, CD27 is a thymic determinant of the balance between interferon-γ- and interleukin 17-producing γδ T cell subsets. Nat. Immunol. 10, 427–436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayday AC, γδ T cells and the lymphoid stress-surveillance response. Immunity 31, 184–196 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Muñoz-Ruiz M, Sumaria N, Pennington DJ, Silva-Santos B, Thymic determinants of γδ T cell differentiation. Trends Immunol. 38, 336–344 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Haas JD, González FHM, Schmitz S, Chennupati V, Föhse L, Kremmer E, Förster R, Prinz I, CCR6 and NK1.1 distinguish between IL-17A and IFN-γ-producing γδ effector T cells. Eur. J. Immunol. 39, 3488–3497 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Schmolka N, Serre K, Grosso AR, Rei M, Pennington DJ, Gomes AQ, Silva-Santos B, Epigenetic and transcriptional signatures of stable versus plastic differentiation of proinflammatory γδ T cell subsets. Nat. Immunol. 14, 1093–1100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds JM, Martinez GJ, Chung Y, Dong C, Toll-like receptor 4 signaling in T cells promotes autoimmune inflammation. Proc. Natl. Acad. Sci. U.S.A. 109, 13064–13069 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenoglio D, Poggi A, Catellani S, Battaglia F, Ferrera A, Setti M, Murdaca G, Zocchi MR, Vδ1 T lymphocytes producing IFN-γ and IL-17 are expanded in HIV-1–infected patients and respond to Candida albicans. Blood 113, 6611–6618 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, Stockinger B, Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12, 255–263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT, Late developmental plasticity in the T helper 17 lineage. Immunity 30, 92–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amado T, Schmolka N, Metwally H, Silva-Santos B, Gomes AQ, Cross-regulation between cytokine and microRNA pathways in T cells. Eur. J. Immunol. 45, 1584–1595 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Haas JD, Nistala K, Petermann F, Saran N, Chennupati V, Schmitz S, Korn T, Wedderburn LR, Forster R, Krueger A, Prinz I, Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in αβ and γδ T cells. PLOS ONE 6, e20171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barros-Martins J, Schmolka N, Fontinha D, Pires de Miranda M, Simas JP, Brok I, Ferreira C, Veldhoen M, Silva-Santos B, Serre K, Effector γδ T cell differentiation relies on master but not auxiliary Th cell transcription factors. J. Immunol. 196, 3642–3652 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D, miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Xue F, Fleming C, Yang J, Ding C, Ma Y, Liu M, Zhang H.-g., Zheng J, Xiong N, Yan J, Differential developmental requirement and peripheral regulation for dermal Vγ4 and Vγ6T17 cells in health and inflammation. Nat. Commun. 5, 3986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas JD, Ravens S, Düber S, Sandrock I, Oberdörfer L, Kashani E, Chennupati V, Föhse L, Naumann R, Weiss S, Krueger A, Förster R, Prinz I, Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity 37, 48–59 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos I-L, Maniou S, Karathanou K, Kalfakakou D, Fevgas A, Dalamagas T, Hatzigeorgiou AG, DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 43, D153–D159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones B, Chen J, Inhibition of IFN-γ transcription by site-specific methylation during T helper cell development. EMBO J. 25, 2443–2452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mercier BC, Ventre E, Fogeron M-L, Debaud A-L, Tomkowiak M, Marvel J, Bonnefoy N, NOD1 cooperates with TLR2 to enhance T cell receptor-mediated activation in CD8 T cells. PLOS ONE 7, e42170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan Y, Seregin SS, Chen J, Chen GY, Nod1 limits colitis-associated tumorigenesis by regulating IFN-γ production. J. Immunol. 196, 5121–5129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Sun X, Huang C, Long X.-r., Lin X, Zhang L, Lv X.-w., Li J, MiR-146a regulates IL-6 production in lipopolysaccharide-induced RAW264.7 macrophage cells by inhibiting Notch1. Inflammation 37, 71–82 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Mei J, Bachoo R, Zhang C-L, MicroRNA-146a inhibits glioma development by targeting Notch1. Mol. Cell. Biol. 31, 3584–3592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro ST, Tesio M, Ribot JC, Macintyre E, Barata JT, Silva-Santos B, Casein kinase 2 controls the survival of normal thymic and leukemic γδ T cells via promotion of AKT signaling. Leukemia 31, 1603–1610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebert MS, Sharp PA, Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bronevetsky Y, Ansel KM, Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 253, 304–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai T-H, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS, MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6, R71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJP, Birolo RS, Moro M, Crosti MC, Gruarin P, Maglie S, Marabita F, Mascheroni D, Parente V, Comelli M, Trabucchi E, De Francesco R, Geginat J, Abrignani S, Pagani M, Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 12, 796–803 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Boldin MP, Yu Y, Liu CS, Ea C-K, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D, miR-146a controls the resolution of T cell responses in mice. J. Exp. Med. 209, 1655–1670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu L-F, Boldin MP, Chaudhry A, Lin L-L, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY, Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142, 914–929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Wang X, Choi IY, Wang Y-C, Liu S, Pham AT, Moon H, Smith DJ, Rao DS, Boldin MP, Yang L, miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J. Clin. Invest. 127, 3702–3716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciumè G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O’Shea JJ, TGF-β and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 13, 587–595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin HY, Oda H, Chen P, Yang C, Zhou X, Kang SG, Valentine E, Kefauver JM, Liao L, Zhang Y, Gonzalez-Martin A, Shepherd J, Morgan GJ, Mondala TS, Head SR, Kim P-H, Xiao N, Fu G, Liu W-H, Han J, Williamson JR, Xiao C, Differential sensitivity of target genes to translational repression by miR-17~92. PLOS Genet. 13, e1006623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cloonan N, Re-thinking miRNA-mRNA interactions: Intertwining issues confound target discovery. Bioessays 37, 379–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY, Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 48, 760–770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal V, Bell GW, Nam J-W, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dweep H, Gretz N, miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 12, 697 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Shaw MH, Reimer T, Sánchez-Valdepeñas C, Warner N, Kim Y-G, Fresno M, Nuñez G, T cell–intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii. Nat. Immunol. 10, 1267–1274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y-G, Park J-H, Shaw MH, Franchi L, Inohara N, Núñez G, The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity 28, 246–257 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nuñez G, Inohara N, An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4, 702–707 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chávez-Arroyo A, Tsai AY, Cevallos SA, Winter MG, Pham OH, Tiffany CR, de Jong MF, Kerrinnes T, Ravindran R, Luciw PA, McSorley SJ, Bäumler AJ, Tsolis RM, NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang K, Kaufman RJ, From endoplasmic-reticulum stress to the inflammatory response. Nature 454, 455–462 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup, The sequence alignment/ map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. miR-146a expression analysis.
Fig. S2. Loss of miR-146a does not affect steady-state γδ T cell subsets.
Fig. S3. Loss of miR-146a does not affect IFN-γ production by γδ27+ T cells or CD4+ TH1 cells upon in vitro polarization.
Fig. S4. Mixed BM and neonatal thymocyte chimeras.
Fig. S5. γδ T cell responses to L. monocytogenes infection.
Fig. S6. Differential Argonaute 2 immunoprecipitation for identification of mRNA targets of miR-146a.
Fig. S7. Nod1 is targeted by miR-146a in γδ T cells.
Fig. S8. Nod1 restriction does not affect IFN-γ production by γδ27+ T cells.
Table S1. Top 25 miscellaneous mRNAs differentially enriched upon Ago2-RNA immunoprecipitation in T cells overexpressing miR-146a.
Table S2. Raw data sets.