Abstract
Background:
Schwann cells (SC) and macrophages play key roles in the response to peripheral nerve injury (PNI). Accurate isolation of such cells is essential for further analyses that can lead to better understanding of the repair process after PNI. Separation of live SC from the injury site without culture enrichment is necessary for targeted gene expression analysis.
New Methods:
Two flow cytometric techniques are presented for rapid enrichment of live SC and macrophages from injured murine peripheral nerve without the need for culture.
Results:
SC were isolated by fluorescent activated cell sorting (FACS) using transgenic expression of eGFP in SC, or by exclusion of other cell types collected from the injury site.
Comparison with Existing Method(s):
Gene expression analyses of peripheral nerve repair have commonly used whole nerve lysates. Isolating SC allows more accurate understanding of their specific role in repair. SC are commonly enriched from nerve by culture, however this changes gene expression patterns and limits the utility for transcriptomic analysis. The surface marker p75-NTR has variable expression in different SC phenotypes and during the course of injury and repair. Using p75-NTR for SC isolation might enrich only a subset of SC. More stably expressed lineage markers for SC are intracellular and not suitable for sorting for gene expression. The methods used here avoid the requirement for surface marker labeling of SC.
Conclusion:
Gene expression analysis of sorted cells from both methods showed successful enrichment of SC. Lineage markers such as Map1b, p75-NTR and S100b were enriched in the sorted SC population. SC sorting by eGFP expression showed improved enrichment, particularly of mature myelinating genes, although this could represent sampling of a subset of SC.
Keywords: Schwann Cell Separation, Flow Cytometry, Peripheral Nerve Injury, Macrophage Isolation, Sciatic Nerve Repair
1. Introduction
Macrophages and Schwann cells (SC) play key roles in the early response to peripheral nerve injury. Teasing out the contribution of and interactions between each cell type will improve our ability to manipulate the early repair process for improved recovery (Stratton et al., 2018). In order to dissect out the contributions of macrophages and SC distinctly, it is necessary to be able to isolate each population. The application of fluorescence-activated cell sorting (FACS) to isolate macrophages for gene expression analysis has been successful in allowing an unbiased gene expression approach to mapping the phenotype of macrophages during peripheral nerve repair (Stratton et al., 2018; Tomlinson et al., 2018). However, a similar approach for SC has been limited by the absence of well characterized SC-specific extracellular markers that are consistently expressed across myelinating and non-myelinating SC phenotypes. Commonly used methods to evaluate the role of SC in peripheral nerve repair include immunohistochemistry, in vitro assays, transgenic mice, and flow cytometric characterization with or without pre-enrichment via culture (Cattin et al., 2015; Roberts et al., 2017; Spiegel and Peles, 2009; Stratton et al., 2018; Wang et al., 2017). These methods are limited in that they do not allow immediate ex vivo evaluation of SC gene expression patterns. Many of the markers used are intracellular, such as the transcription factors Sox10 and Sox2; or markers for subsets of mature or immature SC, such as Necl1 or p75-NTR (Spiegel and Peles, 2009; Stratton et al., 2018). Intracellular labeling requires fixation of the cells and degrades RNA quality. Additionally, pre-enrichment by culture greatly alters gene expression profiles and cell phenotypes. Therefore, there remains a need for a SC enrichment technique that can isolate fresh, live SC, regardless of phenotype, without culture.
We compare two approaches to enriching SC from injured peripheral nerve. First, we performed FACS sorting by exclusion of other expected cell types in the tissue. This method was expected to incorporate all phenotypes of SC at the expense of perhaps including some non-SC. Second, we utilized an S100b-eGFP transgenic mouse line (Zuo, 2004) for positive SC identification, combined with the exclusion markers, to increase specificity of the sort. A number of SC-specific transgenic reporter mouse lines have been described (Laranjeira et al., 2011; Mallon et al., 2002; Vives et al., 2003; Zuo, 2004). We utilized the S100b-eGFP line because S100b is expressed primarily in SC in the peripheral nervous system, and maintenance of expression is anticipated in both myelinating and non-myelinating SC phenotypes (Donato et al., 2009; Liu et al., 2015; Mirsky et al., 2008). Additionally S100b is commonly used as an immunohistochemical marker for SC after peripheral nerve injury (Carroll et al., 1997; Godinho et al., 2013; Jessen and Mirsky, 2005; Mokarram et al., 2012; Raponi et al., 2007), allowing for comparison of similar cells across the two methods of analysis.
2. Methods
2.1. Subject Information
Animal studies were performed in accordance with the PHS Policy on Humane Care and Use of Laboratory Animals, the NIH guide for Care and Use of Laboratory Animals, federal and state regulations, and was approved by the Cornell University Institutional Animal Care and Use Committee (IACUC).
Animals were brought into the research unit and given a 3-day acclimatization period prior to any procedure. Daily record logs of medical procedures were maintained. Rodent cages were replaced weekly. Animals were on a 12/12h light-dark cycle and allowed food and water ad libitum. Group housing (2-5 mice per cage) prior to medical procedures provided socialization. Mice were entered into the study at 9-12 (mean 10) weeks old and both sexes were used.
Hemizygous S100b-eGFP mice (B6;D2-Tg(S100B-EGFP)1Wjt/J, Stock No. 005621), were purchased from Jackson Laboratories and bred to homozygosity. Genotyping was performed (Transnetyx) to screen offspring for the presence of the GFP transgene and mice with highest normalized qPCR expression were crossed to produce a stable homozygous colony. Expression of eGFP was confirmed in all experimental mice by genotyping until a stable homozygous colony was obtained. Control mice were C57Bl/6J (Stock No.000664) which were maintained as a congenic colony.
2.2. Sciatic Nerve Transection Model and Tissue Harvest
Sciatic nerve transection and inert conduit repair was performed as previously described (Tomlinson et al., 2018; Žygelytė et al., 2016). The sequence of surgical procedures and harvests were randomized by mouse strain. . Mice were anesthetized with 3% isoflurane and maintained under anesthesia with 1-2% isoflurane and oxygen. Analgesia was provided by subcutaneous buprenorphine simbadol (0.1mg/kg) injection pre-operatively and 24 hours after surgery. The left sciatic nerve was exposed and transected at mid-femur, proximal to the bifurcation. Proximal and distal nerve stumps were aligned and sutured 1mm into a 5mm long inert silicone nerve conduits with epineurial 10-0 sutures (Ethilon) to create a non-critical 3mm defect. Muscle and cutaneous layers were closed routinely.
Mice were euthanized by pentobarbital overdose 5 or 14 days after repair. The regenerative bridge was harvested within the conduit by transecting the proximal and distal sciatic nerve stumps 1mm from the end of the 5mm conduit. The epineurial sutures were cut and the regenerative bridge was removed from the conduit and placed in a petri dish with 1mL RPMI-1640 (Corning). The nerve was cut into 1mm pieces. The tissues were then transferred to a 50mL conical tube with 10mL of digestion buffer (3mg/mL collagenase type I (Sigma), 1mg/mL hyaluronidase (Sigma), and 0.5mL of 1mM HEPES in RPMI-1640). After 1 hour digestion in a 37°C incubator, tissues were strained through a 70μm mesh strainer (BD Biosciences) to obtain a single cell suspension. The cells were centrifuged at 300g for 10 minutes. The cell pellet was resuspended in 0.5% BSA (Sigma) in DPBS and cells were plated on a v-bottom 96-well plate (Nunc, Thermo Scientific) for antibody staining and FACS.
2.3. Isolation of SC and Macrophages from Injured Sciatic Nerve by FACS
Cells were labeled for 45 minutes at 4°C using species-specific antibodies to label macrophages and other immune cells (Table 1). Cells were washed 2 times after labeling. All wash steps were performed with 0.5% BSA in DPBS and PI was added after labeling as a viability marker. Cells were analyzed using fluorescence-activated cell sorter FACSAriaIII (BD Biosciences) and FACSDiva software (BD Biosciences version 6.1.3). The fluorochromes were excited with the instrument’s 405nm, 488nm, 532nm, and 633nm lasers. The appropriate detection filters were used (Table 2). Compensation beads (UltraComp, eBioscience) were used to set the compensation matrix. Fluorescence was determined by gating against appropriate controls (unstained, fluorescence minus one) on samples prepared in parallel. Gates were set such that less than 1% of positive events were recorded when acquiring the corresponding negative control, and were adjusted as needed to focus on discrete populations. Cells were gated for size, doublet exclusion, and viability.
Table 1:
Antibodies for flow cytometric analysis of injured sciatic nerve samples
| Antibody/ Probe |
Dilution | Fluorochrome | Distributor/Cat No. | Specificity |
|---|---|---|---|---|
| CD16/32 | 1:50 | BV-605 | BD Biosciences/563006 | Pan macrophage |
| F4/80 | 1:400 | PE-Cy7 | eBioscience/25-4801 | Pan macrophage |
| CD11b | 1:200 | Pacific Blue | BioLegend/101224 | Pan macrophage |
| CD14 | 1:100 | PE | eBioscience/12-0141 | Pan macrophage |
| Ly6G | 1:100 | APC | eBioscience/17-9668 | Neutrophils |
| Siglec F | 1:64 | APC | Miltenyi Biotec/130-102-241 | Eosinophils |
| CD19 | 1:400 | APC | eBioscience/17-0193 | B Lymphocytes |
| CD3e | 1:800 | APC | eBioscience/17-0032 | T Lymphocytes |
| Thy1.2 | 1:800 | APC | eBioscience/17/0902 | Fibroblasts |
| CD31 | 1:200 | APC | BD Biosciences/551262 | Endothelial cells |
| Ter119 | 1:200 | APC | eBioscience/17-5921 | Red blood cells |
| PI | 1:200 | eBioscience 00-6990 |
Viability |
Table 2:
Excitation and detection settings for antibodies and conjugates used. Exclusion markers were used for analysis of nerve samples.
| Antibody target | Conjugate | Excitation laser (nm) |
Emission Filters | Mirror |
|---|---|---|---|---|
| CD11b | Pacific Blue | 405 | 450/50 | |
| CD16/32 | BV605 | 405 | 585/42 | 570LP |
| eGFP | n/a | 488 | 515/20 | 505LP |
| CD14 | PE | 532 | 575/25 | |
| F4/80 | PE-Cy7 | 532 | 780/40 | 740LP |
| Propridium iodide | n/a | 532 | 660/20 | 640LP |
| Exclusion markers | APC | 633 | 660/20 |
A dump channel was used to exclude T- and B-lymphocytes (CD3e, CD19), neutrophils (Ly6G), eosinophils (Siglec-F), endothelial cells (CD31), fibroblasts (Thy1.2), and red blood cells (Ter119). Dump cells were sorted as PI− and CD3e+, CD19+, Ly6G+, Siglec-F+, CD31+, Thy1.2+, or Ter119+.
Macrophages were sorted as all viable single cells that were PI− CD3e− CD19− Ly6G− Siglec-F− CD31− Thy1.2− Ter119− F4/80+ CD14+ CD11b+ CD16/32+. Inflammatory macrophages can have reduced F4/80 expression, so if the macrophage population was close to or crossed the lower limit of the gate, co-expression of CD11b and CD16/32 were used to adjust the macrophage gate. Figure 1 shows the full gating strategy. Some samples contained a subset of cells that were F4/80hi and had intermediate fluorescent intensity in the dump channel. These cells were also positive for CD14, CD11b, and CD16/32 and were included in the macrophage gate.
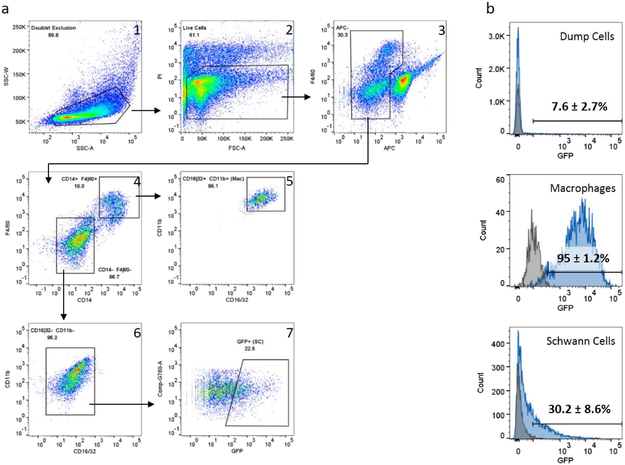
Figure 1 – Gating strategy to sort macrophages and Schwann cells from injured murine sciatic nerve:
The gating tree (a) was set as indicated in: 1) SSC-W/SSC-A (doublet exclusion which excludes cells that might be stuck together), 2) PI/FSC-A (discriminates live/dead and size to exclude debris) to 3) F4/80/APC channel (separates cell populations of interest and eliminates the dump cells by removing eosinophils (Siglec-F), lymphocytes (CD3e, CD19), neutrophils (Ly6G), endothelial cells (CD31), red blood cells (Ter119), and fibroblasts (Thy-1.2)). Macrophages were gated from gate APC-negative as follows: 4) F4/80/CD14 double positive to 5) CD11b/CD16/32 double positive. After final gating, macrophages were single viable APC-F4/80+CD14+CD11b+CD16/32+ cells. Schwann cells were gated from gate APC-negative as follows: 4) F4/80/CD14 double negative to 6) CD11b/CD16/32 double negative to 7) G780-A/GFP. After final gating, Schwann cells were identified in one of 2 ways: by exclusion (single viable APC-F4/80-CD14-CD11b-CD16/32- cells, or by GFP expression (single viable APC-F4/80-CD14-CD11b-CD16/32-GFP+). (b) GFP expression in various cell types harvested from injured sciatic nerve tissue in S100-eGFP mice. Cells from injured sciatic nerve of S100b-eGFP mice (blue) and littermate GFP negative mice (grey) were examined by flow cytometric analysis. GFP expression in three major populations of single viable cells was examined. The dump channel included eosinophils (Siglec-F), lymphocytes (CD3e, CD19), neutrophils (Ly6G), endothelial cells (CD31), red blood cells (Ter119), and fibroblasts (Thy-1.2). Macrophages were single viable Siglec-F-CD3e-CD19-Ly6G-Thy-1.2- F4/80+CD14+CD11b+CD16/32+ cells. Cells that were negative for all markers and positive for GFP were considered Schwann cells.
Schwann cells were sorted using two different methods: by exclusion of other expected cell types in C57BL/6J mice (SCexclusion), or by GFP expression in the S100b-eGFP line (SCGFP). SCexclusion was gated as PI− CD3e− CD19− Ly6G− Siglec-F− CD31− Thy1.2− Ter119− F4/80− CD14− CD11b− CD16/32−. SCGFP was gated as PI− CD3e− CD19− Ly6G− Siglec-F− CD31− Thy1.2− Ter119− F4/80− CD14− CD11b− CD16/32− GFP+. Additionally, the percent of each population (dump channel, macrophages, SCexclusion) that was GFP+ was determined using FlowJo software.
From each subject, a minimum of 10,000 and maximum of 50,000 cells were collected for each of the dump, macrophage and either SCexclusion or SCGFP gates (Figure 1). Prior to sorting, the nozzle, sheath, and sample lines were washed with RNAse Away (Ambion) for 15 minutes then flushed with preservative-free sheath solution (Biosure) for 2-3 minutes to remove RNases. A 100μm ceramic nozzle (BD Biosciences), sheath pressure of 20psi, flow rate < 3 and acquisition rate of <3000 events per second were used as conditions optimized for neuronal cell sorting as previously described (Pruszak et al., 2007). Cells were sorted into 0.5% BSA in DPBS in RNAse-free, lo-bind eppendorf tubes (Zymogen).To assure specificity, purity checks were performed by re-analyzing a subset of sorted cells and only sorts with >80% enrichment were accepted. Sorted cells were lysed with 1:1 solution of RNA lysis buffer (RLT, Qiagen) in RNase-free water (1 μL/4,000 cells, minimum 5μL, for SCexclusion) or in 1:2.5 solution of RLT buffer in DPBS (1 μL/10,000 cells, minimum 5μL, for SCGFP), and stored at −80°C until processed for gene expression analysis.
2.4. Gene Expression Analysis
To assess the SC sorting protocols, dump, macrophage, and SCexclusion cells were isolated from C57BL/6J mice 5 days (n = 3, 2 female, 1 male) and 14 days (n = 5, 2 female, 3 male) after sciatic nerve transection and repair. To determine if SCGFP improved specificity of sorting, dump, macrophage, and SCGFP cells were isolated from S100b-eGFP mice 5 days after sciatic nerve transection and repair (n=8, n=4 male and 4 female). Cells were sorted separately from each individual mouse and whole cell lysates were obtained as described in section 2.3. The cell lysate from each sort was split for analysis on two gene expression panels.
Custom panels of bar-coded probes (nanoString Technologies, Seattle, WA) were designed specific for genes associated with SC and macrophage lineage, phenotype and function (SC and macrophage codesets, 101 genes each, Supplemental Information - Table S1) (Fortina and Surrey, 2008; Geiss et al., 2008; Tomlinson et al., 2018). Samples were analyzed at the Molecular Biology Core Facility at Geisel School of Medicine at Dartmouth, Hanover, NH. NanoString nSolver 4.0 Analysis Software (nanoString Technologies) was used to process raw data. Data from each codeset were processed separately. Background was calculated on the raw data as the average mRNA count across all negative controls plus twice the standard deviation of the negative controls. Genes with expression below background were excluded from further analysis. Six possible housekeeping genes were included on each panel and the genes that were the most stable across samples, including expression above background in all samples and minimal %CV across samples, were used for normalization as previously described (Tomlinson et al., 2018). Data was normalized using the positive control and housekeeping genes. Normalized data were analyzed.
Hierarchical clustering and principal component analysis were performed using JMP Pro 12 software (SAS Institute Inc., Cary, NC) to visualize expression patterns among sorted populations (dump, macrophages, SC), days after injury, and sex. Based on clustering analysis day and sex did not clearly affected separation between the populations, therefore data from day 5 and 14 and both sexes were combined for further analysis and only the single day 5 timepoint was performed for the SCGFP experiment.
Data from each codeset were separately assessed by least square means analysis with Benjamini and Hochberg false discovery rate correction and Tukey post-hoc test to examine expression differences in individual genes among the sorted populations. Log transformation was used to meet the assumptions of the test. Significance was set as p<0.05 throughout. Gene expression data were represented graphically using GraphPad Prism 7 (GraphPad software, Inc) as the median and interquartile range of normalized mRNA count for each group.
2.5. eGFP Expression in Bone Marrow-Derived Macrophages (BMDM)
During SCGFP sorting, GFP expression in macrophages was observed. To further assess leakiness of eGFP expression in macrophages of the S100b-eGFP line, bone marrow was harvested from S100b-eGFP (n=3) and C57BL/6J (n=1) mice. For each mouse, 500,000 cells were seeded onto 3×100mm sterile polystyrene petri dishes (Falcon) with 12mL R10 cell culture medium (RPMI 1640 (Corning) with 10% FBS (ThermoFischer), 1% Penicillin/Streptomycin (ThermoFischer), and 1% HEPES (Sigma Aldrich)). Cells were stimulated to differentiate into macrophages with 10ng/mL macrophage colony stimulating factor (M-CSF, eBioscience). Cells were cultured at 37°C and 5% CO2 for 8 days and were fed every third day.
Ligands known to produce particular extremes of the macrophage polarization spectrum were added to examine eGFP expression in different activation states. Polarization was performed as previously described (Tomlinson et al., 2018), briefly no additional cytokines for M(−); 50ng/mL IFNγ (eBioscience) on day 6 followed by 100ng/mL LPS (Sigma-Aldrich) and 50ng/ml IFNγ (eBioscience) on day 7 for M(IFNγ + LPS) polarization; and 50ng/mL IL-4 on day 7 for M(IL4) polarization. Cells were harvested from the plates on day 8 by incubation with PBS for 30 minutes followed by vigorous trituration.
Bone marrow derived macrophages were labeled with the pan-macrophage marker anti-mouse F4/80 (eBioscience 25-4801) at 1:500 dilution for flow cytometric analysis. Dilutions were determined by titration. Cells were incubated with anti-mouse F4/80 for 45 minutes at 4°C in the dark; washing steps were performed with 0.5% BSA in DPBS. After labeling, cells were resuspended in 0.5% BSA with propidium iodide (PI, eBioscience 00-6990) as a viability marker. Labeled cells were analyzed with FACS Aria III (BD Biosciences) using FACS Diva software (BD Biosciences, version 6.1.3) and final analysis was performed with FlowJo software (TreeStar v10). Fluorochromes were excited with the instrument’s 488nm and 532nm lasers. The appropriate detection filters were used (Table 2). Compensation beads (UltraComp, eBioscience) were used to set the compensation matrix. Fluorescence was determined by gating against appropriate controls (unstained, fluorescence minus one) on samples prepared in parallel. Fully labeled cells from C57BL/6J mice were used for fluorescence minus eGFP. Gates were set such that less than 1% positive events were recorded when acquiring the corresponding negative control. Cells were gated on forward and side scatter for general cell size, forward scatter height and width to exclude doublets, and side scatter and PI to exclude dead cells. Macrophages were defined as all viable single cells that were F4/80+. Percent of macrophages that were GFP+ was calculated for each sample using FlowJo software.
3. RESULTS
3.1. Schwann Cell Isolation by Exclusion of Other Cell Types
Many SC markers, such as S100b and Sox10, are intracellular proteins and require fixation of cells to use them for FACS. Commonly used surface markers, such as p75 neurotrophin receptor (p75-NTR) or Necl1, limit collection to phenotypic subsets such as immature or mature myelinating SC, respectively (Spiegel and Peles, 2009; Stratton et al., 2018). As an alternative that would allow collection of live cells across the spectrum of SC phenotypes, we identified presumptive SC solely by exclusion of other expected cell types (SCexclusion, Figure 1). To assess this method, we used a sciatic nerve transection and empty conduit repair model of injury. After 5 or 14 days of repair, the nerve bridges were harvested and digested, and cells were separated and analyzed by flow cytometry. Dump cells (dump+ = CD3e+, CD19+, Ly6G+, Siglec-F+, CD31+, Thy1.2+, or Ter119+), macrophages (dump− F4/80+ CD14+ CD11b+ CD16/32+), and SCexclusion (cells that were negative for all other markers) were sorted. The SCexclusion population represented 10.4% (6.2-18.2%) of viable single cells in both time points.
Gene expression analysis was performed using nanoString technologies (Geiss et al., 2008; Guerrero et al., 2014) and two custom gene panels (codesets); one focusing on SC markers and one for macrophage markers (Table S1). Sorted cells of individual mice were independently analyzed by nanoString and compared. Gapdh, Actb and Rictor were used as housekeeping genes for the macrophage codeset; and Gapdh and NSE were used as housekeeping genes for the SC codeset. Three samples had content normalization factors >10 (range 10.24 – 20.5) on each codeset, indicating low RNA input and were excluded from further analysis (Supplemental Information - Table S2). The cells collected for these samples were 29,000 – 50,000; however there were samples as low as 10,000 cells with adequate RNA.
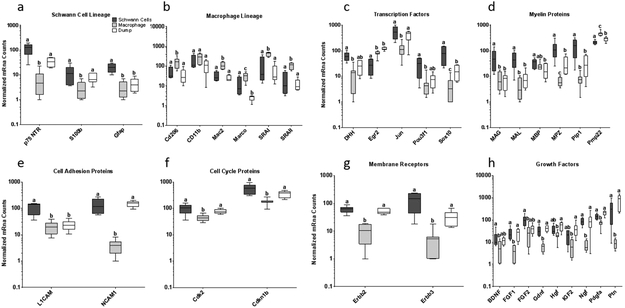
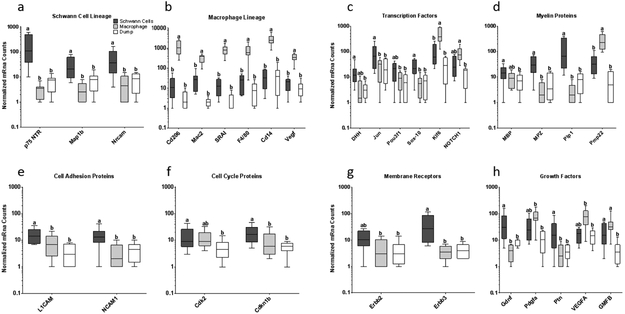
PCA was used to compare gene expression by population and time after injury. Samples clustered more closely by population than by time after injury (Supplemental Information - Figure S1a,c). Because of this, least square means analysis was applied to the pooled data from days 5 and 14. The SCexclusion population demonstrated enrichment of SC related genes compared to the sorted macrophage population; however, separation of the SCexclusion population from the dump population was less consistent. The SC lineage markers p75-NTR and S100b were enriched compared to macrophages, but not dump cells (Figure 2a, p-value<0.05), while Gfap was enriched compared to both macrophages and dump cells. The SCexclusion population had higher Sox10 expression, a master regulator of SC lineage (Britsch et al., 2001), than either the dump or macrophage population (Figure 2c, p-value<0.001). The clearest separation of the SCexclusion population from the dump population was in expression of myelin protein genes (Figure 2d). The macrophage population showed enrichment of macrophage associated genes including Cd206, SRAI, Vegf and Mac2, as expected (Figure 2b, p-value<0.01).
Figure 2 – Schwann cells can be enriched from injured wild type murine sciatic nerve by FACS with exclusion of other cell types:
Gene expression among 3 populations of cells sorted from injured murine sciatic nerve 5 or 14 days after injury. The Dump population included eosinophils (Siglec-F), lymphocytes (CD3e, CD19), neutrophils (Ly6G), endothelial cells (CD31), red blood cells (Ter119), and fibroblasts (Thy-1.2). Macrophages were F4/80+CD14+CD11b+CD16/32+. Presumptive Schwann cells were sorted by excluding cells positive for all of the above markers. Schwann cell lineage markers (a) and other genes associated with Schwann cell function and phenotype (c-g) were enriched in the Schwann cell sort and macrophage associated genes were enriched in the macrophage sort (b). Growth factors relevant to peripheral nerve regeneration had lowest expression in macrophages (h). Least squares model was fit to log transformed mRNA count data and FDR screening was applied. Significance was determined by Tukey post-hoc analysis. Samples with the same Tukey letter are not significantly different.
3.2. Positive Identification of SC by GFP Expression in S100b-eGFP Mice is Complicated by GFP Expression in Macrophages
In the SC isolation by exclusion method described above, it was noted that separation of SC from the dump channel was perhaps not as clear as desired. This could have been because some non-SC were included in the SCexclusion gate. To improve accuracy of the sort, we added positive identification of SC by utilizing transgenic S100b-eGFP mice, which were described to express GFP only in Schwann cells (Zuo, 2004). The same nerve injury model and flow cytometric analysis were applied. Flow cytometric analysis demonstrated GFP expression in 30.2 ± 8.6% (mean ± sd) of the SCexclusion gate (Figure 1b). This GFP expressing population was designated SCGFP and represented a median of 4.1% (range 2.4 – 4.6%) of all viable single cells. However, 95.0 ± 1.2% of macrophages and 7.6 ± 2.7% of dump cells were also GFP positive.
While leaky expression of GFP in macrophages had been reported in some other S100b-eGFP or S100b-YFP lines, it had not been reported in this particular line to the degree observed here (Zuo, 2004).To further assess the finding that macrophages in this mouse line expressed GFP, we analyzed GFP expression in a separate macrophage population, namely bone marrow derived macrophages (BMDM). BMDM expressed GFP under multiple stimulation conditions: M(−), M(IL-4), and M(IFNγ, LPS) (Supplemental Information - Figure S2), indicating macrophage GFP expression in S100b-eGFP mice was not dependent on the local environment of nerve injury or inflammation.
3.3. Schwann Cells and Macrophages Can be Separated from Other Cells in Injured Nerve of S100b-eGFP Mice by FACS
GFP expression in cells other than SC indicated that the S100b-eGFP line could not be used to sort SC by single marker expression. Therefore, the same antibody panel as was used for SCexclusion sorting was applied for the SCGFP sort. The same dump and macrophage gates were used and an additional gate for GFP+ was applied to the prior SCexclusion gate to sort SCGFP cells (Figure 1). Collection and analysis were performed as before. In this experiment, Gapdh and Actb were used as housekeeping genes for the macrophage codeset. Gapdh and Hprt were used as housekeeping genes for the SC codeset. The cells collected for these samples were 20,000 – 50,000 with adequate RNA for each sample. Therefore, no sample had to be excluded from analysis (Supplemental Information - Table S3).
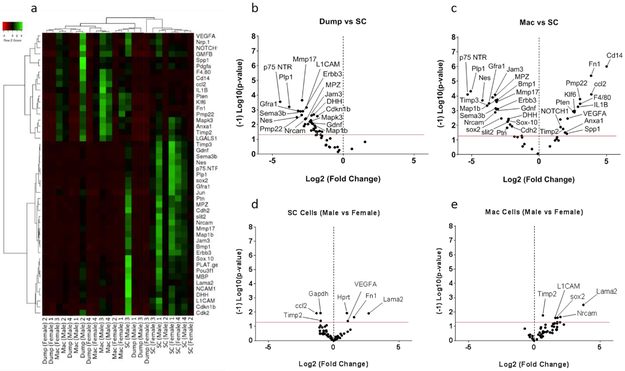
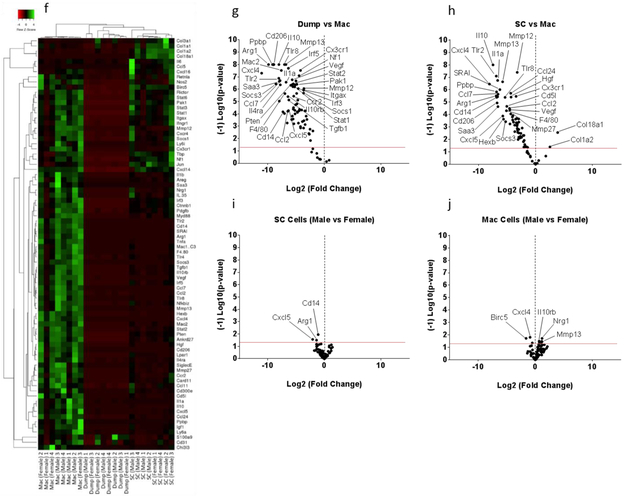
Clustering analysis was performed for both SC (Figure 3a) and macrophage (Figure 3f) codesets. The results indicated distinct clustering by sorted population. Principal component analysis (Supplemental Information - Figure S1b, d) and gene expression comparison (Figure 3d-e and i-j) showed no specific clustering or pattern of gene expression differences in each population by sex. The SCGFP population demonstrated enrichment of SC related genes compared to both the sorted macrophage and dump populations (Figure 3b, c). The macrophage population was appropriately enriched in macrophages, as demonstrated by the higher expression of macrophage specific genes compared to both SC and dump populations (Figure 3g, h).
Figure 3 – Schwann cells and macrophages can be enriched from injured S100-eGFP murine sciatic nerve by FACS:
Clustered gene expression among 3 cell populations from injured murine sciatic nerve 5 days after injury (n=8, 4 male, 4 female) analyzed with NanoString Schwann cell codeset (a-e) and macrophage codeset (f-j). Genes expressed above background in at least one population are analyzed, and the p-values for NanoString data were obtained from nSolver software (p-value < 0.05, red lines). Clear separation of Schwann cells from the macrophage and dump populations is identified by SC (a-c) and macrophage (f-h) lineage markers. Clustering of p75-NTR, Sox 10, Map1b, Nrcam and MPZ genes in isolated SC using eGFP signal compared to macrophages and dump cell population demonstrates the effectiveness of the method. The clustering of the Cd206, Vegf and F4/80 genes in isolated macrophages indicates macrophage enrichment using macrophage specific markers (F4/80, CD14, CD11b, CD16/32). No significant sex effect was detected (d-e and i-j).
Expression of selected genes in the sorted populations is shown in more detail in Figure 4, grouped by gene function. Common SC lineage markers, p75-NTR, Map1b and Nrcam, were expressed at significantly higher levels than in macrophage and dump populations (Figure 4a, p-values<0.0002). In contrast, S100b and Gfap were not consistently expressed above the background of the assay (median and range of 3.37 and 2.37-10 for S100b, and 7 and 4.37-7.62 for Gfap) and comparison between populations could not be made. As with the SCexclusion sort, Sox10 had higher expression in the SCGFP group than either of the other two populations (Figure 4c, p-value<0.002). As expected, the macrophage population showed enrichment of macrophage associated genes including Cd206, SRAI, Vegf, F4/80, Cd14 and Mac2 (Figure 4b, p-values<0.001).
Figure 4 – Schwann cell and macrophage enrichment indicated by cell lineage markers using S100-eGFP mice:
Gene expression among 3 populations of cells sorted from injured murine sciatic nerve 5 days after injury. The Dump population included eosinophils (Siglec-F), lymphocytes (CD3e, CD19), neutrophils (Ly6G), endothelial cells (CD31), red blood cells (Ter119), and fibroblasts (Thy-1.2). Macrophages were F4/80+CD14+CD11b+CD16/32+. Schwann cells were sorted by discarding the cells positive for all of the above markers and including the cells positive for GFP. Schwann cell lineage markers (a) and other genes associated with Schwann cell function and phenotype (c-g) were enriched in the Schwann cell sort and macrophage associated genes were enriched in the macrophage sort (b). Higher expression of p75-NTR, Map1b and Nrcam genes in SC population compare to macrophages and dump, and higher expression of Cd206, Mac2, SRAI, F4/80 and Vegf in macrophages compare to SC and dump indicate an effective separation of SC and macrophages from other cell types. Significance was determined by Tukey post-hoc analysis using p-value < 0.05 (n=8 mice). Samples with the same Tukey letter are not significantly different.
4. Discussion
This study describes two methods for identifying and isolating SC directly from murine peripheral nerve. Most published methods for sorting SC utilize an ex vivo culture step prior to sorting (Maximino et al., 2014; Spiegel and Peles, 2009; Vroemen and Weidner, 2003). These methods are not useful for gene expression analysis because the time in culture likely results in significant gene expression changes that could render results unrepresentative of in vivo expression (Lin et al., 2006; Liu et al., 2015). Magnetic column separation is sometimes used after culture (Masaki et al., 2013; Vroemen and Weidner, 2003); however, this only allows selection based on a single marker and does not give the user visualization of the positive and negative populations to evaluate the validity of the cut-point for separation. Our method allows robust multispectral analysis and visualization in addition to cell sorting.
We attained a reproducible and selective enrichment of SC from other cell populations (macrophages and dump cells) using a FACS method by identifying the GFP+ cells and excluding other cell types. An alternative SC enrichment was also possible by excluding the macrophage and dump populations, and not using a positive selection marker for SC. It is important to note that both methods still required the use of exclusion markers for macrophages at a minimum, and that the S100b-eGFP mouse line cannot be used to isolate SC without additional antibody labeling.
The two methods described here are designed to include SC of multiple phenotypes. While p75-NTR is an available surface marker for SC, its expriosion is primarily limited to immature SC in the healthy adult nervous system, and p75-NTR expression over the course of injury and repair is likely dynamic (Corfas et al., 2004; Jessen and Mirsky, 2005; Meeker and Williams, 2014; Stratton et al., 2018; Taniuchi et al., 1986). The SCexclusion method was designed to be the most inclusive and enrich for all types of SC, while acknowledging that there is likely some contamination with non-SC in the sorted population. The SCGFP method used the S100b marker, which is predicted to be expressed in all adult SC phenotypes (Corfas et al., 2004; Donato et al., 2013; Raponi et al., 2007). Despite this, S100b was not highly expressed in the SC sorted populations. It could be that gene expression is not highly correlated to protein expression under these conditions, or that S100b transcription is downregulated at day 5 after injury. An advantage of using the S100b-eGFP transgenic mouse line over antibody labeling of the SC could be that GFP protein persists after S100b transcription is reduced, allowing continued detection of the SC after injury.
Our data demonstrate that the SCGFP sort improved separation between the dump and SC populations compared to the SCexclusion sort. GFP only identified 30% of the SCexclusion population as SC. It is therefore possible the SCexclusion population included more non-SC, which reduced the pooled gene expression of SC markers toward that of the dump population. Alternatively, the GFP expression was not consistent across all SC phenotypes and the SCGFP sort only identified a subset of SC. However, both markers of myelinating SC (e.g. MPZ, Plp1) and non-myelinating or de-differentiating SC (e.g. c-Jun, L1CAM, p75-NTR) were enriched in the SCGFP sort, suggesting both phenotypes were captured (Jessen and Mirsky, 2008; Martini, 1994; Nieke and Schachner, 1985). Without additional study on an individual cell level, however, we were not able to confirm that the SCGFP population did indeed include all SC phenotypes in the tissue.
The gene expression analysis indicates that separation of SC, macrophage and dump cells is independent of harvest timepoint and subject sex; demonstrating that this method can be used across multiple study designs. In this study, we used a nerve transection model with a non-critical gap repair inside an inert conduit. This experimental approach allowed collection of tissue samples at each timepoint after repair and will also allow the addition of novel immunomodulatory biomaterials to manipulate the microenvironment at the repair site in future work. . At the timepoints examined, 5 and 14 days, developing regenerative bridges were identified in all samples. This approach could be similarly applied to other types of nerve injury including crush, transection and repair and critical length gaps as the method provides excellent sorting of macrophage and SC from the mixed cell populations found at the repair site.
S100b is generally considered a SC specific gene (Donato et al., 2009), and the human S100b promoter in this line’s transgene construct was predicted to result in SC specific eGFP expression. Here we document widespread expression in macrophages as well. GFP was expressed in macrophages independent of phenotype, and was present in the majority of macrophages at the site of peripheral nerve injury. This is in contrast to a prior report of a mouse cross-bred with this S100b-eGFP line where only 9% of GFP+ cells were macrophages (Hayashi et al., 2007). It is a well-known phenomenon that insertion of transgenes can result in ectopic tissue expression, or expression in tissues not predicted by the endogenous expression of the chosen promoter sequence (Brem, 1993; Gabriel et al., 2015; Haruyama et al., 2009; Rossert et al., 1995). In the paper describing the generation of the S100b-eGFP line, there were 7 other lines generated with YFP under the same promoter construct and those lines also expressed fluorescent protein in tissue macrophages (Zuo, 2004). While Zuo and Hayashi et al specifically examined GFP expression in macrophages (Hayashi et al., 2007; Zuo, 2004), more commonly, S100b is used as a SC marker without co-labeling macrophages (Cattin et al., 2015; Dubový et al., 2014; Whitlock et al., 2010). However, our gene expression panels here and RNA sequencing in our prior work (Tomlinson et al., 2018) showed no detectable S100b expression in the macrophages. Additionally, publicly available RNA sequencing data of murine macrophages isolated from peripheral nerve on day 3 and day 8 show S100b FPKM ranging from 0.1 to 5.8, similarly indicating a very low expression in all samples (Stratton et al., 2018).
Therefore, GFP expression in macrophages of the S100b-eGFP line is likely explained by ectopic expression rather than endogenous S100b expression in these macrophages. Our findings reinforce the need to validate key biologic factors in experiments and to utilize redundancy in cell identification methods.
5. Conclusion
Here we present two techniques to enrich SC from other cell populations at the site of peripheral nerve injury. These results indicate that enrichment of SC population is possible using exclusion of other cell types (SCexclusion) and is likely improved by using the GFP signal in S100b-eGFP line (SCGFP). The S100b-eGFP line described here expresses eGFP in SC of the peripheral nervous system and in myeloid-derived macrophages. When using this line of mice in the context of peripheral nerve injury, it is essential to include markers to distinguish between SC and macrophages. Using the markers and the process reported here, live SC and macrophages can be rapidly enriched for further analysis. These methods provide robust techniques for ex vivo isolation of the key cellular contributors to peripheral nerve repair.
Supplementary Material
Highlights.
Schwann cells (SC) and macrophages play key roles in the aftermath of peripheral nerve injury (PNI).
This paper described two methods for accurate separation of live Schwann cells and macrophages from injured murine nerve.
The two different techniques presented in this study provide robust method to isolate SC and macrophages from other cell populations.
The gene expression analysis shows successful enrichment of SC and macrophages in both methods by evaluating the lineage markers such as Nrcam, Map1b, p75 NTR and S100b for SC and Cd206, Mac2 and Vegf for macrophages.
Acknowledgements:
Both Joy E. Tomlinson and Masoud Golshadi are listed as the first author. They both have contributed equally to conduct the experiments. Tomlinson, Golshadi and Cheetham have all designed research, performed research, contributed unpublished reagents/analytic tools, analyzed data and wrote the paper. Donahue and Dong performed research. The authors also want to thank Tim Moore and Megan Bernard for their help.
Funding:
This work was supported by National Institute of Deafness and Communication Disorders (1R01DC017171-01)
Footnotes
Declaration of Interest:
The authors have no conflicts of interests to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brem G, 1993. Inheritance and tissue-specific expression of transgenes in rabbits and pigs. Mol. Reprod. Dev 36, 242–4. 10.1002/mrd.1080360220 [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M, 2001. The transcription factor SoxlO is a key regulator of peripheral glial development. Genes Dev. 15, 66–78. 10.1101/gad.186601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SL, Miller ML, Frohnert PW, Kim SS, Corbett J. a, 1997. Expression of neuregulins and their putative receptors, ErbB2 and ErbB3, is induced during Wallerian degeneration. J. Neurosci 17, 1642–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin A-L, Burden JJ, Van Emmenis L, Mackenzie FE, Hoving JJA, Garcia Calavia N, Guo Y, McLaughlin M, Rosenberg LH, Quereda V, Jamecna D, Napoli I, Parrinello S, Enver T, Ruhrberg C, Lloyd AC, 2015. Macrophage-Induced Blood Vessels Guide Schwann Cell-Mediated Regeneration of Peripheral Nerves. Cell 162,1127–1139. 10.1016/j.cell.2015.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfas G, Velardez MOO, Ko C, Ratner N, Peles E, 2004. Mechanisms and Roles of Axon – Schwann Cell Interactions. October 24, 9250–9260. 10.1523/JNEUROSCI.3649-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL, 2013. Functions of S100 proteins. Curr Mol Med 13, 24–57. [PMC free article] [PubMed] [Google Scholar]
- Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, Brozzi F, Tubaro C, Giambanco I, 2009. S100B ’ s double life : Intracellular regulator and extracellular signal. BBA - Mol. Cell Res 1793, 1008–1022. 10.1016/j.bbamcr.2008.11.009 [DOI] [PubMed] [Google Scholar]
- Dubový P, Klusáková I, Hradilová Svíženská I, 2014. Inflammatory profiling of Schwann cells in contact with growing axons distal to nerve injury. Biomed Res. Int 2014, 691041 10.1155/2014/691041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortina P, Surrey S, 2008. Digital mRNA profiling. Nat. Biotechnol 26, 293–294. 10.1038/nbt0308-293 [DOI] [PubMed] [Google Scholar]
- Gabriel R, Erdelyi F, Szabo G, Lawrence JJ, Wilhelm M, 2015. Ectopic transgene expression in the retina of four transgenic mouse lines. Brain Struct. Funct 10.1007/s00429-015-1128-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol 26, 317–325. 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- Godinho MJ, Teh L, Pollett MA, Goodman D, Hodgetts SI, Sweetman I, Walters M, Verhaagen J, Plant GW, Harvey AR, 2013. Immunohistochemical, Ultrastructural and Functional Analysis of Axonal Regeneration through Peripheral Nerve Grafts Containing Schwann Cells Expressing BDNF, CNTF or NT3. PLoS One 8 10.1371/journal.pone.0069987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero AD, Dong MB, Zhao Y, Lau-Kilby A, Tarbell KV, 2014. IL-2-mediated inhibition of DC development correlates with decreased flt3 expression and increased monocyte-macrophage precursors. Immunology 3, Epub. 10.1111/imm.12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruyama N, Cho A, Kulkarni AB, 2009. Overview: engineering transgenic constructs and mice. Curr. Protoc. Cell Biol Chapter 19, Unit 19.10. 10.1002/0471143030.cb1910s42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Koob JW, Liu DZ, Tong AY, Hunter D. a, Parsadanian A, Mackinnon SE, Myckatyn TM, 2007. A double-transgenic mouse used to track migrating Schwann cells and regenerating axons following engraftment of injured nerves. Exp. Neurol 207, 128–38. 10.1016/j.expneurol.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R, 2008. Negative regulation of myelination: Relevance for development, injury, and demyelinating disease. Glia 56, 1552–1565. 10.1002/glia.20761 [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R, 2005. The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci 6, 671–682. 10.1038/nrn1746 [DOI] [PubMed] [Google Scholar]
- Laranjeira C, Sandgren K, Kessaris N, Richardson W, Potocnik A, Berghe P. Vanden, Pachnis V, 2011. Glial cells in the ENS. J Clin Invest 121 10.1172/JCI58200DS1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DW, Coleman IM, Hawley S, Dumpit R, Gifford D, Kezele P, Hung H, Knudsen BS, Kristal AR, Nelson PS, 2006. Influence of surgical manipulation on prostate gene expression: Implications for molecular correlates of treatment effects and disease prognosis. J. Clin. Oncol 24, 3763–3770. 10.1200/JCO.2005.05.1458 [DOI] [PubMed] [Google Scholar]
- Liu Z, Jin Y-Q, Chen L, Wang Y, Yang X, Cheng J, Wu W, Qi Z, Shen Z, 2015. Specific marker expression and cell state of Schwann cells during culture in vitro. PLoS One 10, e0123278 10.1371/journal.pone.0123278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon BS, Shick HE, Kidd GJ, Macklin WB, 2002. Proteolipid promoter activity distinguishes two populations of NG2-positive cells throughout neonatal cortical development. J. Neurosci 22, 876–885. https://doi.org/22/3/876 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini R, 1994. Expression and functional roles of neural cell surface molecules and extracellular matrix components during development and regeneration of peripheral nerves. J. Neurocytol 23, 1–28. [DOI] [PubMed] [Google Scholar]
- Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A, 2013. Reprogramming Adult Schwann Cells to Stem Cell-like Cells by Leprosy Bacilli Promotes Dissemination of Infection. Cell 152, 51–67. 10.1016/j.cell.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximino JR, de Oliveira GP, Alves CJ, Chadi G, 2014. Deregulated expression of cytoskeleton related genes in the spinal cord and sciatic nerve of presymptomatic S0D1(G93A) Amyotrophic Lateral Sclerosis mouse model. Front. Cell. Neurosci 8, 148 10.3389/fncel.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker R, Williams K, 2014. Dynamic nature of the p75 neurotrophin receptor in response to injury and disease. J. Neuroimmune Pharmacol 9, 615–28. 10.1007/s11481-014-9566-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky R, Woodhoo A, Parkinson DB, Arthur-farraj P, 2008. Novel signals controlling embryonic Schwann cell development, myelination and dedifferentiation 135, 122–135. [DOI] [PubMed] [Google Scholar]
- Mokarram N, Merchant A, Mukhatyar V, Patel G, Bellamkonda RV, 2012. Effect of modulating macrophage phenotype on peripheral nerve repair. Biomaterials 33, 8793–8801. 10.1016/j.biomaterials.2012.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieke J, Schachner M, 1985. Expression of the neural cell adhesion molecules L1 and N-CAM and their common carbohydrate epitope L2/HNK-1 during development and after transection of the mouse sciatic nerve. Differentiation. 30, 141–51. [DOI] [PubMed] [Google Scholar]
- Pruszak J, Sonntag K-C, Aung MH, Sanchez-Pernaute R, Isacson O, 2007. Markers and Methods for Cell Sorting of Human Embryonic Stem Cell-Derived Neural Cell Populations 25, 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C, Deloulme J, 2007. S100B Expression Defines a State in Which GFAP- Expressing Cells Lose Their Neural Stem Cell Potential and Acquire a More Mature Developmental Stage 177, 165–177. 10.1002/glia [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SL, Dun X, Doddrell RDS, Mindos T, Drake LK, Onaitis MW, Florio F, Quattrini A, Lloyd AC, D’Antonio M, Parkinson DB, 2017. Sox2 expression in Schwann cells inhibits myelination in vivo and induces influx of macrophages to the nerve Journal of Cell Science, 10.1242/jcs.210351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossert J, Eberspaecher H, de Crombrugghe B, 1995. Separate cis-acting DNA elements of the mouse pro-alpha 1(1) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol 129, 1421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Peles E, 2009. A novel method for isolating schwann cells using the extracellular domain of Necl1. J. Neurosci. Res 87, 3288–3296. 10.1002/jnr.21985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton JA, Holmes A, Rosin NL, Sinha S, Vohra M, Burma NE, Trang T, Midha R, Biernaskie J, 2018. Macrophages Regulate Schwann Cell Maturation after Nerve Injury. Cell Rep. 24, 2561–2572.e6. 10.1016/j.celrep.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Taniuchi M, Clark HB, Johnson EM, 1986. Induction of nerve growth factor receptor in Schwann cells after axotomy. Proc. Natl. Acad. Sci. U. S. A 83, 4094–8. 10.1073/pnas.83.11.4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JE, Žygelyte E, Grenier JK, Edwards MG, Cheetham J, 2018. Temporal changes in macrophage phenotype after peripheral nerve injury. J. Neuroinflammation 15 10.1186/s12974-018-1219-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives V, Alonso G, Solal AC, Joubert D, Legraverend C, 2003. Visualization of S100B-Positive Neurons and Glia in the Central Nervous System of EGFP Transgenic Mice. J. Comp. Neurol 457, 404–419. 10.1002/cne.10552 [DOI] [PubMed] [Google Scholar]
- Vroemen M, Weidner N, 2003. Purification of Schwann cells by selection of p75 low affinity nerve growth factor receptor expressing cells from adult peripheral nerve. J. Neurosci. Methods 124, 135–143. 10.1016/S0165-0270(02)00382-5 [DOI] [PubMed] [Google Scholar]
- Wang G, Ma Z, Cao L, Yan G, Wang Y, Jin Y, Shen H, Zhang Y, Xu X, Chen X, Shen Z, 2017. A novel method for obtaining highly enriched Schwann cell populations from mature monkey nerves based on in vitro pre-degeneration. Mol. Med. Rep 16, 6600–6607. 10.3892/mmr.2017.7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock EL, Myckatyn TM, Tong AY, Yee A, Yan Y, Magill CK, Johnson PJ, Mackinnon SE, 2010. Dynamic quantification of host Schwann cell migration into peripheral nerve allografts. Exp. Neurol 225, 310–9. 10.1016/j.expneurol.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, 2004. Fluorescent Proteins Expressed in Mouse Transgenic Lines Mark Subsets of Glia, Neurons, Macrophages, and Dendritic Cells for Vital Examination. J. Neurosci. 24, 10999–11009. 10.1523/JNEUROSCI.3934-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žygelytė E, Bernard ME, Tomlinson JE, Martin MJ, Terhorst A, Bradford HE, Lundquist SA, Sledzonia M, Cheetham J, 2016. RetroDISCO: Clearing technique to improve quantification of retrograde labeled motor neurons of intact mouse spinal cords. J. Neurosci. Methods 271 10.1016/j.jneumeth.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.