Abstract
Although antiretroviral therapy (ART) is highly effective at suppressing HIV-1 replication, the virus persists as a latent reservoir in resting CD4+ T cells during therapy. This reservoir forms even when ART is initiated early after infection, but the dynamics of its formation are largely unknown. The viral reservoirs of individuals who initiate ART in chronic infection are generally larger and genetically more diverse than those of individuals who initiate therapy in acute infection, consistent with the hypothesis that the reservoir is formed continuously throughout untreated infection. To determine when viruses enter the latent reservoir, we compared sequences of replication-competent viruses from resting CD4+ T cells from nine women on therapy to viral sequences circulating in blood collected longitudinally prior to therapy. We found that, on average, 71% of the unique viruses from the latent reservoir were most genetically similar to viruses replicating just prior to ART initiation. This proportion is far greater than would be expected if the reservoir formed continuously and was always long-lived. We conclude that ART alters the host environment in a way that allows the formation or stabilization of a majority of the long-lived latent HIV-1 reservoir.
One Sentence Summary:
Most of the long-lived, replication-competent HIV-1 reservoir is formed near the time of therapy initiation.
Introduction
Infection with human immunodeficiency virus type 1 (HIV-1) results in active viral replication in the face of the host immune response, eventually leading to the loss of its primary target cell for replication, CD4+ T cells, and immunodeficiency. The use of multiple potent antiviral drugs stops viral replication and disease progression. However, discontinuation of antiretroviral therapy (ART) results in the rapid rebound of virus, indicating that while therapy suppresses viral replication, HIV-1 is able to persist in an infectious state for years. The best characterized reservoir in individuals on ART is integrated viral DNA in resting memory CD4+ T cells (1–3). One quantitative measure of the reservoir is the number of resting CD4+ T cells that can be induced to produce replication-competent virus after stimulation of the cells in culture, called the quantitative viral outgrowth assay (QVOA). Using this assay, it has been estimated that in people on therapy approximately one in a million resting CD4+ T cells can be induced to produce replication-competent virus, and that this latent reservoir has a half-life of 44 months (4, 5). Given the large number of resting CD4+ T cells in the body, it is impossible to cure HIV-1 by waiting for the infected cells to decay. In addition, clonal expansion of latently infected T cells adds another mechanism for persistence of virus in the body over time (6–13).
In people on ART, greater than 90% of the proviral genomes in resting CD4+ T cells are defective (14, 15). These defective genomes may contribute to continued immune activation and exhaustion (16, 17), but are not the source of the rebound virus if ART is stopped. In contrast, most intact proviruses are theoretically capable of producing replication-competent virus, but the frequency of cells harboring intact proviruses is approximately 30 times higher than the frequency of cells that can be induced to produce virus in a QVOA (15). In addition, the number of resting CD4+ T cells producing outgrowth viruses in QVOA increases with extra rounds of cell stimulation (14), indicating that the typical QVOA using a single round of stimulation underestimates the number of inducible proviruses. Taken together, these results imply that the reservoir of replication-competent proviruses is larger than that measured by standard QVOA. It is currently unknown whether the discrepancy arises because virus expression and outgrowth from resting CD4+ T cells is a noisy stochastic process, or because latency is generated by multiple mechanisms, some of which are not readily reversed in a standard QVOA, or both.
The most widely accepted model of how the reservoir forms involves the infection of a CD4+ T cell as it is transitioning to a resting state (18). However, little is known about when this happens during the course of infection. The reservoir is established even when ART is initiated early (19–21), i.e. virus rebounds with the subsequent discontinuation of therapy even when starting ART soon (e.g. within days) after infection. This suggests that there is early and continuous formation of the reservoir during the period prior to therapy initiation. However, studies of viral DNA have yielded conflicting data about the timing of reservoir formation. One report claimed evidence of continuous introduction of viral DNA into the long-lived reservoir (22), while another report found that most of the viral DNA in the reservoir comes from virus replicating around the time of therapy initiation (23). The timing of reservoir formation is an important question since an understanding of when the reservoir forms could provide new opportunities for limiting its formation. In this study we perform the first quantitative analysis aimed at understanding when replication-competent virus enters the long-lived latent reservoir.
We compared sequences of replication-competent viruses induced from on-ART resting CD4+ T cells (outgrowth viruses, OGVs) to viral sequences in blood collected longitudinally pre-ART. We found that, on average, 71% of OGVs were most genetically similar to viruses replicating just prior to therapy initiation. This proportion is far greater than would be expected if the reservoir were formed continuously. Our results indicate that the initiation of ART alters the host environment to promote latency and/or dramatically extends the half-life of latently infected cells, suggesting new approaches that could be employed at the time of therapy initiation to block formation of a majority of the long-lived CD4+ T cell reservoir.
Results
Cohort and source of viral sequences for the analysis of the latent reservoir
We investigated when replication-competent viruses enter the reservoir by examining viruses induced to replicate from resting CD4+ T cells in the context of QVOA. OGVs were isolated from blood-derived cells of nine women on ART. The women were participants in the CAPRISA 002 cohort, based in KwaZulu-Natal, South Africa, who were originally enrolled into the cohort during acute/primary HIV-1 infection (24). QVOA measures the replication-competent viral reservoir and estimates the number of resting (CD25low, CD69-, HLA-DR-) CD4+ T cells that can produce replication-competent virus following stimulation. This provides a representation of viruses capable of rebound upon ART interruption as opposed to assays that evaluate total viral DNA genomes, the majority of which cannot give rise to replicating virus. The women were ART-naïve for an average of 4.5 years. ART was initiated in accordance with national guidelines in place at the time. The participants had been on suppressive ART for an average of 5.0 years when blood samples were collected for QVOA (Table 1, Fig. 1). Plasma-derived viral RNA genomes were sequenced at multiple time points pre-ART (on average every six months from acute/early infection (Supplementary Data Fig. 1), and these evolving sequences were compared to the sequences of the replication-competent OGVs subsequently grown out of the latent reservoir.
Table 1.
Clinical information for the nine women from the CAPRISA 002 cohort
| Participant ID | Drug regimen* | Time ART naïve (weeks) | Time on ART (weeks) | CD4 count at ART initiation (cells/μl) | CD4 count at QVOA blood draw (cells/μl) | Number of outgrowth viruses | Percent clonal outgrowth viruses | Percent outgrowth viruses from 1 year pre-ART | Percent Outgrowth viruses from within one year of transmission** |
|---|---|---|---|---|---|---|---|---|---|
| CAP188 | EFV/3TC/TDF EFV/FTC/TDF |
247 | 246 | 364 | 430 | 12 | 0 | 50 | 0 |
| CAP206 | EFV/3TC/TDF EFV/FTC/TDF |
274 | 286 | 289 | 978 | 21 | 19 | 78 | 5 |
| CAP217 | EFV/3TC/TDF | 360 | 239 | 287 | 657 | 17 | 12 | 44 | 0 |
| CAP257 | 3TC/NVP/TDF EFV/3TC/TDF EFV/FTC/TDF |
249 | 318 | 170 | 596 | 48 | 17 | 89 | 0 |
| CAP287 | EFV/FTC/TDF | 260 | 223 | 411 | 559 | 6 | 0 | 67 | 17 |
| CAP288 | EFV/FTC/TDF | 210 | 275 | 318 | 860 | 7 | 0 | 100 | 0 |
| CAP302 | EFV/3TC/AZT EFV/3TC/AZT EFV/FTC/TDF |
164 | 270 | 307 | 541 | 8 | 38 | 17 | 17 |
| CAP316 | 3TC/AZT/LPV/r EFV/FTC/TDF |
215 | 215 | 436 | 888 | 11 | 0 | 91 | 0 |
| CAP336 | EFV/3TC/TDF | 142 | 258 | 164 | 558 | 17 | 47 | 100 | 0 |
| Average = | 236 | 259 | 305 | 674 | 16 | 15 | 71 | 4 | |
| Median = | 247 | 258 | 307 | 596 | 12 | 12 | 78 | 0 | |
EFV = Efavirenz, 3TC = Lamivudine, TDF = Tenofovir, NVP = Nevirapine, AZT = Zidovudine, LPV/r = Lopinavir/Ritonavir,
FTC = Emtricitabine
ART = antiretroviral therapy; QVOA = quantitative virus outgrowth assay
Regimen changes indicated below original/prior regimen
Acute/early infection here refers to sampling time points in the first year of study enrolment
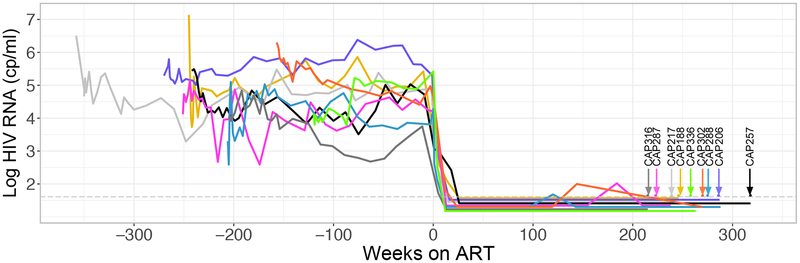
Fig 1. Viral Load and Suppression History of Nine Participants from the CAPRISA 002 Acute Infection Cohort.
The graph shows the viral loads of HIV-1 RNA in the blood pre- and post-therapy, with the time of therapy initiation designated as T=0. Each participant is designated by a different color and the time of blood collection for assessing the reservoir is designated by verticle arrows.
Sequencing plasma virus from throughout untreated infection and OGVs
Phylogenetic trees of sequences from longitudinally sampled viruses generally possess a ladder-like structure as the viral population diverges from its homogeneous founder (25–27) thereby providing a strong phylogenetic and temporal signal for distinguishing populations over time. This allowed us to estimate when each OGV entered the long-lived reservoir by identifying the pre-ART viral sequences that it was most closely related to, and therefore the pre-ART time point when it most likely entered the long-lived reservoir. We sequenced five regions (one in gag, one in nef and three in env), in three viral genes known to have rapid rates of evolution driven by immune responses in the host (28, 29).
An average of nine pre-ART plasma samples were sequenced per participant, using the Illumina MiSeq platform with a Primer ID protocol that allowed us to deeply sample the replicating and evolving viral populations in plasma pre-ART while correcting for sequencing errors and template resampling (30, 31). Phylogenetic trees were constructed for each region that yielded sequences at most pre-ART time points. Two regions were analyzed for one partipicant, and at least three regions were analyzed for the remaining eight particpants. In addition, viral RNA was isolated from the culture supernatant from QVOA wells for analysis of OGV sequences from the replication-competent latent reservoir. Near full-length OGV sequences were generated using the Pacific Biosciences (PacBio) SMRT platform with barcoded primers that likewise allowed for virtually complete error correction; we estimate a residual sequencing error rate of 1 per 50,000 bases based on repeated sequencing of a defective virus produced from a cell clone (8E5 cells; (32)). The sequence of each OGV was determined from a bulk PCR product of the supernatant virus given that the resting CD4+ T cells had been diluted to end point so that a single virus was present in the culture well in most cases. Within the different OGVs from a subject, an average of 15% of the sequences were clonal using a definition in which sequences are clonal if they are identical or differ by 1 base (i.e. 1 difference out of approximately 8850 bases sequenced, likely due to rare sequencing errors and errors within the first round of viral replication in the well; Table 1). Since these OGVs were likely derived from cells that had undergone clonal proliferation in the host (8, 12), we assumed that clonal OGVs represent a single entry into the reservoir followed by subsequent cellular expansion and therefore included only one representative sequence from each putative clonal population in summarizing our findings.
Analysis of the timing of reservoir formation
Phylogenetic analysis revealed that reservoir OGV sequences predominantly clustered in the tree with plasma viruses from the year preceding ART initiation (representative trees for two participants are shown in Fig. 2A–B), although for some participants the OGVs were distributed throughout the tree (two examples are shown in Fig. 3A–B; trees for all participants are shown in Supplementary Data Figs. 2–10). Three methods: patristic distance, clade support and phylogenetic placement (33, 34), were used to estimate when each individual OGV entered the reservoir (pipeline and additional description of the methods are available at https://github.com/veg/ogv-dating). The three methods were applied to each of the multiple regions of the genome sequenced, and the weighted median of these estimates was used to summarize when each OGV entered the reservoir (Fig. 4 for the data shown in Figs. 2 and 3, and Supplementary Data Fig. 11).
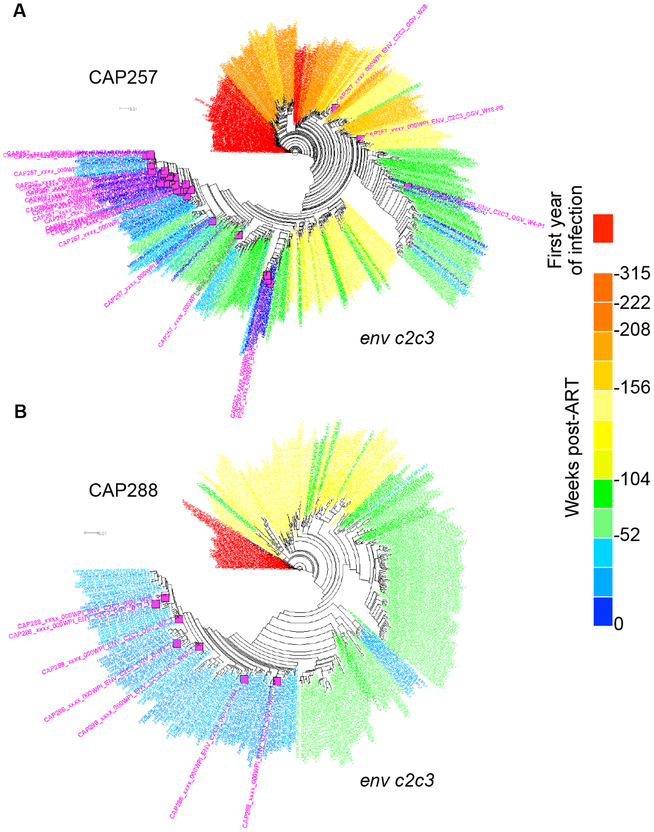
Fig. 2. The Majority of Outgrowth Viruses (OGVs) Are Most Closely Related to Viruses Replicating Near the Time of ART Initiation.
Phylogenetic analyses of one genomic region for each of two participants are shown. Pre-ART sequences are colored red (year after transmission) to blue (year before ART) and outgrowth viruses are shown in pink. The clustering of outgrowth sequences (pink) with sequences from the year before ART (blue) indicates that these participants each had a majority of OGVs with timing assignments for entry into the reservoir in the year preceding ART. (a) CAP257 had 44 unique OGVs of which 89% were most closely related to viruses replicating within one year pre-ART, and 7% to viruses replicating 142 weeks pre-ART. (b) CAP288 had 7 unique OGVs, all (100%) of which were most closely related to viruses replicating within one year pre-ART.
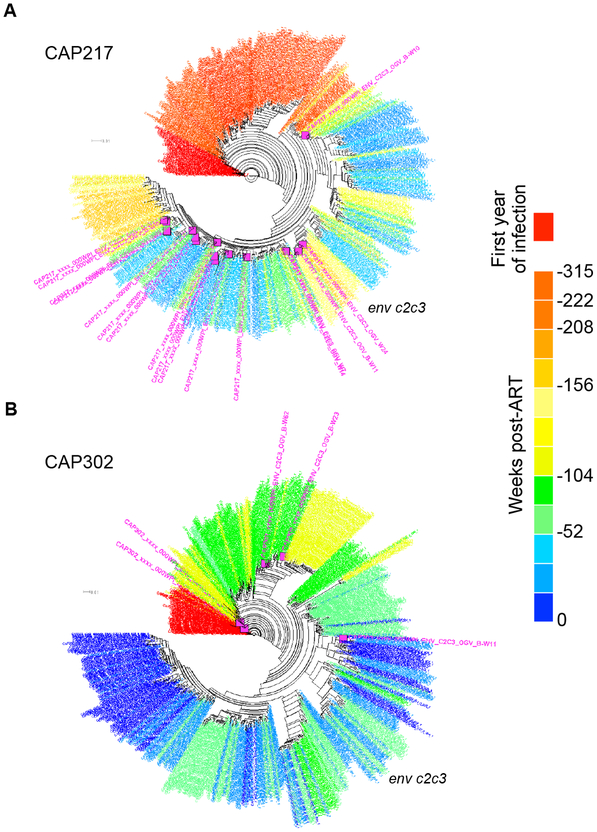
Fig. 3. A Subset of Participants Had Outgrowth Viruses (OGVs) Most Closely Related to Viruses Replicating Throughout Untreated Infection.
Phylogenetic analyses of one genomic region for each of two participants are shown. Selected participants each had OGVs with timing assignments from a wide range of times pre-ART, as illustrated by the clustering of outgrowth sequences (pink) with sequences from different pre-ART time points (red to blue). (a) CAP217 had 17 OGVs, two of which were clones. Of the genetically unique OGVs, 44% were most closely related to viruses replicating within one year pre-ART, and 31% to viruses replicating more than 2 years before ART. (b) CAP302 had 6 unique OGVs of which one was from the year before ART and one was from the year after transmission.
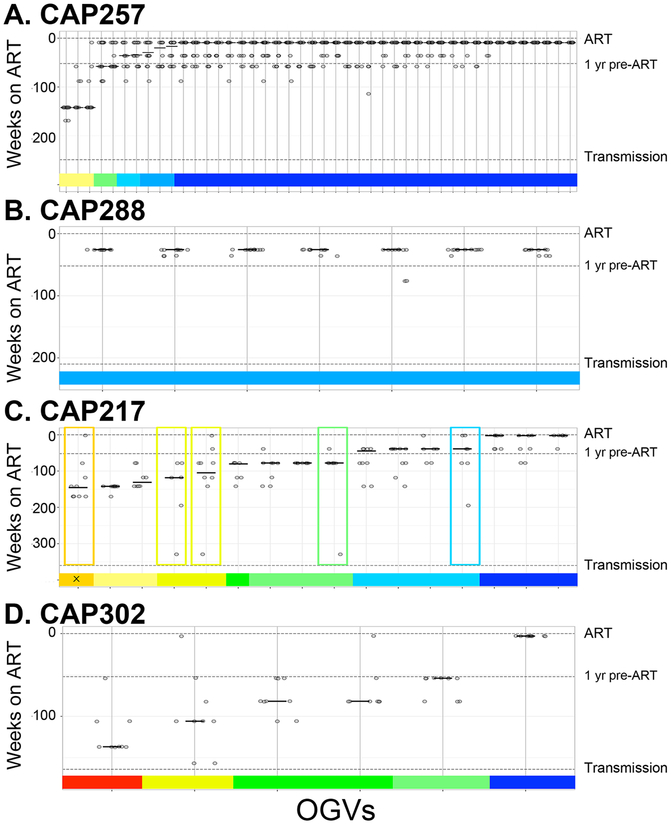
Fig. 4. Timing of Reservoir Entry for OGVs as Estimated by Combining Three Methods.
Three methods (patristic distance, clade support and phylogenetic placement) were used to generate estimates of OGV entry into the reservoir by comparing multiple regions of the OGV genome to that of viral RNA isolated from the plasma pre-ART. (a, b) Estimates are for two participants with OGVs that typically entered the reservoir near the time of ART initiation (CAP257 and CAP288) and (c, d) two with OGVs from throughout untreated infection (CAP217 and CAP302). Each OGV is represented as a vertical line with up to 15 estimates along that line. For each OGV, estimates of reservoir entry were summarized as a weighted median, shown as a horizonal line and represented in a colored bar along the x axis. Boxes indicate OGVs with highly variable estimates (SD > 1 yr). Additional phylogenetic analyses of OGVs with variable estimates found that one (designated with an ‘x’) was in a time-specific lineage that did not correspond to the weighted median. All other OGVs were found in time-specific lineages that corresponded to the weighted median.
The percentage of the OGVs representing viruses replicating in the last year before the initiation of therapy (i.e. “late” virus) was calculated for each of the nine participants (Table 1, Fig. 5A). The median of these values was 78%, and the average percent was 71%. In four of nine women, over 90% of OGVs, and in six of nine women at least 67% of OGVs, entered the reservoir in the year proceeding ART. In contrast, on average only 4% of OGVs entered the reservoir within the first year of infection (Table 1, Fig. 5A).
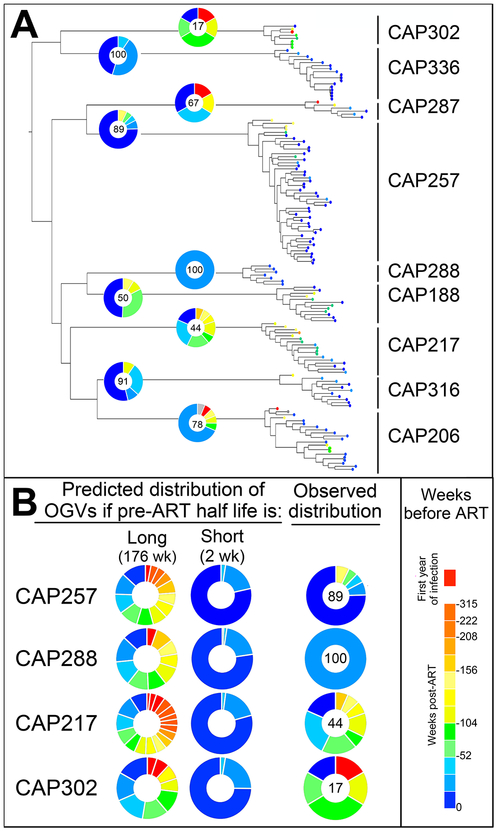
Fig. 5. Skewing of Reservoir Virus to Virus Replicating Near the Time of ART Initiation May be Explained by a Change in the Half-Life of Latently Infected Cells in the Presence of ART.
(a) All outgrowth virus (OGV) sequences are shown in an approximate maximum-likelihood tree. Branch tips are colored according to the estimated time when each OGV entered the reservoir. With OGVs from the year after transmission in red and OGVs from the year before ART in blue. For each participant, the OGV timing distribution is illustrated in a pie chart with the percentage of OGVs from the year pre-ART listed. Overall, 71% of OGVs were produced by cells infected near the time of ART initiation. (b) Two models were considered to explain this pattern. One model assumes that latently infected cells have a long half-life of 176 weeks (44 months) both prior to and during therapy, and predicts that the reservoir will contain variants from throughout untreated infection. The other model assumes that latently infected cells have a short half-life in untreated infection (here chosen as 2 weeks) that then stabilizes to a long half-life during therapy. For the majority of participants (four shown here), the observed data most closely resembles the predicted pattern if latently infected cells have a short half-life prior to ART initiation and then decay slowly after ART initiation.
We estimate that 15% of OGVs were induced from clonally expanded cells (i.e. differed from another OGV by no more than 1 mutation). However, because we sampled a small number of OGVs from some participants, we were only able to identify an OGV as being from an expanded cell clone if that clone was present at a high frequency in the population. In this analysis of 25 clonal OGVs (25 total clones in 10 clone groups), 60% of the clone groups formed with a virus that was replicating in the year prior to ART, with the remaining 40% forming from viruses that had been replicating earlier. There was no statistical difference between the frequency of “late” viruses observed as clones in the OGVs versus their frequency in unique OGVs (Fisher’s exact test P=0.33). Thus there is no evidence (from this limited sampling) that clonal expansion differentially impacted OGVs that entered the reservoir earlier in untreated infection vs. those that entered near the time of ART initiation.
Modeling formation of the long-lived HIV-1 reservoir
Our analyses suggest that cells infected with HIV-1 near the time of ART initiation (or clones of those cells) are more likely to contribute to the long-lived, replication-competent reservoir than cells infected at earlier time points. We considered two simple models to explain our findings. The first model assumes that infected resting CD4+ T cells, which have been observed in untreated infection (35), have a half-life of 44 months (176 weeks), comparable to their half-life on therapy (Fig. 5B, left panel) (4, 5). Under this model the reservoir is expected to contain variants sampled throughout untreated infection with those from later in infection being only slightly more common. The second model assumes that infected resting CD4+ T cells have a short half-life in untreated HIV-1 infection (here set at 2 weeks) that then stabilizes to a longer half-life with the onset of effective ART (Fig. 5B, middle panel). Similar to our observations for two participants with predominantly late reservoir virus (CAP257 and CAP288, Fig. 5B, right panel), this second model predicts that most of the reservoir is seeded by viruses replicating around the time of ART initiation. Thus, the observed high similarity of viruses replicating proximal to ART initiation to those in the latent reservoir can be explained by infected resting CD4+ T cells from early in infection rapidly decaying before they contribute to the long-lived reservoir. However, the second model does not fit the data for the subset of participants (e.g., CAP217 and CAP302) who had virus enter the reservoir over the entire period prior to the initiation of therapy (Fig. 5B, right panel).
Discussion
In this study we have taken advantage of the availability of archived pre-ART viremic samples in the CAPRISA 002 cohort to establish a detailed temporal record of viral evolution in each of nine women enrolled from the time of primary HIV-1 infection. Using this temporal record of viral evolution, we estimated when viruses that had persisted after more than four years on therapy entered the long-lived latent reservoir. In most women the majority of the viruses entered the reservoir around the time of therapy initiation, suggesting that the initiation of ART indirectly changes the host environment by suppressing viral replication to favor the establishment of long-lived cells, some of which are latently infected with replication-competent HIV-1.
Brodin et al. (23) and Jones et al. (22) recently characterized the formation of the long-lived proviral DNA reservoir, which is predominantly defective (14, 15). Our study, which focused on the replication-competent component of the reservoir, is consistent with the observations of Brodin et al. (23) who found that the DNA reservoir in ten individuals was skewed toward variants present in the year preceding ART, although to a lesser extent than identified here. The results of Brodin et al. and our study differ from those of Jones et al. who inferred diversity in the viral DNA reservoir that was reflective of viral evolution over the entire course of infection in two individuals. However, their analysis was based on sequences from a limited number of pre-ART time points (all during chronic infection) and thus required extrapolation based on the model of a molecular clock. In contrast, we were able to sequence virus from acute/early infection onward to the start of therapy, and directly estimate that only a small fraction of the sampled OGV populations were seeded prior to the initiation of therapy. In eight of nine participants there was no evidence of continuous reservoir formation. Only one participant (CAP302) had the replication-competent reservoir seeded primarily with virus from early to mid-stages of the untreated infection (Fig. 3B, Supplementary Data Fig. 8). Overall our data and modeling analysis suggest that much of the long-lived HIV-1 reservoir forms following the initiation of ART which also coincides with rapid declines in HIV-1 viral load, increases in CD4+ T cell counts, and reductions in markers of immune activation. One limitation is that in those subjects where the number of OGVs is small (Table 1) we are able to glean only a coarse picture of reservoir timing. However, even such a broad-strokes picture makes it clear that on average there is preferential entry into the reservoir by viruses replicating around the time of therapy initiation.
It is not clear why only a small fraction of the full-length proviruses from resting CD4+ T cells produce detectable virus in a QVOA (14). It is possible that most proviruses are induced in QVOA, but that the assay is very inefficient at amplifying the small amounts of virus produced by a single resting cell. Alternatively, proviruses may exist in different states of latency with only a small fraction of them inducible in cell culture. Consistent with these possibilities, Ho et al. (14) has shown that within a QVOA repeated rounds of stimulation can give rise to an additional 24% of induced viruses, either pointing to a stochastic nature of the assay or a distinct set of proviruses/cells that are inducible only with multiple rounds of stimulation.
If proviruses exist in different states of latency then it raises the possibility that OGVs recovered by QVOA represent a skewed sample of the viral reservoir, some of which may have been seeded earlier in untreated infection. While it is likely that proviruses experience different degrees of latency, perhaps representing different host mechanisms of gene silencing, the work of Brodin et al. (23) finding that the viral DNA reservoir is also skewed toward variants circulating late in untreated infection indicates that the majority of both the induced and uninduced reservoir is seeded near the time of ART initiation. While we cannot rule out the possibility that a small fraction of the reservoir is seeded early in untreated infection and persists in a state of “deep latency”, current evidence indicates that the majority of the reservoir (both inducible and uninducible) is seeded near the time of ART initiation.
In this study we focused on virus induced to grow from resting CD4+ T cells isolated from the blood of virologically suppressed women. We have not examined the reservior in activated cells (CD25High, CD69+, HLA-DR+), cells in tissues, or cells in HIV-positive men. Also, several studies have found that viral DNA, including intact viral genomes and replication-competent genomes, are found in a wide variety of different CD4+ T cell subsets (13, 36–38). While there is a need to confirm our observations more broadly, they are likely generalizable. For example, Boritz et al. (39) observed that in elite controllers, HIV-infected cells migrate between lymphoid tissue and the blood, suggesting that viral populations at these sites are genetically linked. In addition, analysis of gut tissue biopsies suggests that viral populations in latently infected cells are largely equilibrated between blood and tissue (40), making it less likely that there is a distinct population of latent virus residing within tissue. As mentioned above, the results of Brodin et al. (23), indicate that genetically distinct viruses contained in other cell types such as activated CD4+ T cells would have to represent a small fraction of the long-lived HIV-1 reservoir since they obtained similar results to those reported here analyzing viral DNA present in total peripheral blood cells which would include all subtypes of CD4+ T cells. In addition, while our study of biologically active, inducible virus in the reservoir was done using women, the analogous results of Brodin et al. (23) were obtained in a cohort largely composed of men, suggesting that sex is not a major determinant of when viral sequences enter the long-lived reservoir.
There is ongoing interest as to whether HIV-1 replicates during suppressive antiviral therapy. We were able to account for all OGVs as being phylogenetically close to, and at times identical to, viruses that were replicating before the initiation of therapy, i.e. the OGVs did not represent an outlier group of sequences that had undergone further sequence evolution due to ongoing replication during the average of 5 years on therapy. These results are consistent with those of Brodin et al. (23) and Van Zyl et al. (41) who have failed to find any evidence for viral sequence evolution during suppressive antiretroviral therapy, an observation most compatible with the latent reservoir not being sustained by ongoing viral replication.
Our results point to the long-lived reservoir forming under two very distinct conditions. The majority of the reservoir (on average) forms around the time of therapy initiation and is likely driven by the rapidly changing immune environment within the host that occurs in response to lowering viral loads and decreased antigen drive. These immunologic changes include absolute increases in peripheral CD4+ T cells, reductions in CD4+ T cell turnover (42, 43), and increased frequencies of long-lived memory CD4+ T cells (44). Initiation of ART may therefore promote the increased transition of both infected and uninfected CD4+ T cells from being short-lived effectors to being long-lived memory T cells (45). Similarly, ART-mediated reduction of HIV-1 antigen restores the proliferative ability of HIV-specific CD4+ T cells (46). These changes may directly impact HIV-specific CD4+ T cells by allowing them to become long-lived memory cells capable of proliferation. As this model would predict, HIV-infected people on ART have been shown to have HIV-specific CD4+ T cells that are infected at rates greater than CMV-specific T cells or total memory CD4+ T cells, although the HIV-specific CD4+ T cells represent only a small fraction of the total memory CD4+ T cells and contain only a small fraction of the total viral DNA (47). Together, these findings suggest that profound immunologic changes at the time of ART may allow HIV-infected cells to become long-lived memory cells and form the majority of the stable reservoir.
An additional mechanism is needed to explain how a variable fraction of the reservoir forms in the presence of active viral replication (see examples in Fig. 3). The fraction of OGVs that entered the reservoir at earlier time points prior to therapy initiation was highly variable, with the majority of OGVs from one particpant (CAP 302) seeded earlier in untreated infection. It is not clear what factors contribute to early viral entry into the reservoir. For this particular woman, the infection course was only moderately different from the cohort overall: three years in study prior to therapy, setpoint viral load about two times higher than the mean, nadir CD4+ T cell count only slightly lower than the mean. An important question about the phenomenon of early entry into the reservoir is whether it is continuous or episodic where it might be linked to some other biological events that occur in the person, such as transient exposure to other infectious agents.
Untreated HIV-1 infection is characterized by increased immune activation and dysfunction, which are largely reversed after ART initiation. Most relevant to this study is the loss of CD127+(IL-7 receptor) memory CD4+ T cells during untreated infection (48–50). IL-7 signaling through the CD127 receptor is essential for the transition of effector to memory CD4+ T cells as well as the subsequent maintenance of memory cells (51). With ART initiation, the number of CD127+ memory CD4+ T cells increases (42, 48, 49), most likely due to reductions in inflammation (52). These results suggest a model in which the increase in CD127 expression during ART allows latently infected cells to become long-lived as they now become responsive to IL-7 (Supplementary Data Fig. 12). Consistent with this model in which much of the reservoir should be in cells sensitive to IL-7, administering IL-7 to HIV-infected individuals on ART increases the size of the HIV-1 reservoir (53), and the rate of reservoir decay is slower when IL-7 concentrations are higher (13).
Therapies that limit CD4+ T cell memory generation, perhaps through dampening IL-7/IL-7R signaling in the window from the time of ART initiation to when all viral replication is fully suppressed, could limit reservoir formation. Such an approach would not eliminate all of the reservoir, as it is clear that latency can be seeded at different points during untreated infection. However, blocking the formation of a majority of the reservoir at the time of therapy initiation would greatly reduce the size of the reservoir, lowering the barrier to eradicating HIV-1 from an infected host.
Materials and Methods
Study design
Viral RNA from pre-ART time points ranging from acute/early HIV-1 infection to the time of ART initiation were sequenced across several regions of the viral genome using a deep sequencing protocol. After the women had been on ART for several years they provided a large blood sample that was used to grow out latent HIV-1 from the long-lived viral reservoir. The outgrowth virus sequences were compared to the sequences of the pre-ART longitudinal sampling of replicating virus to determine when the virus in the reservoir had entered the long-lived reservoir prior to the initiation of ART.
Study participants
The CAPRISA 002 cohort is comprised of women from rural and urban KwaZulu-Natal, South Africa, who were identified during the period of acute/primary infection with subtype C HIV-1 and followed longitudinally (24). Participants in the cohort gave blood samples every 3–6 months and were provided antiretroviral therapy (ART) based on the prevailing in-country guidelines. The women were retained in the cohort for up to five years after therapy initiation. We identified nine women in the cohort who had been ART naïve for at least three years prior to initiating therapy, and who had been on ART for a minimum of 4 years. The women provided a 200 ml blood draw while on therapy from which peripheral blood mononuclear cells (PBMCs) were isolated and cryopreserved in liquid nitrogen. Women were excluded based on pregnancy or if they had hemoglobin concentrations lower than 10g/dL. The use of stored samples and the collection and analysis of new samples were done under a protocol approved by the Biomedical Research Ethics Committee of the University of KwaZulu Natal (BE178/150) and the Human Research Ethics Committee of the University of Cape Town (588/2015) in South Africa, and at the University of North Carolina in the United States.
Viral outgrowth assay
Resting CD4+ T cells were isolated from PBMCs by negative selection with CD25 depletion to isolate cell that were CD69-, HLA-DR-, CD25low (Custom kit, Stem Cell). Purified cells were then examined by flow cytometry (LSRII Fortessa) for expression of CD69 (PE, BD), CD25 (APC, BD), CD8 (FITC, BD), CD4 (PerCP-Cy5.5, BD) and viability (aqua live/dead, ThermoFisher). Data were analysed using FlowJo (version 10.4.2).
Resting cells were cultured at 100,000 cells/well with 1.5 μg/ml of highly purified phytohemagglutinin (PHA, Remel-PHA, Thermo Scientific), 60 U/ml of interleukin 2, and irradiated PBMCs from a seronegative donor, and virus outgrowth was facilitated as previously described (4). On days 15 and 19, cultures were tested for HIV-1 p24 capsid protein production using an enzyme-linked immunosorbent assay. Culture supernatants positive for p24 were stored at −80°C.
Sequencing of viral populations in blood plasma pre-ART
Viral RNA copies were extracted from blood plasma using the QIAamp viral RNA Mini kit (Qiagen). The purified RNA was reverse transcribed to cDNA using Superscript III/IV Reverse Transcriptase (Invitrogen). We used the Primer ID method (30) which tags each RNA template through its cDNA primer with a unique 12 nucleotide-long identifier. This allows amplified sequences to be grouped according to their Primer ID tags, from which a consensus sequence can then be derived for each individual template. (template consensus sequence). Multiple cDNA primers were used during the cDNA synthesis step for each sample to generate cDNAs corresponding to multiple regions of the genome and allow multiplexing of the PCR amplicons followed by sequencing. The gene-specific sequence of the cDNA primer was used as an index for the specific region of the genome. Two separate cDNA reactions were performed for each sample to allow efficient use of the available RNA to sequence the different regions of the viral genome. The cDNA products were purified twice using Agencourt RNA Clean XP magnetic beads (Beckman Coulter). Multiplexing during PCR was accomplished by including a common PCR primer binding sequence at the 5’ end of each cDNA primer and a gene-specific forward primer for each amplicon/region. PCR amplification was performed using the KAPA2G Fast Multiplex Mix (Kapa Biosystems) and an equal molar amount of each forward primer and an excess of the universal reverse primer. A second PCR step using the Expand High Fidelity PCR System (Roche) allowed the incorporation of the Illumina MiSeq version 2 indexes and adapters. PCR products were purified using SPRIselect beads (Beckman Coulter), with a 0.6:1 volume ratio of beads to PCR product (to allow for size exclusion of residual primers) following both PCR steps. The final purified amplicon libraries were quantified using the Qubit double-stranded DNA HS assay (Invitrogen, CA), pooled at equimolar ratios and purified again using SPRIselect beads. Illumina MiSeq 2×300 base paired-end sequencing was performed on the multiplexed amplicons.
Sequencing of outgrowth virus populations
Viral RNA was isolated from the QVOA culture supernatant of p24-positive wells and converted to cDNA using Superscript III Reverse Transcriptase and an oligo(dT) primer. The 3’ and 5’ half genomes were amplified in separate PCR reactions using barcoded primers, and the PCR products were gel purified. For CAP257, approximately 35% of the OGVs had only a 3’ half genome amplicon available for analysis. The SMRTbell Template Prep Kit (PacBio) was used to add adaptors to pooled, barcoded amplicons, and the pools of amplicons were then submitted for PacBio sequencing (movie time of 10 hours). Sequences were grouped by barcode, and high quality sequences were analyzed using the PacBio Long Amplicon Analysis (LAA) package. The 3’ and 5’ amplicons for the same virus were joined and visually screened to confirm that open reading frames were intact. Genebank Accession numbers are MN097551–MN097697.
Illumina MiSeq data processing
Raw reads were processed using a custom pipeline written in Python and R programming languages. The MotifBinner2.R program (https://github.com/HIVDiversity/MotifBinner2) carries out quality filtering, merging of overlapping paired-end reads, and implements the Primer ID processing method described by Zhou et al (31). The resulting Primer ID template consensus sequences were processed with an in-house pipeline (https://github.com/ColinAnthony/NGS_processing_pipeline) which removes sequences with degenerate bases or deletions >50 nucleotides long and filters out any contaminant non-target gene sequences.
Phylogenetic and statistical analyses
Hypervariable loop regions of env were manually removed where accurate alignment was not possible. In-frame codon alignments were generated for each genomic region using a custom pipeline (https://github.com/veg/ogv-dating). Briefly, translated protein sequences were used to identify RNA sequences that were in-frame. Sequences that contained frameshifts were aligned to each in-frame sequence using the Smith-Waterman algorithm implemented in the BioExt package (https://github.com/veg/BioExt) and the pairs with the highest score were used to correct frameshifts. MUSCLE (54) was then used to align in-frame amino acid sequences and then generate in-frame, codon-aligned RNA sequences lacking frameshifts. For each pre-ART time point analyzed, identical RNA sequences were replaced with a single representative, and approximately-Maximum-Likelihood trees were constructed using FastTree2 with more rigorous search options (55).
Three methods were used to analyze sequence data from each genomic region and estimate when the OGV entered the reservoir (pipeline and additional description of the methods are available at https://github.com/veg/ogv-dating). First, the tree was rooted to maximize root-to-tip distance versus its time linear regression coefficient. The ‘patristic distance’ method computes the path lengths between an OGV sequence and every other RNA sequence in the tree, finds the minimum distance (d) and assigns the OGV sequence to the time point from which the simple majority of RNA sequences within distance 2d come from. The ‘clade support’ method starts at the OGV sequence, and traverses the tree up to the root until it encounters a well-supported (bootstrap ≥90%) internal node that has at least one RNA sequence descendant. Each OGV sequence was assigned the time point from which the majority of RNA sequences in the clade came from. Third, phylogenetic placement (33, 34) was used to analyze phylogentic trees containing only pre-ART sequences and identify the position in these reference trees where each OGV was most likely to be found and use that new position to identify when the OGV entered the reservoir. Patristic distance and clade support estimates were excluded from subsequent analyses if their tree support value was less than 0.4. Tree support value was estimated by applying the same dating procedure to each RNA sequence, and seeing how often the correct sampling time point was recovered. A support of 0.4 means that the correct time point was recovered for 40% of RNA sequences. Because generating tree support estimates for phylogenetic placement is computationally slow, we did not exclude any estimates generated by phylogenetic placement. Fig. 4 and Supplementary Data Fig. 11 illustrate all of the estimates analyzed for each OGV.
For each OGV, timing of reservoir entry was summarized as the weighed median of estimates generated by the three different methods and the support value for each estimate. As a way of validating this approach, we identified OGVs whose individual estimates had a standard deviation of greater than 1 year. We then visually inspected their phylogentic trees to identify whether the weighted median accurately characterized the position of each OGV in the phylogenetic trees. A total of 25 OGVs (25 of 132 unique OGVs) were identified as having a standard deviation greater than 1 year. We then inspected the phylogenetic trees to identify whether each OGV was found in a time-specific lineage and whether that lineage dated to the same time as the weighted median. This approach revealed that the weighted median accurately characterized 18 of 25 OGVs examined. However, 6 OGVs could be found in time-specific lineages that did not correspond to the weighted median. We therefore revised the estimate of timing of reservoir entry of these 6 OGVs to reflect the age of the time-specific lineage in which they were found. One additional OGV was not located in a time-specific lineage so we revised its entry into the reservoir to be ‘ambiguous’. The majority of the 7 OGVs that were mischaracterized by our automated methods had a nearby group of clonal sequences observed at multiple pre-ART time points which may have been problematic for the methods. Furthermore, 6 of these 7 OGVs were from a single participant (CAP206). Further examination of sequence data for this participant found no evidence of contamination or poor sequence alignment that could explain why our methods failed for this participant.
Supplementary Material
Fig. S1. Longitudinal MiSeq Sampling Depth for Nine Women from the CAPRISA 002 Cohort.
Fig. S2. Timing of Reservoir Outgrowth Viruses for Participant CAP188.
Fig. S3. Timing of Reservoir Outgrowth Viruses for Participant CAP206.
Fig. S4. Timing of Reservoir Outgrowth Viruses for Participant CAP217.
Fig. S5. Timing of Reservoir Outgrowth Viruses for Participant CAP257.
Fig. S6. Timing of Reservoir Outgrowth Viruses for Participant CAP287.
Fig. S7. Timing of Reservoir Outgrowth Viruses for Participant CAP288.
Fig. S8. Timing of Reservoir Outgrowth Viruses for Participant CAP302.
Fig. S9. Timing of Reservoir Outgrowth Viruses for Participant CAP316.
Fig. S10. Timing of Reservoir Outgrowth Viruses for Participant CAP336.
Fig. S11. Estimates of OGV Entry into the Long-lived Reservoir.
Fig. S12. Model of the Relationship Between Viral Suppression and Formation of the Long-lived HIV-1 Reservoir.
Table S1. Data of participant viral loads.
Table S2. Data for timing of outgrowth viruses.
Acknowledgments:
We would like to acknowledge all participants of the CAPRISA 002 acute infection cohort as well as the staff at the Vulindlela and eThekwini Clinical Research Sites, KwaZulu-Natal, South Africa.
Funding: This work was supported by the National Institute of Health(NIH) - South African Medical Research Council (MRC) US-South Africa Program for Collaborative Biomedical Research grants R01 AI115981 to CW and RS, and the Collaboratory of AIDS Researchers for Eradication (UM1 AI126619). The CAPRISA 002 acute infection cohort study has been funded by the South African Department of Science and Technology and the National Research Foundation’s Centre of Excellence in HIV Prevention (Grant # UID: 96354), the South African Department of Health and the South African Medical Research Council Special Initiative on HIV Prevention (Grant #: 96151), the National Institute of Allergy and infectious Disease of the NIH (Grant # AI51794), USAID and CONRAD (USAID co-operative Grant # GP00-08-00005-00, subproject agreement # PPA-09-046), the South African National Research Foundation (Grants # 67385, # 96354), the South African Technology Innovation Agency, and the Fogarty International Center, NIH (Grant # D43TW00231). The work was also supported by the UNC Center For AIDS Research (NIH award P30 AI50410) and the UNC Lineberger Comprehensive Cancer Center (NIH award P30 CA16068).
Footnotes
Competing interests: UNC is pursuing IP protection for Primer ID, and RS is listed as a co-inventor and has received nominal royalties.
Data and materials availability: All sequences will be deposited in public databases and accession numbers provided prior to publication. The near full-length genome sequences will be deposited in GenBank. The MiSeq sequences will be deposited in the Sequencing Read Archive.
References and Notes:
- 1.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, Fauci AS, Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF, Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD, Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, Gay CL, Eron JJ, Margolis DM, Bosch RJ, Archin NM, Precise quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J. Infect. Dis 212, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF, Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4(+) T cells. Nat. Med 9, 727–728 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, Ho YC, Hill AL, Siliciano JD, Siliciano RF, Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. U. S. A 115, E2575–E2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DI, Keele BF, Ho YC, Siliciano JD, Siliciano RF, Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM, Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, Kearney MF, Mellors JW, Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathogens 13, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, Migueles S, Jones RB, Mota T, Joseph SB, Groen K, Pasternak AO, Hwang WT, Sherman B, Vourekas A, Nunnari G, O’Doherty U, Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat. Commun 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F, Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH, Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP, HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med 15, 893–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF, Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF, Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O’Doherty U, Paxinos EE, Fauci AS, Lane HC, Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A 113, 8783–8788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, Capoferri AA, Beg SA, Huang SH, Karandish S, Hao HP, Halper-Stromberg E, Yong PC, Kovacs C, Benko E, Siliciano RF, Ho YC, Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe 21, 494–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan L, Deng K, Gao HB, Xing SF, Capoferri AA, Durand CM, Rabi SA, Laird GM, Kim M, Hosmane NN, Yang HC, Zhang H, Margolick JB, Li LH, Cai WP, Ke RA, Flavell RA, Siliciano JD, Siliciano RF, Transcriptional reprogramming during effector-to-memory transition renders CD4(+) T cells permissive for latent HIV-1 infection. Immunity 47, 766–775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, Chomchey N, Muir R, Haddad EK, Tovanabutra S, Ubolyam S, Bolton DL, Fullmer BA, Gorelick RJ, Fox L, Crowell TA, Trichavaroj R, O’ Connell R, Chomont N, Kim JH, Michael NL, Robb ML, Phanuphak N, Ananworanich J, Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat. Med 24, 923–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrich TJ, Hatano H, Bacon O, Hogan LE, Rutishauser R, Hill A, Kearney MF, Anderson EM, Buchbinder SP, Cohen SE, Abdel-Mohsen M, Pohlmeyer CW, Fromentin R, Hoh R, Liu AY, McCune JM, Spindler J, Metcalf-Pate K, Hobbs KS, Thanh C, Gibson EA, Kuritzkes DR, Siliciano RF, Price RW, Richman DD, Chomont N, Siliciano JD, Mellors JW, Yukl SA, Blankson JN, Liegler T, Deeks SG, HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study. PLoS Med. 14, 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu JY, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DIS, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH, Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones BR, Kinloch NN, Horacsek J, Ganase B, Harris M, Harrigan PR, Jones RB, Brockman MA, Joy JB, Poon AFY, Brumme ZL, Phylogenetic approach to recover integration dates of latent HIV sequences. Proc. Natl. Acad. Sci. U.S.A 115, E8958–E8967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodin J, Zanini F, Thebo L, Lanz C, Bratt G, Neher RA, Albert J, Establishment and stability of the latent HIV-1 DNA reservoir. eLife 5, e18889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, Karim QA, Grobler A, Barnabas N, Iriogbe I, Karim SSA, Team CAIS, Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 Acute Infection Study. PLoS One 3, e1954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J. Virol 83, 3556–3567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun CX, Grayson T, Wang SY, Li H, Wei XP, Jiang CL, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM, Identification and characterisation of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A 105, 7552–7557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI, Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol 73, 10489–10502 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S, HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef Proteins. PLoS One 4, e6687 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frost SDW, Wrin T, Smith DM, Pond SLK, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, Little SJ, Richman DD, Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U.S.A 102, 18514–18519 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R, Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. U.S.A. 108, 20166–20171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S, Jones C, Mieczkowski P, Swanstrom R, Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 denomic RNA populations. J Virol 89, 8540–8555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Folks TM, Powell D, Lightfoote M, Koenig S, Fauci AS, Benn S, Rabson A, Daugherty D, Gendelman HE, Hoggan MD, Biological and biochemical characterization of a cloned Leu-3- cell surviving. J. Exp. Med 164, 280–290 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frost SDW, Murrell B, Hossain A, Silverman GJ, Pond SLK, Assigning and visualizing germline genes in antibody repertoires. Philos. Trans. R. Soc. B-Biol. Sci 370, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsen FA, Kodner RB, Armbrust EV, pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun TW, Carruth L, Finzi D, Shen XF, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, BarditchCrovo P, Siliciano RF, Quantification of latent tissue reservoirs and total body viral load in HIV-1 Infection. Nature 387, 183–188 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM, Quantitation of replication-competent HIV-1 in populations of resting CD4(+) T cells. J. Virol 88, 14070–14077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buzon MJ, Martin-Gayo E, Pereyra F, Ouyang ZY, Sun H, Li JZ, Piovoso M, Shaw A, Dalmau J, Zangger N, Martinez-Picado J, Zurakowski R, Yu XG, Telenti A, Walker BD, Rosenberg ES, Lichterfeld M, Long-Term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J. Virol 88, 10056–10065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S, Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Reports 21, 813–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boritz EA, Darko S, Swaszek L, Wolf G, Wells D, Wu XL, Henry AR, Laboune F, Hu JF, Ambrozak D, Hughes MS, Hoh R, Casazza JP, Vostal A, Bunis D, Nganou-Makamdop K, Lee JS, Migueles SA, Koup RA, Connors M, Moir S, Schacker T, Maldarelli F, Hughes SH, Deeks SG, Douek DC, Multiple origins of virus persistence during natural control of HIV infection. Cell 166, 1004–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, Mohri H, Markowitz M, Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog 8, e1002506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Zyl GU, Katusiime MG, Wiegand A, McManus WR, Bale MJ, Halvas EK, Luke B, Boltz VF, Spindler J, Laughton B, Engelbrecht S, Coffin JM, Cotton MF, Shao W, Mellors JW, Kearney MF, No evidence of HIV replication in children on antiretroviral therapy. J. Clin. Invest 127, 3827–3834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hellerstein MK, Hoh RA, Hanley MB, Cesar D, Lee D, Neese RA, McCune JM, Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J. Clin. Invest 112, 956–966 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohri H, Perelson AS, Tung K, Ribeiro RM, Ramratnam B, Markowitz M, Kost R, Hurley A, Weinberger L, Cesar D, Hellerstein MK, Ho DD, Increased turnover of T lymphocytes in HIV-1 infection and its reduction by antiretroviral therapy. J. Exp. Med 194, 1277–1287 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu WF, Li J, Wu Y, Zhou JK, Zhong JP, Lv QQ, Shao H, Rao HP, CD127 expression in naive and memory T cells in HIV patients who have undergone long-term HAART. Lab. Med 48, 57–64 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Scriba TJ, Tameris M, Smit E, van der Merwe L, Hughes EJ, Kadira B, Mauff K, Moyo S, Brittain N, Lawrie A, Mulenga H, de Kock M, Makhethe L, van Rensburg EJ, Gelderbloem S, Veldsman A, Hatherill M, Geldenhuys H, Hill AVS, Hawkridge A, Hussey GD, Hanekom WA, McShane H, Mahomed H, A Phase IIa Trial of the New Tuberculosis Vaccine, MVA85A, in HIV- and/or Mycobacterium tuberculosis-infected Adults. Am. J. Respir. Crit. Care Med 185, 769–778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNeil AC, Shupert WL, Iyasere CA, Hallahan CW, Mican J, Davey RT, Connors M, High-level HIV-1 viremia suppresses viral antigen-specific CD4(+) T cell proliferation. Proc. Natl. Acad. Sci. U.S.A 98, 13878–13883 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA, HIV preferentially infects HIV-specific CD4+ T cells. Nature 417, 95–98 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Paiardini M, Cervasi B, Albrecht H, Muthukumar A, Dunham R, Gordon S, Radziewicz H, Piedimonte G, Magnani M, Montroni M, Kaech SM, Weintrob A, Altman JD, Sodora DL, Feinberg MB, Silvestri G, Loss of CD127 expression defines an expansion of effector CD8(+) T cells in HIV-infected individuals. J. Immunol 174, 2900–2909 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Koesters SA, Alimonti JB, Wachihi C, Matu L, Anzala O, Kimani J, Embree JE, Plummer FA, Fowke KR, IL-7R alpha expression on CD4(+) T lymphocytes decreases with HIV disease progression and inversely correlates with immune activation. Eur. J. Immunol 36, 336–344 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Dunham RM, Cervasi B, Brenchley JM, Albrecht H, Weintrob A, Sumpter B, Engram J, Gordon S, Klatt NR, Frank I, Sodora DL, Douek DC, Paiardini M, Silvestri G, CD127 and CD25 expression defines CD4(+) T cell subsets that are differentially depleted during HIV infection. J. Immunol 180, 5582–5592 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kondrack RM, Harbertson J, Tan JT, McBreen ME, Surh CD, Bradley LM, Interleukin 7 regulates the survival and generation of memory CD4 cells. J. Exp. Med 198, 1797–1806 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiazyk SAK, Fowke KR, Loss of CD127 expression links immune activation and CD4(+) T cell loss in HIV infection. Trends Microbiol. 16, 567–573 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy JP, Sekaly RP, Chomont N, Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 121, 4321–4329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC, MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP, FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Longitudinal MiSeq Sampling Depth for Nine Women from the CAPRISA 002 Cohort.
Fig. S2. Timing of Reservoir Outgrowth Viruses for Participant CAP188.
Fig. S3. Timing of Reservoir Outgrowth Viruses for Participant CAP206.
Fig. S4. Timing of Reservoir Outgrowth Viruses for Participant CAP217.
Fig. S5. Timing of Reservoir Outgrowth Viruses for Participant CAP257.
Fig. S6. Timing of Reservoir Outgrowth Viruses for Participant CAP287.
Fig. S7. Timing of Reservoir Outgrowth Viruses for Participant CAP288.
Fig. S8. Timing of Reservoir Outgrowth Viruses for Participant CAP302.
Fig. S9. Timing of Reservoir Outgrowth Viruses for Participant CAP316.
Fig. S10. Timing of Reservoir Outgrowth Viruses for Participant CAP336.
Fig. S11. Estimates of OGV Entry into the Long-lived Reservoir.
Fig. S12. Model of the Relationship Between Viral Suppression and Formation of the Long-lived HIV-1 Reservoir.
Table S1. Data of participant viral loads.
Table S2. Data for timing of outgrowth viruses.