Summary
Epigenetic control of gene expression contributes to dynamic responsiveness of cellular processes that include cell cycle, cell growth and differentiation. Mitotic gene bookmarking, retention of sequence-specific transcription factors at target gene loci, including the RUNX regulatory proteins, provide a novel dimension to epigenetic regulation that sustains cellular identity in progeny cells following cell division. Runx transcription factor retention during mitosis coordinate physiological control of cell growth and differentiation in a broad spectrum of biological conditions, and is associated with compromised gene expression in pathologies that include cancer.
Keywords: RUNX, mitotic bookmarking, epigenetic control, gene expression
Introduction
Epigenetic regulation of gene expression is essential; it enables cells to accommodate dynamic changes that take place during a variety of cellular processes that include cell cycle, cell growth, and cell differentiation(Chi et al. 2010; Sarkies and Sale 2012; Ptashne 2013; Cerase et al. 2014; Attar and Kurdistani 2014). DNA methylation and histone modifications are the most studied and well understood epigenetic mechanisms that contribute to spatial and temporal regulation of gene expression(Strahl and Allis 2000; He and Lehming 2003; Chi et al. 2010; Rivera and Bennett 2010; Schübeler 2015). We used RUNX proteins as model for lineage commitment and maintenance to identify mitotic gene bookmarking – retention of sequence specific phenotypic transcription factors on target genes during mitosis – as a novel epigenetic mechanism that ensures maintenance of cellular identity across cell generations and coordinates cell growth and differentiation (Zaidi et al. 2003; Young et al. 2007a; Young et al. 2007b; Ali et al. 2008; Bakshi et al. 2008; Pande et al. 2009; Ali et al. 2010; Zaidi et al. 2010; Ali et al. 2012; Zaidi et al. 2014; Lopez-Camacho et al. 2014). Numerous independent studies (Xing 2005; Sarge and Park-Sarge 2005; Dey et al. 2009; Blobel et al. 2009; Zhao et al. 2011; Kadauke et al. 2012; Arampatzi et al. 2013; Caravaca et al. 2013; Kadauke and Blobel 2013; Lake et al. 2014; Zaret 2014; Lodhi et al. 2014; Wong et al. 2014; Lodhi et al. 2016; Lerner et al. 2016; Festuccia et al. 2016) have since confirmed that mitotic gene bookmarking is a prevalent epigenetic mechanism in a number of biological models and under physiological and pathological conditions. In this chapter, we present an overview of mitotic bookmarking as a key mechanistic, epigenetic, dimension of RUNX control for multiple cellular processes.
Mitotic Bookmarking: A Historical Perspective
In the 1980s, Weintraub, Groudine, and Struhl found that a limited number of nuclease accessible sites on the condensed mitotic chromatin persist through the cell cycle (Struhl 1981; Groudine and Weintraub 1982; Weintraub 1985). In the 1990s, John and Workman proposed that these inheritable hypersensitive sites are suggested that these accessible sites provide a platform to place “bookmarks” for rapid activation of genes following mitosis (John and Workman 1998). This model explained observations by Levens and colleagues as well as by Wu and colleagues that promoters of the Myc, hsp70i, and β Globin genes, contain nuclease accessible sites that persist through mitosis (Martínez-Balbás et al. 1995; Michelotti et al. 1997). Wu and colleagues also examined several sequence-specific transcription factors during mitosis and found that these transcription factors are displaced from the condensed mitotic chromatin (Martínez-Balbás et al. 1995). In 2003, our group identified the osteogenic master regulator RUNX2 as the first sequence specific bookmark that remained associated with target genes through mitosis (Zaidi et al. 2003). Subsequent studies of RUNX family of phenotypic transcription factors from our group (Zaidi et al. 2003; Young et al. 2007a;Young et al. 2007b; Ali et al. 2008; Bakshi et al. 2008; Pande et al. 2009; Ali et al. 2010; Zaidi et al. 2010; Ali et al. 2012; Zaidi et al. 2014; Lopez-Camacho et al. 2014) and studies by other groups examining various transcription factors (Xing 2005; Sarge and Park-Sarge 2005; Dey et al. 2009; Blobel et al. 2009; Zhao et al. 2011; Kadauke et al.2012; Arampatzi et al. 2013; Caravaca et al. 2013; Kadauke and Blobel 2013; Lake et al.2014; Zaret 2014; Lodhi et al. 2014; Wong et al. 2014; Lodhi et al. 2016; Lerner et al. 2016; Festuccia et al. 2016) have identified mitotic bookmarking as a key epigenetic mechanism for regulation of genes that coordinate cell growth and lineage maintenance following mitosis.
Characteristics of Mitotic Bookmarks
Studies over the past decade have contributed to an emerging view of mitotic bookmarking and have revealed shared characteristics of mitotic bookmarks across biological models:
Properties of Genes that are Bookmarked During Mitosis
Condensation of chromosomes during mitosis is a key event that leads to brief pause in transcription (Hartl et al. 1993; Gottesfeld and Forbes 1997) and displacement of some sequence-specific transcription factors from their target genes (Martínez-Balbás et al. 1995). While reconfiguration of cellular architecture during mitosis requires coordination of several independent mechanisms (Pines 2006), studies have established that phosphorylation of histone H3 on serine residues at positions 10 and 28 plays a key role in condensation of mitotic chromosomes (Kouzarides 2007; Margueron and Reinberg 2010). These phosphorylation events are mediated by the mitotic Aurora B kinase and results in displacement of several regulatory and structural proteins that include Heterochromatin Protein 1 (HP1), RNA Polymerase II, and chromatin remodeler B cell-specific Moloney murine leukemia virus integration site 1 (BMI1) from mitotic chromosomes (Sabbattini et al. 2007). It is noteworthy that earlier studies in the 1980s by the Weintraub, Groudine and Struhl groups showed that mitotic chromosomes harbor inheritable nuclease-accessible regions that are inheritable (Struhl 1981; Groudine and Weintraub 1982; Weintraub 1985). Consistent with these observations, more recent studies, using nuclease accessibility assays combined with genome-wide approaches, have shown that the state of open chromatin remains preserved during mitosis, although it appears to be remodeled at the level of individual genes and regulatory elements (Hsiung et al. 2015). Studies focused on understanding posttranslational modifications of histone proteins show that mitotic chromosomes retain several histone modifications(Wang and Higgins 2013). Two of these modifications (Kouzarides 2007) – H3K4me3, a modification associated with transcriptional activation, and H3K27me3, a modification linked with transcriptional silencing – have been shown to be retained on promoters of genes that are rapidly reactivated following mitosis (Grandy et al. 2016). Bivalency of some genes, i.e., marking of gene regulatory regions with both activating and repressive histone marks, is emerging as a key mechanism to not only retain cellular memory but also provide necessary plasticity in human embryonic stem cells, as well as in cancer cells.
In addition to histone modifications, variants of histone proteins and their nucleosomal distribution within the regulatory regions of certain genes during mitosis appear to contribute to transcriptional memory (Weber and Henikoff 2014). The histone variants H3.3 and H2A.Z are well-studied examples (Ng and Gurdon 2008; Kelly et al. 2010). For example, the H3.3 variant is predominantly distributed in actively transcribed genes during interphase, and this distribution is preserved during mitosis, indicating that incorporation of histone H3.3 in regulatory regions of genes that are reactivated immediately after mitosis may be a key hallmark of mitotically bookmarked genes. Consistent with a role of histone H3.3 in maintaining cellular memory, Gurdon and colleagues have found that incorporation of histone H3.3 into the Myogenic Differentiation 1 (MyoD1) gene promoter can maintain cellular transcription memory of the gene through 24 cell divisions (Ng and Gurdon 2007; Ng and Gurdon 2008). The histone H2A.Z variant appears to be retained during mitosis. Interestingly, genes that are active in the G2 phase of the cell cycle contain nucleosomes at 1+ position. These nucleosomes contain the H2A.Z variant, and slide onto transcription start site (TSS) during mitosis, thus silencing genes. It has been proposed that this sliding of H2A.Z-containing nucleosome at 1+ position may contribute to marking genes that require rapid reactivation following mitosis (Kelly et al. 2010). Together, these observations point to a central role for histone modifications and variants in making gene loci accessible in the condensed chromatin environment of mitotic chromosomes.
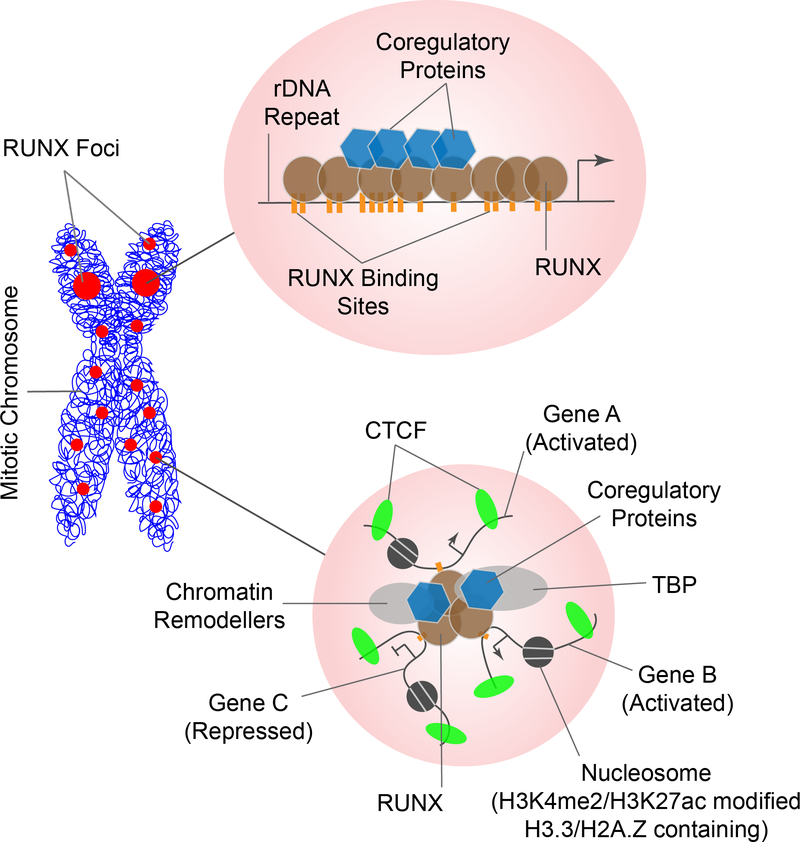
Studies using RUNX phenotypic transcription factors as a model to examine mitotic gene bookmarking have revealed two classes of genes that are bookmarked by RUNX proteins during mitosis (Zaidi et al. 2010): 1) Highly repetitive genes containing dozens of RUNX binding sites, thus providing a natural amplification of RUNX signal that is easily visualized by immunofluorescence microscopy. Ribosomal RNA genes are one example of such genes. Each of the rDNA repeats – that, in turn, is tandemly organized as 200–300 copies on each of the 5 acrocentric chromosomes in humans – contains more than 40 binding sites for RUNX proteins. Collectively, these repeats provide a concentration of high affinity binding sites for RUNX proteins, resulting in allelic visualization of RUNX nuclear foci that colocalize with RNA Polymerase I machinery and regulate the expression of rRNA genes post-mitotically (Young et al. 2007a; Ali et al. 2008). 2) Single copy genes that must be coordinately regulated to maintain cellular identity as well as proliferative and growth potential. For example, in osteoblasts, SMAD 2 and 4 genes – two key effectors of osteogenic BMP signaling – are localized on chromosome 18 and are bookmarked by RUNX2 during mitosis (Young et al. 2007b; Zaidi et al. 2011). This observation suggests that a subset of genes that are bookmarked by a transcription factor during mitosis may be organized in a “mitotic enhancers” to coordinate post-mitotic gene expression (Figure 1). It remains to be seen whether, in addition to containing multiple genes and transcriptional regulators, these “mitotic enhancer” also share other properties of the interphase enhancers, such as CTCF binding and presence of H3K4me2 and H3K27ac histone modifications.
Figure 1: Retention of RUNX proteins with target genes on mitotic chromosomes reveals two distinct mitotic microenvironments.
RUNX proteins (shown in red circles) associate with target genes on mitotic chromosomes (shown in blue line). More than a decade of studies has revealed two distinct target genes that are occupied by RUNX proteins during mitosis: 1) Genes that normally have several hundred copies in human genome, with each copy carrying several dozen RUNX binding sites (depicted as orange vertical lines in the top left circle). These genes provide a physiological amplification of RUNX occupancy on mitotic chromosomes (organized as large, allelic foci that can be identified by immunofluorescence microscopy). A key example is ribosomal RNA genes that are intimately linked with cell growth, and – together with a subset of regulatory proteins (blue hexagons) – are regulated by RUNX proteins (brown circles). 2) Single copy genes that carry fewer RUNX binding sites, but are localized on the same chromosome (depicted in bottom left circle). These genes may also be occupied by CTCF proteins (green ovals), with specialized nucleosomes (dark gray circles) containing histone variants H3.3 and/or H2A.Z, and may be coordinately activated (e.g., Genes A and B, usually linked with phenotype maintenance) and repressed (e.g., Gene C, often linked with cell proliferation) by RUNX proteins. The collective outcome of mitotic gene bookmarking by RUNX proteins is a coordinate regulation of cell growth and proliferation as well as lineage maintenance in post-mitotic cells.
Properties of Transcription Factors that Function as Mitotic Bookmarks
It is increasingly apparent that, in addition to interacting with target genes in a sequence specific manner, a mitotic bookmark, i.e., a transcription factor that occupies a subset of its target genes during mitosis, is usually a phenotypic regulatory protein. Studies over the past decade have demonstrated that more than 20 transcription factors and chromatin regulatory proteins including many lineage determining factors are retained on mitotic chromosomes. Examples include proteins involved in genome organization (e.g., the chromatin remodeler BRD4 and global enhancer binding protein CTCF), as well as lineage-restricted transcription factors that include the basic helix-loop-helix myogenic regulatory factors in muscle cell differentiation, CCAAT/enhancer-binding protein α in the adipocyte differentiation program, FoxA1 in liver cells, GATA1 and Runx1 in hematopoietic lineage differentiation, and Runx2 in osteoblast differentiation (Zaidi et al. 2003; Xing 2005; Sarge and Park-Sarge 2005; Young et al. 2007a; Young et al. 2007b; Ali et al. 2008; Bakshi et al. 2008; Pande et al. 2009; Dey et al. 2009; Blobel et al. 2009; Ali et al. 2010; Zaidi et al. 2010; Zhao et al. 2011; Ali et al. 2012; Kadauke et al. 2012; Arampatzi et al. 2013; Caravaca et al. 2013; Kadauke and Blobel 2013; Zaidi et al. 2014; Lake et al. 2014; Zaret 2014; Lodhi et al. 2014; Wong et al. 2014; Lopez-Camacho et al. 2014; Lodhi et al. 2016; Lerner et al. 2016; Festuccia et al. 2016). Each of these transcription factors occupy a subset of their target genes during mitosis in their respective lineages. Interference with mitotic bookmarking by these proteins results in deregulation of target genes following mitosis and compromised lineage identity. Interestingly, transcription factors that dictate lineage commitment of mesenchymal stem cell into muscle (MyoD), adipocytes (C/EBPa), or osteoblasts (RUNX2) not only bookmark RNA Pol II regulated genes during mitosis, but also the ribosomal RNA genes that are transcribed by RNA Pol I (Young et al. 2007a; Ali et al. 2008). Ribosomal RNA (rRNA) genes that are intimately linked with cellular growth potential provide a model system to study mechanistic ramifications of mitotic gene bookmarking. These tandemly repeated genes are located on five acrocentric chromosomes in humans and are naturally amplified regulatory units with binding sites for numerous transcription factors in addition to RUNX proteins; these include Myc, MyoD, and C/EBPa and RUNX proteins (Young et al. 2007a; Ali et al. 2008). In undifferentiated mesenchymal cells, rRNA genes are occupied by Myc, which is a transcriptional activator of these genes, and an upregulator of genes that are involved in cell proliferation. When mesenchymal stem cells are differentiated into myoblasts, adipocytes or osteoblasts, they exit cell cycle and Myc is replaced by MyoD, C/EBPa or RUNX2, respectively, each of which bookmarks and downregulates rRNA genes, as well as genes that are involved in cell cycle regulation. Concomitantly, these phenotypic proteins mitotically bookmark genes that are expressed immediately after mitosis and are critical for commitment to and maintenance of their respective lineages. (Ali et al. 2008). These findings highlight an important mechanistic aspect of mitotic bookmarking during lineage commitment i.e., coordination of cell proliferation, cell growth and cell identity. Genome-wide experimental approaches involving endogenous transcription factors in biologically relevant systems will provide further mechanistic insights into functional relevance of mitotic gene bookmarking in maintaining epigenetic cell memory in progeny cells.
Mitotic Bookmarking by Oncogenes as a Mechanism for Maintenance of Disease Phenotype
Recent studies also suggest that mitotic gene bookmarking has an important role in the onset, progression, and perpetuation of disease (Zaidi et al. 2014). A key example is provided by the leukemic fusion protein AML1-ETO (also known as RUNX1T1) that blocks myeloid cell differentiation and enhances proliferative potential (Bakshi et al. 2008). Interestingly, the leukemic AML1-ETO fusion protein mitotically bookmarks rRNA genes, as well as genes controlling cell proliferation and myeloid cell differentiation. Functionally, AML1-ETO upregulates rRNA and cell proliferation-related genes, but downregulates genes that mediate myeloid cell differentiation, promoting and/or supporting the transformed phenotype. Another recent example of cancer-related mitotic gene bookmarking is the mixed lineage leukemia protein (MLL). MLL is a chromatin-remodeling factor that is associated with leukemia and regulates transcription by recruiting chromatin modifying machinery to target genes (Gilliland et al. 2004). The mitotic retention of MLL with target genes favors rapid post-mitotic reactivation of target gene transcription required for the onset and progression of MLL post- mitotically (Blobel et al. 2009; Follmer et al. 2012). Another example of a link between human disease and mitotic gene bookmarking is provided by the examination of hepatocyte nuclear factor HNFβ1 (Lerner et al. 2016). HNFβ1 is frequently mutated in Congenital Abnormalities of Kidney and Urogenital Tract. Many of these mutations disrupt DNA binding activity of HNFβ1, and compromise its gene bookmarking capabilities. Whether disruption of mitotic gene bookmarking by HNFβ1 contributes to the observed congenital abnormalities remains to be seen. It will be informative to establish whether mitotic bookmarking of disease/cancer-related genes is a shared trait of all oncogenic proteins that interact with target genes in a sequence-specific manner.
Concluding Remarks
It is increasingly apparent that mitotic bookmarking is a central epigenetic mechanism that is essential for maintenance of cellular memory through cell divisions. Emerging evidence indicate that phenotypic transcription factors mitotically bookmark a subset of target genes and that this bookmarking plays a key role in coordination of cell proliferation, growth and differentiation. Importantly, mitotic gene bookmarking by oncogenes in cancer cells appear to be necessary for maintenance of the tumor phenotype. There are several open ended questions that must be addressed to acquire further comprehensive mechanistic insights into mitotic gene bookmarking by RUNX proteins and transcription factors in a broad biological context: 1) Do genes that are localized on the same chromosome and are regulated by the same transcription factors are also bookmarked during mitosis for coordinate transcriptional regulation post-mitotically? 2) What extracellular signals regulate the switch between mitotic bookmarking of a gene by global transcriptional activators (e.g., Myc) in undifferentiated cells and by phenotypic transcription factors as cells commit to a specific lineage? 3) What are the contributions of coregulatory proteins to reactivation of a bookmarked gene post-mitotically? 4) Can the accumulation of transcription factors on mitotic chromosomes be therapeutically targeted in dividing cells? 5) Does mitotic bookmarking play any role in asymmetrically dividing cells? Additional studies will be necessary to functionally link mitotic gene bookmarking and maintenance of cellular memory within in relation to biological control and pathology.
References
- Ali SA, Dobson JR, Lian JB, et al. (2012) A Runx2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. Journal of Cell Science. doi: 10.1242/jcs.100909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SA, Zaidi SK, Dacwag CS, et al. (2008) Phenotypic transcription factors epigenetically mediate cell growth control. Proceedings of the National Academy of Sciences 105:6632–6637. doi: 10.1073/pnas.0800970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SA, Zaidi SK, Dobson JR, et al. (2010) Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proceedings of the National Academy of Sciences 107:4165–4169. doi: 10.1073/pnas.1000620107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arampatzi P, Gialitakis M, Makatounakis T, Papamatheakis J (2013) Gene-specific factors determine mitotic expression and bookmarking via alternate regulatory elements. Nucleic Acids Research. doi: 10.1093/nar/gks1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attar N, Kurdistani SK (2014) Layered regulation of the epigenome and its links to cellular metabolism and physiology.
- Bakshi R, Zaidi SK, Pande S, et al. (2008) The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. Journal of Cell Science 121:3981–3990. doi: 10.1242/jcs.033431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, et al. (2009) A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Molecular Cell 36:970–983. doi: 10.1016/j.molcel.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, et al. (2013) Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes & Development 27:251–260. doi: 10.1101/gad.206458.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerase A, Smeets D, Tang YA, et al. (2014) Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci USA 111:2235–2240. doi: 10.1073/pnas.1312951111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG (2010) Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10:457–469. doi: 10.1038/nrc2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Nishiyama A, Karpova T, et al. (2009) Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell 20:4899–4909. doi: 10.1091/mbc.E09-05-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N, Dubois A, Vandormael-Pournin S, et al. (2016) Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat Cell Biol. doi: 10.1038/ncb3418 [DOI] [PubMed] [Google Scholar]
- Follmer NE, Wani AH, Francis NJ (2012) A Polycomb Group Protein Is Retained at Specific Sites on Chromatin in Mitosis. PLoS Genetics 8:e1003135. doi: 10.1371/journal.pgen.1003135.s005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland DG, Jordan CT, Felix CA (2004) The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program 80–97. doi: 10.1182/asheducation-2004.1.80 [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ (1997) Mitotic repression of the transcriptional machinery. Trends in Biochemical Sciences 22:197–202. [DOI] [PubMed] [Google Scholar]
- Grandy RA, Whitfield TW, Wu H, et al. (2016) Genome-Wide Studies Reveal that H3K4me3 Modification in Bivalent Genes Is Dynamically Regulated during the Pluripotent Cell Cycle and Stabilized upon Differentiation. Mol Cell Biol 36:615–627. doi: 10.1128/MCB.00877-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M, Weintraub H (1982) Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell 30:131–139. [DOI] [PubMed] [Google Scholar]
- Hartl P, Gottesfeld J, Forbes DJ (1993) Mitotic repression of transcription in vitro. The Journal of Cell Biology 120:613–624. doi: 10.1083/jcb.120.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Lehming N (2003) Global effects of histone modifications. Briefings in Functional Genomics and Proteomics 2:234–243. [DOI] [PubMed] [Google Scholar]
- Hsiung CCS, Morrissey CS, Udugama M, et al. (2015) Genome accessibility is widely preserved and locally modulated during mitosis. Genome Research 25:213–225. doi: 10.1101/gr.180646.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Workman JL (1998) Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays 20:275–279. doi: [DOI] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA (2013) Mitotic bookmarking by transcription factors. Epigenetics & Chromatin 6:1–1. doi: 10.1186/1756-8935-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Udugama MI, Pawlicki JM, et al. (2012) Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150:725–737. doi: 10.1016/j.cell.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, et al. (2010) H2A.Z Maintenance during Mitosis Reveals Nucleosome Shifting on Mitotically Silenced Genes. Molecular Cell 39:901–911. doi: 10.1016/j.molcel.2010.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin Modifications and Their Function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Lake RJ, Tsai P-F, Choi I, et al. (2014) RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking. PLoS Genetics 10:e1004204–15. doi: 10.1371/journal.pgen.1004204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner J, Bagattin A, Verdeguer F, et al. (2016) Human mutations affect the epigenetic/bookmarking function of HNF1B. Nucleic Acids Research 44:8097–8111. doi: 10.1093/nar/gkw467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi N, Ji Y, Tulin A (2016) Mitotic Bookmarking: Maintaining Post-Mitotic Reprogramming of Transcription Reactivation. Curr Mol Bio Rep 2:10–15. doi: 10.1007/s40610-016-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi N, Kossenkov AV, Tulin AV (2014) Bookmarking promoters in mitotic chromatin: poly(ADP-ribose)polymerase-1 as an epigenetic mark. Nucleic Acids Research 42:7028–7038. doi: 10.1093/nar/gku415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Camacho C, van Wijnen AJ, Lian JB, et al. (2014) CBFβ and the leukemogenic fusion protein CBFβ-SMMHC associate with mitotic chromosomes to epigenetically regulate ribosomal genes. J Cell Biochem 115:2155–2164. doi: 10.1002/jcb.24892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11:285–296. doi: 10.1038/nrg2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Dey A, Rabindran SK, et al. (1995) Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83:29–38. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Sanford S, Levens D (1997) Marking of active genes on mitotic chromosomes. Nature 388:895–899. doi: 10.1038/42282 [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB (2008) Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10:102–109. doi: 10.1038/ncb1674 [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB (2007) Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol 10:102–109. doi: 10.1038/ncb1674 [DOI] [PubMed] [Google Scholar]
- Pande S, Ali SA, Dowdy CR, et al. (2009) Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol 218:473–479. doi: 10.1002/jcp.21630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J (2006) Mitosis: a matter of getting rid of the right protein at the right time. Trends in Cell Biology 16:55–63. doi: 10.1016/j.tcb.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Ptashne M (2013) Epigenetics: Core misconcept. Proceedings of the National Academy of Sciences. doi: 10.1073/pnas.1305399110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera RM, Bennett LB (2010) Epigenetics in humans: an overview. Current Opinion in Endocrinology, Diabetes and Obesity 17:493–499. doi: 10.1097/MED.0b013e3283404f4b [DOI] [PubMed] [Google Scholar]
- Sabbattini P, Canzonetta C, Sjoberg M, et al. (2007) A novel role for the Aurora B kinase in epigenetic marking of silent chromatin in differentiated postmitotic cells. The EMBO Journal 26:4657–4669. doi: 10.1038/sj.emboj.7601875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge O-K (2005) Gene bookmarking: keeping the pages open. Trends in Biochemical Sciences 30:605–610. doi: 10.1016/j.tibs.2005.09.004 [DOI] [PubMed] [Google Scholar]
- Sarkies P, Sale JE (2012) Cellular epigenetic stability and cancer. Trends in Genetics 28:1–10. doi: 10.1016/j.tig.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Schübeler D (2015) Function and information content of DNA methylation. Nature 517:321–326. doi: 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Strahl B, Allis C (2000) The language of covalent histone modifications. [DOI] [PubMed]
- Struhl G (1981) A gene product required for correct initiation of segmental determination in Drosophila. Nature 293:36–41. [DOI] [PubMed] [Google Scholar]
- Wang F, Higgins JMG (2013) Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends in Cell Biology 23:175–184. doi: 10.1016/j.tcb.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Weber CM, Henikoff S (2014) Histone variants: dynamic punctuation in transcription. Genes & Development 28:672–682. doi: 10.1101/gad.238873.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H (1985) Assembly and propagation of repressed and depressed chromosomal states. Cell 42:705–711. [DOI] [PubMed] [Google Scholar]
- Wong MM, Byun JS, Sacta M, et al. (2014) Promoter-Bound p300 Complexes Facilitate Post-Mitotic Transmission of Transcriptional Memory. PLoS ONE 9:e99989. doi: 10.1371/journal.pone.0099989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H (2005) Mechanism of hsp70i Gene Bookmarking. Science 307:421–423. doi: 10.1126/science.1106478 [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, et al. (2007a) Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445:442–446. doi: 10.1038/nature05473 [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang X, et al. (2007b) Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci USA 104:3189–3194. doi: 10.1073/pnas.0611419104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Grandy RA, Lopez-Camacho C, et al. (2014) Bookmarking target genes in mitosis: a shared epigenetic trait of phenotypic transcription factors and oncogenes? Cancer Research 74:420–425. doi: 10.1158/0008-5472.CAN-13-2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino M, et al. (2011) Bookmarking the genome: maintenance of epigenetic information. Journal of Biological Chemistry 286:18355–18361. doi: 10.1074/jbc.R110.197061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, et al. (2010) Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet 11:583–589. doi: 10.1038/nrg2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SM, et al. (2003) Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci USA 100:14852–14857. doi: 10.1073/pnas.2533076100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS (2014) Genome Reactivation after the Silence in Mitosis: Recapitulating Mechanisms of Development? Developmental Cell 29:132–134. doi: 10.1016/j.devcel.2014.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Nakamura T, Fu Y, et al. (2011) Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature Publishing Group 13:1295–1304. doi: 10.1038/ncb2341 [DOI] [PMC free article] [PubMed] [Google Scholar]