Abstract
Differentiation of insulin-producing β cells from induced pluripotent stem cells (iPSCs) derived from patients with diabetes promises to provide autologous cells for diabetes cell replacement therapy. However, current approaches produce such patient iPSC-derived β (SC-β) cells with poor function in vitro and in vivo. Here, we used CRISPR/Cas9 to correct a diabetes-causing pathogenic variant in Wolfram Syndrome 1 (WFS1) in iPSCs derived from a patient with Wolfram Syndrome (WS). After differentiation with our recent 6-stage differentiation strategy, corrected WS SC-β cells performed robust dynamic insulin secretion in response to glucose and reversed pre-existing streptozocin-induced diabetes when transplanted into mice. Single-cell transcriptomics showed that corrected SC-β cells displayed increased insulin and decreased expression of genes associated with endoplasmic reticulum stress. CRISPR/Cas9 correction of a diabetes-inducing gene variant thus allows for robust differentiation of autologous SC-β cells that can reverse severe diabetes in an animal model.
One Sentence Summary:
Patient stem cell-derived β cells CRISPR/Cas9-corrected for a diabetes-causing gene variant in WFS1 restore glucose homeostasis when transplanted into diabetic mice.
Introduction
Derivation of induced pluripotent stem cells (iPSCs) from patients followed by differentiation into disease-relevant cell types holds great promise for in vitro disease modeling, drug screening, and autologous cell replacement therapy for multiple diseases (1, 2). Diabetes mellitus is caused by the death or dysfunction of insulin-producing β cells within the pancreas. Although insulin injections are often used to replace this lost function (3), long-term complications can arise (4). Alternatively, transplantation of cadaveric allogeneic islets containing β cells has been performed successfully, demonstrating the feasibility of a cell therapy approach that is however limited due to low donor numbers and the need for immunosuppressant drugs (5-7).
Stem-cell derived β cells (SC-β cells) differentiated from iPSCs derived from patients with diabetes would provide a source of autologous replacement cells (8), but the lack of robust physiological function of these cells has been an unmet need in the field (9). Specifically, prior reports using patient iPSCs have generated pancreatic or endocrine progenitors lacking β cell identity (10-14). Recently we and others have developed differentiation strategies with human embryonic stem cells (hESCs) to generate functional non-progenitor SC-β cells in vitro as an alternative source of replacement cells (15-17). Although these and similar approaches have been used in vitro to generate iPSC- or nuclear transfer stem cell-derived β cells from patients with Type 1 (18, 19), Type 2 (20), and neonatal diabetes (21, 22), these cells have showed only modest function in vitro and in vivo. In particular, unlike with primary β cells, these SC-β cells derived from patients with diabetes required long times after transplantation (12-19 wk) to functionally mature and normalize blood glucose in modestly diabetic mice or had a high failure rate, being unable to achieve normoglycemia or having formation of overgrowths. In addition, they were not transplanted into mice with pre-existing diabetes and in vitro dynamic glucose-stimulated insulin secretion (GSIS) was not tested. To overcome these limitations, we recently developed a differentiation protocol that leverages a previously unknown role of the cytoskeleton in pancreatic fate choice to produce highly functional SC-β cells across multiple cell lines (23).
Single pathogenic gene variants that cause diabetes can be corrected in iPSCs (21, 22) using CRISPR/Cas9 gene editing (24). One applicable condition is Wolfram Syndrome (WS), a rare autosomal recessive disorder caused by pathogenic variants in the WFS1 gene (25, 26), which cause chronic endoplasmic reticulum (ER) stress that stimulates the unfolded protein response and eventually causes the death of β cells (27-29). Individuals with WS develop diabetes in childhood along with other ailments, including optic nerve atrophy and neurodegeneration (26). ER stress is a common feature shared with all forms of diabetes and other diseases (30-35). There is currently no effective treatment for WS or ER stress-related diseases (26).
Here we combined our recently devised differentiation strategy (23) with CRIPSR/Cas9 gene editing of WFS1 in patient-derived iPSCs to generate autologous, gene-corrected SC-β cells from patients with WS. We assessed the differentiation efficacy and glucose-stimulated insulin secretion of the gene-corrected, patient-derived SC-β cells as compared to unedited cells both in vitro and in vivo when transplanted into a mouse model of diabetes. We also used single-cell RNA sequencing (scRNA-seq) to further characterize these cells and followed up these observations with assessment of ER and mitochondrial stress. Our work highlights that CRISPR/Cas9 correction of patient-derived iPSCs can be used to generate cells with utility in diabetes cell replacement therapy.
Results
Gene-corrected WS SC-β cells display dynamic glucose stimulated insulin secretion and express β cell markers
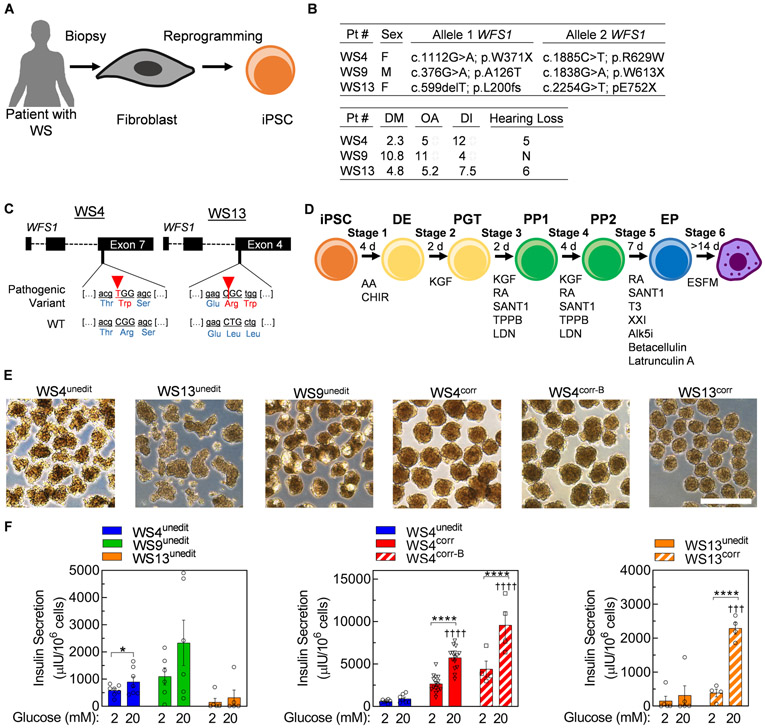
We started our study with iPSC lines reprogrammed from fibroblasts of two patients with WS, previously (36) (WU.WOLF-09 and WU.WOLF-13, designated WS9unedit and WS13unedit, respectively, in this study) and also generated iPSCs from skin fibroblasts of one additional, previously described patient with Wolfram Syndrome (25) (WU.WOLF-04, designated WS4unedit in this study). We used CRISPR/Cas9 to correct the pathogenic variants in the WFS1 gene for patients WU.WOLF-04 and WU.WOLF-13, generating multiple corrected iPSC clones designated WS4corr, WS4corr-B, and WS13corr (Fig. 1A-C, fig. S1, table S1). We attempted to adapt the unedited iPSC lines from all three patients to suspension culture for propagation and differentiation to generate SC-β cells as previously reported (17). However, we were unable to establish these suspension cultures due to extensive cell death and low cell yields. Due to these challenges, we did not continue to attempt suspension differentiations for corrected clones and instead used our recent differentiation protocol based on cytoskeletal modulation (23). We differentiated WS iPSCs into SC-β cells with small molecules and growth factors to recapitulate pancreatic development using attachment culture and aggregated the final cell population into islet-like clusters for assessment (Fig. 1D-E). Differentiation of unedited iPSC lines from all three patients produced cells capable of secreting insulin (Fig. 1F). However, our three CRISPR/Cas9-corrected cells lines (WS4corr, WS4corr-B, and WS13corr) were more glucose responsive, secreting 6.4±1.4x, 10.6±1.3x, and 7.2±0.6x more insulin at 20 mM glucose compared to their unedited isogenic and patient-matched controls, respectively (Fig. 1F). These data indicate that Wolfram Syndrome-causing pathogenic variants in WFS1 contribute to defects in GSIS that can be corrected with gene editing.
Fig. 1. CRISPR/Cas9 correction of WFS1 generates functional WS SC-β cells in vitro.
(A) Schematic summary of iPSC generation from patients with WS. (B) Information on the 3 patients, including the genetic location of autosomal recessive pathogenic variants in WFS1 and age of symptom onset. N=none, M=male, F=female, DM=diabetes mellitus, OA=optic atrophy, DI=diabetes insipidus. (C) Gene variants in WFS1 in WS4unedit and WS13unedit iPSCs targeted for CRISPR/Cas9 correction. (D) Schematic of differentiation protocol mimicking embryonic development of pancreatic β cells. (E) Bright field images of stage 6 clusters produced from all cell lines in this study. Scale bar, 500 μm. (F) Static GSIS functional assessment of WS4unedit (n=7), WS9unedit (n=6), and WS13unedit (n=5), WS4corr (n=15) and WS4corr-B (n=4), and WS13corr (n=4) stage 6 cells in an in vitro static GSIS assay. Data with WS4unedit and WS13unedit cells are regraphed to help with comparisons. Y axes differ between panels. *p<0.05, ****p<0.0001 by one-way paired t-test. †††p<0.001, ††††p<0.0001 by two-way unpaired t-test comparing to high glucose in unedited cells. iPSC, induced pluripotent stem cell; DE, definitive endoderm; PGT, primitive gut tube; PP, pancreatic progenitor; EP, endocrine progenitor. Act A, activin A; CHIR, CHIR99021; KGF, keratinocyte growth factor; RA, retinoic acid; LDN, LDN193189; T3, triiodothyronine; Alk5i, Alk5 inhibitor type II; ESFM, enriched serum-free medium.
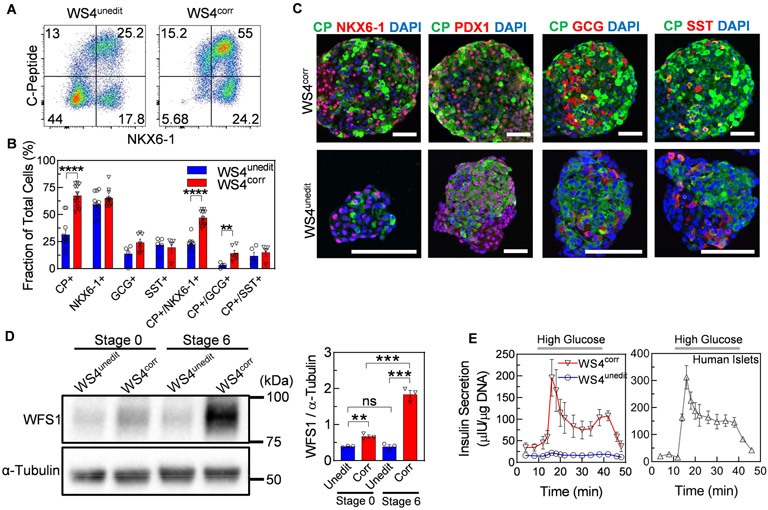
We performed further characterization of the islet-like clusters from patient WS4 expressing α, β, and δ cell hormones with and without CRISPR/Cas9 correction (fig. S2A). WS4unedit, WS4corr, and WS4corr-B all produced C-peptide+ cells that co-stained with β cell transcription factors NKX6-1 and PDX1 (Fig. 2A-C, fig. S2B-D). However, SC-β cell generation, as indicated by C-peptide+/NKX6-1+ cells, was lower for WS4unedit (22±2%) cells compared to WS4corr (47±2%) and WS4corr-B (59.1±1%) cells (Fig. 2A-C, fig. S2B-D). The fractional yield of SC-β cells from corrected clones was similar to our previously reported SC-β cells derived from donors without diabetes (non-diabetic SC-β cells) (17, 23). The early stages of differentiation generated cells with similar relative gene expression and fraction of cells expressing key progenitor markers (fig. S3). We began to detect differences in differentiation efficiency at stage 5, when endocrine cell induction predominantly occurs (15), and these differences amplified during stage 6 (Fig. 2C). The mechanistic cause of WS4unedit SC-β cell differentiation defects is unknown, but other reports have shown organelle calcium and oxidative stress influence the differentiation of other cell types (37, 38). WFS1 gene and protein expression were detectable across all stages of differentiation. However, the detrimental effects of WFS1 pathogenic variants on differentiation towards SC-β cells appeared mostly during later stages of differentiation when WFS1 expression increased (Fig. 2D, fig. S4). We observed lower expression of WFS1 protein in WS4unedit compared to WS4corr cells (Fig. 2D). In prior reports, variant WFS1 protein and RNA were shown to be unstable and undergo proteasomal degradation, cellular depletion, and non-sense-mediated decay (39, 40).
Fig. 2. In vitro characterization of unedited and corrected β cells from iPSCs derived from an individual with WS.
(A) Representative flow cytometry dot plots and (B) quantified fraction of cells expressing or co-expressing pancreatic β cell or islet markers for WS4unedit (n=4-7) and WS4corr (n=4-8) stage 6 cells. **p<0.01, ****p<0.0001 by two-way unpaired t-test. (C) Immunostaining of sectioned WS4corr and WS4unedit stage 6 clusters stained for β cell or islet markers. Scale bar, 100 μm. (D) Western blot (left) and quantified intensity (right) of WFS1 protein in stage 0 and stage 6 WS4corr (n=3) and WS4unedit (n=3) cells. **p<0.01, ***p<0.001 by two-way unpaired t-test. (E) Dynamic human insulin secretion of WS4corr (n=5) and WS4unedit (n=4) stage 6 cells and primary human islets (17). Clusters were perfused with 2 mM glucose except where indicated by high glucose (20 mM). CP, C-peptide; GCG, glucagon; SST, somatostatin.
Lack of WFS1 can cause β cell dysfunction (27, 29, 32). Indeed, although both WS4unedit and WS4corr SC-β cells responded to glucose by secreting insulin in a static assay (Fig. 1F), only WS4corr SC-β cells demonstrated robust first- and second-phase insulin secretion (Fig. 2E). WS4corr SC-β cells secreted 8.9±2.1x more insulin in dynamic GSIS assays compared to WS4unedit SC-β cells at peak (first phase) insulin secretion in 20 mM glucose. WS4corr SC-β cells were functionally similar to non-diabetic SC-β cells and human islets (Fig. 2E), as previously reported (17). Even accounting for the differences in differentiation efficacy between WS4unedit and WS4corr iPSC lines, WS4corr SC-β cells still showed superior static and dynamic GSIS (fig. S5). Collectively, these data demonstrate that CRISPR/Cas9 correction of the WFS1 pathogenic variant in WS iPSCs increased SC-β cell differentiation efficacy and function, leading to greater and near islet-like dynamic insulin secretion in vitro in response to high glucose.
Gene-corrected WS SC-β cells correct blood glucose in mice with pre-existing diabetes
To evaluate the potential of corrected WS SC-β cells for cell replacement therapy, we transplanted 5x106 WS4unedit stage 6 cells, WS4corr stage 6 cells, and cadaveric human islet cells under the kidney capsule of mice previously rendered diabetic by streptozotocin (STZ) injection to destroy endogenous mouse β cells (Fig. 3A, fig. S6). Three sham mice spontaneously died, two at 14 wk and one at 25 wk, presumably due to their diabetes. After transplantation with WS4corr SC-β cells, blood glucose dropped <200 mg/dL within 1 wk (Fig. 3B), with an average blood glucose concentration of 81±5 mg/dL from 2-27 wk. The grafts at two weeks contained cells mostly expressing C-peptide but there were also minority populations of glucagon+ and somatostatin+ cells (Fig. 3C, fig. S6G). We performed glucose tolerance tests 9 d and 10 wk after transplantation, with WS4corr SC-β cells demonstrating improved glucose tolerance compared to transplanted WS4unedit cells and control sham mice (Fig. 3D-E). Transplanted human islets performed similarly albeit more slowly compared to WS4corr cells in mice, resulting in normoglycemia at 2 wk, improved glucose tolerance at 9 days, and demonstrating in vivo GSIS at 2 wk post transplantation (Fig. 3B, 3D-E). For the 6-month observation period, both mice transplanted with WS4unedit SC-β cells and sham mice were unable to achieve glycemic control, whereas the WS4corr SC-β cells maintained blood glucose normalization. We collected mouse serum 2 and 10 wk after transplantation, and although all transplantation groups had detectable human insulin, WS4corr had higher concentrations than WS4unedit, 5.8±1.5x and 14.4±2.8x at 2 and 10 wk respectively, and human islets had higher concentrations than WS4unedit, 4.4±1.5x at 2 wk, (Fig. 3F). This was consistent with our in vitro observations of increased insulin secretion and glucose responsiveness of WS4corr compared to WS4unedit SC-β cells in vitro. Serum insulin concentrations doubled from 2 to 10 wk in the WS4corr SC-β cells, suggesting the SC-β cells were maturing in vivo (Fig. 3F). Mice transplanted with WS4unedit SC-β cells also had a higher human proinsulin-to-insulin ratio in the serum, suggesting the presence of ER stress and problems with insulin processing (Fig. 3G). We performed nephrectomy of two live mice 12 wk after their transplantation with WS4corr SC-β cells, which resulted in a return to hyperglycemia within 1 wk, indicating that prior glycemic control was due to the transplanted WS4corr SC-β cells (Fig. 3H). We observed that kidneys receiving WS4unedit cells contained undesirable large overgrowths that were not present in any kidneys with WS4corr cells (fig. S6H). The ability of WS4corr SC-β cell-transplanted mice to reverse diabetes demonstrates that CRISPR/Cas9 correction of differentiated iPSCs from a patient with diabetes is a promising source of autologous β cells for diabetes therapy.
Fig. 3. Transplantation of gene edited patient-derived β cells into mice reverses preexisting diabetes.
(A) Schematic of diabetes induction with streptozotocin (STZ), transplantation of stage 6 cells containing WS SC-β cells, and nephrectomy of the transplanted mice. (B) Blood glucose measurements before and after STZ treatment, and after transplantation with SC-β cells or human islets. Five groups were studied: Diabetic mice without a transplant (STZ, No Txp; n=7; black), diabetic mice transplanted with WS4unedit stage 6 cells (STZ, WS4unedit Txp; 5x106 cells; n=6; blue), diabetic mice transplanted with human islets (STZ, HI Txp; 4000 IEQ; n=4; dark grey), diabetic mice transplanted with WS4corr stage 6 cells (STZ, WS4corr Txp; 5x106 cells; n=10; red), and non-diabetic mice with a sham transplant (No STZ, No Txp; n=5; light grey). (C) Immunostaining of sectioned kidney explanted from a mouse that had received WS4corr stage 6 cells 2 wk prior. Scale bar, 100 μm. (D) Glucose tolerance test (GTT) 9 d and 10 wk after Txp. Five groups were studied: STZ, No Txp (n=7 at 9 d and 10 wk; black); STZ, WS4unedit Txp (n=6 at 9 d; n=5 at 10 wk; blue); STZ, HI Txp (n=4 at 9 d; dark grey); STZ, WS4corr Txp (n=10 at 9 d; n=9 at 10 wk; red); and No STZ, No Txp (n=5 at 9 d and 10 wk; light grey). (E) Area under the curve (AUC) quantification of GTT data. **p<0.01, ***p<0.001 by Mann-Whitney two-tailed nonparametric test. (F) In vivo GSIS secretion 2 and 10 wk after transplantation for STZ, WS4unedit Txp (n=6 and 5 at 2 and 10 wk; blue); STZ, WS4corr Txp (n=10 and 9 at 2 and 10 wk; red); and STZ, HI Txp (n=4; dark grey) mice at 0 and 60 min after 2 g/kg glucose injection. ns=not significant, **p<0.01 by one-way paired t-test. †p<0.05, †††p<0.001 by two-way unpaired t-test compared to WS4unedit 60 min measurements. (G) Molar ratio of serum human proinsulin to insulin for STZ, WS4unedit Txp (n=5; blue) and STZ, WS4corr Txp (n=9; red) 60 min after 2 g/kg glucose injection 10 wk after transplantation. ***p<0.001 by two-way unpaired t-test. (H) Blood glucose measurements before and after nephrectomy of two STZ, WS4corr Txp (orange) mice compared to remaining non-nephrectomized STZ, WS4corr Txp (n=6; red). CP, C-peptide; GCG, glucagon; SST, somatostatin.
Single-cell RNA sequencing of differentiated WS iPSCs reveals SC-β cell, pancreatic endocrine, and non-pancreatic populations
To compare SC-β cells generated from WS4corr and WS4unedit iPSCs while accounting for differences in differentiation efficacy (Fig. 2A), we performed scRNA-seq using the 10x Genomics platform. We sequenced a total of 7,215 and 3,640 stage 6 single cells differentiated from WS4unedit and WS4corr hiPSCs, respectively, averaging 50,996 reads per cell. After quality control (fig. S7), an average of 2,861 genes were detected per cell. We used dimensionality reduction and unsupervised clustering to evaluate our scRNA-seq data and identified 8 cell populations in WS4corr and WS4unedit (Fig. 4A) based on the top upregulated genes in each cluster matched to exocrine and endocrine cell types distinguished in published pancreatic transcriptome data (table S2) (43).
Fig. 4. Single cell transcriptional analysis reveals WS4corr and WS4unedit SC-β cell populations and off-targets.
(A) tSNE projection from unsupervised clustering of transcriptional data from scRNA-seq of WS4unedit and WS4corr stage 6 cells. (B) Calculated percentages of defined cluster populations for WS4unedit and WS4corr stage 6 cells. (C) Heat map of key β cell population gene markers (insulin [INS], chromogranin A [CHGA], SPINK1, ID3) with low/none (grey), medium (yellow), and high (red) expression. NP1, neural progenitor 1; NP2, neural progenitor 2; NP3, neural progenitor 3; PH, polyhormonal; EC, enterochromaffin.
We identified SC-β cell populations in stage 6 cells differentiated from both WS4corr and WS4unedit iPSCs (Fig. 4A). Although SC-β cells were the largest population in differentiated WS4corr cells (42%), they were a minority population in differentiated WS4unedit cells (11%) (Fig. 4B). The vast majority (91%) of cells within WS4corr stage 6 cells were identified as pancreatic endocrine (SC-β, SC-α, SC-δ), whereas pancreatic endocrine (SC-β, polyhormonal) were a minority (16.5%) for WS4unedit cells (Fig. 4A-B). The majority of WS4unedit stage 6 cells were either pancreatic exocrine or non-pancreatic cells (Fig. 4A-B), with many cells expressing non-β cell markers such as SPINK1 and ID3 (Fig. 4C, fig. S7C, fig. S8A), suggesting that the WFS1 pathogenic variants carried by these cells caused misdirection of cell fate choice to off-targets with our differentiation protocol. Gene expression of the off-target markers was detectable as early as stage 2 with real-time PCR (fig. S8B), suggesting the non-SC-β cell off-target cells were likely expanding as differentiation progressed to reduce the fraction of on-target pancreatic cells. The effects of these off-target populations on the function of the SC-β cell population, particularly in terms of transplantation efficacy, is unknown. Both WS4corr and WS4unedit pancreatic endocrine populations had cells with multiple detected hormones (insulin, glucagon, somatostatin) (Fig. 4C, fig. S7C), as has been observed with endocrine stem cell differentiations and human islets (44-46). Taken together, scRNA-seq enabled identification of SC-β cells from WS4corr and WS4unedit iPSCs differentiated to stage 6, in addition to several unexpected off-targets from WS4unedit cells, illustrating the greatly improved SC-β cell differentiation efficacy enabled by CRISPR/Cas9 correction in WS iPSCs.
CRISPR/Cas9 gene editing modulated SC-β cell gene expression
scRNA-seq enabled investigation of the transcriptome specifically in SC-β cells from the heterogeneous stage 6 cell population in both WS4corr and WS4unedit lines. This eliminated dilution effects on analysis of the bulk population that could occur due to the presence of non-SC-β cell populations and markers expressed by multiple cell types, such as NKX6-1 expressed by both pancreatic progenitors and β cells. We first evaluated expression of β cell markers within the SC-β cell population, as our prior work had shown that inflammatory stress could reduce key β cell proteins (18). INS, CHGA, and GCG expression within SC-β cells was reduced, whereas SST expression increased, in WS4unedit compared to WS4corr SC-β cells (Fig. 5A, table S3A). Additionally, gene expression of transcription factors important to β cell identity (NKX6-1, ISL1, PDX1) and GCK, a necessary β cell functional gene, were similar in WS4unedit compared to WS4corr SC-β cells. Real-time PCR measurements of the total stage 6 populations for WS4unedit, WS4corr, and WS4corr-B cell lines showed similar reductions in INS and CHGA transcripts (Fig. 5B, fig. S9A). Some β cell markers such as NKX2-2 were lower in WS4unedit compared to WS4corr and WS4corr-B stage 6 cells (Fig. 5B, fig. S9A), however this was likely due to lower differentiation yields of WS4unedit SC-β cells. Immunostaining confirmed that C-peptide+ cells co-expressed many β cell proteins, including PDX1, CHGA, ISL1, NKX6-1, and NEUROD1 (Fig. 5C, fig. S9B). These data indicate that SC-β cells with CRISPR/Cas9 correction of WFS1 were mostly similar in terms of β cell marker expression compared to unedited patient cells except notably for INS, which showed greatly reduced transcript abundance. Furthermore, both WS4unedit and WS4corr SC-β cells co-expressed many β cell protein markers, similar to prior reports on non-diabetic SC-β cells (17, 23).
Fig. 5. CRISPR/Cas9 correction of WFS1 improves β cell gene expression in differentiated cells.
(A) Violin plots detailing log-normalized gene expression of β cell and islet markers in the WS4unedit (blue) and WS4corr (red) SC-β cell populations defined in Fig. 4. Log fold change and p-values for violin plots are available in table S3A. (B) Real-time PCR analysis of the total stage 6 population measuring expression of β cell and islet genes for WS4unedit (n=6-13; blue) and WS4corr (n=7-14; red). **p<0.01, ***p<0.001, ****p<0.0001 by Man-Whitney two-tailed nonparametric test. (C), Immunostaining of single-cell dispersed WS4corr and WS4unedit stage 6 cells stained for indicated pancreatic and β cell markers. Scale bar, 50 μm.
CRISPR/Cas9 gene editing decreased SC-β cell ER and mitochondrial stress
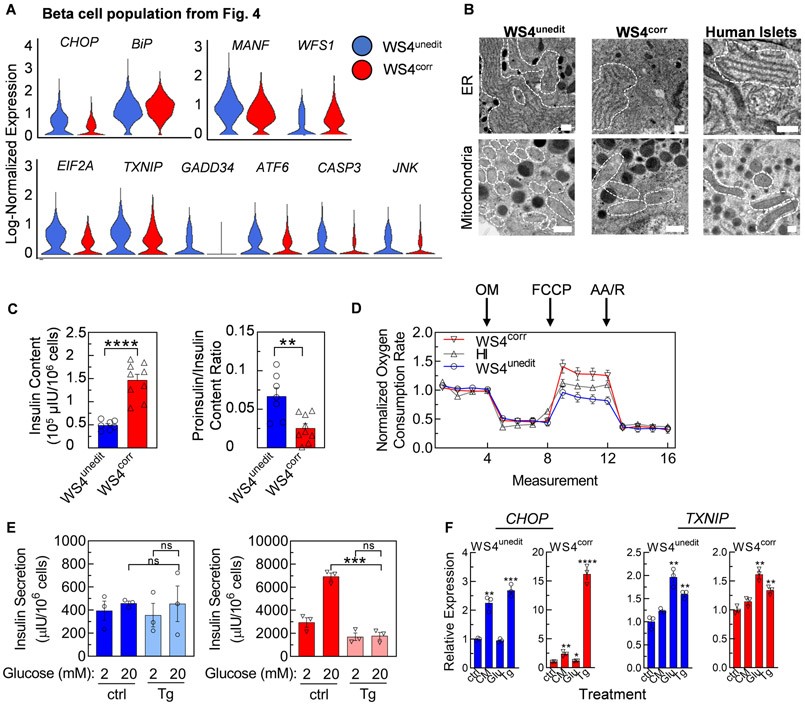
As pathogenic variants in WFS1 cause ER stress in β cells (27-29), we next evaluated the expression of stress markers within WS4corr and WS4unedit SC-β cells using our scRNA-seq data. ER stress can affect many pathways in cell metabolism, including activation of the unfolded protein response, mitochondrial stress, and apoptosis. We observed increased expression of many adaptive ER stress (eIF2a, MANF), terminal ER stress (CHOP), mitochondrial stress (TXNIP), and apoptotic (CASP3) markers within WS4unedit compared to WS4corr SC-β cells (Fig. 6A, table S3B). WFS1 expression was elevated in WS4corr compared to WS4unedit SC-β cells (Fig. 6A), consistent with mouse and immortalized cell line studies (28, 29) and transcript and protein measurements of the entire stage 6 population. Discerning definitive trends with real-time PCR analysis for a wide range of markers in the bulk stage 6 population was difficult (fig. S10A), likely due to the dilution effects of differing SC-β cell differentiation efficacy. Adaptive ER (eIF2a, MANF) and mitochondrial (TXNIP) stress markers are expressed across most cell types but are typically more highly expressed in high protein-producing and glucose-responsive cells (47), confounding study in this system.
Fig. 6. CRISPR/Cas9 correction of WFS1 reduces WS SC-β cell stress.
(A) Violin plots detailing log-normalized expression of stress genes in the WS4unedit (blue) and WS4corr (red) SC-β cell populations defined in Fig. 4. Log fold-change and adjusted p-values for violin plots available in table S3B. (B) Representative transmission electron microscopy images of ER (top) and mitochondria (bottom) for WS4unedit, WS4corr SC-β cells, and human islets. White dotted lines outline the ER and mitochondria in the cell cytoplasm. Scale bar, 500 nm. (C) Human insulin content (left) and proinsulin/insulin content ratio (right) of WS4unedit (n=7; blue) and WS4corr (n=9; red) stage 6 cells. **p<0.01, ****p<0.0001 by two-way unpaired t-test. (D) Mitochondrial respiration of WS4unedit stage 6 (n=11; blue), WS4corr stage 6 (n=9; red), and human islets (n=6; dark grey) represented as percentage of baseline oxygen consumption rate measurements. Respiration was interrogated by measuring changes in relative OCR after injection with oligomycin (OM), FCCP, and antimycin A (AA)/rotenone (R). (E) Static GSIS functional assessment of stage 6 cells treated with DMSO or thapsigargin (Tg). n=3. *** p<0.001 by two-way unpaired t-test. ns, not significant. (F) Real-time PCR analysis of bulk stage 6 population measuring expression of stress genes after treatment with cytokine mixture (CM), high glucose (Glu), or Tg. n=4. **p<0.01, ***p<0.001 by two-way unpaired t-test compared to ctrl. ctrl=control.
To investigate the failure of WS4unedit SC-β cells to properly function, we used transmission electron microscopy (TEM) to observe the morphology of SC-β cell ER and mitochondria (Fig. 6B, fig. S11A). ER and mitochondria for WS4corr cells appeared normal and healthy compared to human islets. WS4unedit cell ER were swollen and expanded in the cytoplasm, measuring 4.9±1.2x (p<0.05) and 3.0±0.8x (p<0.01) larger than WS4corr cell and human islets, respectively, which can occur in response to ER stress (32). The WS4unedit mitochondria appear fragmented and undergoing fission, measuring 1.9±0.3x (p<0.001) and 2.0±0.3x (p<0.0001) smaller than WS4corr and human islets, respectively, consistent with mitochondria exhibiting dysfunction in response to chronic ER stress (48-50). Our SC-β cells contained a variety of granules in various stages of maturation, as previously reported (15, 16, 51). As the ER is critical for proper insulin processing, we measured the insulin content and proinsulin-to-insulin ratio of stage 6 cells. We observed that WS4unedit cells had lower insulin content and a higher proinsulin-to-insulin ratio than WS4corr cells (Fig. 6C, fig. S11B). Combined with the observed increases in unfolded protein response markers and unhealthy ER morphology shown by TEM, these data support WS4unedit SC-β cells have defects in insulin processing that persist for months after transplantation. Mitochondria are critical for proper glucose sensing, and therefore we assessed their function by measuring the oxygen consumption rate (OCR) after injection of interrogating compounds (52). We found that WS4unedit cells had a lower OCR after injection with carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), a mitochondrial oxidative phosphorylation uncoupler, compared to WS4corr cells (p<0.0001; two-way ANOVA), WS4corr-B, and human islets (p<0.001; two-way ANOVA) (Fig. 6D, fig. S11C). There was no difference in OCR when interrogating the cells with Oligomycin or Antimycin A (AA) and rotenone injections, which inhibits ATP synthase and mitochondrial electron transport chain, respectively. This indicates WS4unedit cells have reduced maximal mitochondrial respiratory capacity, as seen in other reports of monogenic diabetes (53) and consistent with stressed and unhealthy mitochondria (Fig. 6A-B). The difference was not observed with undifferentiated WS4corr and WS4unedit iPSCs (fig. S11C), indicating this phenotype was restricted to certain cell types, such as SC-β cells. Taken together, SC-β cells harboring pathogenic variants in WFS1 displayed ER and mitochondrial dysfunction, which was restored with CRISPR/Cas9 gene correction.
To further investigate the effects of cellular stress on WS SC-β cells, we treated WS4unedit and WS4corr stage 6 cells with thapsigargin, an inhibitor of sarco/endoplasmic reticulum Ca2+ ATPase (SERCA), to induce ER stress. We performed static GSIS and whereas insulin secretion was not affected in WS4unedit cells, likely because these cells already had low secretion and function, insulin secretion in WS4corr cells was strongly negatively affected by this compound, decreasing insulin secretion by 3.9±0.7x and eliminating glucose-responsiveness (Fig. 6E). Using bulk stage 6 population analysis with real-time PCR and western blot, we also observed that both WS4unedit and WS4corr cells responded to thapsigargin treatment by increasing known stress markers (Fig. 6F, fig. S12, fig. S13). These data indicate that the enhanced functional and reduced ER stress transcriptional benefits of CRISPR/Cas9 gene correction can be reversed with chemical-induced ER stress.
Discussion
Here we applied a technological strategy of combining iPSCs derived from a patient with WS, CRISPR/Cas9 correction of the disease-causing pathogenic gene variant, and our recent robust differentiation protocol to generate SC-β cells that were able to successfully and rapidly rescue mice with severe pre-existing diabetes. Correction of WFS1 drastically improved SC-β cell differentiation efficacy and function in the SC-β cells, which displayed robust first- and second-phase insulin secretion. scRNA-seq and direct assessment of the ER and mitochondria revealed reduction of stress in these organelles and a reduction in apoptotic markers after CRISPR/Cas9 correction.
Prior work with WS patient-derived iPSCs in the context of diabetes did not utilize gene therapy nor generate SC-β cells. Instead, this work focused on earlier stages of development by generating and studying pancreatic progenitors and polyhormonal endocrine cells that lack bone fide β cell features, such as high insulin per cell, dynamic function, or diabetes reversal in mice upon transplantation (12). More recent work with patient-derived iPSCs or nuclear transfer stem cells having other forms of diabetes has instead generated SC-β cells, but these cells are still missing important features of mature β cells, most noteworthy being continued inferior function (18-22). There is also limited data comparing differentiation of nuclear transfer stem cells (19) and iPSCs (15-18, 21, 22) to SC-β cells, so more studies comparing these two potential autologous cell sources is needed, particularly as the immunogenicity of these cells is unclear (8). In terms of cell therapy, only prevention, not reversal, of diabetes in mice using SC-β cells derived from patients with diabetes has been reported. This required long maturation times in vivo (12-19 wk) and has only been successful with limited mouse numbers, and often with the inability to return blood glucose to normoglycemia (18, 19, 22). Overgrowths and other abnormal structures have also been observed in some of these reports (19, 22). In contrast, we showed reversal of severe pre-existing diabetes in all mice with corrected WS SC-β cell transplantation, with no observed teratoma or cystic structure formation by use of our differentiation strategy. We did observe overgrowths in the WS4unedit SC-β cell transplants that likely resulted from the detected proliferating off-target mesenchyme and progenitors.
Our study demonstrates that CRISPR/Cas9 correction of a diabetes-inducing gene variant allows for robust differentiation of autologous SC-β cells capable of reversing severe pre-existing diabetes. Application of our strategy for treating diabetes in mice may enable development of autologous cell replacement therapy in patients with diabetes. By using patient-derived cells that have been gene corrected to fix diabetic variants and produce robustly functional cells, the need for the patient to take immunosuppressant drugs could be avoided for many forms of diabetes (8). Alternative strategies for encapsulating cells for immune protection are currently being explored with allogeneic products but still require improvements for efficacy and safety (54, 55). Instead, autogenic SC-β cell therapy is possible and would benefit from combination with transplantation using macroporous scaffolds to better enable retrievability (56, 57) or other cells or materials to promote engraftment, survival, and function (58-62).
There are some important limitations to note in this study and in its implications. We primarily studied cells from one patient with WS. While these results are promising, differences in clinical presentation of WS is known, for example, with age of diabetes diagnosis ranging from 3 wk to 16 yr after birth (63). Further in-depth study with other patients is warranted to better represent heterogeneity in WS and to expand to other monogenetic forms of diabetes. A concern in iPSC culture (64) and in CRISPR/Cas9 (65) is the induction of off-target mutations, which likely necessitates careful karyotyping and genomic sequencing before translation to actual clinical use. In addition, while the SC-β cells generated in this study display many of the features of bone fide primary β cells, they are still not fully mature in terms of gene expression and function (23). Improvements in differentiation protocols to fully mature SC-β cells would help enable clinical translation of this technology to patients with WS.
Another major feature of our corrected WS SC-β cells is their ability to display robust dynamic function in vitro, with first- and second-phase insulin secretion. This feature is weak, absent, or not demonstrated in other diabetic SC-β cell strategies (18, 20-22), indicating that our observation of robust improvements in function with CRISPR/Cas9 would have been difficult with alternate approaches. Demonstrating dynamic function in patient SC-β cells is important for the study of diabetes, as dynamic function is commonly lost early on in the disease (66-69).
Human WS patient-derived β cells are not normally available for study due to the rarity of the disease and death of β cells during disease progression (70), and our ability to differentiate these cells from patient iPSCs facilitates in-depth modeling of this disease. Current common models of WS, including Wfs1 knockout mice and insulinoma lines, are of limited value in the study of WS due to species differences (21, 27-29, 31, 32). Of importance, Wfs1 knockout mice have only mild diabetes, in contrast with WS patients having insulin-dependent diabetes (31, 71). Understanding of human WS progression is lacking, making development of effective therapies difficult, and although the scope of this report is focused on one WS patient, we hope it enables more in-depth mechanistic study. Our human iPSC-derived model of WS validated the presence of elevated ER stress, activation of the unfolded protein response, and defects in insulin processing (39, 40). The proportion of endocrine subtype markers was affected in our model, with relatively lower expression of glucagon and C-peptide/insulin and higher somatostatin with the WFS1 pathogenic variant. Focused study of the influence of WFS1 and ER stress on differentiation could provide new insights. A major limitation of this study is the depth that we were capable of investigating molecular control and signaling of cellular stress, predominantly because of the purity differences of the SC-β cell population based on WFS1 pathogenic state and compared to human islets. The use of single-cell RNA sequencing enabled examination of gene expression differences, however sorting SC-β cells would be an ideal option for more robust study. Patient-derived SC-β cells can serve as a human patient-specific model of WS for further study of molecular events in β cell failure, drug screening, and a source of autologous cells for therapy (8).
Single-cell sequencing and in vitro assays demonstrated that WS4unedit SC-β cells display ER stress. ER stress is found in other types of diabetes, including Type 1 and 2, neonatal diabetes, maturity onset diabetes of the young (MODY), and many other disorders (30-35). Future studies investigating WS and other forms of diabetes are now more rigorously possible via our platform for the discovery of new biology, therapeutic compounds, and replacement cells. WS, neonatal diabetes, and MODY are all monogenic forms of diabetes, and through gene therapy using CRISPR/Cas9, the pathogenic variant in specific genes causing diabetes in these patients can be corrected. Our approach, leveraging CRISPR/Cas9 and a robust differentiation protocol, allows for rigorous study of these forms of diabetes for disease modeling and drug screening. In the future, we suspect our advanced strategy combining patient iPSCs, gene editing, and differentiation to high functioning SC-β cells and other cell types will produce a viable, personalized cell source for cell therapy in patients with diabetes and other degenerative disorders (72).
Materials and Methods
Study design
The objective of this study was to analyze and transplant insulin-producing β cells into mice with pre-existing diabetes to determine the translational potential of differentiated iPSCs from a patient with diabetes, specifically WS, after CRISPR/Cas9 correction of the pathogenic diabetes-causing variant. We used CRISPR/Cas9 on three iPSC lines derived from patients with WS to generate three corrected iPSC lines, all of which were differentiated to SC-β cells. We transplanted WS4corr SC-β cells, WS4unedit SC-β cells, and cadaveric human islet β cells into nine, six, and four STZ-treated diabetic mice, respectively. STZ-treated diabetic and nondiabetic mice without transplants served as transplantation controls. Mouse groups were assigned randomly, and the study was not blinded. Transplanted mice were monitored through blood glucose measurements and blood serum collection, then sacrificed for ex vivo analysis. Nephrectomy surgery was performed on WS4corr transplanted mice to confirm transplanted SC-β cells were the source of glucose tolerance and nondiabetic blood glucose concentrations. For all in vitro analyses, at least 3 differentiations were used. Sample size was not determined with a power calculation, and specific sample cells are defined for each dataset. Data collection was stopped at pre-determined, arbitrary times. No data was excluded. Further methods details are available in the Supplementary Materials.
hiPSC line generation
This work as performed in accordance with the ESCRO Committee at Washington University in St. Louis. WS9unedit and WS13unedit iPSC lines were previously published as Wolf-2010-9 and Wolf-2010-13, respectively (36). WS4unedit iPSC line was generated from a previously described WS patient (WU.WOLF-04) (25) as previously described (36) by the Genetic Engineering and iPSC Center (GEiC) at Washington University in St. Louis from skin fibroblast with Sendai viral reprogramming (Life Technologies).
CRISPR/Cas9 gene correction
CRISPR/Cas9 gene correction of the WFS1 pathogenic variants in the WS4 and WS13 iPSC lines was performed at GEiC at Washington University in St. Louis. Guide RNAs were generated to target the WFS1 variants and validated using next generation deep sequencing analysis (NGS) (73). YH421.WFS1.sp14 and YS422.WFS1.sp11 was selected for homology directed repair using a single stranded DNA oligo (ssODN) as a template to target the point mutation in WFS1 on allele 2 for WS4 iPSCs and allele 1 for WS13 iPSCs, respectively. The designated allele to correct was determined based on the highest CRISPR/Cas9 gRNA specificity. Cells with successful CRISPR/Cas9 correction of WFS1 were termed WS4corr, and cells in which WFS1 retained the point mutation were termed WS4unedit, as confirmed by targeted NGS (73). The top 5 off-target sequences were also confirmed with NGS. gRNA sequences used are listed in table S1.
Stem cell-derived β cell differentiation
Undifferentiated cells were seeded at 5.2-7.3x105 cells/cm2 and differentiated performed as outlined in tables S4-6 to generate SC-β cells (23). Cells were aggregated 7-9 d into stage 6 on an OrbiShaker (Benchmark) at 100 RPM for assessment. Islets were purchased from Prodo Labs and cultured in islet media (table S6) for comparison 24 hr after shipment arrival. Bright field images were captured with a Leica DMi1.
Glucose-stimulated insulin secretion
For static GSIS, approximately 30 stage 6 clusters were placed in transwells (Corning), equilibrated for 1 hr in 2 mM glucose KRB (table S6), and challenged with sequential 1-hr 2 and 20 mM KRB treatments. For dynamic GSIS, clusters were loaded into a cell chamber, perfused with buffer at 100 μL/min, and equilibrated in 2 mM glucose KRB for 90 min. The samples were then challenged for 12 min at 2 mM, 24 min at 20 mM, and 12 min at 2 mM glucose KRB, collecting effluent every 2-4 min. Human insulin was quantified with ELISA and data normalized to cell counts from Vi-Cell XR or DNA quantification using Quant-iT Picogreen dsDNA assay kit (Invitrogen).
Mouse transplants
Animal studies were performed in accordance with Washington University IACUC. Mice were randomly designated for STZ treatment and transplantation groups. Mouse number per group was selected to allow for statistical significance based on our prior studies (15, 17, 18). Surgical procedures and follow up studies were performed by unblinded individuals. Male 7 wk old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories, rendered diabetic with injection of 45 mg/kg STZ (R&D) for 5 d, with diabetes confirmed after 8 d. Anaesthetized mice were injected with 5x106 WS4unedit stage 6 cells, 5x106 WS4corr stage 6 cells, 5x106 (4000 IEQ) islet cells, or saline under the kidney capsule. Islet transplantation was performed in a separate cohort. Animals were monitored up to 6 months. Blood glucose was measured with a Contour Blood Glucose Monitoring System (Bayer). Glucose tolerance and in vivo GSIS assays were performed by fasting mice for 4 hr and injecting with 2 g/kg glucose. Serum hormones were quantified using Human Ultrasensitive Insulin ELISA kit (ALPCO Diagnostics), Mouse C-peptide ELISA (ALPCO Diagnostics), and human proinsulin ELISA (Mercodia). 12-wk after transplantation, live nephrectomy was performed on 2 anaesthetized transplanted mice.
Statistical analysis
Statistical analysis was calculated with GraphPad Prism. Data was tested for normal (Gaussian) distribution using Shapiro-Wilk normality test. One- and two-sided unpaired and paired t-tests and one- and two-way ANOVA with Tukey’s or Dunnett’s tests were used for data sets with a normal distribution. Otherwise, Mann-Whitney two-tailed nonparametric tests were used. Statistical tests are specified in figure legends. p<0.05 was considered statistically significant. Data is shown as mean±s.e.m unless otherwise noted. The sample size, n, indicates the total number of biological replicates.
Supplementary Material
Fig. S1. Additional patient and cell line information.
Fig. S2. Additional WS4 stage 6 and human islet analysis.
Fig. S3. Differentiation progression and efficiency for WS4corr and WS4unedit lines.
Fig. S4. WFS1 expression during SC-β cell differentiation in WS4corr and WS4unedit lines.
Fig. S5. Glucose-stimulated insulin secretion normalized to β cell population.
Fig. S6. Additional analysis of WS4corr and WS4unedit SC-β cell transplantations in diabetic mice.
Fig. S7. Additional analysis of WS4corr and WS4unedit SC-β cell scRNA-seq.
Fig. S8. Additional analysis of WS4corr and WS4unedit SC-β cell scRNA-seq for off-targets.
Fig. S9. Additional analysis of differences in SC-β cell beta and islet markers for WS4corr clones and WS4unedit lines.
Fig. S10. Additional analysis of ER stress gene expression.
Fig. S11. Additional analysis of TEM and mitochondrial respiration.
Fig. S12. Treatment of SC-β cells with chemical stressors.
Fig. S13. Stress marker measurements of WS4corr-B and human islets.
Table S1. CRISPR sequences
Table S2. Additional analysis of WS4corr and WS4unedit SC-β cell scRNA-seq and population upregulated genes.
Table S3. Log fold change values between WS4corr and WS4unedit SC-β cells for markers in Figure 5A and 6A.
Table S4. Differentiation protocol
Table S5. Differentiation factor list
Table S6. Media and buffer formulations
Table S7. Antibody list
Table S8. Primers used for real-time PCR
Data file S1. Individual-level data for all figures
Acknowledgments:
We thank Mason Simmons, Anurima Sharma, William Buchsner, and John Bramley for assistance. Cell line generation and correction was performed by the Washington University (WU) Genome Engineering and iPSC Center (GEiC). Microscopy was performed through the WU Center for Cellular Imaging. Processing of electron microscopy samples was performed at the Harvard Medical School Electron Microscope Core Facility. We also thank all the members of the WU Wolfram Syndrome Study and Research Clinic for their support (https://wolframsyndrome.dom.wustl.edu) and all the participants in the Wolfram syndrome International Registry and Clinical Study, Research Clinic, and Clinical Trials for their time and efforts.
Funding: This work was primarily supported by the NIH (R01DK114233 to J.R.M.), JDRF Career Development Award (5-CDA-2017-391-A-N to J.R.M.), WU Center of Regenerative Medicine to J.R.M., and startup funds from WU School of Medicine Department of Medicine to J.R.M. This work was also partly supported by the NIH (DK112921, TR002065, TR002345 to F.Urano; T32DK108742 to support K.G.M.; R25GM103757 to support L.V.C.; T32DK007120 to support N.J.H.) and from Unravel Wolfram Syndrome Fund, Silberman Fund, Stowe Fund, Ellie White Foundation for Rare Genetic Disorders, Eye Hope Foundation, Snow Foundation, Feiock Fund to F.Urano. This work was further supported by Manpei Suzuki Diabetes Foundation and JSPS Overseas Research Fellowship to S.M. CRISPR/Cas9 work was supported by the Children’s Discovery Institute to J.R.M. Seahorse analysis and microscopy was supported by the NIH (P30DK020579 to the WU Diabetes Research Center). Sequencing work was performed by the NIH (NIH P30CA91842 and UL1TR000448 to WU Genome Technology Access Center in the Department of Genetics; UL1TR002345 to the WU Institute of Clinical and Translational Sciences).
Footnotes
Competing interests: L.V.C., N.J.H., and J.R.M. are inventors on patent application (PCT/US19/32643; METHODS AND COMPOSITIONS FOR GENERATING CELLS OF ENDODERMAL LINEAGE AND BETA CELLS AND USES THEREOF) for material described in this manuscript.
Data and materials availability: All data associated with this study are in the paper or supplementary materials. Single cell RNA sequencing raw data is available in GEO under accession number GSE139535. iPSC lines are available under an MTA upon request to the corresponding authors.
References and Notes:
- 1.Karagiannis P, Takahashi K, Saito M, Yoshida Y, Okita K, Watanabe A, Inoue H, Yamashita JK, Todani M, Nakagawa M. J. P. r., Induced pluripotent stem cells and their use in human models of disease and development. 99, 79–114 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. J. c., Induction of pluripotent stem cells from adult human fibroblasts by defined factors. 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Atkinson MA, Eisenbarth GS, Michels AW, Type 1 diabetes. Lancet 383, 69–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathan DM, Long-term complications of diabetes mellitus. N Engl J Med 328, 1676–1685 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Shapiro AM, Lakey JR, Ryan EA, Korbutt GS, Toth E, Warnock GL, Kneteman NM, Rajotte RV, Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med 343, 230–238 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Shapiro AM, Ricordi C, Hering BJ, Auchincloss H, Lindblad R, Robertson RP, Secchi A, Brendel MD, Berney T, Brennan DC, Cagliero E, Alejandro R, Ryan EA, DiMercurio B, Morel P, Polonsky KS, Reems JA, Bretzel RG, Bertuzzi F, Froud T, Kandaswamy R, Sutherland DE, Eisenbarth G, Segal M, Preiksaitis J, Korbutt GS, Barton FB, Viviano L, Seyfert-Margolis V, Bluestone J, Lakey JR, International trial of the Edmonton protocol for islet transplantation. N Engl J Med 355, 1318–1330 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Scharp DW, Lacy PE, Santiago JV, McCullough CS, Weide LG, Falqui L, Marchetti P, Gingerich RL, Jaffe AS, Cryer PE, et al. , Insulin independence after islet transplantation into type I diabetic patient. Diabetes 39, 515–518 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Millman JR, Pagliuca FW, Autologous Pluripotent Stem Cell-Derived beta-Like Cells for Diabetes Cellular Therapy. Diabetes 66, 1111–1120 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Odorico J, Markmann J, Melton D, Greenstein J, Hwa A, Nostro C, Rezania A, Oberholzer J, Pipeleers D, Yang L, Cowan C, Huangfu D, Egli D, Ben-David U, Vallier L, Grey ST, Tang Q, Roep B, Ricordi C, Naji A, Orlando G, Anderson DG, Poznansky M, Ludwig B, Tomei A, Greiner DL, Graham M, Carpenter M, Migliaccio G, D’Amour K, Hering B, Piemonti L, Berney T, Rickels M, Kay T, Adams A, Report of the Key Opinion Leaders Meeting on Stem Cell-derived Beta Cells. Transplantation 102, 1223–1229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA, Generation of pluripotent stem cells from patients with type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America 106, 15768–15773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thatava T, Kudva YC, Edukulla R, Squillace K, De Lamo JG, Khan YK, Sakuma T, Ohmine S, Terzic A, Ikeda Y, Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Mol Ther 21, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang L, Hua H, Foo K, Martinez H, Watanabe K, Zimmer M, Kahler DJ, Freeby M, Chung W, LeDuc C, Goland R, Leibel RL, Egli D, beta-cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes 63, 923–933 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua H, Shang L, Martinez H, Freeby M, Gallagher MP, Ludwig T, Deng L, Greenberg E, Leduc C, Chung WK, Goland R, Leibel RL, Egli D, iPSC-derived beta cells model diabetes due to glucokinase deficiency. The Journal of clinical investigation 123, 3146–3153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, Goland RS, Leibel RL, Solomon SL, Benvenisty N, Sauer MV, Egli D, Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature 510, 533–536 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Pagliuca FW, Millman JR, Gurtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA, Generation of functional human pancreatic beta cells in vitro. Cell 159, 428–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, Yang YH, Johnson JD, Kieffer TJ, Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature biotechnology 32, 1121–1133 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Velazco-Cruz L, Song J, Maxwell KG, Goedegebuure MM, Augsornworawat P, Hogrebe NJ, Millman JR, Acquisition of Dynamic Function in Human Stem Cell-Derived beta Cells. Stem cell reports 12, 351–365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millman JR, Xie C, Van Dervort A, Gurtler M, Pagliuca FW, Melton DA, Generation of stem cell-derived beta-cells from patients with type 1 diabetes. Nat Commun 7, 11463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui L, Danzl N, Campbell SR, Viola R, Williams D, Xing Y, Wang Y, Phillips N, Poffenberger G, Johannesson B, Oberholzer J, Powers AC, Leibel RL, Chen X, Sykes M, Egli D, beta-Cell Replacement in Mice Using Human Type 1 Diabetes Nuclear Transfer Embryonic Stem Cells. Diabetes 67, 26–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Sterr M, Ansarullah, Burtscher I, Böttcher A, Beckenbauer J, Siehler J, Meitinger T, Häring H-U, Staiger H, Cernilogar FM, Schotta G, Irmler M, Beckers J, Wright CVE, Bakhti M, Lickert H, Point mutations in the PDX1 transactivation domain impair human β-cell development and function. Molecular metabolism, (2019). [DOI] [PMC free article] [PubMed]
- 21.Balboa D, Saarimaki-Vire J, Borshagovski D, Survila M, Lindholm P, Galli E, Eurola S, Ustinov J, Grym H, Huopio H, Partanen J, Wartiovaara K, Otonkoski T, Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma S, Viola R, Sui L, Cherubini V, Barbetti F, Egli D, beta Cell Replacement after Gene Editing of a Neonatal Diabetes-Causing Mutation at the Insulin Locus. Stem cell reports 11, 1407–1415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogrebe NJ, Augsornworawat P, Maxwell KG, Velazco-Cruz L, Millman JR, Targeting the cytoskeleton to direct pancreatic differentiation of human pluripotent stem cells. Nature biotechnology, (2020). [DOI] [PMC free article] [PubMed]
- 24.Hsu PD, Lander ES, Zhang F, Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall BA, Permutt MA, Paciorkowski AR, Hoekel J, Karzon R, Wasson J, Viehover A, White NH, Shimony JS, Manwaring L, Austin P, Hullar TE, Hershey T, W. U. W. S. Grp, Phenotypic characteristics of early Wolfram syndrome. Orphanet J Rare Dis 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urano F, Wolfram Syndrome: Diagnosis, Management, and Treatment. Current diabetes reports 16, 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y, Urano F, WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. The Journal of biological chemistry 280, 39609–39615 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R, Hayashi E, Ishihara H, Oka Y, Permutt MA, Urano F, Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. The Journal of clinical investigation 120, 744–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D, Tokita A, Satake C, Tashiro F, Katagiri H, Aburatani H, Miyazaki J, Oka Y, WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum Mol Genet 15, 1600–1609 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Araki E, Oyadomari S, Mori M, Impact of endoplasmic reticulum stress pathway on pancreatic beta-cells and diabetes mellitus. Exp Biol Med (Maywood) 228, 1213–1217 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, Murray J, Schmidt RE, Herrera PL, Permutt MA, Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia 48, 2313–2321 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Fonseca SG, Gromada J, Urano F, Endoplasmic reticulum stress and pancreatic beta-cell death. Trends in endocrinology and metabolism: TEM 22, 266–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding HP, Ron D, Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes 51 Suppl 3, S455–461 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS, Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science (New York, N.Y 306, 457–461 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Zhong J, Rao X, Xu JF, Yang P, Wang CY, The role of endoplasmic reticulum stress in autoimmune-mediated beta-cell destruction in type 1 diabetes. Exp Diabetes Res 2012, 238980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu S, Kanekura K, Hara T, Mahadevan J, Spears LD, Oslowski CM, Martinez R, Yamazaki-Inoue M, Toyoda M, Neilson A, A calcium-dependent protease as a potential therapeutic target for Wolfram syndrome. Proceedings of the National Academy of Sciences 111, E5292–E5301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombardi AA, Gibb AA, Arif E, Kolmetzky DW, Tomar D, Luongo TS, Jadiya P, Murray EK, Lorkiewicz PK, Hajnoczky G, Murphy E, Arany ZP, Kelly DP, Margulies KB, Hill BG, Elrod JW, Mitochondrial calcium exchange links metabolism with the epigenome to control cellular differentiation. Nat Commun 10, 4509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mody N, Parhami F, Sarafian TA, Demer LL, Oxidative stress modulates osteoblastic differentiation of vascular and bone cells. Free Radic Biol Med 31, 509–519 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Hofmann S, Bauer MF, Wolfram syndrome-associated mutations lead to instability and proteasomal degradation of wolframin. FEBS letters 580, 4000–4004 (2006). [DOI] [PubMed] [Google Scholar]
- 40.de Heredia ML, Cleries R, Nunes V, Genotypic classification of patients with Wolfram syndrome: insights into the natural history of the disease and correlation with phenotype. Genet Med 15, 497–506 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EMJC, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. J. N. b., Integrating single-cell transcriptomic data across different conditions, technologies, and species. 36, 411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning E. J. J. C. s., A single-cell transcriptome atlas of the human pancreas. 3, 385–394. e383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arda HE, Li L, Tsai J, Torre EA, Rosli Y, Peiris H, Spitale RC, Dai C, Gu X, Qu K, Wang P, Wang J, Grompe M, Scharfmann R, Snyder MS, Bottino R, Powers AC, Chang HY, Kim SK, Age-Dependent Pancreatic Gene Regulation Reveals Mechanisms Governing Human beta Cell Function. Cell metabolism 23, 909–920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segerstolpe A, Palasantza A, Eliasson P, Andersson EM, Andreasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, Smith DM, Kasper M, Ammala C, Sandberg R, Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell metabolism 24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen MBK, Azad A, Ingvorsen C, Hess K, Hansson M, Grapin-Botton A, Honore C, Single-Cell Gene Expression Analysis of a Human ESC Model of Pancreatic Endocrine Development Reveals Different Paths to beta-Cell Differentiation. Stem cell reports 9, 1246–1261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arunagiri A, Haataja L, Cunningham CN, Shrestha N, Tsai B, Qi L, Liu M, Arvan P, Misfolded proinsulin in the endoplasmic reticulum during development of beta cell failure in diabetes. Ann N Y Acad Sci 1418, 5–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maechler P, Wollheim CB, Mitochondrial function in normal and diabetic beta-cells. Nature 414, 807–812 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Rötig A, Cormier V, Chatelain P, Francois R, Saudubray J, Rustin P, Munnich A, Deletion of mitochondrial DNA in a case of early-onset diabetes mellitus, optic atrophy, and deafness (Wolfram syndrome, MIM 222300). The Journal of clinical investigation 91, 1095–1098 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cagalinec M, Liiv M, Hodurova Z, Hickey MA, Vaarmann A, Mandel M, Zeb A, Choubey V, Kuum M, Safiulina D, Role of mitochondrial dynamics in neuronal development: mechanism for wolfram syndrome. PLoS biology 14, e1002511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nair GG, Liu JS, Russ HA, Tran S, Saxton MS, Chen R, Juang C, Li ML, Nguyen VQ, Giacometti S, Puri S, Xing Y, Wang Y, Szot GL, Oberholzer J, Bhushan A, Hebrok M, Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived beta cells. Nature cell biology 21, 263–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millman JR, Doggett T, Thebeau C, Zhang S, Semenkovich CF, Rajagopal R, Measurement of Energy Metabolism in Explanted Retinal Tissue Using Extracellular Flux Analysis. J Vis Exp, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardenas-Diaz FL, Osorio-Quintero C, Diaz-Miranda MA, Kishore S, Leavens K, Jobaliya C, Stanescu D, Ortiz-Gonzalez X, Yoon C, Chen CS, Haliyur R, Brissova M, Powers AC, French DL, Gadue P, Modeling Monogenic Diabetes using Human ESCs Reveals Developmental and Metabolic Deficiencies Caused by Mutations in HNF1A. Cell stem cell 25, 273–289 e275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomei AA, Villa C, Ricordi C, Development of an encapsulated stem cell-based therapy for diabetes. Expert Opin Biol Ther 15, 1321–1336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scharp DW, Marchetti P, Encapsulated islets for diabetes therapy: history, current progress, and critical issues requiring solution. Adv Drug Deliv Rev 67-68, 35–73 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Pedraza E, Brady AC, Fraker CA, Molano RD, Sukert S, Berman DM, Kenyon NS, Pileggi A, Ricordi C, Stabler CL, Macroporous three-dimensional PDMS scaffolds for extrahepatic islet transplantation. Cell Transplant 22, 1123–1135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song J, Millman JR, Economic 3D-printing approach for transplantation of human stem cell-derived beta-like cells. Biofabrication 9, 015002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Najjar M, Manzoli V, Abreu M, Villa C, Martino MM, Molano RD, Torrente Y, Pileggi A, Inverardi L, Ricordi C, Hubbell JA, Tomei AA, Fibrin gels engineered with pro-angiogenic growth factors promote engraftment of pancreatic islets in extrahepatic sites in mice. Biotechnology and bioengineering 112, 1916–1926 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Sackett SD, Tremmel DM, Ma F, Feeney AK, Maguire RM, Brown ME, Zhou Y, Li X, O’Brien C, Li L, Burlingham WJ, Odorico JS, Extracellular matrix scaffold and hydrogel derived from decellularized and delipidized human pancreas. Sci Rep 8, 10452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tremmel DM, Odorico JS, Rebuilding a better home for transplanted islets. Organogenesis 14, 163–168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang K, Chaimov D, Patel SN, Liang JP, Wiggins SC, Samojlik MM, Rubiano A, Simmons CS, Stabler CL, 3-D physiomimetic extracellular matrix hydrogels provide a supportive microenvironment for rodent and human islet culture. Biomaterials 198, 37–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Augsornworawat P, Velazco-Cruz L, Song J, Millman JR, A hydrogel platform for in vitro three dimensional assembly of human stem cell-derived islet cells and endothelial cells. Acta Biomater, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrett TG, Bundey SE, Macleod AF, Neurodegeneration and diabetes: UK nationwide study of Wolfram (DIDMOAD) syndrome. Lancet 346, 1458–1463 (1995). [DOI] [PubMed] [Google Scholar]
- 64.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee JH, Loh YH, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Izpisua Belmonte JC, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein LS, Zhang K, Somatic coding mutations in human induced pluripotent stem cells. Nature 471, 63–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD, High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caumo A, Luzi L, First-phase insulin secretion: does it exist in real life? Considerations on shape and function. Am J Physiol Endocrinol Metab 287, E371–385 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Del Prato S, Tiengo A, The importance of first-phase insulin secretion: implications for the therapy of type 2 diabetes mellitus. Diabetes/metabolism research and reviews 17, 164–174 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Seino S, Shibasaki T, Minami K, Dynamics of insulin secretion and the clinical implications for obesity and diabetes. The Journal of clinical investigation 121, 2118–2125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB, Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell 105, 745–755 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Hilson JB, Merchant SN, Adams JC, Joseph JT, Wolfram syndrome: a clinicopathologic correlation. Acta Neuropathol 118, 415–428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ishihara H, Takeda S, Tamura A, Takahashi R, Yamaguchi S, Takei D, Yamada T, Inoue H, Soga H, Katagiri H, Tanizawa Y, Oka Y, Disruption of the WFS1 gene in mice causes progressive beta-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet 13, 1159–1170 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Abreu D, Urano F, Current Landscape of Treatments for Wolfram Syndrome. Trends Pharmacol Sci, (2019). [DOI] [PMC free article] [PubMed]
- 73.Bell CC, Magor GW, Gillinder KR, Perkins AC, A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genomics 15, 1002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, Ryu JH, Wagner BK, Shen-Orr SS, Klein AM, Melton DA, Yanai I, A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst, (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Additional patient and cell line information.
Fig. S2. Additional WS4 stage 6 and human islet analysis.
Fig. S3. Differentiation progression and efficiency for WS4corr and WS4unedit lines.
Fig. S4. WFS1 expression during SC-β cell differentiation in WS4corr and WS4unedit lines.
Fig. S5. Glucose-stimulated insulin secretion normalized to β cell population.
Fig. S6. Additional analysis of WS4corr and WS4unedit SC-β cell transplantations in diabetic mice.
Fig. S7. Additional analysis of WS4corr and WS4unedit SC-β cell scRNA-seq.
Fig. S8. Additional analysis of WS4corr and WS4unedit SC-β cell scRNA-seq for off-targets.
Fig. S9. Additional analysis of differences in SC-β cell beta and islet markers for WS4corr clones and WS4unedit lines.
Fig. S10. Additional analysis of ER stress gene expression.
Fig. S11. Additional analysis of TEM and mitochondrial respiration.
Fig. S12. Treatment of SC-β cells with chemical stressors.
Fig. S13. Stress marker measurements of WS4corr-B and human islets.
Table S1. CRISPR sequences
Table S2. Additional analysis of WS4corr and WS4unedit SC-β cell scRNA-seq and population upregulated genes.
Table S3. Log fold change values between WS4corr and WS4unedit SC-β cells for markers in Figure 5A and 6A.
Table S4. Differentiation protocol
Table S5. Differentiation factor list
Table S6. Media and buffer formulations
Table S7. Antibody list
Table S8. Primers used for real-time PCR
Data file S1. Individual-level data for all figures