Abstract
Despite being discovered over 60 years ago, the precise role of phospholipase D (PLD) is still being elucidated. PLD enzymes catalyze the hydrolysis of the phosphodiester bond of glycerophospholipids producing phosphatidic acid and the free headgroup. PLD family members are found in organisms ranging from viruses, and bacteria to plants, and mammals. They display a range of substrate specificities, are regulated by a diverse range of molecules, and have been implicated in a broad range of cellular processes including receptor signaling, cytoskeletal regulation and membrane trafficking. Recent technological advances including: the development of PLD knockout mice, isoform-specific antibodies, and specific inhibitors are finally permitting a thorough analysis of the in vivo role of mammalian PLDs. These studies are facilitating increased recognition of PLD’s role in disease states including cancers and Alzheimer’s disease, offering potential as a target for therapeutic intervention.
Keywords: Phospholipase D, Phosphatidic acid, Lipid signaling, Membrane transport
1. Introduction
Hanahan and Chaikoff discovered phospholipase D (PLD) in 1947 as a factor in carrot extracts capable of causing lipid and membrane degradation. They characterized it as a phospholipid-specific phosphodiesterase, able to release free choline from phosphatidylcholine (PtdCho), using water as a nucleophilic acceptor, to produce phosphatidic acid (PtdOH or PA) [1–3]. PLDs have since been described in organisms as diverse as viruses, bacteria, plants and mammals, and the enzymes have been broadly divided into members of a ‘superfamily’ containing the canonical catalytic motif HxKxxxxD (HKD), and non-HKD enzymes with a broader range of substrate specificities. However, bacterial cardiolipin synthases (CLSs), phosphatidylserine synthases (PSSs) and endonucleases are now also known to contain HKD motifs. As such, perhaps a better definition for PLD enzymes is that of proteins possessing the ability to hydrolyze glycerophospholipids, pre-dominantly PtdCho, at the headgroup phosphodiester bond to produce the free headgroup and PtdOH.
PLD activity and PtdOH regulate a remarkable range of cellular processes including: vesicle trafficking, endocytosis, exocytosis, secretion [4–13], cytoskeletal rearrangement [14], phagocytosis [15,16], neuronal- and cardiac stimulation [17–23], matrix metalloproteinase (MMP) production [24], the oxidative respiratory burst in neutrophils [25], plant stress responses ([26,27], pathogen resistance [28], the pathogenic actions of bacteria, and spider venom [29–32], pluripotency, stem cell reprograming, and apoptosis [33]. PLD is also implicated in a number of diseases including: inflammation, diabetes [34] oncogenesis [35], Alzheimer’s disease (AD) [36–41], thrombotic disease [42,43], hypertension [44], multiple sclerosis [45], and viral infection [46].
2. Early biochemical studies of mammalian PLD
The first PLD activities were identified in plants and bacteria, however given the mammalian PLD focus of this review, and since this early history has been covered elsewhere, the reader is referred to the following articles [1,47–50]. Importantly though, the early plant studies demonstrated PLD catalyzes transphosphatidylation reactions in the presence of glycerol or short chain primary aliphatic -alcohols (such as butan-1-ol). Being stronger nucleophilic acceptors than water, the natural acceptor, they are used preferentially by the enzyme with a preference of over 1000-fold [17,51]. The resultant phosphatidylalcohol products of these reactions are metabolically stable, and unlike PtdOH are poor substrates for lipid phosphate phosphatases (LPPs [previously called phosphatidate phosphohydrolases or PAPs]) [52,53]. Their stability, presumed functional inactivity, and ability to block PtdOH production resulted in widespread use of phosphatidylalcohol formation both as a marker of PLD activity, and as a way of blocking PLD mediated PtdOH production through alternate production of phosphatidyi-alcohols.
Initially considered absent from animal tissue, interest in PLD grew following demonstration that rat-brain solubilized enzyme preparations released choline, and ethanolamine from PtdCho, and phosphatidylethanolamine (PtdEth) respectively [54,55]. Later studies confirmed PtdOH increases were direct results of PLD activity, and not of the phospholipase C (PLC)/ Diacylglycerol (DAG) kinase pathway, or de novo synthesis [56,57]. Attempts to purify PLD from mammalian tissues resulted in identification of multiple isoforms with differing: pH optima, and responses to: Ca2+ and Mg2+, oleate, phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), and guanosine 5′-(gamma-thio) triphosphate (GTPγS) [58–65]. PLD partially purified from HL60 membranes was potently activated by GTPγS, could be stimulated by the small GTPase ADP-ribosylation factor (Arf), and had an absolute requirement for PtdIns(4,5)P2 [66,67]. Similar activities were subsequently discovered in rat brain [68], alongside a second oleate-dependent activity [69]. Further studies revealed that the GTP-stimulated activation of PLD in neutrophils was dependent on Rho, another small GTPase [70], and a pig brain membrane PLD was identified that could be stimulated by both small GTPases [71]. PLD activities were subsequently described in most mammalian cell types [72], and in many sub-cellular organelles including plasma membrane, nucleus, Golgi, and endoplasmic reticulum (ER) [73–77]. These activities could be stimulated by a wide variety of agonists including: serum, growth factors, phorbol esters, N-formyl-methionyl-leucyl-phenylalanine (FMLP), adenosine triphosphate (ATP), epinephrine, vasopressin and GTPγS [56,57,78–81].
3. Cloning of the first PLD-encoding genes
Lack of DNA or protein sequences hindered initial PLD studies but this problem was surmounted following purification of PLD from cabbage (Brassica oleracea), and castor bean (Ricinus communis) [50,82,83]. The N-terminal amino acid sequence of the later enabling cDNA clone isolation, from an endosperm cDNA library, encoding a 92-kDa protein [50]. Expression of the cloned enzyme in E.coli, facilitated demonstration that hydrolysis and transphosphatidylation were performed by the same enzyme [50]. Identification of plant PLD sequences paved the way for identification of PLD-encoding genes in other organisms, initially in the yeast Saccharomyces cerevisiae [84,85]. Sequencing of the yeast SP014 PLD facilitated identification of homologous mammalian expressed sequence tag (EST) clones and subsequently full-length mammalian PLD1 isoforms were identified from a human HeLa cell cDNA library [34], mouse [86] and rat [87]. PLD1 encoded a predominantly membrane associated, PtdCho-specific, 1072 amino acid (aa), trans-phosphatidylation-competent protein which could be stimulated by PtdIns(4,5)P2, and GTPγS-loaded Arf1 [88]. Shortly after PLD1’s identification, recognition of similar but distinct sequences in EST databases revealed a second mammalian PLD isoform: PLD2. Isolation of PLD2 cDNA from a mouse embryo cDNA library revealed a 932aa protein with 51% homology to PLD1 including particular homology within the central catalytic core [89–91]. PLD1 and PLD2 have a similar substrate preference, preferring mono- or di-unsaturated PtdCho, and generate structurally identical PtdOH species [92]. They both possessing four highly conserved catalytic domains I-IV with catalytic HKD motifs in domains II and IV. They also both contain N-terminal PX- and PH-do- mains and a polybasic PtdIns(4,5)P2-binding region. The isoforms vary considerably between conserved domain-II, and -III, with human PLD1 (but not PLD2) possessing a variable thermolabile loop region that appears to be inhibitory, since its deletion increases basal activity threefold [93]. The region is cleaved by caspases during apoptosis conferring, heightened GTPase sensitivity and elevated activity [94–96]. PLD1 has multiple splice variants, with the isoform that was initially discovered being designated PLD1a. The PLD1b splice variant lacks 38aa/43aa in the loop region between the catalytic domains but possesses a similar regulatory and catalytic profile [34,88]. PLD1a, and 1b variants lacking the C-terminal 113aa, designated PLD1a2 and -b2, were also identified, possessing dramatically reduced (8–12% of wild-type) activity [97]. PLD1c contains an mutation close to the N-terminus generating an inactive, truncated product of unknown function, which is expressed in a number of tissues including brain, and postulated to be an inhibitor of endogenous PLD activity [91]. PLD2 also has multiple splice variants, the first variant to be identified being designated PLD2a. PLD2b possesses an 11aa C-terminal deletion, and PLD2c contains a 56 bp insertion resulting in a truncated protein of unknown biological significance, since it lacks the catalytic HKD motifs, and is catalytically inactive [91]. A comparison of PLD1 and PLD2 structure is illustrated in Fig. 1.
Fig. 1.
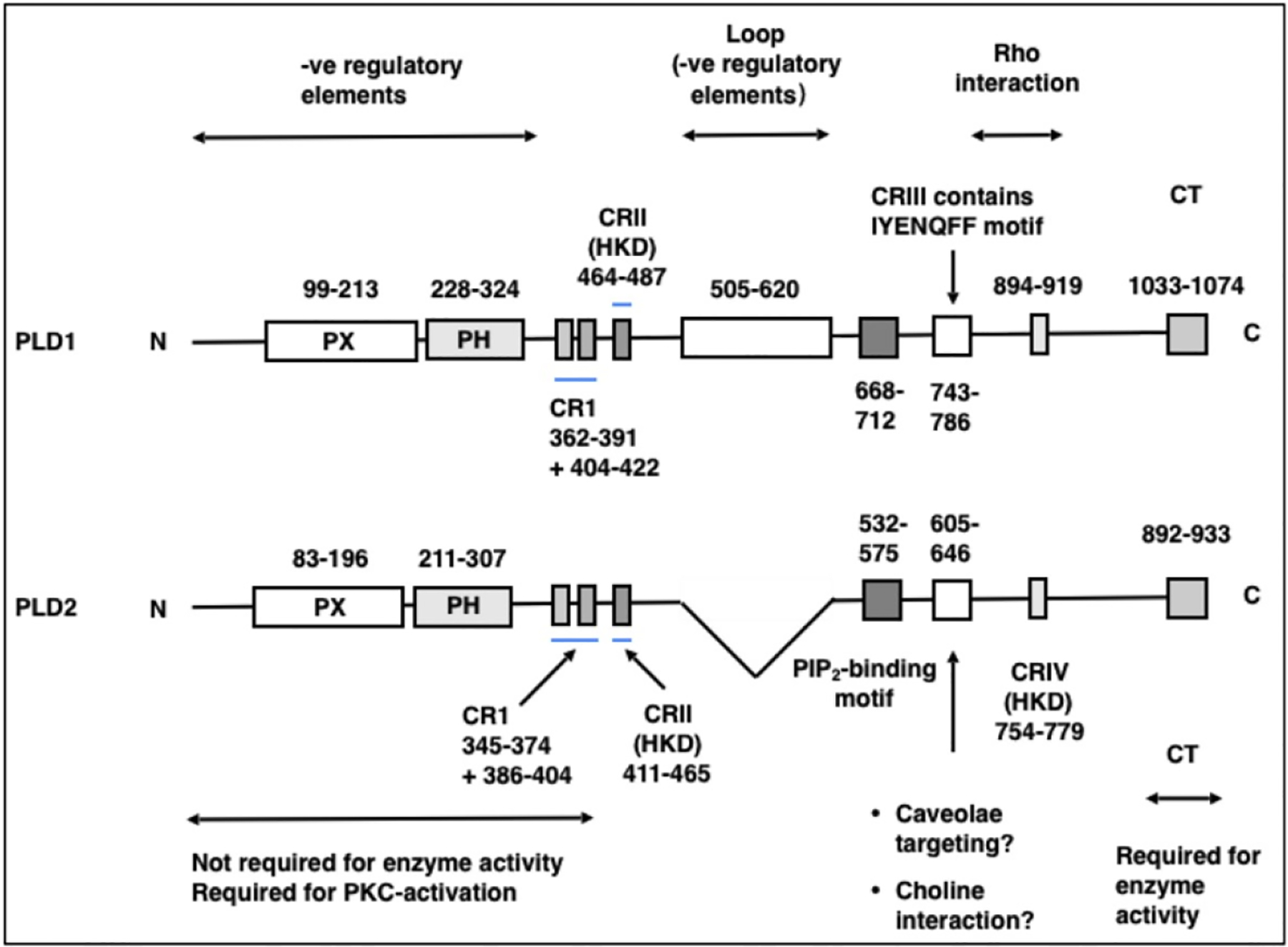
A detailed comparison of PLD1 and PLD2 structure. PLD1 and PLD2 display 51% sequence homology and share a conserved structure with 4 conserved PLD regions (CR), of which CRII and IV contain the catalytic sequence HKD. They both contain PX- and PH-domains and a PIP2 binding motif. PLD1 contains a loop sequence between CRII and CRIII absent in PLD2. Known and potential region-specific functions are indicated.
4. Mammalian PLD structure
4.1. The PLD catalytic domain and potential catalytic mechanisms
Sequencing of PLD proteins identified four conserved domains (I-IV) [98], domains II, and IV containing the charged catalytic motif HxK (x)4D (HKD) [48,99]. Substitution mutations within the HKD motifs inactivate mammalian PLD1, PLD2, and yeast Spo14 [93,97,100,101]. Two catalytic models of PLD activity have been proposed: 1) each catalytic motif functions independently; 2) each forms half of the active site [98]. Evidence supports the latter, since: a) mutation of single HKD motifs ablates activity [100], and b) individually expressed N-, and C- terminal PLD1 fragments [102,103] are catalytically inactive, but activity is reconstituted upon co-expression. In addition, several crystal structures of ‘PLD superfamily’ members reveal a conserved fold agreeing with this model, including that of the Salmonella typhimurium endonuclease Nuc, which contains a single HKD motif, and dimerizes to form an active site [104]. Other structural studies using S. typhimurium (strain PMF) PLD [105,106], and the human tyrosyl-DNA phosphodiesterase (Tdp1) family member also validate this model. Currently no resolved mammalian PLD structures exist however, a PLD2 3D-model has been proposed, combining homology, and ab initio 3D structural modeling methods, and docking conformations [107]. While theoretical, this model superimposed on bacterial PLD crystal structures, broadly agrees with biochemical studies showing that the N-, and C-terminal HKD motifs form a catalytic pocket, accommodating the PtdCho headgroup [107].
Radiolabel exchange experiments using a plant PLD, suggested a two-step “ping pong” catalysis reaction, involving a covalent phosphatidyl-enzyme intermediate, followed by hydrolysis/transphosphatidylation [48,108]. Later, a combination of labeling [109], and crystallization studies [104] using the bacterial Nuc enzyme, revealed a histidine from one HKD motif forms the catalytic nucleophile attacking the phosphate of the phosphodiester bond. This forms a phosphor-enzyme intermediate which is then hydrolyzed by water in the second half of the reaction, the other HKD motif acting as an acid, protonating the leaving group [104]. Radiolabeling studies using Nuc and another family member: the Yersinia, pestis murine toxin, validated this model [110], while subsequent crystallization of S. typhimurium PLD, and Tdp1 indicated the N-terminal motif likely provides the nucleophilic histidine [105,106,111,112]. Recent mammalian PLD2 molecular threading data also supports this model with the N-, and C-terminal histidines (H442/H756) of the HKD motifs forming a catalytic pocket, accommodating the PtdCho head group (but not that of PtdEth or PtdSer), facilitating nucleophilic attacks by H442, and water, aided by H756 [107]. The motifs lysine and aspartic acid residues are also implicated in catalysis [100,113,114], with postulated roles including intra-molecular interactions outside the active site [105,106], or interaction with-, and charge neutralization of-, the substrate [104]. Six amino acids downstream of the second HKD motif, within domain IV of mammalian PLD1 and PLD2 is the IGSANIN motif. This is essential for mammalian PLD1 catalysis, and palmitoylation, with mutation ablating activity, and causing abnormal, diffuse localization [115]. Variants in the catalytic sequences exist in a number of family members, but they generally adhere to the consensus motif HxKx(4)Dx(6)GGxD/N for domain II, and HxKx(4)Dx(6)GS/TxN for domain IV [116].
4.2. Lipid binding domains of mammalian PLDs
Mammalian PLD1, and PLD2 possess N-terminal phox homology (PX)- and pleckstrin homology (PH) domains and an additional poly-basic PtdIns(4,5)P2 binding site [17,117]. The interplay of lipid-binding sites likely confers localization and function, and is implicated in sub-cellular cycling [118] (see Fig. 2).
Fig. 2.
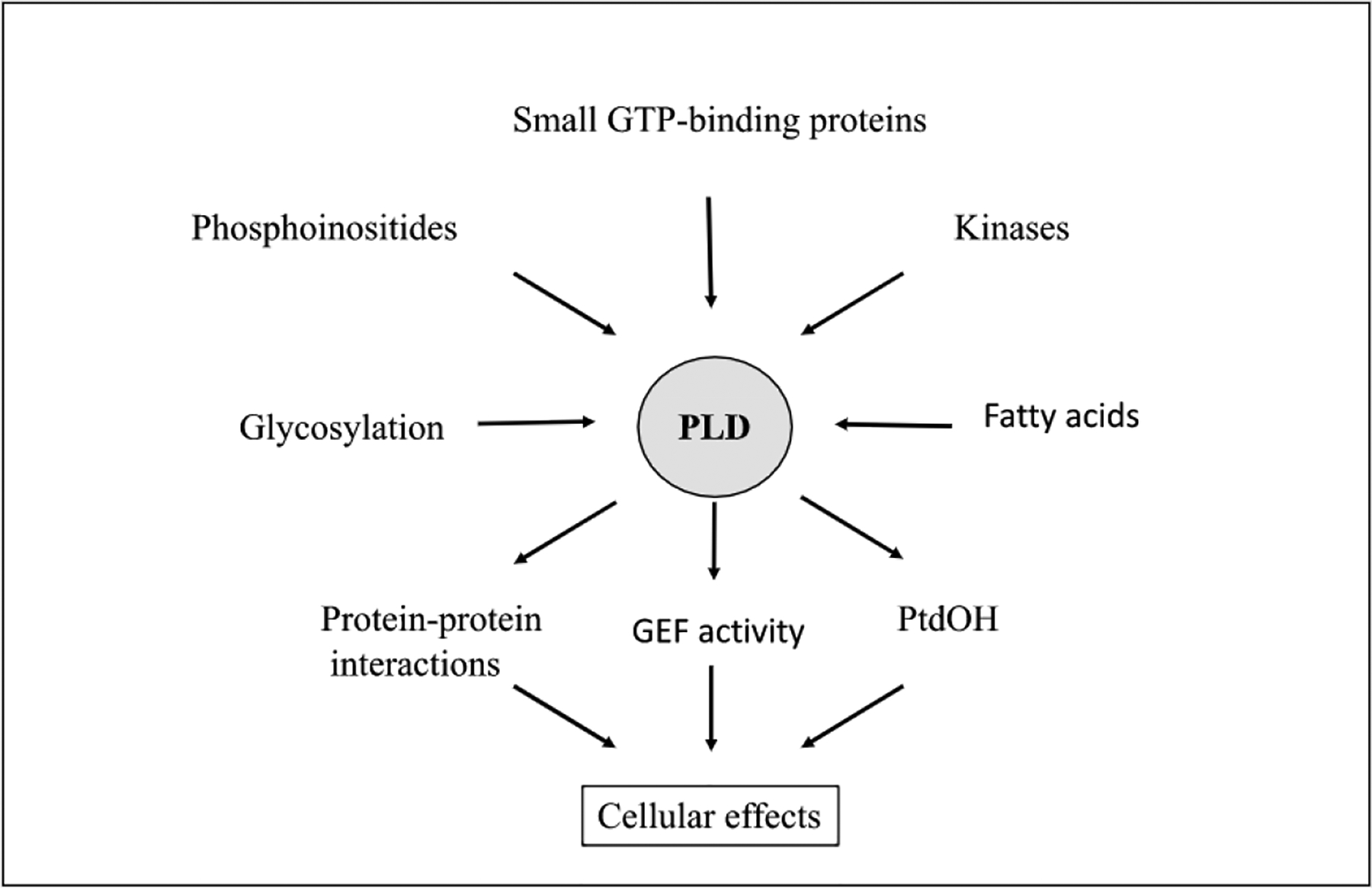
PLD regulation and function. PLD activity is regulated by a multitude of factors including proteins and lipids. Its activity results in a diverse range of biological outcomes through production of PtdOH, GEF-activity, and protein-protein interactions.
PH-domains are best known as high-affinity, high-specificity phosphoinositide (PIP) binding domains, although it is now apparent that less than 10% of them perform this function (as low as 3% in yeast) [119], such binding requiring phosphorylation at two adjacent sites on the PIP e.g. PtdIns(4,5)P2, PtdIns(3,4)P2 and PtdIns(3,4,5)P3 [120]. Membrane targeting roles once thought to be characteristic of these domains now appear restricted to this high-affinity binding subset [121], although at least one PH-domain is reported displaying PIP-binding independent membrane localization [119]. Other PH-domain subsets exist, including PIP-binders with weaker affinities, and specificities [122,123], and those postulated to be involved in coincidence detection [120,124,125]. Surface plasmon resonance (SPR) studies indicate PLD1 binds PtdIns(4,5)P2, PtdIns(3,4)P2, and PtdIns3P, with the isolated PH-domain having significantly higher affinity for PtdIns(4,5)P2 [126]. However, since PtdIns(4,5)P2 levels are higher than agonist stimulated PtdIns(3,4)P2 levels and PLD activity can be measured in the absence of PI3-kinase activity, the relevance of PtdIns (3,4)P2, and PtdIns3P interactions in vivo are unclear [126,127]. Immuno-affinity chromatography using large-unilamellar vesicles and the purified PLD2 PH-domain indicate a 10-fold PtdIns(4,5)P2 binding preference over PtdIns4P, PtdOH, or PtdSer though it bound these lipids to a lesser degree [128].
PLD PH-domain point mutations or deletions have variously reported effects on in vitro activity ranging from complete inhibition to no affect, under conditions of PtdIns(4,5)P2 excess. A number of reports suggest PH-domain point mutation, or deletion in PLD1 [93,101], or PLD2 [93,101,128,129] does not affect in vitro activity. However, conflicting data exists for PLD1 [47,126], and the PH -domain is required for in vivo PLD2 activity [128]. PH-domain mutations in PLD1 and PLD2 result in aberrant localizations, indicating a membrane-targeting role [126,128]. The isolated PLD1 PH-domain localized weakly to perinuclear vesicles, translocating weakly to the plasma membrane upon PMA-stimulation, indicating a potential function in exocytosis [118]. The PLD1 PH-domain is palmitoylated at cysteines 240, and 241, loss of palmitoylation resulting in translocation from peri-nuclear vesicles to plasma membrane [118,130]. The PLD1 PH-domain is additionally required for efficient return from the cell surface to endosomes, and for lipid raft entry, a palmitoylation-dependent process, which appears critical for internalization [118]. The PLD PH-domains are now also recognized as protein binding domains. The PLD1 PH-domain interacts with the μ2 subunit of adaptor protein 2 (AP2), facilitating AP2 membrane recruitment, and subsequent endocytosis of epidermal growth factor receptor (EGFR) [131]. The PLD2 PH-domain binds aldolase [132], the kinase domain of c-Src [133], and appears to play an important role in interaction with the small GTPase Rac2 [103].
Several studies demonstrated PH-domain mutation, or removal of the PLD1 or PLD2 N-termini (including the PH-domain) didn’t impair in vitro activity, or affect concentration dependent PtdIns(4,5)P2 activation, indicating the presence of another PtdIns(4,5)P2 binding site [93,101,128,129]. A conserved aromatic amino acid sequence: the “polybasic motif’, between domains III, and IV of mammalian PLD1, PLD2 and the yeast Spo14 PLD, is absent in bacterial and plant PLDs not regulated by PtdIns(4,5)P2. Substitution of these residues in mammalian PLD2 dramatically impaired PtdIns(4,5)P2 binding and stimulation of in vitro catalytic activity [129]. Furthermore, a peptide corresponding to the polybasic motif bound PtdIns(4,5)P2 with high affinity [128], and while PLD2, and Spo1 4 mutated within the polybasic motif display normal localization, they are non-functional in vivo [128,129]. In contrast, in PLD1, the polybasic motif acts in concert with other PLD domains to mediate localization. In COS-7 cells over-expressing tagged PLD1, the PtdIns(4,5)P2-interaction site was sufficient for mediating PMA-stimulated translocation to the plasma membrane, however the PH-, and PX-domains were required for endosomal return [118].
PX-domains are found in the N-termini of human, yeast, and nematode PLD [98,134]. These domains frequently display binding preferences for PtdIns3P, and its derivatives, which, are enriched at the early endosome [135,136]. PLD1 is activated by PtdIns(3,4,5)P3 in vitro [88,126,137,138], which specifically interacts with arginine 179 of the PLD1 (but not PLD2) PX-domain. Mutation of this residue inhibits platelet-derived growth factor (PDGF)-stimulated PLD1 membrane recruitment [137,139]. At the plasma-membrane, the PX-domain is required for further compartmentalization, with threonine 147 of PLD1 being one of three residues required for maximum PMA-stimulation, and which is reversibly phosphorylated by PKC at this location, driving PLD1 into caveolin-enriched microdomains, [140,141]. The PLD1 PX- domain is also required for efficient return from the cell surface, binding PtdInsSP to target it to endocytic vesicles [118]. Sequence alignment of the PLD1- and PLD2 PX-domains with the NADPH oxidase p47-phox, suggest they contain a ‘second ion-binding pocket’. In p47- phox the conserved region enables it to bind anionic phospholipids such as PtdSer and PtdOH while it’s other pocket binds PtdIns(3,4)P2 [142]. Consistent with this, the PLD1 PX-domain can bind both lipids, raising the possibility that PLD activity could be regulated by its own product. Simultaneous occupation of both pockets resulted in a synergistic increase in membrane binding with electrostatic potential calculations suggesting occupation of the second pocket by PtdOH, PtdSer, or other ionic lipids may initiate binding to the membrane, followed by interaction with the other pocket with PtdIns(3,4,5)P3 [139].
Non-lipid binding PLD PX-domain roles also exist including the guanine nucleotide exchange activity (GEF) activity of the PLD2 PX-domain, with specificity for the small GTPases Rac2 and Ras [143–146]. Additionally, the PLD1 and PLD2 PX-domains bind several proteins in a lipase independent manner. These include dynamin, for which they act as GTPase activating proteins (GAPs) stimulating GTPase activity, and accelerating EGFR endocytosis [131,147]). The PLD2 PX-domain additionally binds: 1) the PKCζ kinase domain, stimulating in vitro kinase activity [148]; 2) the PLCγ1 Src homology 3 (SH3) domain, an EGF-stimulated interaction, that potentiates PLCγ activity [149]; 3) Munc-18–1 which inhibits PLD activity in an EGF-reversible manner [150]; 4) Syk kinase, promoting its activation, and downstream mast cell activation [151]; 5) Collapsin Response Mediator Protein (CRMP), an interaction that additionally requires the PLD2 C-terminus, and that acts to inhibit neuronal PLD2 [152].
4.3. The mammalian PLD N- and C-termini
The poorly conserved mammalian PLD N-termini are modification tolerant, and deletion in PLD1 or PLD2 does not majorly impact catalytic activity [93,101,102,153]. The PLD1 N-terminus however, is required for PKC-stimulation [93,102,153,154], and appears to contain negative regulatory elements since deletion of the N-terminal 325 amino acids enhances Arf-, and Rac1-dependent activities [93]. Similarly, removal of the PLD2 N-terminus dramatically increases Arf-stimulation [101]. The PLD C-termini display greater conservation, and are less modification tolerant [93,101,155]. Deletion of the PLD1 C-terminus [93], or single amino acid mutations in this region abolish activity [155]. The PLD1a2, and −1b2 splice-variants lack the C-terminal 113aa, instead possessing distinct 10aa sequences conferring partial activity (8–12% of PLD1a/1b). They display altered localization, being absent from the endosome, indicating a C-terminal sub-cellular targeting role [97]. The C-termini also contain Rho family interaction sites, with additional postulated roles in stabilizing active site conformation [155].
4.4. Other mammalian PLD structural motifs
Motif III consists of the highly conserved IYIENQFF motif [117,156]. Its exact function remains unclear, but mutations in this sequence reduce PLD activity [100]. IYIENQFF is postulated to interact with PtdCho via the choline headgroup’s methyl group [117,156] and it is also suggested to increase the catalytic rate, or bind caveolin [117]. The amino acid residues leucine 405 and glycine 412 of the mammalian PLD1 domain I also have important functions for activity. Confusingly however, their mutation, but not deletion, seriously impacts activity [93]. Mammalian PLD1 and the C. elegans PLD isoform contain a loop region between domains II, and III. While the role in C. elegans is unlear, in human PLD1 it appears to be the region least well conserved between human and rodent isoforms, the site of alternate splicing [88], an autoinhibitory region [93], and a site of proteolytic cleavage [95].
4.5. The tissue and cellular distribution of mammalian PLD1, and PLD2
Early biochemical studies revealed PLD activities in a wide range of mammalian tissues and cell lines. Following the cloning of PLD1 and PLD2 isoform-specifïc expression data emerged, most of which reported mRNA levels until the advent of high-affinity antibodies [48,72]. PLD1b [91,157,158] and PLD2a are the most abundantly expressed splice variants [91,158,159] and most studies agree that PLD1 and PLD2 are ubiquitously expressed and present in almost all tissues and cell lines with the potential exception of peripheral leukocytes, lymphocytes [160,161], and renal tissue which reportedly lack PLD1 [162]. Isoform-specific tissue expression patterns exist: PLD1 is enriched in human heart, brain, pancreas, uterus, and intestine, and PLD2 is enriched in brain, placenta, lung, thymus, prostate, and uterine tissue [90,163]. Highly sensitive PLD2 brain expression analysis, using Locked Nucleotide Amplification (LNA), indicates that expression in this organ is not ubiquitous [164]. Most functional regions of the brain expressed PLD2 with levels being detectable in the pyramidal cells of the cornu ammonis (CA) regions of the hippocampus, the Purkinje cells in the cerebellum and the mitral cells of the olfactory bulb [164], However, in contrast to previous studies using antibody staining [165], PLD2 was absent from glia rich regions such as the fimbria, arbor vitae of the cerebellum, the internal capsule running through the caudoputamen, or the corpus callosum [164].
Early sub-cellular localization studies were limited to subcellular fractionation and activity measurements [166]. Later, studies addressing individual isoform localization were mainly limited to overexpression of tagged proteins in transformed cell lines, due to lack of specific antibodies. These yielded confusing data, arguing for, and against localization to practically every known cellular location [48,134]. Thankfully trends emerged, and more recent reports using specific antibodies are helping to clarify the situation. Continuing discrepancies as to the precise nature of their localizations are likely a reflection of the different imaging strategies, cell lines and tissues used [167,168].
The majority of localization studies have used overexpressed, tagged-proteins, and overexpressed PLD1 is frequently described as cytoplasmic [169], perinuclear [89,118,169], and vesicular [118,170–173], with co-localization studies placing it at early- and late endosomes [7,89,97,118,168–170,174], lysosome [7,170,174], autophagosomes [170], ER [89], secretory granuoles [7,175], Golgi [89,118,168] and actin cytoskeleton [176]. Overexpressed PLD1 has also been observed at the plasma membrane [7,169,175,177], including caveolin-rich microdomains [140], lamellipodia and ruffles [178] and to move to the plasma membrane following cell-stimulation with some [7,118,175,178], but not all agonists [89,175]. Over-expressed PLD1 has also observed in some studies within the nucleus [170,173] and overexpression studies have proved critical in determining how the proteins localization at this site is determined. PLD1 possesses a nuclear localization sequence (NLS), mutation of which abolishes nuclear import [173]. Following apoptosis-induced caspase cleavage the NLS lies within a C-terminal fragment (CF-PLD1) which moves exclusively to the nucleus. The NLS of wild-type PLD1 and CF-PLD1 interact with importin-β which mediates nuclear import [95,170,173]. Four hydrophobic residues (L647, V648, 1649, 1670) within the PLD1 N-terminal HKD motif, determine PLD1 nuclear localization, their mutation causing translocation from vesicles, and an increase in PLD1-importin-β interaction with subsequent nuclear-translocation [170]. Staining with specific antibodies localizes endogenous PLD1 to many of the same localizations as the overexpressed protein including the cytoplasm/ cytoplasmic puncta [12,179], Golgi [9,12], TGN [179], nucleus [12,95,179], a perinuclear region [179], a vesicular localization [170], and plasma membrane [177]. Notably however, these studies do not detect endogenous PLD1 in the lysosomes, endosomes or secretory granuoles. Careful studies employing combined cell fractionation, immunofluorescence and immuno-EM have confirmed the endogenous localization of PLD1 in multiple cell lines as being within the Golgi and nucleus, and that overexpression of PLD1 forces the protein from the Golgi causing it to mislocalize [12]. This report is consistent with some overexpression studies reporting PLDs absence from the Golgi [7,174]. This study also noted much lower levels of plasma membrane PLD1 staining (if any in some cell types) than many studies employing overexpression [12]. Other studies looking at the endogenous protein have also called into question the extent of PLD1 at the plasma membrane and have suggested that in contrast to the overexpressed protein it is absent from caveolin-rich microdomains [180].
Many PLD2 localization studies have also employed overexpressed tagged-protein, and have reported it in cytosol [181], vesicles [171], and in particular the plasma membrane [89,168,171,172,181], including in caveolin-rich micro domains [180], and membrane ruffles [171] in unstimulated cells. PLD2 has also been observed to translocate to plasma membrane ruffles [484], filopodia, and sub-PM vesicles [89] following stimulation. The endogenous protein has also been observed at the plasma membrane in multiple cell lines [11,168,179,182], although some studies suggest this is much less extensive than with overexpression and limited to discrete areas such as ruffles or regions of active recycling [11]. Endogenous PLD2 may also localize to caveolae since it co-localizes with caveolin-1 [11] and is present in caveolin rich fractions [180,182,183]. Endogenous PLD2 is also observed in cytoplasmic puncta [11,179,182], within the perinuclear area [11,179], at the TGN [179] and at the rims of the Golgi, an area distinct from that of endogenous PLD1 suggesting a potential different function within this organelle [11]. Endogenous PLD2 has also been observed to translocate to the nucleus following treatment with the Arf inhibitor brefeldin A, suggesting a potential nuclear signaling role under certain circumstances [11].
4.6. The PLD1, and PLD2 knockout mice
Given the range of cellular processes in which PLD1 or PLD2 have been implicated, the observation that mice lacking either (or both) isoforms are viable and appear overtly normal [42,184] surprised many in the field, but offered the hope that PLD inhibitors may be valuable therapeutic agents without major health risks. Absence of either isoform fails to elicit compensatory elevation of expression of the other [42,184], and both PLD1−/−, and PLD2−/− -knockout mice are viable, bom at expected mendelian ratios, develop normally, appear healthy, are fertile, and display overtly normal behavior. However, mouse phenotypes associated with PLD1/ PLD2 absence do exist. While red-and white blood cell, and platelet counts appear normal in PLD1−/− mice, platelet volume was increased slightly resulting in increased surface abundance of platelet surface receptors, including GPV, CD9 and βl and β3 integrins [42]. PLD1−/− mice also exhibit reduced αIIbβ3-dependent platelet activation, rendering them resistant to major pathological hemostasis events such as strokes and pulmonary embolisms [42]. Furthermore PLD1−/− mice, display macroautophagy defects [185], which could potentially affect cancer cell-intrinsic metabolic pathways [186], and display decreased tumor formation and angiogenesis [43]. PLD-deficient mice (single or double PLD1/PLD2 knockout) display impaired brain development, having reduced brain growth at 14–27 days post partum [187]. They also display impaired cognitive function in social (PLD1 KO and PLD2 KO), and object recognition (PLD2 KO, and double KO). Brain microdialysis revealed severely reduced hippocampal acetylcholine (Ach) release following behavioral stimulation in PLD1−/− and PLD2−/− single knockout animals [187]. Since choline is a precursor for Ach production, this may be a consequence of decreased choline production due to reduced PLD activity. These observations may have relevance for cognitive dysfunction observed in fetal alcohol syndrome and Alzheimer’s disease (AD) [187]. Consistent with a role in this disease, PLD2−/− mice displayed protection against the synaptotoxic, and memory-impairing actions of β-amyloid in a transgenic AD model [39,188]. PLD1 and PLD2 have also been implicated in suppression of appetite, and appear to protect against overweight [189]. In addition, PLD1 and PLD2 knockout mice consume more food than controls, and also display reduced energy expenditure, elevated body weight, and increased adipose tissue [189]. PLD1 and PLD2 knockout mice both displayed elevated free-fatty acids and are insulin and glucose intolerant [189]. Complicating interpretation of the knockout mouse phenotypes, PLDs may possess non-phospholipase roles, including scaffolding [190,191] and their absence therefore could potentially disrupt multi-protein complexes. Knockdown could also induce compensatory mechanisms including those increasing PtdOH [190].
4.7. The discovery of additional mammalian PLD isoforms
While the vast majority of mammalian PLD research has focused on PLD1 and PLD2, other potential mammalian isoforms have been reported in the literature [190]. These proteins namely PLD3 (also called Sam-9 or HUK4); PLD4; PLD5 and PLD6 (also called Mitochondrial PLD, MitoPLD, Zucchini, or Zuc), contain HKD domains or variants thereof, but have been termed non-classical PLDs, lacking the PX- and PH-domains, and -classical PLD activity required to convert PtdCho to PtdOH [192–194]. As such, these proteins remain largely outside the scope of this current article and will only be mentioned briefly. It is now clear that PLD3 and PLD4 are single-stranded nucleic acid exonucleases regulating endosomal nucleic-acid sensing, rather than PtdCho-hydrolyzing PLDs [195], PLD5 is likely catalytically inactive due to poor conservation of the catalytic domains, lacking the histidine and lysine of the first catalytic motif and the histidine of the second [116,196]. PLD5 KO mice exhibit no significant abnormalities [197], however in humans PLD5 is linked to a profibrotic uterine phenotype that occurs during childbearing years. PLD5 polymorphisms may also be associated with an increased risk of tumor progression in multiple cutaneous and uterine leiomyomatosis syndrome [198], PLD6 encodes a single HKD-motif containing protein, which binds mitochondrial outer surfaces via an N-terminal trans-membrane tail [199]. Initial reports studying the mouse isoform stated that PLD6 hydrolyzes cardiolipin to PtdOH and displayed no in vitro nuclease activity towards RNA or DNA [199–201]. Recent studies however, clearly demonstrate that PLD6 is not a lipase/ phosophodiesterase, but rather a single strand specific RNase, essential for primary biogenesis of piRNAs (piwi-interacting RNAs), and suppression of transposon expression, with an important role in maintaining genomic integrity of germline cells [202–205].
4.8. Redundancy of mammalian PLD isoforms
The existence of multiple PLD isoforms within individual organisms raises the possibility of functional redundancy. Redundancy in plants is well documented [28], however the situation in other organisms is less clear [190], The differing localizations of mammalian PLD1 and PLD2 in some cell types suggest non-redundant functions, as does the fact there is no compensatory expression increase observed in knockout mice lacking a single PLD isoform [42,184]. Furthermore, while inactivation of either isoform decreased macrophage phagocytosis, no further decrease occurred upon dual disruption [206]. Nor is PLD1’s regulation of reactive oxygen species (ROS) levels in neutrophils affected by absence of PLD2 [184]. In addition, while stable expression of PLD1 or PLD2 in HEK293 cells, resulted in elevation of tissue plasminogen activator (tPA)-stimulated p38 MAPK phosphorylation, only those expressing PLD1 demonstrated increased cyclooxygenase-2 (Cox-2), and interleukin-8 (IL-8) expression [207]. Other studies, however argue for full-, or partial redundancy of PLD1 and PLD2. These include those demonstrating dual occupation of the same sub-cellular localizations, and the fact that inhibition of mouse embryonic fibroblast (MEF) dorsal membrane-ruffling by the dual PLD1/2 inhibitor FIPI, requires inhibition or deletion of both isoforms [208]. Evidence of partial redundancy also exists, from studies in which lack of PLD1 (but not PLD2) reduced platelet activation [42], and lack of both isoforms elicited a stronger phenotype [209].
4.9. The identification of isoform-specific mammalian PLD inhibitors
Until the advent of specific PLD inhibitors, primary alcohols (primarily butan-1-ol) were the only method to inhibit the production of PLD derived PtdOH, with secondary and tertiary alcohols used as controls [51,79,184,210], Problems associated with the technique became evident when it was revealed that the alcohol concentrations required to substantially block PtdOH production were toxic, and that routinely used concentrations gave incomplete inhibition [184,211–215]. Concerns of off-target effects also grew, and with the advent of RMAi, and small molecular inhibitors, it became evident, that many biological effects associated with primary alcohols could not be attributed to PLD inhibition [190,210,212,215,216]. In particular, the PLD-specific inhibitor: 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI) didn’t have any effect on a number of biological processes which were inhibited by butan-1-ol, and that had previously been attributed to PLD [212,215]. Similarly, a number of ethan-1-ol sensitive events, failed to be replicated with more reliable techniques, such as a genetic deletion of PLD [210]. The impact of these revelations was enormous. After over 100 publications demonstrating an essential PLD role in fMLP peptide signaling-activated superoxide production [216] Sato, and colleagues demonstrated that the dual PLD1/ PLD2 inhibitor 5-fluoro-2-indoyl des-chlorohalopemide (FIPI) had no effect on this process, or on PtdOH production. PtdOH increases stimulated by fMLP were instead being produced by DAG kinase mediated phosphorylation of PLC-produced DAG. Clearly, alcohol was inhibiting superoxide production through an unknown PLD-independent mechanism. To emphasize just how big an impact this finding had on the field, over 4800 PLD articles had been published prior to this finding, many of which employed primary alcohols [190]. Furthermore, later studies indicated that primary alcohols aren’t equally effective ‘inhibitors’ for all family members, for example they poorly inhibit Pseudomonas aeruginosa PLD (PldA)-catalyzed PtdOH production [217]. It has also been demonstrated that some organisms such as fungi can also use the secondary alcohols, traditionally employed as controls in these experiments, for transphosphatidylation [218,219]. In addition, primary, and secondary alcohols are also reported to actually stimulate plant PLD activity [220] and the effects of primary alcohols have been mimicked by higher concentrations of secondary alcohols in algae [221,222]. Despite these caveats, inhibition of PtdOH production by primary alcohols continues to be employed within the PLD field. As such, and given the historical reliance on this technique, reports employing this technique have been included in this review, where possible with more recent reports validating the findings. It is perhaps best that all findings previously based upon this technique should be revisited, using genetic knockdown, siRNA (or similar), or specific inhibition. The identification of highly specific PLD inhibitors has enabled functional analysis without the concerns of off-target effects, inhibitor dose, or time course, associated with previous methods [184]. A wide-range of PLD inhibitors are reported, though many are currently poorly characterized [47]. A 2007 Novartis screen to isolate PLD2 inhibitors identified the psychotropic agent halopemide (also called R34301) [223], which inhibits both PLD1 (IC50 21 nM in cells and IC50 220 nM in vitro), and PLD2 (IC50 300 nM in cells and IC50 310 nM in vitro) [224]. The drug had previously been through clinical trials in which plasma concentrations sufficient for human PLD1 inhibition were used, demonstrating the clinical viability of PLD1 inhibition [225]. Following halopemide’s identification, hundreds of analogs were synthesized and assayed in attempts to identify isoform specific inhibitors [47,224,226–228]. Notable inhibitors included FIPI, which inhibited PLD1 (IC50 1 nM in cells, and IC50 9.5 nM in vitro) and PLD2 (IC50 44 nM in cells and IC50 17 nM in vitro) with increased potency [224], FIPI acts by interacting with the second HKD motif and as such inhibits the catalytic sites of both PLD1 and PLD2 [229]. Isoform specific inhibitors also exist including: the PLD1 specific inhibitor: VU0155069 (approximately 163-fold selectivity over PLD2), and the PLD2 inhibitor: VU0155072 (approximately 10-fold selectivity over PLD1) [223,224,230–232]. Further optimization via medicinal chemistry approaches greatly improved isoform specificity and potency, resulting in the highly selective PLD1 inhibitor: VU0359595 (IC50 3.7 nM, 1700-fold over PLD2), and an improved PLD2 inhibitor: VU0364739 (IC50 20 nM, 75-fold preference over PLD1) [47,233–235].
4.10. Mammalian PLD1 and PLD2 regulation
Mammalian PLD enzymes are tightly regulated by multiple mechanisms including post-translational modification, cofactor availability, molecular interactions and signal-induced sub-cellular translocation (Fig. 2) [47,163]. PLD1, and PLD2 are differentially regulated with PLD2 being generally less agonist responsive. For example, PLD2 transfected cells are stimulated only 2-fold by phorbol 12-myristate 13- acetate (PMA), versus 10–15 fold for PLD1. Such observations led to suggestion that PLD2 is basally active (unlike PLD1) in unstimulated cells [89]. This was confirmed by the observation that PLD2 (but not PLD1) knockdown resulted in a reduction of basal PLD activity. PLD1 in vitro basal activity is activated directly by small G proteins and PKC, in conjunction with PtdIns(4,5)P2 [88,236,237]. PLD2 displays constitutively high activity (1000-fold greater than PLD1) in the presence of PtdIns(4,5)P2, but is insensitive to PLD1 activators such as Arf, Rho and PKC in vitro [34,89,236].
4.11. Regulation of mammalian PLD by phosphoinositides and other lipids
Early mammalian PLD studies identified a rat brain PLD activity which, was oleate-dependent and preferred neutral pH. It was inhibited by negatively-charged-phospholipid-binding aminoglycoside antibiotics, including neomycin, in a manner reversed by adding back PtdIns(4,5)P2 [59,68,238–241]. Later, in vitro PLD assays using HL60 cell membranes revealed a PtdIns(4,5)P2 requirement for choline release from PtdCho [66], and that in the absence of sodium oleate, the partially purified oleate-dependent activity could also be stimulated by this phospholipid [68,69]. Cloning of mammalian PLD1 [34,138], and PLD2 [89,90,242] isoforms later revealed an obligate PtdIns(4,5)P2 requirement for the activity of these isoforms. Several proteins involved in PtdIns(4,5)P2 production, sequestration, and dephosphorylation are implicated in PLD regulation. Over-expression of PtdIns4P 5-kinase increases PLD activity in mammalian cells, and both PLD1 and PLD2 bind to PtdIns4P 5-kinase Iα [243]. PLD2 activity (but not that of PLD1) is increased when co-expressed with PtdIns4P 5-kinase Iα and it is inhibited by over-expression of a catalytically inactive PtdIns4P 5-kinase Iα mutant [243]. PtdIns4P 5-kinase Iα activity is stimulated by PtdOH, providing a potential, positive feedback loop, or the possibility that one PLD isoform could stimulate another through stimulation of kinase activity. Dephosphorylation of PtdIns(4,5)P2, or its sequestration, may also regulate PLD since the inositol phosphate 5-phosphatase synaptojanin [244,245] and the PtdIns(4,5)P2-, and actin-binding protein gelsolin [246] inhibit PLD in vitro. Numerous studies demonstrate roles for the PH-domains and polybasic motifs of PLD1, and PLD2 in interaction with PtdIns(4,5)P2. These lipid-binding domains play an important role in localization of these isoforms, and are reported to contribute to varying degrees in their obligate PtdIns(4,5)P2 requirement for activity (see section 4.2). Other phosphoinositides also stimulate PLD activities, including PtdIns(3,4,5)P3, a weak activator of the purified enzyme, which demonstrates greater activation in partly purified preparations [247]. PtdIns(3,4,5)P3 stimulates human- [88,126] and rat PLD1 [138], activating and binding human PLD1 half as efficiently as PtdIns(4,5)P2, suggesting a critical role of the phosphate position on the inositol ring [126]. Surface plasmon resonance analysis demonstrated only limited PtdIns(3,4,5)P3 binding to intact PLD1 and none to the isolated PH- domain [126,247]. In contrast, the isolated PLD1 PX-domain bound PtdIns(3,4,5)P3 with high affinity in a vesicle binding study. Binding required lysine (119,121) and arginine (179), and the PLD1 PX-domain also displayed modest affinity PtdIns3P as well as for PtdInsSP [139]. In addition to binding of these phosphoinositides, the PLD1 PX-domain can simultaneously bind PtdOH or PtdSer, and potentially other ionic lipids due to the presence of a second distinct lipid-binding pocket, dual occpacy of these pockets increases membrane association and likely plays a role in spatiotemporal PLD1 regulation [139] .The in vivo role of PI-3-kinase in regulation of PLD activity is complicated by the fact that while PI-3-kinase inhibitors such as wortmannin inhibit cellular PLD activity, they are non-specific and affect multiple other pathways [248]. In addition, PI-3-kinase may regulate PLD through indirect mechanisms, including lipid-binding proteins such as the cytohesin [249] and Tiam-1 [250] exchange factors regulating Arf-, and Rho, whose activity is controlled by PtdIns(3,4,5)P3-dependent PH-domains.
Purification of multiple PLD activities from plants and mammals has indicated stimulation by unsaturated fatty acids [63,163,251]. These include one of the earliest purified mammalian PLDs, from porcine lung, which was stimulated by unsaturated oleic (18:1), linoleic (18:2), and arachidonic (20:4) fatty acids [63]. Study of oleate-dependent PLD activity has been hampered by the failure to conclusively identify the oleate-stimulated protein and it is likely that multiple isoforms can be oleate-stimulated depending on the assay system. For example, oleate stimulated PLD1 activity in Huh-7 cells through PI-3 kinase-mediated activation of Arf and Rho [252], while oleate stimulated PLD2 but not PLD1 in RBL-2H3 mast cells [172]. Several other studies suggested PLD2 is oleate-stimulated, and consistent with the early studies this isoform is enriched in lung [90,163], PLD activity was oleate-stimulated in Jurkat T-cells, only expressing PLD2, but not HL60’s uniquely expressing PLD1 [90]. Similarly, oleate-stimulated activity in L1210 cells solely expressing PLD2, but not in U937 cells only expressing PLD1 [253]. Oleate (18:1) also stimulated purified PLD2 (but not PLD1) in vitro, as did the other unsaturated fatty acids linoleate (18:2) and arachidonate (20:4), but not saturated fatty acids such as myristate (14:0), palmitate (16:0), stearate (18:0), or arachidate (20:0) [253].
5. Post-translational modification of mammalian PLDs
5.1. Palmitoylation, and ubiquitination
Both mammalian PLD1, and PLD2 are palmitoylated with important consequences for localization and regulation. The PLD1 [115,130], and PLD2 [254] PH-domains are palmitoylated on cysteine residues: a modification requiring a full-length, catalytically competent protein [255]. Mutation of cysteines −240 and −241 of human PLD1 didn’t significantly alter in vitro activity, but decreased activity in vivo. This was associated with decreased perinuclear-localization, and a concomitant increase in plasma-membrane localization [130]. Mutation of corresponding rat PLD1 residues decreased basal activity and membrane localization, and also reduced levels of PLD1 serine and threonine phosphorylation [256]. Palmitoylation of rat PLD1 required association of the N- and C-termini of the PLD protein [256]. In addition to palmitoylation, PLD is also regulated by ubiquitination. The PLD1 (but not PLD2) PH-domain is multi-monoubiquitinated in a manner requiring catalytic activity and palmitoylation. Ubiquitination targets PLD to the proteasome for degradation likely serving to reduce lipase activity [231].
5.2. Phosphorylation
PLD1 and PLD 2 are phosphorylated on serine (S)-, tyrosine (Y)-, and threonine (T) residues [141,163,257–259]. Confusingly, basal in vitro and in vivo PLD activities are frequently insensitive to mutation of phosphorylation sites, suggesting phosphorylation alters localization and substrate/ regulator access, rather than activity. Many observations are also complicated by the failure of studies to address the possibility of intermediate kinases.
Casein kinase II, a serine-threonine-selective kinase, phosphorylates PLD1 (S911) [260] and PLD2 (on multiple sites in vitro) [261], but does not affect catalytic activity. The kinase complexes with both proteins and is suggested to stimulate U87 glioblastoma cell proliferation through a PLD-dependent mechanism [261]. PLD1 S505 is phosphorylated by AMP-activated protein kinase (AMPK), a modification required for in vivo glucose-stimulated PLD activity [262]. Similarly, cyclin-dependent kinase 5 (Cdk5)-mediated PLD2- S134 phosphorylation regulates EGF-stimulated insulin secretion in rat insulinoma cells [263]. PLD1 T147 is phosphorylated by the p90 ribosomal S6 kinase, an event required for stimulation of PLD activity in PC12 neuroendocrine cells by K+, a process in which PLD is stimulated following K+-mediated depolarization of membranes, and subsequent increase in cellular Ca2+ [264]. Association of the rat PLD1 N- and C-termini occurs in vivo, through conserved residues in the HKD domains. Association is required for palmitoylation of cysteine residues C240 and C241, which in turn is essential for widespread serine / threonine phosphorylation, possibly by direction of rat PLD1 to a kinase containing membrane fraction [103,256]. Serine/ threonine phosphorylation does not appear to be required rat-PLD1 activity, or for its stimulation by PKC, or small GTP-binding proteins but did localize it exclusively to the membrane fraction [103]. An inhibitory role for phosphorylation may exist for PLD2. PLD2 over-expressed in HeLa cells was phosphorylated on serine and threonine residues, following treatment with the phosphatase inhibitor okadaic acid. This resulted in a concomitant inhibition of PtdIns(4,5)P2_ stimulated PLD2 which was not due to alteration of PtdIns(4,5) P2 affinity [265].
Tyrosine phosphorylation also regulates PLD activity. The tyrosine phosphatase inhibitor vanadate increases PLD activity in HL60 granulocytes [266] and both fMLP-stimulated PLD activity and tyrosine phosphorylation are increased in neutrophils, in a manner inhibited by tyrosine kinase inhibitors [267]. PLD2 is tyrosine phosphorylated and confusingly dephosphorylation of PLD2 by tyrosine phosphatases is reported to both increase [268], and decrease [269] in vitro activity. PLD2 is tyrosine phosphorylated following EGF-stimulation [270–272], and since the mutations Y11A [272], or Y296F [270] increase EGF-stimulated PLD activity, phosphorylation of the residues appears to down-regulate activity. The tyrosine kinases Janus kinase 3 (JAK3), and Src phosphorylate PLD2 at Y415, and Y511 respectively [270]. Phosphorylation of Y415 stimulates PLD2 activity, while phosphorylation of Y511 is inhibitory [270]. Tyrosine phosphorylation of the PLD2 PX-domain residues Y169 and Y179 regulate SH2 domain-mediated binding to the adaptor protein Grb2 (Growth factor receptor bound protein 2). This in turn, recruits the proline rich C-terminus of the Ras GEF SOS (son of sevenless), via the Grb2 SH3-domain. This stimulates Ras, coupling EGF-stimulation to Ras activation, and increasing PLD2 activity. Y169 and Y179 act as Grb2 docking sites, with a level of redundancy. Mutation of either residue reduces Grb2 binding, while dual mutation abolishes it. The two residues appear to regulate distinct functions however, Y179 being dispensible for PLD2 activity while mutation of Y169F ablates it [273]. Furthermore,while both residues serve to bind Grb2, a Y179F but not a Y169F mutant could stimulate Ras activity. Transient over-expression of PLD2 Y179F additionally upregulates p21, Ras, Erk activity and increases cell proliferation, indicating that Y179 of PLD2 has a negative roll in Ras-mediated regulation of cell proliferation [273]. Interaction with Grb2 through PLD2 Y169, additionally plays an important role in regulation of PLD2 phospholipase activity, and localization. Cos 7 cells constitutively expressing Grb2 shRNA had significantly reduced PLD2 activity and PLD2 over-expressed in these cells localized to the cytoplasm and to a peri-nuclear localization to a lesser extent. Following co-transfection with an shRNA resistant Grb2 construct, PLD2 relocalized to the perinuclear/ Golgi region upon EGF-stimulation. Endogenous PLD2 and Grb2 also co-localized in Huvec cells [274], Akt (protein kinase B) also phosphorylates PLD2 on threonine 175 and mutation of this residue (T175A) inhibits the PLD2 Y179F mutant from stimulating Ras, suggesting these residues allow PLD2 to fine-tune Ras signaling [275]. The phosphorylation state of PLDs may have relevance to disease. Tyrosine kinases are frequently upregulated in cancer, and a proteomic study examining global phosphotyrosine changes in cells overexpressing constitutively-active, transforming nucleophosmin-anaplastic lymphoma-kinase indicated increased phosphorylation of PLD 1 (Y711), and PLD2 (Y573) [276].
5.3. Regulation of mammalian PLDs by protein kinase C
Phorbol esters including Phorbol myristate acetate (PMA), which mimic the PKC-product DAG, stimulate PLD activity in a range of cell lines and tissues [277,278]. PLD1 is activated by both PKCα, and PKCδ in vitro [279], and while the high basal activity of PLD2 appears unresponsive to PKC in some systems [89,101,280] in others it is inhibited by PKC-inhibitors, or stimulated by PKC, or PMA [163,281–283]. Over-expressed and endogenous PKCα and PKCδ co-immunoprecipitate with over-expressed PLD1 and PLD2, in SF9 cells, in a PMA potentiated manner [283]. Similarly, endogenous PKCα and over-expressed PLD1 co-immunoprecipitate in COS-1 cell lysates, and endogenous PLD1, and PKCα co-immunoprecipitate in NIH3T3 cell lysates [284]. Purified PKCα and PLD1 interact directly [178,284], and SPR indicates this occurs in the absence of exogenous phorbol ester, ATP, or GTPγS [178]. The PLD1 PKC-binding site is N-terminal to the PX-domain [153], while PKCα requires both its regulatory and catalytic domains and the phenylalanine residue 663. The interaction is independent of PKC-kinase activity, occuring in the absence of ATP in vitro. Consistent with this, truncated PKC devoid of the catalytic domain activates PLD [247,286].
PKC phosphorylates PLD but the function in vivo is unclear, in many cases having little to no effect on activity [140,277]. Furthermore, many observations appear highly dependent on cell system, stimulus, and PKC-isoform, and the possibility exists of intermediate proteins [163]. In neutrophil membranes, PKCα stimulation of PLD requires ATP and is inhibited by staurosporine, a nonselective kinase inhibitor [257], suggesting PKCα directly phosphorylates PLD, or phosphorylates an intermediate PLD activator. However, in fibroblast membranes, PKC stimulated PLD1 activity through direct protein-protein interaction and not phosphorylation [277]. Proteomic analysis revealed PMA-stimulated PKC phosphorylates PLD1 on S2, S561, and T147, mutation of which results in slightly decreased PMA-stimulated PLD activity in vivo, but not in vitro [141]. Although dispensable for activity, S2 phosphorylation is required for receptor-stimulated actin association [259], PLD2 is phosphorylated at S134, S146, S243, T72, T99, T100, and T252 following COS-7 cell PMA-stimulation [258]. S243 and T252 were the predominantly phosphorylated residues but their mutation did not inhibit PMA-stimulated PLD activity [258]. Some studies suggest PKC phosphorylation of PLD is required for cell-surface receptor mediated PLD stimulation. For example, PLD1 T147 phosphorylation and activity increased rapidly following COS-7 cell EGF-stimulation, which is ablated by expression of dominant-negative PKCα, or mutation of the phosphorylation site [281]. In contrast, other studies suggest PKC-mediated PLD phosphorylation down-regulates PLD activity. For example, PMA rapidly stimulated PLD1 [285] and PLD2 [282] activities in COS-7 cells, but PLD phosphorylation only increased after a much longer period of stimulation, and this correlated with decreased activity [279,282,285]. Furthermore, phosphorylation of PLD2 serine and threonine residues following COS-7 cell PMA-stimulation was not required for PLD2 activity, since a kinase-dead PKC mutant acted as a more potent activator. Nor was it required for PMA-potentiated PLD2-PKC interaction, but rather served to down-regulate PLD2 activity [282]. Further evidence of PKC mediated PLD inhibition comes from the observation that the PKC-inhibitor staurosporine prolongs PMA-stimulated PLD activity [285], and PKCα/β inhibitors block phosphorylation, but not PMA-stimulated activity [282]. Furthermore, PLD inhibition by DGKζ occurs through a mechanism that appears to involve it complexing with PKCα [234,287,288]. The dual, positive and negative regulation of PLD by PKC-mediated phosphorylation has led to suggestion that different PKC isoforms perform these capacities, activated by different agonists [163]. For example, EGF may only activate positive regulators like PKCα, while PMA may activate multiple isoforms, including negative PLD1 regulators such as PKCδ [281,289]. Further complicating matters, individual PKC isoforms may behave differently depending on the PLD isoform. As an additional level of regulation, PKC also regulates PLD by influencing its expression levels. PMA selectively upregulated PLD1 but not PLD2 expression in HCT116 colorectal cancer cells, with upregulation being ablated following pre-treatment with PKC-inhibitors [280]. Regulation of PLD by PKC may also link PLD to Phospholipase C (PLC) signaling. PLD activation is frequently accompanied by stimulation of both PLC, and PKC is suggested as a potential link between these pathways [247], since PKC inhibition downstream of PLC blocks PLD stimulation [290].
6. Regulation of mammalian PLDs by small GTPases
6.1. PLD regulation by Arf proteins
Ras superfamily GTPases were the first proteins demonstrated to directly activate PLD. The first to be described was Arf, which was identified as a cytosolic factor capable of activating PLD in HL60 cell membranes, that was essential for GTPγS-dependent stimulation [66]. The Arf inhibitor Brefeldin A, or over-expression of dominant negative Arf1, or Arf6, block PLD-stimulation [178,247,291]. Furthermore, numerous studies demonstrate in vitro stimulation of PLD activity by Arf1-[66,67,279,292], and both in vitro [66], and in vivo stimulation of PLD activity by Arf6 [178]. Early studies suggested Arf only stimulated PLD1 [89] however, later studies indicated a 2-fold stimulation of PLD2 above its high basal activity, with GTPγS-activated Arf [90,130], compared with 4–6 fold stimulation of PLD1 [292].
ARF proteins are reported to bind PtdIns(4,5)P2 [295], and PtdIns P2 stimulates both ARF guanine nucleotide exchange, and ARF GAP activity [296]. A relationship between Arf6, PLD2 and PtdIns(4)P- 5-kinase Ια is proposed [181], and several studies have indicated ARF-mediated PLD-stimulation is dependent on PtdIns(4,5)P2, inciting speculation that Arf indirectly activates PLD by PtdIns(4,5)P2-dependent membrane phospholipid head group rearrangement [47,297]. However, other studies report Arf-mediated PLD activation in the absence of PtdIns(4,5)P2 [47,293,294], suggesting alternate or additional mechanisms of activation. PLD1b co-localizes with Arf6 at the plasma membrane in stimulated RBL-2H3 cells [178,247], and Arf1 in COS-1/−7 cell Golgi [298]. PLD1b bound Arf1 in an SPR study [178] and both Arf1 [299], and Arf6 [178] co-immunoprecipitate with PLD1. Since Arf activates N-terminally truncated PLD1 [292] and PLD2 [101] the Arf-binding site is likely elsewhere on the protein. The Arf N-terminus is believed to contain the PLD-interaction site, and Arf1 lacking the N-terminal 17aa no longer stimulates PLD1 activity [300,301]. Arf is myristoylated in this region and although PLD activation does not require this modification, stimulation is enhanced by it [66,69]. Arf myristoylation only appears to be required for certain PLD functions, being required for fMLP, but not GTPγS-stimulated PLD activity [302,303]. Synergistic stimulation is also observed when Arf is combined with other Rho family GTPases and PKC [292], and PLD1b co-localizes with Arf6, Rac1, and PKCγ following plasma membrane translocation in vivo [178,247].
6.2. PLD regulation by Rho family members
Pretreatment with the Rho GTPase inhibitors Clostridium difficile- or C. botulinum C3 toxin blocks PLD activation [304], and RhoA, Cdc42, Rac1, and Rac2 stimulate PLD activity in vitro [71,178,247,279,305–307]. Rho, Cdc42, and Rac1 are proposed to be PLD 1-selective binding activators, stimulating substrate-binding affinity [292]. Direct binding of Rac1 to PLD1 has been demonstrated by SPR [178]. PLD1 lacks Cdc42 and Rac-interactive binding (CRIB)-, or Rho-binding Rho effector (REM) motifs [308], with Rho instead interacting via residues in the PLD1 C-terminus, within domains III and IV [100,237,309,310]. Rac2−/− mice also show reduced PLD1 activity suggesting this isoform may also regulates PLD1 [311]. Rac2 regulates PLD2 through a dual, positive and negative, biphasic-mechanism during leukocyte chemotaxis [311]. GTP-bound Rac2 binds PLD2 through two poorly conserved Cdc42 and Rac-interactive binding (CRIB) motifs in the region of the PLD2 PH-domain [311–313]. PLD2 acts as a Rac2 GEF switching Rac2 from a GDP-bound inactive state to a GTP-bound active state, through a mechanism which requires the Rac2-binding, PLD2 CRIB motifs [146,313]. PLD also influences Rac activity through PtdOH-production aiding dissociation of the Rho-specific guanine nucleotide dissociation inhibitor (Rho GDI), facilitating Rac 1/2 plasma membrane association [314]. As such, a mechanism has been proposed in which rapid activation of a Rac2-PLD2 positive feedback loop occurs during onset of chemotaxis. This is followed by a second phase in which PLD2 activity is decreased, potentially by competitive binding of the PLD2 PH-domain region between GTP-bound Rac2 and PtdIn4,5P2, or by limition of PLD2’s association with the membrane [311–313]. PtdOH additionally recruits and stabilizes interaction of the Rho family GEFs DOCK1/2 (dedicator of cytokinesis 1/2), and Tiam1 (T-lymphoma invasion and metastasis-inducing protein 1) to the plasma membrane, through their C-terminal PtdOH-binding polybasic motifs [208,313,315,316]. The Ras-GEF Sos (son of sevenless) also binds PtdOH and is recruited by PLD2 leading to Ras activation [273,317].
Other small GTPases play roles in PLD regulation and activity including Rheb (Ras-homology enriched in brain), which activates PLD1 in vitro [318] and RalA (Ras-related protein A), which increases PLD1 activity in vitro, and co-immunoprecipitates with PLD1, but not PLD2 [299,319]. The RalA interaction site maps to a site independent of Arf interaction, allowing synergistic PLD1 activation [299].
6.3. Proteolysis of mammalian PLD, and apoptotic signaling
Mammalian PLD1 and PLD2 are substrates for caspase cleavage, a process linked to apoptotic signaling [47,96], Both isoforms are cleaved in vitro, and in vivo by caspases 3 and −8, with PLD1 additionally cleaved by caspase 7 [94,95]. Analysis of Alzheimer’s patient brain tissue demonstrated caspase 3 activation and caspase-proteolyzed PLD1 fragments [95], Caspase 3 cleaves PLD1 at aspartic acid residues D41, D545 and D581 in vitro, and at D545 in vivo where this is the dominant cleavage site [95,96], D545 cleavage cuts PLD1 within the loop region yielding a 56-kDa C-terminal fragment (CF-PLD1), which localizes to the nucleus via an exposed nuclear localization sequence (NLS), and a 60-kDa cytosolic N-terminal fragment (NF-PLD1) [95,96,173], Full-length PLD1 protects against apoptosis via suppression of p53 signaling, but in vivo PLD1 activity is suppressed by the dominant negative action of NF-PLD1, resulting in p53 de-repression and induction of apoptotic signaling [96], The PLD1c splice variant which possesses an early stop codon at amino acid 513, is expressed in human brain and may also function in a pro-apoptotic mechanism, similar to NF-PLD1 [91]. CF-PLD1 generated during apoptosis is imported into the nucleus via association of its NLS with importin-β [95,170,173]. Residues in the N-terminal HKD motif target PLD1 to vesicles, and mutation studies have indicated that when the residues are absent the protein’s association with importin-β increases [320]. Full-length PLD1 also possesses a NLS and is partially imported into the nucleus [96,173] where it mediates PKCα, and extracellular signal-regulated kinase (ERK) activation, a function which CF-PLD1 is unable to perform [173]. The ability of caspase-cleaved fragments to interact with each other, or the intact enzyme appeared likely given that separate PLD1 N-and C-terminal fragments can associate to form an active enzyme [102]. It is now known that the caspase-cleaved fragments interact via hydrophobic residues within the catalytic motifs and that this inhibits CF-PLD1’s nuclear import [170,320].
Like PLD1, the PLD2 isoform is also anti-apoptotic, with PLD2 inhibition or knockdown increasing apoptotic signaling. This likely occurs via induction of anti-apoptotic protein expression (Bcl-2 and Bcl-XI), and down-regulation of pro-apoptotic proteins (Egr-1, and PTEN) [47], Caspase-3 cuts PLD2 at multiple sites upstream of the PX domain (13–28aa), although the function of this is unclear, resulting in no significant changes in molecular weight, localization, apoptotic signaling, or catalytic activity [94,96].
7. Mammalian PLD function
Despite years of study, the precise nature of mammalian PLD function is still being elucidated. While largely believed to generate localized PtdOH pools, the observation that PLD can interact with over 30 different proteins has invited speculation of additional non-enzyme functions, including scaffolding roles [190,191]. PtdOH comprises about 1–4% of total cellular lipid [321] and plays both structural, and signaling roles. Complicating the study of PLD function is the fact that PtdOH can also be generated by de novo synthesis through: 1) sequential acylation of glycerol-3-phosphate [322–324], an important step in the Kennedy pathway for generation of glycerophospholipids [325]; 2) through the acylation of lysophosphatidic acid (LPA) by LPA acyltransferase (LPAAT), or 3) phosphorylation of DAG by DAG-kinase (DGK) [47]. For example, despite PLD activity being stimulated immediately following purinergic (Ρ2Υ6) G-protein-coupled receptor (GPCR) stimulation, the majority of PtdOH generated is via DGKζ [234]. The small negatively charged headgroup and resultant conical-shape of PtdOH intrinsically induce negative membrane curvature. This is believed to lower the activation energy for fission of membrane vesicles, and for their fusion with cell membranes [326], Importantly, PtdOH acts as a signaling node, recruiting binding proteins including the GEFs: DOCK2 and SOS, which activate Rac1 and Ras, respectively [191,315,317], and the mammalian target of rapamycin (mTOR) [327]. PtdOH also mediates translocation and activation of the Raf-1 (c-Raf) serine/ threonine kinase [328–331]. Multiple cAMP-specific phosphor-diesterase (PDE) 4 family members are also regulated by PtdOH, with PDE4A5; PDE4D3 and PDE4EB1 being stimulated by PtdOH in vitro [332], and PDE4D3 binding PtdOH directly [333]. PDE4A1, while not activated in vitro by PtdOH [332] requires PtdOH-binding through its N-terminus for membrane localization [298]. These interactions may indicate a PLD role in terminating G-protein coupled receptor signaling by metabolism of the second messenger cAMP [163].
While PtdOH production may represent a general PLD function, precise spatial and temporal regulation of this activity likely contributes to isoform specific functionalities. Mammalian PLD1, and PLD2 are implicated in a wide range of overlapping and isoform specific functions (see Fig. 3). PLD1 is implicated in vesicular trafficking [334,335] and exocytosis [175,336,337]. PLD1 and PLD2 are both implicated in anti-apoptosis [47,338], and PLD2 in endocytic recycling [339–341], PC 12 cell differentiation [342], actin-based membrane dynamics [181] and LI (neuronal cell adhesion molecule L1 or L1CAM)-dependent neurite outgrowth [343]. A PLD2 role as a guanine nucleotide exchange factor for Rac2 is now also recognized [146] (see Tables 1 and 2).
Fig. 3.
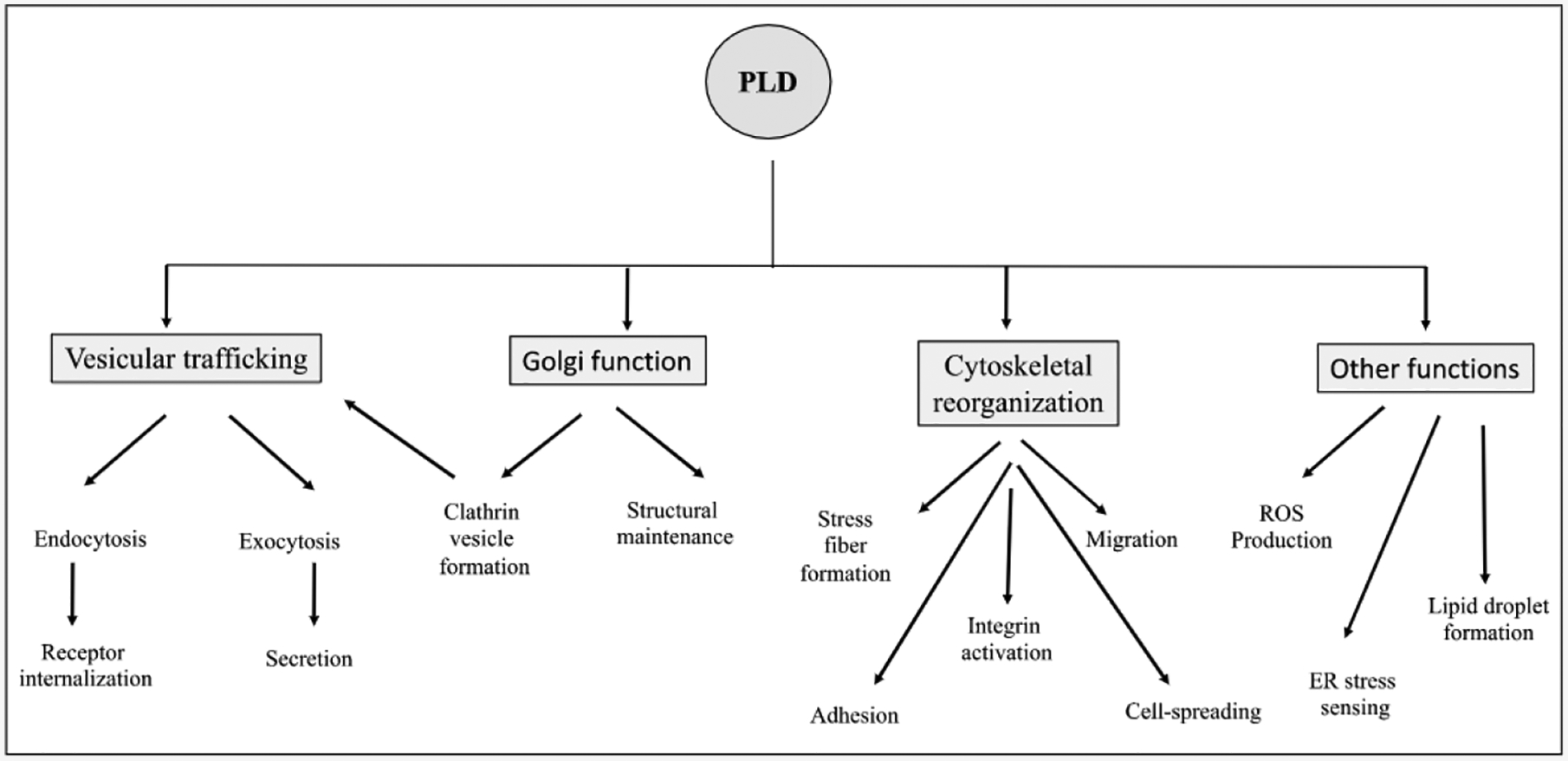
Examples of mammalian PLD function. Mammalian PLDs are involved in a vast array of cellular processes, notably Vesicular trafficking, Golgi-function and cytoskeletal regulation.
Table 1.
Functions and disease relevance of mammalian PLD1.
| Cancer (Melanoma), metastasis, angiogenesis. | [43,224,481] |
| Pathological hemostasis (e.g. strokes and pulmonary embolisms, autophagy, proteinopathies). | [42,185] |
| Brain development. | [187] |
| Behavioral stimulation response, cognitive function, social recognition. | |
| Receptor internalization. | [353] |
| Secretion/ exocytosis. | [373–377,381] |
| Actin Stress fiber formation, adhesion/ cell spreading. | [391,392,394] |
| ROS production. | [184] |
| Lipid droplet formation. | [439,440] |
| Integrin activation. | [42] |
| Diabetes. | [558,560] |
A large number of studies have identified important roles for PLD1 in cellular and physiological processes as diverse as endocytosis, lipid droplet formation, cytoskeletal regulation, cognitive function and social recognition. PLD1 has also been implicated in numerous diseases including diabetes, strokes and cancer. Numbers in parentheses refer to reference number in manuscript bibliography.
Table 2.
Functions and disease relevance of mammalian PLD2.
| Cancers including renal, breast, colorectal, bladder and lung. Tumor transformation, tumor metastasis and invasion. Angiogenesis. | [458,464,482,489,491,494] |
| Alzheimer’s disease (AD), brain development, behavioral stimulation response, cognitive function, social and object recognition. | [39,187,188] |
| Receptor internalization, recycling and trafficking. | [41,97,168,334,339–341,353,354,364] |
| Golgi/ TGN clathrin-coated vesicle formation, Golgi, and lysosome structural maintenance. | [8,10,179,384] |
| Adhesion. | [392,394] |
| Blood pressure regulation, thrombotic disease. | [44,168,196,452] |
A large number of studies have indicated a diverse range of important cellular and physiological PLD2 functions including receptor internalization, Golgi function, cellular adhesion, behavioral stimulation response and blood pressure regulation. PLD2 has also been linked to a number of human diseases including Alzheimer’s disease and cancer. Numbers in parentheses refer to reference number in manuscript bibliography.
7.1. The role of mammalian PLDs in receptor signaling/ trafficking
PLD activity is stimulated by many receptor agonists including the receptor tyrosine kinase (RTK) agonists EGF [163,344] and PDGF [345], as well as by the cannabinoid type 1- [346]; D2 dopamine- [347]; endothelin-1 (ET-1) [345]; P2Y purinergic- [234,348–350]; formyl peptide- [41]; angiotensin II- (AngII) [345] and μ-opiod- [341] GPCRs. In turn, PLD activity and PtdOH are implicated in multiple aspects of receptor trafficking, including PtdOH’s role in recruiting cellular machinery required to facilitate receptor endocytosis/ exocytosis events [351], and induction of membrane curvature [5,134,352]. PLD activity is implicated in EGFR [353]; μ opiod- [341,354]; mGluR1a- (metabotropic glutamate) [355], angiotensin II type 1- [168] and FcyRI-receptor internalization [97,334]. The plasma membrane localizations of PLD2, and stimulated-PLD1, ideally posit them to influence vesicular trafficking at this location and both mammalian PLD1, [353,356,357], and PLD2 [168,339–341,358] regulate receptor endocytosis. EGF-receptor (EGFR) internalization and degradation are enhanced by overexpression of either isoform, and are inhibited by expression of catalytically inactive PLD1/2 mutants, the presence of primary alcohols, or inhibition of the RalA, or PKC PLD-activators [353,356,357]. PLD1 opposes the action of hypoxia-inducible factor 1α (HIF1α), a negative regulator of EGFR endocytosis [357]. Following EGF-stimulation, PLD1 promotes degradation of HIF1α, in a manner requiring the PLD1 PH-domain. This increases expression of the Rab-effector Rabaptin 5 and accelerates EGFR internalization [357]. PLD2 co-immunoprecipitates with EGFR [272], and PLD2 over-expression increases EGFR mRNA and protein-levels, stabilizing mRNA and preventing it from decay, and inhibiting intemalized-EGFR degradation [359]. Dynamin plays a vital role in endocytosis and both PLD1 and PLD2 bind to its GTP-bound form stimulating GTPase activity through PLD PX-domain dependent GAP activity [147]. Expression of either PLD1- or PLD2’s isolated PX-domain stimulated EGFR endocytosis, similar to PLD1 or PLD2 overexpression [353,360]. Knockdown of both PLD isoforms with siRNA decreased EGFR endocytosis in a manner rescued by catalytically inactive PLD expression [147], suggesting PLD influences clathrin-endocytosis independently of PtdOH, or downstream DAG production. In addition to EGFR, PLD2 regulates endocytosis of the transferrin-receptor [339] and GPCRs [341,361,363] such as the angiotensin-[168], metabotropic glutamate −1 and −5 [355], β2-adrenergic-, M3 muscarinic- and μ-opioid receptors [341,354,362,364]. PLD proteins likely regulate the endocytic process on multiple levels, for example PLD derived PtdOH may induce negative membrane curvature at neck of endocytosing vesicles, while the PLD PX-domain acts to stimulate dynamin, facilitating vesicle scission [365]. In the case of the μ-opiod receptor, whose C-terminus interacts with PLD2, receptor endocytosis requires conversion of PLD2 derived PtdOH to DAG [341,354].
In addition to roles in endocytosis, PLD is also linked to Golgi/ TGN clathrin-coated vesicle formation [179] as well as COPI vesicle budding and tubulation [10]. PLD is also linked to clathrin-independent endocytic recycling, a process which requires Arf6 activation to recycle membrane proteins back to the PM [364,366–369]. Consistent with this, expression of an Arf6 effector domain mutant (N48R), which is unable to activate PLD activity, but is otherwise functional in terms of GEF/ GAP regulation and ability to activate PtdIns4P 5-kinase, caused an endosomal-tubule build-up in HeLa cells. This phenotype is consistent with a recycling defect, and is similar to that observed following inhibition of PtdOH production with butan-1-ol [211,352,370]. The Arf6 mutant (N48R) induced recycling block could be relieved by PMA treatment, (PLD being activated by PKC, bypassing the requirement for Arf6) [371]. Propranolol treatment which blocks PtdOH to DAG conversion, broke tubular recycling endosomes into vesicles, but didn’t relieve blockage of vesicle fusion with the plasma membrane, suggesting a PtdOH-derived DAG role in vesicle fusion [365]. PLD2 has been linked to recycling, depletion of this isoform but not PLD1 inhibiting transferrin receptor recycling from the endocytic recycling compartment (ERC), but not receptor internalization in HeLa cells [339]. PLD2-mediated recycling required EFA6 (exchange factor for Arf6) an ARF6 GEF, which coordinatinates both this process, and membrane ruffling [372].
PLD is also required for exocytosis including: nascent secretory vesicle release from the TGN [175,373–377], exocytosis of myeloperoxidase (MPO) [378,379], IgE-receptor degranulation [380], andPLD1-regulated secretion of von Willebrand factor [381]. The exocytotic mechanism is reported to involve secretory carrier-associated membrane protein 2 (SCAMP2) interaction with Arf6 and PLD1, linking them to exocytotic fusion pore formation [382], in which the serine/ threonine kinase ribosomal protein s6 kinase (RSK2) and PtdOH are essential [199,383]. However, many studies linking PLD activity with the exocytotic process have been called in to question since the departure for using primary alcohols to block PtdOH production. More recent studies using the PLD1/2 inhibitor FIPI and PLD KO mice have indicated that PLD is not required for degranulation/ secretion from mast cells, or neutrophils [210,212].
In addition to roles in vesicular formation, PLD2 plays an important role in in structural maintenance of the Golgi, and lysosome, and PLD2 (but not PLD1) specific inhibitors [179], primary alcohols [8,179], PLD2 siRNA [10] or catalytically inactive PLD2 mutants [384], cause dramatic morphological changes to these organelles.
7.2. PLD’s role in cell motility and cytoskeletal regulation
PLD activity and PtdOH are linked to mammalian cytoskeletal regulation and remodeling, and are key regulators of cell motility [13,14,89,385–389]. Mammalian PLD1 and PLD2 are both implicated in these processes. PLD activation stimulates actin stress fiber formation in fibroblasts [390] and endothelial cells [14], a PLD1 mediated event [391], while both PLD1 [392–394] and PLD2 [393] are involved in adhesion, cell spreading [395] and membrane ruffling [11,358] in leukocytes. The role of PLD in motility is evolutionarily conserved, being required for motility of algal monospores [396,397] and the slime mold Dictyostelium both of which are inhibited by primary alcohols, the latter being reversible by addition of exogenous PtdOH [398].
PLD’s small GTPase regulators, such as Rho, Rac, and Arf6 have well established roles in actin rearrangement, cell motility, and wound healing [247,399]. Consequently, it is perhaps unsurprising that many of PLDs roles in these phenomena are tightly linked to GTP-binding protein activities, for example Arf6-stimulated MDCK cell migration occurs through Rac- and PLD activation [399], and antigen-stimulated mast cells exhibit Arf6-, and PLD2-dependent membrane ruffles [171]. Many PLD-mediated roles in cytoskeletal modulation and cell-motility occur through downstream effectors, and in addition to PLD activity being regulated by small GTPases, PLD has also been observed to act upstream of the small GTPase Rac, a major regulator of cytoskeletal rearrangement required for cell-spreading and migration. PLD-derived PtdOH targets and anchors Rac1-GTP to the plasma membrane [400], through the Rac C-terminal polybasic motif. Rac mutated in this region cannot translocate, nor initiate integrin-activated, downstream-activation of p21-activated kinase (PAK) proteins, which have known roles in cytoskeletal rearrangement, motility and filiapodia formation [400]. The role of PLD2 in stimulation of membrane ruffling and subsequent motility have been closely linked to its relationship with Rac2, and the Grb2 (Growth receptor bound 2) adaptor protein [144,388,401]. The PLD2 PX-domain residues Y169, and Y179 interact with the Grb2 SH2 domain [274]. This interaction is reported to have major implications for PLD2 biology, being required for both PLD2 phospholipase activity, and it’s ability to re-localization to COS-7 cell Golgi, from the cytoplasm, following EGF-stimulation [274]. PLD2 and Grb2 co-localize in the actin-rich membrane-ruffles of macrophages, and play a role in the formation of these structures in resting and stimulated cells. PLD2 derived PtdOH was necessary but insufficient for the formation of these structures, requiring Rac-2 (Or Rac-1 to a lesser degree) for complete membrane ruffling induction [144]. Rac-2 is an activator of ARP2/3 which polymerizes actin and the three components coordinate to stimulate actin polymerization and membrane ruffling [144]. A model has been proposed in which PLD2 is targeted and activated at sites of membrane ruffling by Grb2, stimulating local PtdOH production. Rac2 a known PtdOH binding protein [580], associates with these proteins at the ruffle site enhancing membrane ruffle formation. In addition, Grb2 also links PLD2 to regulation of the actin nucleating Arp2/3 complex by promoting interaction of PLD2 with the Wiscott-Aldrich syndrome protein (WASP), which binds and activates Arp2/3, and is recruited to the plasma membrane during this process, enhancing phagocytic cup formation [401]. PLD2-and Grb2 also complex with the protein tyrosine phosphatase 1b (PTP1b), which serves to stimulate PLD2 activity [268]. PTP1b’s role in cell adhesion and motility is complex, and seemingly dependent on cell type, and regulatory context, both positively, and negatively regulating these processes [402]. In general, PTP1b appears to positively affect cell motility associated with integrin-dependent signaling stimulation, and negatively regulates motility by antagonizing growth factor receptor tyrosine kinase signaling [402]. PLD2-, and to a lesser extent PLD1-derived PtdOH also bind and activate ribosomomal 6 kinase (S6K) mediating actin polymerization, and facilitating motility [388,389,403,404]. Mammalian PLD is also linked to cytoskeletal regulation and remodeling through PtdIns4P 5-kinase, a protein which both produces PtdIns(4,5)P2 stimulating PLD activity, and is itself regulated by PtdOH [181,392]. The type Ιγ PtdIns4P 5-kinase isoform is implicated in cell motility, interacting with talin, and regulating focal adhesions [405,406]. Expression of dominant negative phosphatidylinositol PtdIns4P 5-kinase Iγb inhibited over-expressed PLD2’s ability to stimulate cellular adhesion [392]. Antibodies against β1 and β2 integrins blocked PtdOH-, and PtdIns(4,5)P2-stimulated adhesion inviting postulation that PLD-derived PtdOH stimulates PtdIns4P 5-kinase Iγb regulating integrins though PtdIns(4,5)P2 production [392].
In addition to interacting with the cytoskeleton through binding partners and downstream effectors, PLDs also directly interact with cytoskeletal components. Furthermore, while PLD activity promotes cytoskeletal rearrangement, PLD itself can be regulated by cytoskeletal elements. Mammalian PLD activity is regulated bidirectionally by actin, with monomeric actin inhibiting activity and filamentous activity promoting it [407]. Stimulated PLD1 associates with actin, binding both monomeric and filamentous forms [407], and stimulated PLD2 translocates to filopodia [89], and membrane ruffles [171,181], co-localizing with cytoskeletal elements [92] including actin [176]. PLD2 activity is inhibited following actin binding, a process reversed by Arf1 in vitro [176].
7.3. The role of mammalian PLD in respiratory burst
Mammalian PLDs are implicated in pathogen induced reactive oxygen species (ROS) production, during which the membrane-bound nicotinamide adenine dinucleotide phosphate (NADPH) -oxidase is activated during respiratory burst, producing superoxide radicals in immune cells used by the host in pathogen defense [408]. Historically, many studies implicated PLD in this process [409], since primary alcohols blocked ROS production [184,378,410]. However, demonstration that: 1) the dual PLD1/ PLD2 inhibitor FIPI didn’t affect respiratory burst stimulated by the GPCR fMLP, 2) that PtdOH generated in this process is through PLC/ DAG kinase, 3) that primary alcohols inhibit superoxide production through a mechanism independent of PLD activity [216], and 4) PLD−/− neutrophils could also generate fMLP induced superoxide [210], resulted in PLDs role in this process being called into question. However, more recent experiments using bone marrow-derived primary neutrophils isolated from the PLD2−/− knockout mouse, combined with use of isoform specific inhibitors, have demonstrated a role for PLD1 in regulation of phorbol ester-stimulated, adhesion-dependent, chemoattractant-, and Fcγ-receptor-stimulated ROS production, indicating a role for at least this isoform in these processes [184]. The precise mechanism by which PLD regulates ROS production however still remains to be elucidated, and is somewhat confusing since the response to FMLP is enhanced by removing PLD activity, compared to the results with PMA indicating that PLD activity may only regulate specific ROS pathways [184].
7.4. Mammalian PLD and mTOR activation
The target of rapamycin (TOR) is a serine/ threonine kinase, which is highly conserved throughout the eukaryotes. It controls cellular metabolism and growth, in response to nutrients, growth factors and stress [411]. PLD and PtdOH have been implicated in mammalian TOR (mTOR) activation, and a PLD requirement has been demonstrated in serum, phenylephrine, and mechanically induced mammalian target of rapamycin (mTOR) signaling [327,412–415]. Following cell entry, rapamycin complexes with FK506-binding protein of 12 kDa (FKBP12) and binds the mTOR FKBP12-rapamycin binding (FRB) domain, inhibiting mTOR signaling [416–418]. PtdOH also interacts with this region and may compete for binding, being expected to increase resistance to rapamycin inhibition [327,419]. However, many studies addressing the role of PLD activity in mTOR regulation historically employed primary alcohols, and they should be viewed in the context of the caveats now known to be associated with that methodology. Furthermore, despite numerous studies implicating PtdOH’s role in the mechanical induction of mTOR signaling, the role of PLD in this process is debated as the time courses of mechanically induced PLD activation and PtdOH production don’t correspond [420]. PLD activity is elevated for the first 15 min following mechanical induction, before returning to, or decreasing below control levels. In contrast, PtdOH levels increase over the first 30 min ultimately reaching an elevated steady state. Specifically, PLD activity at 15–30 min post induction is 27% lower than in an unstimulated muscle control, while PtdOH levels are elevated 138% and 187% above the control at 15- and 30 min respectively [413]. These data do not rule out the possibility that PLD is involved in the early response to mechanical induction, but do indicate that PLD activity cannot account for the majority of the PtOH response. Moreover, the observation that the dual mammalian PLD1/ PLD2 inhibitor FIPI blocked agonist-induced PLD activation, but didn’t prevent mechanically induced PtdOH or mTOR signaling indicates a major role for these isoforms in mTOR induction is unlikely [415]. This is supported by the observation that while knockout of both PLD1 and PLD2 result in viable mice [42,184], knockout of mTOR is lethal [421,422]. As such, it seems increasingly likely that the PtdOH source required for mTOR activation primarily comes from sources other than PLD, primarily the LPAAT pathway [423]. However, interpretation of these findings is complicated by the observation that inhibition of PLD, can result in a compensatory elevation of PtdOH from undetermined sources [351]. It has also been speculated that the LPAAT pathway provides PtdOH required for nutrient sensing by mTOR, while PLD provides local PtdOH in response to growth factors, insulin and stress [423]. Since mTOR localizes to the lysosome under conditions of amino acid sufficiency [424], it has been postulated that PLD (particularly PLD1) may contribute to activation at this localization, particularly since the LPAAT pathway produces PtdOH at the ER, which is then moved by vesicular trafficking to other cellular locals [425]. Furthermore mTOR forced to localize to the lysosome in the absence of amino acids, could still be activated in the presence of the small GTPase Rheb [426], a known activator of PLD1 [318]. PLD has also been postulated to play a role in mTOR signaling in the context of cancer. Dysregulation of signaling pathways regulating mTOR are postulated to be the most common in a cancer setting and mTOR is critical for cancer cell survival [423,427]. The dysregulated pathways include the phosphatidylinositol-3 kinase/ AKT/ Rheb pathway [428]. PLD activity is upregulated in many cancers and is substantially increased in cells depleted of nutrients, particularly in cells harboring Ras mutations [423,429,430]. Mutation of Ras results in an increased requirement for exogenously supplied lipid due to compromised activity of stearoyl-CoA desaturase-1, resulting in an inability of newly synthesized fatty acids to be desaturated, an essential requirement for membrane-targetted phospholipids [431,432]. Importantly, PLD appears to supply PtdOH in the correct format for stabilization of mTOR. Dipalmitoyl -PtdOH containing two saturated fatty acids caused the mTORC2 complex to fall apart, in contrast to PtdOH containing palmitate (saturated) and oleate (mono-unsaturated), which stabilized mTORC1 and mTORC2 under conditions of PLD suppression [419]. As PLD produces its PtdOH from membrane PtdCho, the composition of the PtdOH produced would be more similar to the later condition, consisting of a saturated and unsaturated fatty acid. Elevation of PLD activity in the context of Ras mutation is therefore proposed to aid escape from a defult apopototic program, and since PLD is required for cell migration in the absence of serum, may also help them move to a more favorable environment [423,433].
7.5. PLD and lipid droplet formation
ARF-regulated PLD activity is linked to cell-free lipid droplet (LD) assembly [434], and assembly of very-low-density lipoproteins (VLDLs) [435,436], a process involving lipid droplet formation in the microsomal lumen [437]. PLD is also implicated in stimulation of LD-formation by fatty acids, particularly oleate. Fatty acids stimulate some PLD activities, and oleate is reported to stimulate LD-formation through activation of PLD and PI3kinase [172,252,253]. The PLD1 isoform (but not PLD2) is present in LDs, and increased PLD1 expression promotes LD formation, while PLD1 siRNA inhibits it [438,439]. PLD1 activity in LDs is reported to be stimulated by Arf1 [439], and in cell-free LD assembly PLD1 additionally required the extracellular signal-regulated kinase 2 (ERK2) [434,438,440]. PLD1 (but not PLD2) and ERK2 were essential factors for both basal and insulin-stimulated cytosolic lipid droplet production. Inhibition of ERK2 ablated PLD1’s effect on lipid droplet formation, but did affect PLD1 activity. As such, PLD1 was proposed to act upstream of ERK2 [440]. ERK2 increases phosphorylation of the dynein cytoskeletal motor protein, increasing its translocation to adipose-differentiation related protein (ADRP)-containing LDs. Dynein plays a critical role in LD-formation, and microinjection of dynein antibodies strongly inhibits this process [438], Excessive triglyceride accumulation in ADRP-containing LDs, especially in liver, and skeletal muscle, is associated with metabolic disorders including insulin resistance and type 2 diabetes [441,442], strong risk factors for cardiovascular disease. Lipid droplets are also linked to [443] arteriosclerosis and ADRP is the major LD associated protein in foam cells (lipid-loaded macrophages) associated with atherosclerotic lesions [444]. Consistent with a role in LD-formation, PLD activities are linked to obesity, diabetes [189] and to thrombotic disease [42,43,209].
8. Mammalian PLD isoforms and human health
As techniques for studying PLDs have improved, they have linked PLD activity to a number of disease states including thrombosis [42,43,209], Alzheimer’s disease [37–41], multiple sclerosis [206,445] and a number of human cancers [162,446,447] (see Fig. 4). A finding perhaps unsurprising given the wide range of cellular processes in which PLD activities are implicated. The lack of severe phenotypes displayed by PLD knockout mice, and the apparent lack of overt toxicity of PLD1/2 inhibitors offers the hope that PLD specific inhibition may provide therapeutic potential [190].
Fig. 4.
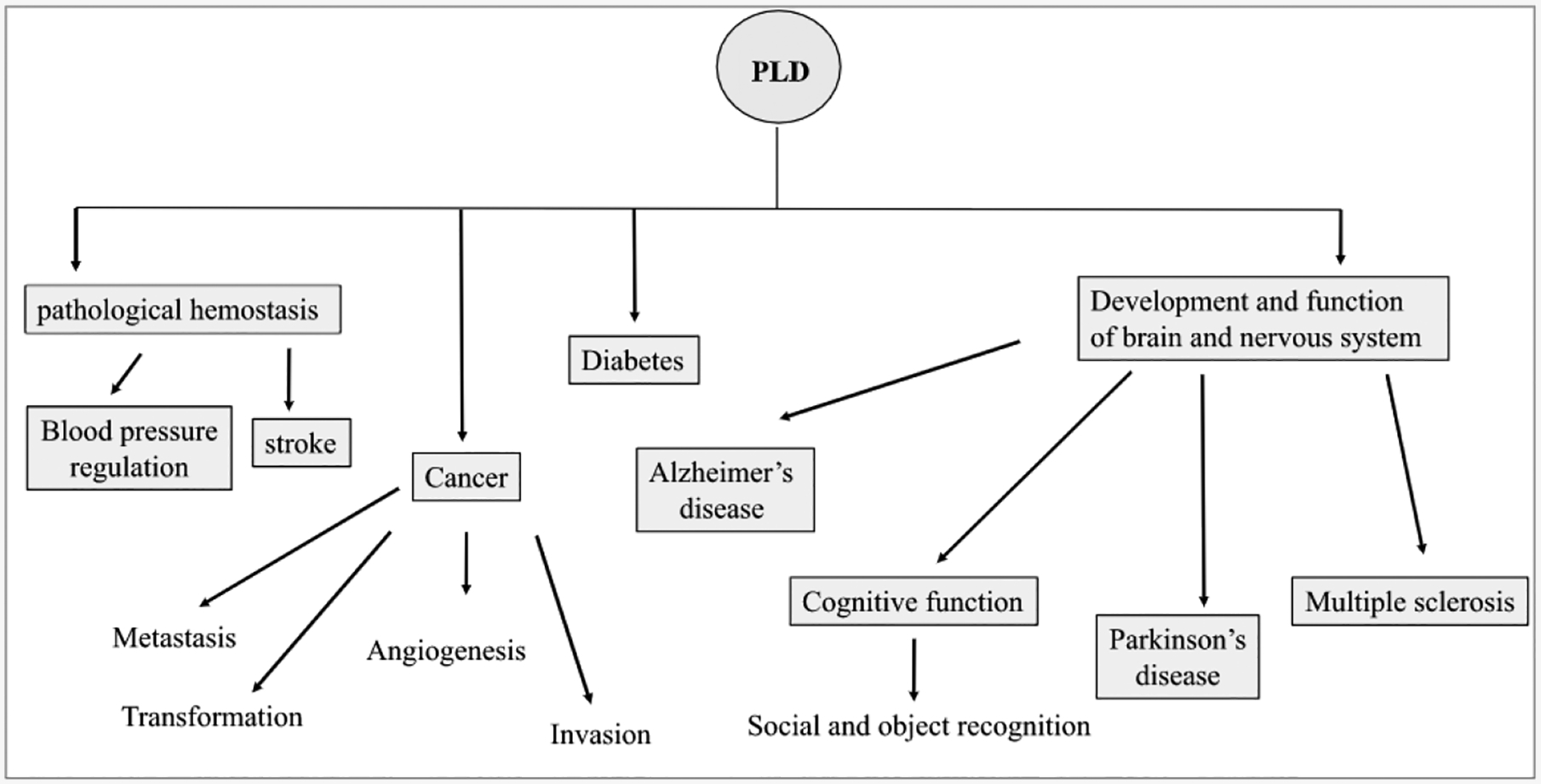
The role of mammalian PLDs in disease. Mammalian PLDs have been implicated in a number of physiological processes and diseases, including brain function and development, Alzheimer’s disease, cancer, diabetes and stroke.
8.1. PLD in vascular disease
Mammalian PLDs are linked to a number of vascular disease states including clotting disorders and atherosclerosis. Platelet aggregation is essential for hemostasis, and adhesion and activation of platelets at sites of vascular injury plays an essential role in limiting blood loss. These processes also form the mechanism underlying myocardial infarction and stroke [42], PLD1 and PLD2 are both expressed in platelets where they localize to ‘granule-like’ organelles. PLD1 being found through the platelet while PLD2 localizes to the platelet periphery, both isoforms redistributing to the plasma membrane upon activation and stimulation [42,448]. PLD1 is implicated in platelet activation, and stable thrombus formation under high shear forces, thrombi being unstable in PLD1’s absence, under rapid flow [42]. A key stage in platelet activation is activation of the integrin αIIb β3 adhesion receptor, facilitating adhesion and aggregation. Mice lacking PLD1 display integrin αIIbβ3 activation, resulting in decreased fibrinogen binding and vascular thrombi formation [42]. PLD1’s role in this process is unclear but decreased integrin αIIb βIII activation confers protection in stroke, pulmonary embolism and aortic thrombosis models [42,43]. Consistent with a role in regulation of the clotting process PLD activity is also linked to Von Willebrand disease, the most common inherited bleeding disorder, characterized by reduced blood-clotting which results from deficiency of the procoagulant and proinflammatory Von Willebrand clotting factor [449]. PLD1, (but not PLD2) regulates histamine-induced secretion of the prothrombotic Von Willebrand clotting factor from endothelial cells, which is inhibited by butan-1-ol or PLD1 RNAi [381]. The PLD2 isoform has also been linked to thrombus formation in a mouse, with both isoforms needing to be deleted for protection from FeCl3-induced arteriolar thrombosis [209]. Dual deletion was also required to inhibit release of a granules, important factors in platelet biology with roles in clotting, and atherosclerosis [209]. PLD2 is also linked to blood pressure regulation, a key factor in thrombotic disease, facilitating endocytosis of the GPCR angiotensin [168] and subsequent release of the blood-pressure regulating hormone aldosterone in adrenal cortex cells [450,451]. The PLD2−/− mouse displays increased blood pressure and decreased cardiac function due to decreased endothelial nitric oxide synthase (eNOS), resulting in decreased nitric oxide (NO), an important vasodilatory factor [581]. The decrease in eNOS appears to be a consequence of reduced free cholesterol in the PLD2−/− mouse, resulting in an increase in HMG-CoA reductase, the rate limiting enzyme in cholesterol synthesis, and a negative regulator of eNOS [581]. Human PLD2 PX-domain small nucleotide polymorphisms (SNPs) were identified in a screen to identify polymorphisms associated with hypertension. R172C correlated with patients suffering from hypertension, while R98C was associated with decreased blood pressure and reduced hypertension risk [44]. Analysis of the R172C mutation has determined that this mutation likely results in partial loss of function, or altered function rather than complete PLD2 deficiency [581]. The role of mammalian PLDs in blood pressure regulation and thrombus formation makes them attractive drug targets for treatment of thrombotic disease, and inhibition of both PLD activities with the PLD1/2 inhibitor FIPI reduced occlusive thrombus formation following chemical injury in mice [452]. This was not associated with intracerebral hemorrhaging or increased duration of tail bleeding, suggesting pharmacological PLD-inhibition may provide a safe anti-thrombotic therapeutic strategy for ischemic stroke and arterial thrombosis [43,190]. However, it should be noted that hemostasis in mouse is less dependent on platelet activation than humans, and this remains to be clinically tested [453]. Furthermore, application may be limited, since it was only effective when delivered prior to thrombosis, indicating inhibition may be useful for prophylactic treatment for high risk individuals, but less beneficial following thrombotic events such as stroke [190].
PLD is also implicated in atherosclerosis. During the atherosclerotic process exaggerated phagocytosis of lipids within the arterial wall leads to the accumulation of foam cells. Phagocytosis occurs through the uptake of low density lipoprotein (LDL). Foam cells containing LDL form atherosclerotic plaques which then act to restrict blood supply potentially resulting in stroke or peripheral arterial disease. Studies employing macrophages from PLD-deficient mice and specific PLD inhibitors have demonstrated that macrophage phagocytosis of cholesterol, and foam cell formation are less efficient in the absence of PLD2, demonstrating a role for this isoform in the formation of atherosclerotic plaques. PLD2 interacts with the Grb2-adaptor protein, actin, and WASP during macrophage phagocytosis of aggregated oxidized LDL (Agg-Ox-LDL) in the presence of CD36, and is proposed to act within this complex in the formation of the phagocytic cup, facilitating Agg-Ox-LDL uptake. PLD2 (but not PLD1), Grb2 and WASP are upregulated in diseased artery tissues indicating the potential in vivo relevance of these observations [454].
PLD activity has also been implicated in recovery following ischemia (reduced blood flow), which can occur as a result of multiple causes including vascular disease. In particular, PLD1 has been demonstrated to play an important role in tumor necrosis factor-α-mediated inflammation and scar formation following mouse myocardial ischemia, and reperfusion. Mice lacking PLD1 exhibited increased infarct size and reduced ventricular function, likely as a result of reduced tumor necrosis factor-α release, and altered interstitial collagen deposition [445].
8.2. PLD in viral pathogenesis
Host PLD activities are linked to pathogenesis associated with a number of human viruses [46,455–457]. Human PLD2 was identified in a whole genome RNAi screen for host genes required for influenza virus infection [457], a finding confirmed in cell culture models using isoform-selective PLD inhibitors [46], Influenza infection stimulates PLD activity and PLD co-localizes with influenza virus during infection. PLD inhibition delayed viral entry and reduced viral titers with specific PLD2 inhibition correlating with significant increases in transcription of innate antiviral effectors. Reduction in viral titer following PLD2 inhibition was dependent on Rig-I (retinoic acid-inducible gene-1), IRF3 (Interferon regulatory factor 3) and MxA (myxovirus resistance gene A). Together these data suggest PLD facilitates rapid endocytosis of influenza virus, permitting viral escape from innate immune detection and effectors capable of limiting lethal infection. As such, PLD2 inhibition could potentially be used proactively, or subsequent to infection to limit disease progression [46]. PLD inhibitors also suppressed HIV-1 replication in activated T cells [455,456], and are proposed to block Ras/ERK signaling, activating the c-Myc transcription factor required to produce dNTPs required for viral replication during T cell activation [455,456]. These findings offer potential for therapeutic intervention for HIV-1 and other viral infections dependent on host nucleotide bio-synthesis [455].
8.3. PLD in cancer
Aberrant PLD/PtdOH signaling is observed in numerous human cancers, including those of breast, ovary, kidney and colon [162,446,447]. A range of PLD abnormalities have been observed including PLD mutation [458,459], increased abundance, and increased activity [47,162,447,458–464]. PLD is upregulated in cells transformed by oncogenes including v-Src [465], v-Raf [466], v-Ras [467] and V-Fps [468], and PLD activity was required for Η-Ras transformation of Rat-2 fibroblasts [463]. PLD activity in cancer cells is linked to: increased metastasis and invasiveness [433,469–471], proliferative signaling [163,317,330,472], growth suppressor evasion [433,473–476], and resistance to cell death [477]. These findings are perhaps unsurprising, given the role of PLD activity in cancer-relevant processes such as cell motility, cytoskeletal rearrangement, [330], MMP-secretion [35,47,478] and regulation of mTOR [327,412–415]. While a consensus is emerging for PLD playing a pro-oncogenic role many older studies relied on inhibition of PtdOH production by primary alcohols and should be viewed with caution [163,190,479].
Mammalian PLD1 and PLD2 are both linked to cancer biology and isoform specific up-regulation is reported in a number of cancers including PLD1 in osteosarcoma [480], PLD2 in renal cancer [162], oleate-dependent PLD activity in breast cancer [460] and oleate-, and Arf-dependent activities in colon cancer [481]. PLD1 and PLD2 are linked to the prometastatic phenotype [482] with PLD1 in particular being implicated in metastasis and angiogenesis [43]. Isoform-specific PLD inhibitors decrease cancer cell invasion, migration, metastasis and angiogenesis [43,224,433,469], and specific inhibition of either PLD1, or PLD2, or both proteins inhibited invasion of highly metastatic breast cancer cells [224]. Furthermore, treatment of mice with the PLD1/ PLD2 dual inhibitor FIPI suppressed tumor growth and metastasis, and reduced growth of subcutaneously implanted B16F10 mouse melanoma cells into B16F10 tumors by 50% in PLD1−/− but not PLD2−/− mice [43]. PLD1 is required for metastatic tumor seeding and tumor vascularization with deficiency additionally impacting endothelial cell signaling [42]. Tumor platelet-interactions form an important stage in metastatic seeding, experimental metastasis being almost completely inhibited in a platelet-depleted host [483–486]. PLD1 deficiency impairs these interactions, reducing platelet-coating of tumor cells following stimulation by platelet-activating factors like PAR4-activating peptide or thrombin [43]. PLD1−/− platelets, also display impaired αIIbβ3 integrin activation [42], and reduced metastasis was observed in PLD1−/− mice after blocking αIIbβ3, suggesting a PLD role in facilitating αIIbβ3 mediated tumor cell-platelet contact [43]. Platelets release transforming growth factor-β1 (TGF β1) locally activating tumor cell signaling, facilitating the prometastatic phenotype [483], a process in which PLD is implicated [487].
In addition, to differences in platelet biology, PLD1-deficient mice possess endothelial cells with differences in angiogenic signaling pathways. The cells display: reduced basal phosphorylation of Akt at serine 473, and absence of VEGF-induced phosphorylation of this residue, they also exibit a 50% reduction of ERK1/2 basal phosphorylation and diminished VEGF-induced p38 phosphorylation [43]. Consistent with PLD1’s role in angiogenesis, the zebrafish PLD1 (zPLD1) isoform which is 64–68% identical to human and rodent PLD1 proteins at the amino acid level, is also required for angiogenesis, and both butan-1-ol and PLD1 morpholinos impair intersegmental blood vessel development [488].
Mammalian PLD2 is implicated in transformation, tumor metastasis and invasion [35,429,482]. Elevated PLD2 expression is observed in colorectal cancer patients [464,489] and a PLD2 polymorphism (C1814T) was identified in this group which remains of unclear significance since it does not effect PLD activity [458]. PLD2 is linked to in vivo EL4 lymphoma metastasis, with over-expression of catalytically inactive PLD2 reducing liver metastases [490], PLD2 is also linked invasion and metastasis of breast cancer [224,491]. Invasion of highly metastatic breast cancer cells is inhibited by a PLD2 specific inhibitor [224] and stable silencing of PLD2 in highly invasive breast cancer cells resulted in tumors with a mildly invasive capacity. In contrast, PLD2 over-expression in mildly invasive cells increased tumorgenicity following implantation into SCID (severe combined immunodeficient) mice. Furthermore, implantation of a micro-osmotic pump providing the PLD2-specific inhibitor NOPT reduced primary tumor volume and onset, and reduced axillary tumors by 50%, while a PLD 1/2-specific PLD inhibitor resulted in no axillary tumors suggesting an additional PLD1 role in this process [491]. PLD activity, including that of the PLD2 isoform has been linked to the epithelial-mesenchymal-transitition (EMT), an important stage in breast cancer metastasis [492]. PLD2 is regulated by the zinc-finger transcription factors Slug (SNAI2) and Snail (SNAI1), which play key roles in EMT, cell survival and invasion. Their activity is reported to account for elevated PLD2 activity in breast cancers of larger size and poor prognosis. PLD2 is positively regulated by Slug which activates PLD2 gene expression, and negatively regulated by Snail which competitively binds the PLD2 promoter. Both PLD2 and Slug levels are elevated in highly aggressive cells, while Snail levels are reduced. An inverse pattern of expression is observed in cells displaying lower aggression [492]. Following the production of PtdOH, Snail activity is negated and Snail expression reduced in the highly invasive setting, while expression of the Slug positive regulator is increased [492]. PLD in breast cancer is also regulated by MicroRNAs (miRs). Breast cancer cells displaying low aggression, express low levels of PLD and high levels of four miRs, which decrease PLD-translation. Three of these (miR 203, −887, and −3619) target PLD2, while one targets PLD1 (miR 182). Combined expression of miR 887, and miR 3619 abolishes over 90% of PLD2 activity. These tumor-suppressor like miRs are down-regulated in postEMT, highly-aggressive cells, which display high levels of both PLD1 and PLD2, and tumor aggressiveness could be reversed by miR transfection. The junction proteins vimentin and E-cadherin play key roles in miR regulation. E-cadherin triggers miR expression in pre- EMT cells, down-regulating PLD activity, while vimentin reduces expression in invasive cells post-EMT transition, leading to higher PLD activation [493].
PLD2 is linked to vascular endothelial growth factor (VEGF) signaling [494], a key mediator of angiogenesis, embryonic development and pathophysiological repair [495–498]. VEGF is dramatically upregulated during pathogenic angiogenic events including diabetic retinopathy and cancer [499,500], PLD2 deficiency in immortalized human umbilical vein endothelial (iHUVEC) cells decreased VEGF mediated cell-survival, proliferation, migration, and tubulation [494]. Furthermore, ex vivo aortic sprouting assays using PLD2−/− KO mouse tissue revealed PLD2 absence inhibits ex vivo angiogenesis [494]. Retinal angiogenesis was also decreased in the absence of PLD2, as was both tumor growth and tumor angiogenesis in Lewis lung carcinoma cells subcutaneously implanted in PLD2 knockout mice versus wild type [501]. PLD2 is also linked to angiogenesis in clear cell renal carcinoma (ccRCC). PLD1 and PLD2 are both upregulated in ccRCC, with increased PLD levels corresponding with higher tumor stage and grade. Knockdown of PLD2 suppressed proliferation and invasion of ccRCC in vitro and growth, and invasion of tumors in a nude mouse xenograft model. Importantly elevated PLD2 expression was also found to be significantly associated with poor prognosis in 67 ccRCC patients. Expression of angiogenin (ANG), a protein involved in angiogenesis, invasion and metastasis is elevated in a number of human cancers, and inhibition of ANG significantly suppressed tumor invasion. PtdOH produced by PLD2 increases ANG expression, and treatment of renal cell carcinoma cells with PLD2 shRNA downregulated ANG mRNA, indicating a PLD2 role in regulating angiogenesis through regulation of ANG levels [502]. In addition to a role in angiogenesis, PLD2 is is implicated in cancer cell migration and invasion [433,503], a role which is perhaps unsurprising since the isoform is linked to motility in numerous cell types including phagocytes [386,387,504,505], epithelial cells [506,507], and fibroblasts [508,509]. PLD2’s role in increased proliferation of cancer cells appears tightly linked to the products of the oncogenes JAK (Janus kinase) and Fes (the human counterpart of feline sarcoma retrovirus protein associated with leukemia and sarcoma). These tyrosine kinases display elevated expression associated with tumor-growth, angiogenesis and metastasis. JAK and Fes physically interact, but binding appears to be reduced in transformed cells. PLD2 is phosphorylated on Y415, Y169 and Y179 following cell-stimulation, with JAK3 phosphorylating PLD2 at Y415 in vitro [270,503]. Fes interacts with these phosphorylated residues differentially binding to PLD2 on Y415 in non-transformed cells, and to Y415, and Y169 in transformed cells, at least in the later-case through the Fes SH2-domain [503]. JAK3 and PLD2 also interact although the binding remains to be fully characterized [503]. In transformed cells PLD2, JAK and Fes are over-expressed and the activity of all three proteins is elevated. The kinases activate PLD2 leading to increased PtdOH which further activates Fes (but not JAK3) forming a positive feedback loop [503]. In non-transformed cells, Fes negatively feeds back to JAK3 but this appears to be diminished in transformed cells, in which binding of the two proteins is reduced [503]. Silencing of any of the three proteins with siRNA reduced proliferation of highly proliferative breast cancer cells, and silencing of all three had a synergistic silencing effect [503], Another aspect of cancer biology in which PLD2 plays an important role, is in anti-tumor immunity. Growth of tumors formed by subcutaneously transplanted cancer cells is enhanced in PLD2 knockout mice and is at least partly ascribable to the absence of PLD2 in bone marrow. Cytotoxic CD8+T cells were significantly reduced in tumors within the PLD2 knockout animal and division of cultured CD8+T cells was significantly suppressed indicating an important PLD2 role in T-lymphocyte proliferation [510].
8.4. PLD in neuronal physiology and pathology
PLD is expressed in brain tissue [511,512] and linked to both normal, and pathological brain function [39]. PLD isoforms mediate signaling, and trafficking of brain receptors including those of mito-genic growth factors [187,513], and opiod-, muscarinic-, and metabotropic glutamate receptors [341,354,355,514]. They play key roles in cell proliferation during brain development [187,513,515,516]; and regulate neurite outgrowth, particularly axonal sprouting [165,343,517,518]. For example, stimulation of astrocytes with the muscarinic-receptor agonist carbachol results in axonal elongation and hippocampal neurite outgrowth, through PLD-dependent release of fibronectin, plasminogen, and laminin [519].
PLD1 and PLD2 are highly expressed in brain, particularly within the white matter. PLD1 expression changes only slightly during rat development (E15-P49), while PLD2 is upreguiated in white and gray matter during the early postnatal stage [520]. Both isoforms are implicated in hippocampal mossy fiber sprouting, and neuronal plasticity [165]. PLD2 is also implicated in signaling by the neuronal L1 cell adhesion molecule (L1 CAM), and acts downstream of the L1-MAP Kinase pathway that regulates neunte outgrowth [343,518], Highly sensitive analysis of PLD2 brain expression, using Locked Nucleotide Amplification (LNA) based in situ hybridization indicated PLD2 is expressed in hippocampus, cerebellum and olfactory bulb [164]. Loss of PLD2 from these regions could therefore impact function reflected in mouse behavior. Consistent with this, PLD2−/− mice display learning and memory deficiencies, and reduced hippocampal acetylcholine release following behavioral stimulation [187]. PLD2−/− mice also display behavioral abnormalities demonstrating decreased olfaction and potential anosmia, the inability to smell [164]. PLD2−/− mice display abnormal cerebellar architecture, with ectopic Purkinje cells visible in the arbor vitae (cerebellar white matter) or clustered on the molecular layer surface, instead of being sandwiched between the molecular- and granular layers. Lipidomic analysis of the PLD2−/− brain indicates that the quantitatively normal PtdOH-pool is qualitatively and regionally abnormal, with a general shift from shorter 32: and 34: PtdOH species to longer 36: and 38: species [39,164].
PLD2 [521,522] and the PLD family member PLD5 have both been linked to autism suggesting a potential common role for PLD family proteins in autism risk [522]. PLD2 was linked to autism in an Australian study which discovered an autism-linked SNP (rs4141463) within an intron of the mono-ADP ribosylhydrolase 2 (MACROD2) gene, a region linked to regulation of PLD2 expression [521,522]. MACROD2 copy number variants are associated with schizophrenia [523], brain infarct [524], brain volume in multiple sclerosis [525] and deletion is reported in attention deficit hyperactivity disorder (ADHD) [526]. The in vivo relevance of this finding however, is currently unclear and another study looking at a European cohort failed to find an association between MACROD2 and autism, suggesting the finding may be population dependent [527].
PLD is implicated in Alzheimer’s disease (AD) [37–41], PLD activity being increased in AD brain homogenates [36,528]. Many facets of AD pathogenesis are mediated by cerebral Amyloid β (Aβ) accumulation, resulting from sequential cleavage of amyloid precursor protein (APP) by β-, and γ-secretases [39,529–532]. Aβ-stimulated PLD activity correlated with release of lactate dehydrogenase, a cell death indicator, suggesting amyloid neurotoxic actions are PLD mediated [47,533]. PLD activity was increased by APP in P19 mouse embryonic carcinoma cells [534] and oligomeric Aβ in cultured neurons [39]. PLD is also implicated in regulation of APP trafficking, as well as trafficking of pre-senilin-1 (PS1), a component of the γ-secretase which mediates APP cleavage to generate Aβ [36–41,188,528,535–539]. Both the PLD1 and PLD2 isoforms are linked to AD [37,39,188]. Studies using cell culture models suggest PLD1 is AD protective, and negatively regulates Aβ formation [37]. PLD1 (but not PLD2) is also implicated in APP- and PS1 trafficking, with important implications for APP metabolism and Aβ secretion [37–39]. PLD2 is activated by Aβ in cultured neurons, and reduction of PLD2 levels blocked Aβ-induced PLD activation [188]. PLD2 is predominantly localized to the cell-surface: a major site of Aβ action [168]. Oligomeric Aβ causes a partial internalization of PLD2 from the plasma membrane [188], in an extracellular Ca2+ dependent manner, re-localization being blocked by cPLA2 inhibition. This is consistent with studies indicating PLD2 activation occurs downstream of Ca2+ entry and cPLA2 stimulation [188]. PLD activity is elevated in a transgenic mouse AD model (SwAPP) and PLD2 deletion in this background blocked Aβ42 oligomer synaptotoxicity. This resulted in synaptic protection, memory deficit rescue, and neuronal protection, despite significant levels of Aβ [188]. Liquid chromatography mass-spectrometry (LC-MS) analysis of ethanol-injected SwAPP mice versus control animals indicated increased PLD activity in the SwAPP AD background, with increases in phosphatidylethanol (PEtOH) (32:1, 34:2, and 34:1) and PtdOH 34:2 within the brain. PtdOH 34:2 accumulation was PLD-dependent and was not observed in SwAPP PLD2 knockout mice [188], Profiling of PLD2−/− knockout mouse brain lipids in an independent study revealed a similar decrease in shorter 32: and 34: PtdOH species and an increase in larger 36: and 38: species, suggesting compensation by other PtdOH producing enzymes [164], The PtdOH 34:2 species was previously observed accumulating in D. mela-nogaster photoreceptors following PLD over-expression, which resulted in receptor degeneration, consistent with this lipid species playing a pathogenic role [540]. Taken together these data potentially suggest PLD2: 1) mediates signaling downstream of Aβ; (2) regulates availability of putative Aβ receptors at synapses, or 3) alters Aβ to binding to putative receptors [188].
PLDs are implicated in the biology of synucleins a group of poorly characterized proteins that are enriched in nerve endings, and implicated in both AD and Parkinson’s disease (PD). They are present in AD-associated amyloid plaques, and accumulate in PD Lewy bodies, with mutations in synuclein genes predisposing the PD condition [541–545]. The neurotoxic α-synuclein peptide co-immunoprecipitates with PLD1 and PLD2 [546], and α, β and γ synuclein inhibit PLD2 in vitro [545,547]. Activation of PLD2 by the muscarinic receptor in human dopaminergic cells is proposed to involve loss of PLD2-inhibition by α-synuclein [548]. Two α-synuclein point mutations associated with early-onset PD have been assayed for the ability to inhibit PLD2 activity in vitro [543–545]. While α-synuclein A30P displayed equal inhibition to wild-type α-synuclein, — A53T’s ability to inhibit PLD2 was significantly enhanced [545]. Inhibition required residues in exon 4 and 6 of α-synuclein, a region required for the proteins a helical structure. Deletion or mutation of these regions rendered the protein unable to inhibit PLD2. This ability could also be blocked by phosphorylation of serine 129, or tyrosine 125, or 136 within α-synuclein exons 5 and 6 [545]. However, translation of in vitro findings to our understanding of the in vivo PLD-synuclein relationship is complicated, by contrasting reports that α-synuclein does not inhibit PLD activity [549], and that PLD does not contribute to α-synuclein mediated brain lesions [546].
PLD is also linked to fetal alcohol spectrum disorder (FASD) resulting from alcohol consumption during pregnancy [550]. A range of effects are observed within the spectrum depending on timing and extent of consumption during the developmental process [551]. PLD has long been known to produce phoshatidyl-alcohols in the presence of primary alcohols but the in vivo relevance of this has remained unclear. The consequence of transphosphatidylation are two-fold: 1) cellular phosphatidyl-alcohol accumulation; 2) inhibition of downstream PtdOH dependent pathways. There is a sparsity of evidence linking phosphatidyl-alcohol accumulation to in vivo toxicity. However, accumulation was associated with disruption of membrane function, including a reduction in Ins(1,4,5)P3 levels [552] and reduced binding of 3H Ins(1,4,5)P3 to cerebellar membranes [553]. Phosphatidyl-alcohol persisted in the brains of ethanol fed rats with a half-life of 8–10 h [553].
PLD activity is also linked to multiple sclerosis (MS) a disabling neurological autoimmune disease in which the immune system attacks the myelin nerve sheath. PLD1 reduces immune responses [206,445] and PLD1 ablation reduced symptoms in allergic encephalomyelitis (EAE) the murine MS model [45]. The CNS is ordinarily protected from lymphocyte incursions from the periphery preventing uncontrolled inflammation and tissue damage. However, during diseases of the CNS such as MS, lymphocytes readily cross the blood-brain barrier and induce demyelination and axonal damage. Inhibition of lymphocyte trafficking is therefore a major focus of research into treatment of CNS disorders. PLD1 is expressed in the lymphocytes of MS patients, during autoimmune CNS inflammation, but not ini the lymphocytes of healthy individuals. Absence of PLD1 reduced chemokine static adhesion of lymphocytes to the endothelial adhesion molecules vascular cell adhesion molecule 1 (VCAM1) and Intercellular adhesion molecule 1 (ICAM1) in vitro with concomitant decreased motility in cell migration, and blood brain barrier models [45]. Absence of PLD1 resulted in attenuated disease severity in EAE mice indicating the relevancy of these findings to recruitment of the CNS in vivo.
8.5. PLD in insulin signaling and diabetes
Insulin stimulates mammalian PLD activity and numerous studies link PLD-derived PtdOH with the glucose uptake mechanism [213,291,554–556], In-tum, mammalian PLD also regulates insulin signaling by regulating multiple aspects of vesicular-trafficking, [555], and potentially through regulation of mTOR (see previous section), a critical component of insulin signaling. Although the precise role of PLD in insulin signaling through mTOR and associated pathways is still being elucidated. Mammalian mTOR exists in two complexes defined by the nature of the their primary accessory proteins, namely mTORC1, containing raptor (regulatory associated protein of Tor) and mTORC2, containing rictor (rapamycin insensitive component of TOR). The mechanisms by which these proteins regulate insulin signaling are still being fully resolved, but mTOR is known to activate AKT by phosphorylating it at ser473 enhancing insulin signaling [419,557]. Serum activation of PLD results in phosphorylation of AKT ser473, however this appears to be independent of mTORC2 [557].
PLD1 and PLD2 are both implicated in insulin signaling and PLD1 and PLD2 deficient mice exhibit insulin intolerance, characteristic of a pre-diabetic state [189]. PLD1, and PLD2 interact with PEA15 (phos-phoprotein enriched in diabetes/ astrocytes [also called PED]) [558], and alterations in the expression level are linked to diabetes and other disease states including Alzheimer’s disease, polycystic ovary syndrome, cardiovascular disease, and cancer (PEA15 is a tumor suppressor) [559]. PLD1 interaction is dependent on a D- (also called DED [death effector domain])-peptide binding motif in the PEA15 N-terminus, and the D4-motif of PLD1 (which overlaps with its Rho activation site) [560,561]. The interaction is independent of PEA15 phosphorylation [562], and increases PLD1 activity [561]. This is potentially a result of increased PLD stability, since both PLD1- and PLD2 protein levels increase following PEA15 co-expression, suggesting it may act as a PLD chaperone [561]. PEA15 is upregulated in type 2 diabetics and may contribute to insulin resistance in glucose uptake. Cellular glucose transport at the plasma membrane occurs through the facilitative glucose transporter (Glut) family [563]. Glut1 resides at the plasma membrane and is thought to perform basal transport, while Glut4 is responsible for insulin-stimulated transport [564]. PEA15 overexpression in skeletal muscle, a major site of diabetic insulin resistance, increases cell-membrane Glut1 expression, and inhibits insulin-stimulated glucose transport, and Glut4 cell-surface recruitment [565]. Expansion of the Glut1 cell surface compliment, prevents further insulin-stimulated Glut1 translocation, and in cells uniquely expressing this transporter, may account for the lack of insulin-stimulated glucose transport, and increased basal glucose transport, which decreases insulin effectiveness, increasing insulin-resistance. In cells expressing both receptors a significant decrease in insulin responsiveness is additionally observed as Glut4 translocation to the plasma membrane is blocked [565]. Primary alcohols inhibit Glut-4 translocation and glucose uptake [566,567], and PLD1 RNAi, or catalytically inactive PLD1 mutant expression reduced plasma membrane Glut4 through delayed docking of Glut4 containing vesicles at the exocytic site [213]. Conversely, increased PLD-expression, increased insulin-stimulated Glut4 translocation [213,568]. In cell culture, or transgenic animal systems PEA15 over-expression impairs insulin-mediated glucose regulation in a PEA15-PLD1 interaction-dependent manner [560]. PEA15 over-expression also causes PLD-dependent PKCα-activation, preventing subsequent activation of ΡKξ [569,570], which is a major Glut4 membrane translocation regulator [571]. Disruption of the PEA15-PLD1 interaction in vitro results in reduced PKCα activity and restoration of insulin sensitivity [560]. Furthermore, insulin sensitivity was restored in PEA15 over-expressing mice, following adenoviral expression of the PLD1 D4 PEA15-binding-domain. This disrupted the PEA15-PLD1 interaction, reduced PKCα activity and increased ΡΚCξ activation thereby enhancing insulin sensitivity [572]. PEA15 is also implicated in other aspects of the diabetic phenotype, PEA15 over-expression increasing mouse creatine levels and urine volume, and elevating transforming growth factor-α1 (TGFβ1) in kidney, and serum [573]. Pharmacological PLD- or PKCβ inhibition resulted in reduced TGFβ1 and the extra-cellular matrix component fibronectin, which could potentially result in diabetes-associated renal dysfunction and kidney damage [573]. TGFβ1 induces PEA15-mediated autophagy, and induces PEA15 transcription [574], Defects in autophagy are linked to the etiology of many diseases including type two diabetes (T2D), as well as cancer and neurodegenerative disease [579]. In T2D, impaired pancreatic beta cell function and development of insulin resistance has been linked to autophagy. PLD1 is linked to autophagy modulation, and butan-1-ol, RNAi, PLD inhibitors and knockout of PLD1 in a mouse model all decreased macroautophagy [185]. Autophagy and the endocytic process are tightly linked [575] and it is perhaps unsurprising that PLD is implicated in regulation of autophagy given its links to endocytic trafficking [575–577]. PLD1’s role in autophagy is required for cancer cell survival under conditions of prolonged glucose insufficiency [578].
9. Discussion
Since the initial discovery of PLD activity in plant tissue, activities have been detected across the genera. Improvements in scientific methodology have since resulted in cloning of PLD genes, identification of signature motifs, and an improved understanding of PLD structure and the catalytic mechanism. Improved detection of endogenous PLDs, has reduced dependency on over-expression constructs, allowing, with ever increasing accuracy, the precise localization of PLDs in vivo. The advent of RNA interference and in particular the evolution of isoform-specific PLD inhibitors to inhibit PLD expression or activities has facilitated a departure from our dependence on over-expression of mutant PLDs, and perhaps most importantly from use of primary alcohols to inhibit PtdOH production. These advances have greatly improved our understanding of the cellular functions of PLD enzymes. More recently technological advances including TALEN and CRISPR have enabled manipulation of DNA with increased ease, and fidelity, reducing off target effects, and promise to clarify the situation even further. The development and careful analysis of PLD knockout animal models, has vastly improved our understanding of the role of PLDs in vivo and are allowing for the first-time a thorough analysis of PLDs role in the context of whole organisms. As a result of these technological advances, PLD activities are now convincingly being linked to numerous diseases including Alzheimer’s disease, thrombosis and numerous cancers. As our comprehension of PLDs role in the pathology of these diseases increases, and our understanding of PLDs potential for drugability grows, the hope that PLD proteins may provide drugable targets is becoming closer to reality.
Acknowledgments
This work was supported by funding from the Robert A. Welch Foundation (BE0017), and a grant from the National Institutes of Health (GM131804, both awarded to Vytas A. Bankaitis, and strategic funding from the BBSRC (BB/P013384/1) to Michael J. O. Wakelam. The authors would also like to acknowledge Max Lönnfors for editing assistance and discussion.
References
- [1].Hanahan DJ, Chaikoff IL, A new phospholipide-splitting enzyme specific for the ester linkage between the nitrogenous base and the phosphoric acid grouping, J. Biol. Chem 169 (3) (1947) 699–705. [PubMed] [Google Scholar]
- [2].Hanahan DJ, Chaikoff IL, The phosphorus-containing lipides of the carrot, J. Biol. Chem 168 (1) (1947) 233–240. [PubMed] [Google Scholar]
- [3].Hanahan DJ, Chaikoff IL, On the nature of the phosphorus-containing lipides of cabbage leaves and their relation to a phospholipide-splitting enzyme contained in these leaves, J. Biol. Chem 172 (1) (1948) 191–198. [PubMed] [Google Scholar]
- [4].Dohke Y, Fujita-Yoshigaki J, Sugiya H, Furuyama S, Hara-Yokoyama M, Involvement of phospholipase D in the cAMP-regulated exocytosis of rat parotid acinar cells, Biochem. Biophys. Res. Commun 299 (4) (2002) 663–668. [DOI] [PubMed] [Google Scholar]
- [5].Roth MG, Molecular mechanisms of PLD function in membrane traffic, Traffic 9 (2008) 1233–1239. [DOI] [PubMed] [Google Scholar]
- [6].Morris AJ, Regulation of phospholipase D activity, membrane targeting and intracellular trafficking by phosphoinositides, Biochem. Soc. Symp 74 (2007) 247–257. [DOI] [PubMed] [Google Scholar]
- [7].Brown FD, Thompson N, Saqib KM, Clark JM, Powner D, Thompson NT, et al. , Phospholipase D1 localises to secretory granules and lysosomes and is plasma- membrane translocated on cellular stimulation, Curr. Biol 8 (14) (1998) 835–838. [DOI] [PubMed] [Google Scholar]
- [8].Siddhanta A, Backer JM, Shields D, Inhibition of phosphatidic acid synthesis alters the structure of the Golgi apparatus and inhibits secretion in endocrine cells, J. Biol. Chem 275 (16) (2000) 12023–12031. [DOI] [PubMed] [Google Scholar]
- [9].Sweeney DA, Siddhanta A, Shields D, Fragmentation and re-assembly of the Golgi apparatus in vitro. A requirement for phosphatidic acid and phosphatidylinositol 4,5-bisphosphate synthesis, J. Biol. Chem 277 (4) (2002) 3030–3039. [DOI] [PubMed] [Google Scholar]
- [10].Yang JS, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, et al. , A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance, Nat. Cell Biol 10 (10) (2008) 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Freyberg Z, Bourgoin S, Shields D, Phospholipase D2 is localized to the rims of the Golgi apparatus in mammalian cells, Mol. Biol. Cell 13 (11) (2002) 3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Freyberg Z, Sweeney D, Siddhanta A, Bourgoin S, Frohman M, Shields D, Intracellular localization of phospholipase D1 in mammalian cells, Mol. Biol. Cell 12 (4) (2001) 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gomez-Cambronero J, Phosphatidic acid, phospholipase D and tumorigenesis, Adv. Biol. Regul 54 (2014) 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cross MJ, Roberts S, Ridley AJ, Hodgkin MN, Stewart A, Claesson-Welsh L, et al. , Stimulation of actin stress fibre formation mediated by activation of phospholipase D, Curr. Biol 6 (5) (1996) 588–597. [DOI] [PubMed] [Google Scholar]
- [15].Kusner DJ, Hall CF, Jackson S, Fc gamma receptor-mediated activation of phospholipase D regulates macrophage phagocytosis of IgG-opsonized particles, J. Immunol 162 (4) (1999) 2266–2274. [PubMed] [Google Scholar]
- [16].Suchard SJ, Hinkovska-Galcheva V, Mansfield PJ, Boxer LA, Shayman JA, Ceramide inhibits IgG-dependent phagocytosis in human polymorphonuclear leukocytes, Blood 89 (6) (1997) 2139–2147. [PubMed] [Google Scholar]
- [17].Frohman MA, Morris AJ, Phospholipase D structure and regulation, Chem. Phys. Lipids 98 (1–2) (1999) 127–140. [DOI] [PubMed] [Google Scholar]
- [18].Dhalla NS, Xu YJ, Sheu SS, Tappia PS, Panagia V, Phosphatidic acid: a potential signal transducer for cardiac hypertrophy, J. Mol. Cell. Cardiol 29 (11) (1997) 2865–2871. [DOI] [PubMed] [Google Scholar]
- [19].Bocckino SB, Wilson PB, Exton JH, Phosphatidate-dependent protein phosphorylation, Proc. Natl. Acad. Sci. U. S. A 88 (14) (1991) 6210–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park JB, Kim JH, Kim Y, Ha SH, Yoo JS, Du G, et al. , Cardiac phospholipase D2 localizes to sarcolemmal membranes and is inhibited by alpha-actinin in an ADP- ribosylation factor-reversible manner, J. Biol. Chem 275 (28) (2000) 2129S–21301. [DOI] [PubMed] [Google Scholar]
- [21].Henry RA, Boyce SY, Kurz T, Wolf RA, Stimulation and binding of myocardial phospholipase C by phosphatidic acid, Am. J. Phys 269 (2 Pt 1) (1995) C349–C358. [DOI] [PubMed] [Google Scholar]
- [22].Xu YJ, Panagia V, Shao Q, Wang X, Dhalla NS, Phosphatidic acid increases intracellular free Ca2+ and cardiac contractile force, Am. J. Phys 271 (2 Pt 2) (1996) H651–H659. [DOI] [PubMed] [Google Scholar]
- [23].Xu YJ, Yau L, Yu LP, Elimban V, Zahradka P, Dhalla NS, Stimulation of protein synthesis by phosphatidic acid in rat cardiomyocytes, Biochem. Pharmacol 52 (1996) 1735–1740. [DOI] [PubMed] [Google Scholar]
- [24].Reich R, Blumenthal M, Liscovitch M, Role of phospholipase D in laminin-induced production of gelatinase A (MMP-2) in metastatic cells, Clin. Exp. Metastasis 13 (1995) 134–140. [DOI] [PubMed] [Google Scholar]
- [25].Waite KA, Wallin R, Qualliotine-Mann D, McPhail LC, Phosphatidic acid-mediated phosphorylation of the NADPH oxidase component p47-phox. Evidence that phosphatidic acid may activate a novel protein kinase, J. Biol. Chem 272 (24) (1996) 15569–15578. [DOI] [PubMed] [Google Scholar]
- [26].Bargmann BO, Munnik T, The role of phospholipase D in plant stress responses, Curr. Opin. Plant Biol 9 (5) (2006) 515–522. [DOI] [PubMed] [Google Scholar]
- [27].Uraji M, Katagiri T, Okuma E, Ye W, Hossain MA, Masuda C, et al. , Cooperative function of PLDdelta and PLDalphal in abscisic acid-induced stomatal closure in Arabidopsis, Plant Physiol. 159 (1) (2012) 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Johansson ON, Fahlberg P, Karimi E, Nilsson AK, Ellerstrom M, Andersson MX, Redundancy among phospholipase D isoforms in resistance triggered by recognition of the Pseudomonas syringae effector AvrRpml in Arabidopsis thaliana, Front. Plant Sci 5 (2014) 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee S, Lynch KR, Brown recluse spider (Loxosceles reclusa) venom phospholipase D (PLD) generates lysophosphatidic acid (LPA), Biochem. J 391 (Pt 2) (2005) 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lajoie DM, Cordes MH, Spider, bacterial and fungal phospholipase D toxins make cyclic phosphate products, Toxicon 108 (2015) 176–180. [DOI] [PubMed] [Google Scholar]
- [31].Lajoie DM, Zobel-Thropp PA, Kumirov VK, Bandarian V, Binford GJ, Cordes MH, Phospholipase D toxins of brown spider venom convert lysopho- sphatidylcholine and sphingomyelin to cyclic phosphates, PLoS One 8 (8) (2013) e72372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].van Dijk MC, Postma F, Hilkmann H, Jalink K, van Blitterswijk WJ, Moolenaar WH, Exogenous phospholipase D generates lysophosphatidic acid and activates Ras, Rho and Ca2+ signaling pathways, Curr. Biol 8 (7) (1998) 386–392. [DOI] [PubMed] [Google Scholar]
- [33].Jiang Y, Du M, Wu M, Zhu Y, Zhao X, Cao X, et al. , Phosphatidic acid improves reprogramming to pluripotency by reducing apoptosis, Stem Cells Dev. 25 (1) (2016) 43–54. [DOI] [PubMed] [Google Scholar]
- [34].Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, et al. , Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family, J. Biol. Chem 270 (50) (1996) 29640–29643. [DOI] [PubMed] [Google Scholar]
- [35].Foster DA, Xu L, Phospholipase D in cell proliferation and cancer, Mol. Cancer Res 1 (11) (2003) 789–800. [PubMed] [Google Scholar]
- [36].Kanfer JN, Singh IN, Pettegrew JW, McCartney DG, Sorrentino G, Phospholipid metabolism in Alzheimer’s disease and in a human cholinergic cell, J. Lipid Mediat. Cell Signal 14 (1–3) (1996) 361–363. [DOI] [PubMed] [Google Scholar]
- [37].Cai D, Zhong M, Wang R, Netzer WJ, Shields D, Zheng H, et al. , Phospholipase D1 corrects impaired betaAPP trafficking and neurite outgrowth in familial Alzheimer’s disease-linked presenilin-1 mutant neurons, Proc. Natl. Acad. Sci. U. S. A 103 (6) (2006) 1936–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu Y, Zhang YW, Wang X, Zhang H, You X, Liao FF, et al. , Intracellular trafficking of presenilin 1 is regulated by beta-amyloid precursor protein and phospholipase D1, J. Biol. Chem 284 (18) (2009) 12145–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oliveira TG, Di Paolo G, Phospholipase D in brain function and Alzheimer’s disease, Biochim. Biophys. Acta 1801 (8) (2010) 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Singh IN, McCartney DG, Kanfer JN, Amyloid beta protein (25–35) stimulation of phospholipases A, C and D activities of LA-N-2 cells, FEBS Lett. 365 (2–3) (1995) 125–128. [DOI] [PubMed] [Google Scholar]
- [41].Brandenburg LO, Konrad M, Wruck C, Koch T, Pufe T, Lucius R, Involvement of formyl-peptide-receptor-like-1 and phospholipase D in the internalization and signal transduction of amyloid beta 1–42 in glial cells, Neuroscience 156 (2) (2008) 266–276. [DOI] [PubMed] [Google Scholar]
- [42].Elvers M, Stegner D, Hagedorn I, Kleinschnitz C, Braun A, Kuijpers ME, et al. , Impaired alpha(IIb)beta(3) integrin activation and shear-dependent thrombus formation in mice lacking phospholipase D1, Sci. Signal 3 (103) (2010) ral. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo JA, et al. , Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis, Sci Signal 5 (249) (2012) ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hong KW, Jin HS, Lim JE, Cho YS, Go MJ, Jung J, et al. , Non-synonymous single-nucleotide polymorphisms associated with blood pressure and hypertension, J. Hum. Hypertens 24 (11) (2010) 763–774. [DOI] [PubMed] [Google Scholar]
- [45].Gobel K, Schuhmann MK, Pankratz S, Stegner D, Herrmann AM, Braun A, et al. , Phospholipase D1 mediates lymphocyte adhesion and migration in experimental autoimmune encephalomyelitis, Eur. J. Immunol 44 (8) (2014) 2295–2305. [DOI] [PubMed] [Google Scholar]
- [46].Oguin TH 3rd, Sharma S, Stuart AD, Duan S, Scott SA, Jones CK, et al. , Phospholipase D facilitates efficient entry of influenza virus, allowing escape from innate immune inhibition, J. Biol. Chem 289 (37) (2014) 25405–25417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Selvy PE, Lavieri RR, Lindsley CW, Brown HA, Phospholipase D: enzymology, functionality, and chemical modulation, Chem. Rev 111 (10) (2011) 6064–6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McDermott M, Wakelam MJ, Morris AJ, Phospholipase D, Biochem. Cell Biol 82 (1) (2004) 225–253. [DOI] [PubMed] [Google Scholar]
- [49].Waite M, The Phospholipases, 1 ed, Springer, USA, 1987. [Google Scholar]
- [50].Wang X, Xu L, Zheng L, Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L, J. Biol. Chem 269 (32) (1994) 20312–20317. [PubMed] [Google Scholar]
- [51].Yang SF, Freer S, Benson AA, Transphosphatidylation by phospholipase D, J. Biol. Chem 242 (3) (1967) 477–484. [PubMed] [Google Scholar]
- [52].Wakelam MJ, Hodgkin M, Martin A, The measurement of phospholipase D-linked signaling in cells, Methods Mol. Biol 41 (1995) 271–278. [DOI] [PubMed] [Google Scholar]
- [53].Tang X, Benesch MG, Brindley DN, Lipid phosphate phosphatases and their roles in mammalian physiology and pathology, J. Lipid Res 56 (11) (2015) 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Saito M, Kanfer J, Phosphatidohydrolase activity in a solubilized preparation from rat brain particulate fraction, Arch. Biochem. Biophys 169 (1) (1975) 318–323. [DOI] [PubMed] [Google Scholar]
- [55].Kanfer JN, The base exchange enzymes and phospholipase D of mammalian tissue, Can. J. Biochem 58 (12) (1980) 1370–1380. [DOI] [PubMed] [Google Scholar]
- [56].Bocckino SB, Blackmore PF, Wilson PB, Exton JH, Phosphatidate accumulation in hormone-treated hepatocytes via a phospholipase D mechanism, J. Biol. Chem 262 (31) (1987) 15309–15315. [PubMed] [Google Scholar]
- [57].Pai JK, Siegel MI, Egan RW, Billah MM, Phospholipase D catalyzes phospholipid metabolism in chemotactic peptide-stimulated HL-60 granulocytes, J. Biol. Chem 262 (25) (1988) 12472–12477. [PubMed] [Google Scholar]
- [58].Tettenborn CS, Mueller GC, 12-O-tetradecanoylphorbol-13-acetate activates phosphatidylethanol and phosphatidylglycerol synthesis by phospholipase D in cell lysates, Biochem. Biophys. Res. Commun 155 (1) (1988) 249–255. [DOI] [PubMed] [Google Scholar]
- [59].Chalifa Y, Mohn H, Liscovitch M, A neutral phospholipase D activity from rat brain synaptic plasma membranes. Identification and partial characterization, J. Biol. Chem 265 (29) (1990) 17512–17519. [PubMed] [Google Scholar]
- [60].Hurst KM, Hughes BP, Barritt GJ, The roles of phospholipase D and a GTP-binding protein in guanosine 5’-[gamma-thio] triphosphate-stimulated hydrolysis of phosphatidylcholine in rat liver plasma membranes, Biochem. J 272 (3) (1990) 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kanaho Y, Kanoh H, Saitoh K, Nozawa Y, Phospholipase D activation by platelet- activating factor, leukotriene B4, and formyl-methionyl-leucyl-phenylalanine in rabbit neutrophils. Phospholipase D activation is involved in enzyme release, J. Immunol 146 (10) (1991) 3536–3541. [PubMed] [Google Scholar]
- [62].Nishida A, Shimizu M, Kanaho Y, Nozawa Y, Yamawaki S, Characterization of phospholipase D in a cell-free system of cultured cells derived from rat frontal cortex, Brain Res. 595 (1) (1992) 12–16. [DOI] [PubMed] [Google Scholar]
- [63].Okamura S, Yamashita S, Purification and characterization of phosphatidylcholine phospholipase D from pig lung, J. Biol. Chem 269 (49) (1994) 31207–31213. [PubMed] [Google Scholar]
- [64].Exton JH, Phospholipase D: enzymology, mechanisms of regulation, and function, Physiol. Rev 77 (2) (1997) 303–320. [DOI] [PubMed] [Google Scholar]
- [65].Exton JH, Phosphatidylcholine breakdown and signal transduction, Biochim. Biophys. Acta 1212 (1) (1994) 26–42. [DOI] [PubMed] [Google Scholar]
- [66].Brown HA, Gutowski S, Moomaw CR, Slaughter C, Stemweis PC, ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity, Cell 75 (6) (1993) 1137–1144. [DOI] [PubMed] [Google Scholar]
- [67].Cockcroft S, Thomas GM, Fensome A, Geny B, Cunningham E, Gout I, et al. , Phospholipase D: a downstream effector of ARF in granulocytes, Science 263 (5146) (1994) 523–526. [DOI] [PubMed] [Google Scholar]
- [68].Liscovitch M, Chalifa V, PertHe P, Chen CS, Cantley LC, Novel function of phosphatidylinositol 4,5-bisphosphate as a cofactor for brain membrane phospholipase D, J. Biol. Chem 269 (34) (1994) 21403–21406. [PubMed] [Google Scholar]
- [69].Massenburg D, Han JS, Liyanage M, Patton WA, Rhee SG, Moss J, et al. , Activation of rat brain phospholipase D by ADP-ribosylation factors 1,5, and 6: separation of ADP-ribosylation factor-dependent and oleate-dependent enzymes, Proc. Natl. Acad. Sci. U. S. A 91 (24) (1994) 11718–11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bowman EP, Uhlinger DJ, Lambeth JD, Neutrophil phospholipase D is activated by a membrane-associated Rho family small molecular weight GTP-binding protein, J. Biol. Chem 268 (29) (1993) 21509–21512. [PubMed] [Google Scholar]
- [71].Brown HA, Gutowski S, Kahn RA, Stemweis PC, Partial purification and characterization of Arf-sensitive phospholipase D from porcine brain, J. Biol. Chem 270 (25) (1995) 14935–14943. [DOI] [PubMed] [Google Scholar]
- [72].Meier KE, Gibbs TC, Knoepp SM, Ella KM, Expression of phospholipase D isoforms in mammalian cells, Biochim. Biophys. Acta 1439 (2) (1999) 199–213. [DOI] [PubMed] [Google Scholar]
- [73].Balboa MA, Insel PA, Nuclear phospholipase D in Madin-Darby canine kidney cells. Guanosine 5’−0-(thiotriphosphate)-stimulated activation is mediated by RhoA and is downstream of protein kinase C, J. Biol. Chem 270 (50) (1995) 29843–29847. [DOI] [PubMed] [Google Scholar]
- [74].Ktistakis NT, Brown HA, Stemweis PC, Roth MG, Phospholipase D is present on Golgi-enriched membranes and its activation by ADP ribosylation factor is sensitive to brefeldin A, Proc. Natl. Acad. Sci. U. S. A 92 (11) (1995) 4952–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ktistakis NT, Brown HA, Waters MG, Stemweis PC, Roth MG, Evidence that phospholipase D mediates ADP ribosylation factor-dependent formation of Golgi coated vesicles, J. Cell Biol 134 (2) (1996) 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Provost JJ, Fudge J, Israelit S, Siddiqi AR, Exton JH, Tissue-specific distribution and subcellular distribution of phospholipase D in rat: evidence for distinct RhoA- and ADP-ribosylation factor (ARF)-regulated isoenzymes, Biochem. J 319 (Pt 1) (1995) 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Whatmore J, Morgan CP, Cunningham E, Collison KS, Willison KR, Cockcroft S, ADP-ribosylation factor 1-regulated phospholipase D activity is localized at the plasma membrane and intracellular organelles in HL60 cells, Biochem. J 320 (Pt 3) (1996) 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bocckino SB, Wilson PB, Exton JH, Ca2+ — mobilizing hormones elicit phosphatidylethanol accumulation via phospholipase D activation, FEBS Lett. 225 (1–2) (1987) 201–204. [DOI] [PubMed] [Google Scholar]
- [79].Pai JK, Siegel MI, Egan RW, Billah MM, Activation of phospholipase D by chemotactic peptide in HL-60 granulocytes, Biochem. Biophys. Res. Commun 150 (1) (1987) 355–364. [DOI] [PubMed] [Google Scholar]
- [80].Besterman JM, Duronio V, Cuatrecasas P, Rapid formation of diacylglycerol from phosphatidylcholine: a pathway for generation of a second messenger, Proc. Natl. Acad. Sci. U. S. A 83 (18) (1986) 6785–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Daniel LW, Waite M, Wykle RL, A novel mechanism of diglyceride formation. 12-O-tetradecanoylphorbol-13-acetate stimulates the cyclic breakdown and resynthesis of phosphatidylcholine, J. Biol. Chem 261 (20) (1986) 9128–9132. [PubMed] [Google Scholar]
- [82].Abousalham A, Riviere M, Teissere M, Verger R, Improved purification and biochemical characterization of phospholipase D from cabbage, Biochim. Biophys. Acta 1158 (1) (1993) 1–7. [DOI] [PubMed] [Google Scholar]
- [83].Dyer JH, Ryu SB, Wang X, Multiple forms of phospholipase D following germination and during leaf development of Castor bean, Plant Physiol. 105 (2) (1994) 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J, Phospholipase D signaling is essential for meiosis, Proc. Natl. Acad. Sci. U. S. A 92 (26) (1995) 12151–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Rudge SA, Cavenagh MM, Kamath R, Sciorra VA, Morris AJ, Kalin RA, et al. , ADP- Ribosylation factors do not activate yeast phospholipase Ds but are required for sporulation, Mol. Biol. Cell 9 (8) (1998) 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Colley WC, Altshuller YM, Sue-Ling CK, Copeland NG, Gilbert DJ, Jenkins NA, et al. , Cloning and expression analysis of murine phospholipase D1, Biochem. J 326 (Pt 3) (1997) 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Park SK, Provost JJ, Bae CD, Ho WT, Exton JH, Cloning and characterization of phospholipase D from rat brain, J. Biol. Chem 272 (46) (1997) 29263–29271. [DOI] [PubMed] [Google Scholar]
- [88].Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, et al. , Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C- alpha, J. Biol. Chem 272 (6) (1997) 3860–3868. [DOI] [PubMed] [Google Scholar]
- [89].Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, et al. , Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization, Curr. Biol 7 (3) (1997) 191–201. [DOI] [PubMed] [Google Scholar]
- [90].Lopez I, Arnold RS, Lambeth JD, Cloning and initial characterization of a human phospholipase D2 (hPLD2). ADP-ribosylation factor regulates hPLD2, J. Biol. Chem 273 (21) (1998) 12846–12852. [DOI] [PubMed] [Google Scholar]
- [91].Steed PM, Clark KL, Boyar WC, Lasala DJ, Characterization of human PLD2 and the analysis of PLD isoform splice variants, FASEB J. 12 (13) (1998) 1309–1317. [DOI] [PubMed] [Google Scholar]
- [92].Pettitt TR, McDermott M, Saqib KM, Shimwell N, Wakelam MJ, Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells, Biochem. J 360 (Pt 3) (2001) 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sung TC, Zhang Y, Morris AJ, Frohman MA, Structural analysis of human phospholipase D1, J. Biol. Chem 274 (6) (1999) 3659–3666. [DOI] [PubMed] [Google Scholar]
- [94].Riebeling C, Bourgoin S, Shields D, Caspase cleavage of phospholipase D1 in vitro alters its regulation and reveals a novel property of the “loop” region, Biochim. Biophys. Acta 1781 (8) (2008) 376–382. [DOI] [PubMed] [Google Scholar]
- [95].Jang YH, Ahn BH, Namkoong S, Kim YM, Jin JK, Kim YS, et al. , Differential regulation of apoptosis by easpase-mediated cleavage of phospholipase D isozymes, Cell Signal. 20 (12) (2008) 2198–2207. [DOI] [PubMed] [Google Scholar]
- [96].Jang YH, Namkoong S, Kim YM, Lee SJ, Park BJ, Min DS, Cleavage of phospholipase D1 by caspase promotes apoptosis via modulation of the p53-dependent cell death pathway, Cell Death Differ. 15 (11) (2008) 1782–1793. [DOI] [PubMed] [Google Scholar]
- [97].Hughes WE, Parker PJ, Endosomal localization of phospholipase D1a and 1b is defined by the C-termini of the proteins, and is independent of activity, Biochem. J 356 (Pt 3) (2001) 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ponting CP, Kerr ID, A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues, Protein Sci. 5 (5) (1996) 914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Exton JH, Phospholipase D, Ann. N. Y. Acad. Sci 905 (2000) 61–68. [DOI] [PubMed] [Google Scholar]
- [100].Sung TC, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, et al. , Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity, EMBO J. 16 (15) (1997) 4519–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sung TC, Altshuller YM, Morris AJ, Frohman MA, Molecular analysis of mammalian phospholipase D2, J. Biol. Chem 274 (1) (1999) 494–502. [DOI] [PubMed] [Google Scholar]
- [102].Xie Z, Ho WT, Exton JH, Association of N- and C-terminal domains of phospholipase D is required for catalytic activity, J. Biol. Chem 273 (52) (1998) 34679–34682. [DOI] [PubMed] [Google Scholar]
- [103].Xie Z, Ho WT, Exton JH, Association of the N- and C-terminal domains of phospholipase D. Contribution of the conserved HKD motifs to the interaction and the requirement of the association for Ser/Thr phosphorylation of the enzyme, J. Biol. Chem 275 (32) (2000) 24962–24969. [DOI] [PubMed] [Google Scholar]
- [104].Stuckey JA, Dixon JE, Crystal structure of a phospholipase D family member, Nat. Struct. Biol 6 (3) (1999) 278–284. [DOI] [PubMed] [Google Scholar]
- [105].Leiros I, Hough E, D’Arrigo P, Carrea G, Pedrocchi-Fantoni G, Secundo F, et al. , Crystallization and preliminary X-ray diffraction studies of phospholipase D from Streptomyees sp, Acta Crystallogr. D Biol. Crystallogr 56 (Pt 4) (2000) 466–468. [DOI] [PubMed] [Google Scholar]
- [106].Leiros I, Secundo F, Zambonelli C, Servi S, Hough E, The first crystal structure of a phospholipase D, Structure 8 (6) (2000) 655–667. [DOI] [PubMed] [Google Scholar]
- [107].Mahankali M, Alter G, Gomez-Cambronero J, Mechanism of enzymatic reaction and protein-protein interactions of PLD from a 3D structural model, Cell. Signal 27 (1) (2015) 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Stanacev NZ, Stuhne-Sekalec L, On the mechanism of enzymatic phosphatidylation. Biosynthesis of cardiolipin catalyzed by phospholipase D, Biochim. Biophys. Acta 210 (2) (1970) 350–352. [DOI] [PubMed] [Google Scholar]
- [109].Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE, Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate, Proc. Natl. Acad. Sci. U. S. A 95 (16) (1998) 9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Rudolph AE, Stuckey JA, Zhao Y, Matthews HR, Patton WA, Moss J, et al. , Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member, J. Biol. Chem 274 (17) (1999) 11824–11831. [DOI] [PubMed] [Google Scholar]
- [111].DR Davies, Interthal H, Champoux JJ, Hoi WG, The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1, Structure 10 (2) (2002) 237–248. [DOI] [PubMed] [Google Scholar]
- [112].DR Davies, Interthal H, Champoux JJ, Hoi WG, Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures, J. Mol. Biol 324 (5) (2002) 917–932. [DOI] [PubMed] [Google Scholar]
- [113].Secundo F, Carrea G, D’Arrigo P, Servi S, Evidence for an essential lysyl residue in phospholipase D from Streptomyces sp. by modification with diethyl pyrocarbonate and pyridoxal 5-phosphate, Biochemistry 35 (30) (1996) 9631–9636. [DOI] [PubMed] [Google Scholar]
- [114].Iwasaki Y, Horiike S, Matsushima K, Yamane T, Location of the catalytic nucleophile of phospholipase D of Streptomyces antibioticus in the C-terminal half domain, Eur. J. Biochem 264 (2) (1999) 577–581. [DOI] [PubMed] [Google Scholar]
- [115].Manifava M, Sugars J, Ktistakis NT, Modification of catalytically active phospholipase D1 with fatty acid in vivo, J. Biol. Chem 274 (2) (1999) 1072–1077. [DOI] [PubMed] [Google Scholar]
- [116].Yoshikawa F, Banno Y, Otani Y, Yamaguchi Y, Nagakura-Takagi Y, Morita N, et al. , Phospholipase D family member 4, a transmembrane glycoprotein with no phospholipase D activity, expression in spleen and early postnatal microglia, PLoS One 5 (11) (2010) e13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Frohman MA, Sung TC, Morris AJ, Mammalian phospholipase D structure and regulation, Biochim. Biophys. Acta 1439 (2) (1999) 175–186. [DOI] [PubMed] [Google Scholar]
- [118].Du G, Altshuller YM, Vitale N, Huang P, Chasserot-Golaz S, Morris AJ, et al. , Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs, J. Cell Biol 162 (2) (2003) 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, et al. , Genome- wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains, Mol. Cell 13 (5) (2004) 677–688. [DOI] [PubMed] [Google Scholar]
- [120].Lemmon MA, Pleckstrin homology (PH) domains and phosphoinositides, Biochem. Soc. Symp 74 (2007) 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, et al. , Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast, EMBO J. 17 (18) (1998) 5374–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].H Takeuchi, Kanematsu T, Misumi Y, Sakane F, Konishi H, Kikkawa U, et al. , Distinct specificity in the binding of inositol phosphates by pleckstrin homology domains of pleckstrin, RAC-protein kinase, diacylglycerol kinase and a new 130 kDa protein, Biochim. Biophys. Acta 1359 (3) (1997) 275–285. [DOI] [PubMed] [Google Scholar]
- [123].Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, et al. , Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains, J. Biol. Chem 273 (46) (1998) 30497–30508. [DOI] [PubMed] [Google Scholar]
- [124].Levine TP, Munro S, Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components, Curr. Biol 12 (9) (2002) 695–704. [DOI] [PubMed] [Google Scholar]
- [125].Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, et al. , FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns (4)P, Nat. Cell Biol 6 (5) (2004) 393–404. [DOI] [PubMed] [Google Scholar]
- [126].Hodgkin MN, Masson MR, Powner D, Saqib KM, Ponting CP, Wakelam MJ, Phospholipase D regulation and localisation is dependent upon a phosphatidylinositol 4,5-biphosphate-specific PH domain, Curr. Biol 10 (1) (2000) 43–46. [DOI] [PubMed] [Google Scholar]
- [127].Cross MJ, Hodgkin MN, Plumb JA, Brunton VG, Stewart A, MacAully G, et al. , Inhibition of phospholipid signalling and proliferation of Swiss 3T3 cells by the wortmannin analogue demethoxyviridin, Biochim. Biophys. Acta 1362 (1) (1997) 29–38. [DOI] [PubMed] [Google Scholar]
- [128].Sciorra VA, Rudge SA, Wang J, McLaughlin S, Engebrecht J, Morris AJ, Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes, J. Cell Biol 159 (6) (2002) 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Sciorra VA, Rudge SA, Prestwich GD, Frohman MA, Engebrecht J, Morris AJ, Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes, EMBO J. 18 (21) (1999) 5911–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Sugars JM, Cellek S, Manifava M, Coadwell J, Ktistakis NT, Fatty acylation of phospholipase B1 on cysteine residues 240 and 241 determines localization on intracellular membranes, J. Biol. Chem 274 (42) (1999) 30023–30027. [DOI] [PubMed] [Google Scholar]
- [131].Lee CS, Kim KL, Jang JH, Choi YS, Suh PG, Ryu SH, The roles of phospholipase D in EGFR signaling, Biochim. Biophys. Acta 1791 (9) (2009) 862–868. [DOI] [PubMed] [Google Scholar]
- [132].Kim JH, Lee S, Kim JH, Lee TG, Hirata M, Suh PG, et al. , Phospholipase D2 directly interacts with aldolase via its PH domain, Biochemistry 41 (10) (2002) 3414–3421. [DOI] [PubMed] [Google Scholar]
- [133].Ahn BH, Kim SY, Kim EH, Choi KS, Kwon TK, Lee YH, et al. , Transmodulation between phospholipase D and c-Src enhances cell proliferation, Mol. Cell. Biol 23 (2003) 3103–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Liscovitch M, Czarny M, Fiucci G, Tang X, Phospholipase D: molecular and cell biology of a novel gene family, Biochem. J 345 (Pt 3) (2000) 401–415. [PMC free article] [PubMed] [Google Scholar]
- [135].Sato TK, Overduin M, Emr SD, Location, location, location: membrane targeting directed by PX domains, Science 294 (5548) (2001) 1881–1885. [DOI] [PubMed] [Google Scholar]
- [136].Xu Y, Hortsman H, Seet L, Wong SH, Hong W, SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P, Nat. Cell Biol 3 (7) (2001) 658–666. [DOI] [PubMed] [Google Scholar]
- [137].Lee JS, Kim JH, Jang IH, Kim HS, Han JM, Kazlauskas A, et al. , Phosphatidylinositol (3,4,5)-trisphosphate specifically interacts with the phox homology domain of phospholipase D1 and stimulates its activity, J. Cell Sci 118 (Pt 19) (2005) 4405–4413. [DOI] [PubMed] [Google Scholar]
- [138].Min DS, Park SK, Exton JH, Characterization of a rat brain phospholipase D isozyme, J. Biol. Chem 273 (12) (1998) 7044–7051. [DOI] [PubMed] [Google Scholar]
- [139].Stahelin RV, Ananthanarayanan B, Blatner NR, Singh S, Bruzik KS, Murray D, et al. , Mechanism of membrane binding of the phospholipase D1 PX domain, J. Biol. Chem 279 (52) (2004) 54918–54926. [DOI] [PubMed] [Google Scholar]
- [140].Kim Y, Han JM, Han BR, Lee KA, Kim JH, Lee BD, et al. , Phospholipase D1 is phosphorylated and activated by protein kinase C in caveolin-enriched micro-domains within the plasma membrane, J. Biol. Chem 275 (18) (2000) 13621–13627. [DOI] [PubMed] [Google Scholar]
- [141].Kim Y, Han JM, Park JB, Lee SD, Oh YS, Chung C, et al. , Phosphorylation and activation of phospholipase D1 by protein kinase C in vivo: determination of multiple phosphorylation sites, Biochemistry 38 (32) (1999) 10344–10351. [DOI] [PubMed] [Google Scholar]
- [142].Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, et al. , Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction, EMBO J. 21 (19) (2001) 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Jeon H, Kwak D, Noh J, Lee MN, Lee CS, Suh PG, et al. , Phospholipase D2 induces stress fiber formation through mediating nucleotide exchange for RhoA, Cell. Signal 23 (8) (2011) 1320–1326. [DOI] [PubMed] [Google Scholar]
- [144].Mahankali M, Peng HJ, Cox D, Gomez-Cambronero J, The mechanism of cell membrane ruffling relies on a phospholipase D2 (PLD2), Grb2 and Rac2 association, Cell. Signal 23 (8) (2011) 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Henkels KM, Mahankali M, Gomez-Cambronero J, Increased cell growth due to a new lipase-GEF (phospholipase D2) fastly acting on Ras, Cell. Signal 25 (1) (2013) 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Mahankali M, Henkels KM, Alter G, Gomez-Cambronero J, Identification of the catalytic site of phospholipase D2 (PLD2) newly described guanine nucleotide exchange factor activity, J. Biol. Chem 287 (49) (2012) 41417–41431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lee CS, Kim IS, Park JB, Lee MN, Lee HY, Suh PG, et al. , The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis, Nat. Cell Biol 8 (5) (2006) 477–484. [DOI] [PubMed] [Google Scholar]
- [148].Kim JH, Kim JH, Ohba M, Suh PG, Ryu SH, Novel functions of the phospholipase D2-Phox homology domain in protein kinase Czeta activation, Mol. Cell. Biol 25 (8) (2005) 3194–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Jang IH, Lee S, Park JB, Kim JH, Lee CS, Hur EM, et al. , The direct interaction of phospholipase C-gamma 1 with phospholipase D2 is important for epidermal growth factor signaling, J. Biol. Chem 278 (20) (2003) 18184–18190. [DOI] [PubMed] [Google Scholar]
- [150].Lee HY, Park JB, Jang IH, Chae YC, Kim JH, Kim IS, et al. , Munc-18–1 inhibits phospholipase D activity by direct interaction in an epidermal growth factor-reversible manner, J. Biol Chem 279 (16) (2004) 16339–16348. [DOI] [PubMed] [Google Scholar]
- [151].Lee JH, Kim YM, Kim NW, Kim JW, Her E, Kim BK, et al. , Phospholipase D2 acts as an essential adaptor protein in the activation of Syk in antigen-stimulated mast cells, Blood 108 (3) (2006) 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Lee S, Kim JH, Lee CS, Kim JH, Kim Y, Heo K, et al. , Collapsin response mediator protein-2 inhibits neuronal phospholipase D(2) activity by direct interaction, J. Biol. Chem 277 (8) (2002) 6542–6549. [DOI] [PubMed] [Google Scholar]
- [153].Park SK, Min DS, Exton JH, Definition of the protein kinase C interaction site of phospholipase D, Biochem. Biophys. Res. Commun 244 (2) (1998) 364–367. [DOI] [PubMed] [Google Scholar]
- [154].Zhang Y, Altshuller YM, Hammond SM, Hayes F, Morris AJ, Frohman MA, Loss of receptor regulation by a phospholipase D1 mutant unresponsive to protein kinase C, EMBO J. 18 (22) (1999) 6339–6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Liu MY, Gutowski S, Stemweis PC, The C terminus of mammalian phospholipase D is required for catalytic activity, J. Biol. Chem 276 (8) (2001) 5556–5562. [DOI] [PubMed] [Google Scholar]
- [156].Qin C, Wang X, The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains, Plant Physiol. 128 (3) (2002) 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Katayama K, Kodaki T, Nagamachi Y, Yamashita S, Cloning, differential regulation and tissue distribution of alternatively spliced isoforms of ADP-ribosylation-factor-dependent phospholipase D from rat liver, Biochem. J 329 (Pt 3) (1998) 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Millar CA, Jess TJ, Saqib KM, Wakelam MJ, Gould GW, 3T3-L1 adipocytes express two isoforms of phospholipase D in distinct subcellular compartments, Biochem. Biophys. Res. Commun 254 (3) (1999) 734–738. [DOI] [PubMed] [Google Scholar]
- [159].Di Fulvio M, Gomez-Cambronero J, Phospholipase D (PLD) gene expression in human neutrophils and HL-60 differentiation, J. Leukoc. Biol 77 (6) (2005) 999–1007. [DOI] [PubMed] [Google Scholar]
- [160].Kinsky SC, Loader JE, Benedict SH, Phorbol ester activation of phospholipase D in human monocytes but not peripheral blood lymphocytes, Biochem. Biophys. Res. Commun 162 (2) (1989) 788–793. [DOI] [PubMed] [Google Scholar]
- [161].Bradshaw CD, Ella KM, Qi C, Sansbury HM, Wisehart-Johnson AE, Meier KE, Effects of phorbol ester on phospholipase D and mitogen-activated protein kinase activities in T-lymphocyte cell lines, Immunol. Lett 53 (2–3) (1996) 69–76. [DOI] [PubMed] [Google Scholar]
- [162].Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y, et al. , Increased activity and intranuclear expression of phospholipase D2 in human renal cancer, Biochem. Biophys. Res. Commun 278 (1) (2000) 140–143. [DOI] [PubMed] [Google Scholar]
- [163].Bruntz RC, Lindsley CW, Brown HA, Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer, Pharmacol. Rev 66 (4) (2014) 1033–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Vermeren MM, Zhang Q, Smethurst E, Segonds-Pichon A, Schrewe H, Wakelam MJ, The phospholipase D2 knock out mouse has ectopic Purkinje cells and suffers from early adult-onset anosmia, PLoS One 11 (9) (2016) e0162814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang Y, Huang P, Du G, Kanaho Y, Frohman MA, Tsirka SE, Increased expression of two phospholipase D isoforms during experimentally induced hippo-campal mossy fiber outgrowth, Glia 46 (1) (2004) 74–83. [DOI] [PubMed] [Google Scholar]
- [166].Edwards YS, Murray AW, Accumulation of phosphatidylalcohol in cultured cells: use of subcellular fractionation to investigate phospholipase D activity during signal transduction, Biochem. J 308 (Pt 2) (1995) 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Cockcroft S, Signalling roles of mammalian phospholipase D1 and D2, Cell. Mol. Life Sci 58 (11) (2001) 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].G Du, Huang P, Liang BT, Frohman MA, Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis, Mol. Biol. Cell 15 (3) (2004) 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Lucocq J, Manifava M, Bi K, Roth MG, Ktistakis NT, Immunolocalisation of phospholipase D1 on tubular vesicular membranes of endocytic and secretory origin, Eur. J. Cell Biol 80 (8) (2001) 508–520. [DOI] [PubMed] [Google Scholar]
- [170].Jang YH, Do SM, The hydrophobic amino acids involved in the interdomain association of phospholipase D1 regulate the shuttling of phospholipase D1 from vesicular organelles into the nucleus, Exp Mol Med 44 (10) (2012) 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].O’Luanaigh N, Pardo R, Fensome A, Allen-Baume V, Jones D, Holt MR, et al. , Continual production of phosphatidic acid by phospholipase D is essential for antigen-stimulated membrane ruffling in cultured mast cells, Mol. Biol. Cell 13 (8) (2002) 3730–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Sarri E, Pardo R, Fensome-Green A, Cockcroft S, Endogenous phospholipase D2 localizes to the plasma membrane of RBL-2H3 mast cells and can be distinguished from ADP ribosylation factor-stimulated phospholipase D1 activity by its specific sensitivity to oleic acid, Biochem. J 369 (Pt 2) (2003) 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Jang YH, Do SM, Nuclear localization of phospholipase D1 mediates the activation of nuclear protein kinase C(alpha) and extracellular signal-regulated kinase signaling pathways, J. Biol. Chem 286 (6) (2011) 4680–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Toda K, Nogami M, Murakami K, Kanaho Y, Nakayama K, Colocalization of phospholipase D1 and GTP-binding-defeetive mutant of ADP-ribosylation factor 6 to endosomes and lysosomes, FEBS Lett. 442 (2–3) (1999) 221–225. [DOI] [PubMed] [Google Scholar]
- [175].Choi WS, Kim YM, Combs C, Frohman MA, Beaven MA, Phospholipases D1 and D2 regulate different phases of exocytosis in mast cells, J. Immunol 168 (11) (2002) 5682–5689. [DOI] [PubMed] [Google Scholar]
- [176].Lee S, Park JB, Kim JH, Kim Y, Kim JH, Shin KJ, et al. , Actin directly interacts with phospholipase D, inhibiting its activity, J. Biol. Chem 276 (30) (2001) 28252–28260. [DOI] [PubMed] [Google Scholar]
- [177].Vitale N, Caumont AS, Chasserot-Golaz S, Du G, Wu S, Sciorra VA, et al. , Phospholipase D1: a key factor for the exocytotic machinery in neuroendocrine cells, EMBO J. 20 (10) (2001) 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Powner DJ, Hodgkin MN, Wakelam MJ, Antigen-stimulated activation of phospholipase D1b by Raci, ARF6, and PKCalpha in RBL-2H3 cells, Mol. Biol. Cell 13 (4) (2002) 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Brito de Souza L, Pinto da Silva LL, Jamur MC, Oliver C, Phospholipase D is involved in the formation of Golgi associated clathrin coated vesicles in human parotid duct cells, PLoS One 9 (3) (2014) e91868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Czarny M, Lavie Y, Fiucci G, Liscovitch M, Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-182–101, J. Biol. Chem 274 (5) (1999) 2717–2724. [DOI] [PubMed] [Google Scholar]
- [181].Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, et al. , Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation, Cell 99 (5) (1999) 521–532. [DOI] [PubMed] [Google Scholar]
- [182].Zheng X, Bollinger Bollag W, Aquaporin 3 colocates with phospholipase d2 in caveolin-rich membrane microdomains and is downregulated upon keratinocyte differentiation, J. Invest Dermatol 121 (6) (2003) 1487–1495. [DOI] [PubMed] [Google Scholar]
- [183].Czarny M, Fiucci G, Lavie Y, Banno Y, Nozawa Y, Liscovitch M, Phospholipase D2: functional interaction with caveolin in low-density membrane microdomains, FEBS Lett. 467 (2–3) (2000) 326–332. [DOI] [PubMed] [Google Scholar]
- [184].Norton LJ, Zhang Q, Saqib KM, Schrewe H, Macura K, Anderson KE, et al. , PLD1 rather than PLD2 regulates phorbol-ester-, adhesion-dependent and Fc{gamma}- receptor-stimulated ROS production in neutrophils, J. Cell Sci 124 (Pt 12) (2011) 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Dall’Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan RB, et al. , The phospholipase D1 pathway modulates macroautophagy, Nat. Commun 1 (2010) 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Rabinowitz JD, White E, Autophagy and metabolism, Science 330 (6009) (2010) 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Burkhardt U, Stegner D, Hattingen E, Beyer S, Nieswandt B, Klein J, Impaired brain development and reduced cognitive function in phospholipase D-deficient mice, Neurosci. Lett 572 (2014) 48–52. [DOI] [PubMed] [Google Scholar]
- [188].Oliveira TG, Chan RB, Tian H, Laredo M, Shui G, Staniszewski A, et al. , Phospholipase d2 ablation ameliorates Alzheimer’s disease-linked synaptic dys-function and cognitive deficits, J. Neurosci 30 (49) (2010) 16419–16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Trujillo Viera J, El-Merahbi R, Nieswandt B, Stegner D, Sumara G, Phospholipases D1 and D2 suppress appetite and protect against overweight, PLoS One 11 (6) (2016) e0157607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Frohman MA, The phospholipase D superfamily as therapeutic targets, Trends Pharmacol. Sci 36 (3) (2015) 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Jang JH, Lee CS, Hwang D, Ryu SH, Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners, Prog. Lipid Res 51 (2) (2012) 71–81. [DOI] [PubMed] [Google Scholar]
- [192].Munck A, Bohm C, Seibel NM, Hashemol Hosseini Z, Hampe W, Hu-K4 is a ubiquitously expressed type 2 transmembrane protein associated with the endoplasmic reticulum, FEBS J. 272 (7) (2005) 1718–1726. [DOI] [PubMed] [Google Scholar]
- [193].Pedersen KM, Finsen B, Cells JE, Jensen NA, Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain, J. Biol. Chem 273 (47) (1998) 31494–31504. [DOI] [PubMed] [Google Scholar]
- [194].Otani Y, Yamaguchi Y, Sato Y, Furuichi T, Ikenaka K, Kitani H, et al. , PLD$ is involved in phagocytosis of microglia: expression and localization changes of PLD4 are correlated with activation state of microglia, PLoS One 6 (11) (2011) e27544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Gavin AL, Huang D, Huber C, Martensson A, Tardif V, Skog PD, et al. , PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing, Nat Immunol. 19 (9) (2018) 942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Nelson RK, Frohman MA, Physiological and pathophysiological roles for phospholipase D, J. Lipid Res 56 (12) (2015) 2229–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Gardin A, White J, The sanger mouse genetics programme: high throughput characterisation of knockout mice, Acta Ophthalmol. 89 (2011) 1755–3768. [Google Scholar]
- [198].Aissani B, Wiener H, Zhang K, Multiple hits for the association of uterine fibroids on human chromosome 1q43, PLoS One 8 (3) (2013) e58399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA, A common lipid links Mfh-mediated mitochondrial fusion and SNARE-regulated exocytosis, Nat. Cell Biol 8 (11) (2006) 1255–1262. [DOI] [PubMed] [Google Scholar]
- [200].Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, et al. , piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling, Dev. Cell 20 (3) (2011) 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Ha EE, Frohman MA, Regulation of mitochondrial morphology by lipids, Biofactors 40 (4) (2014) 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Nureki O, Is zucchini a phosphodiesterase or a ribonuclease? Biom. J 37 (6) (2014) 369–374. [DOI] [PubMed] [Google Scholar]
- [203].Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, et al. , Structure and function of Zucchini endoribonuclease in piRNA biogenesis, Nature 491 (7423) (2012) 284–287. [DOI] [PubMed] [Google Scholar]
- [204].Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ, The structural bio-chemistry of Zucchini implicates it as a nuclease in piRNA biogenesis, Nature 491 (7423) (2012) 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O, Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nue, RNA 18 (12) (2012) 2128–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Ali WH, Chen Q, Delgiomo KE, Su W, Hall JC, Hongu T, et al. , Deficiencies of the lipid-signaling enzymes phospholipase D1 and D2 alter cytoskeletal organization, macrophage phagocytosis, and cytokine-stimulated neutrophil recruitment, PLoS One 8 (1) (2013) e55325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Grab LT, Kearns MW, Morris AJ, Daniel LW, Differential role for phospholipase D1 and phospholipase D2 in 12-O-tetradecanoyl-13-phorbol acetate-stimulated MAPK activation, Cox-2 and IL-8 expression, Biochim. Biophys. Acta 1636 (1) (2004) 29–39. [DOI] [PubMed] [Google Scholar]
- [208].Sanematsu F, Nishikimi A, Watanabe M, Hongu T, Tanaka Y, Kanaho Y, et al. , Phosphatidic acid-dependent recruitment and function of the Rac activator DOCK1 during dorsal ruffle formation, J. Biol. Chem 288 (12) (2013) 8092–8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Thielmann I, Stegner D, Kraft P, Hagedorn I, Krohne G, Kleinschnitz C, et al. , Redundant functions of phospholipases D1 and D2 in platelet alpha-granule release, J. Thromb, Haemost 10 (11) (2012) 2361–2372. [DOI] [PubMed] [Google Scholar]
- [210].Sato T, Hongu T, Sakamoto M, Funakoshi Y, Kanaho Y, Molecular mechanisms of N-formyl-methionyl-leucyl-phenylalanine-induced superoxide generation and de-granulation in mouse neutrophils: phospholipase D is dispensable, Mol. Cell. Biol 33 (1) (2013) 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Skippen A, Jones DH, Morgan CP, Li M, Cockcroft S, Mechanism of ADP ribosylation factor-stimulated phosphatidylinositol 4,5-bisphosphate synthesis in HL60 cells, J. Biol. Chem 277 (8) (2002) 5823–5831. [DOI] [PubMed] [Google Scholar]
- [212].Yanase Y, Carvou N, Frohman MA, Cockcroft S, Reversible bleb formation in mast cells stimulated with antigen is Ca2+/calmodulin-dependent and bleb size is regulated by ARF6, Biochem. J 425 (1) (2009) 179–193. [DOI] [PubMed] [Google Scholar]
- [213].Huang P, Altshuller YM, Hou JC, Pessin JE, Frohman MA, Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1, Mol. Biol. Cell 16 (6) (2005) 2614–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Huang P, Frohman MA, The potential for phospholipase D as a new therapeutic target, Expert Opin. Ther, Targets 11 (5) (2007) 707–716. [DOI] [PubMed] [Google Scholar]
- [215].Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, et al. , 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis, Mol. Pharmacol 75 (3) (2009) 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Kanaho Y, Sato T, Hongu T, Funakoshi Y, Molecular mechanisms of fMLP-induced superoxide generation and degranulation in mouse neutrophils, Adv. Biol. Regul 53 (1) (2013) 128–134. [DOI] [PubMed] [Google Scholar]
- [217].Spencer C, Brown HA, Biochemical characterization of a Pseudomonas aeruginosa phospholipase D, Biochemistry 54 (5) (2015) 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].McLain N, Dolan JW, Phospholipase D activity is required for dimorphic transition in Candida albicans, Microbiology 143 (Pt 11) (1997) 3521–3526. [DOI] [PubMed] [Google Scholar]
- [219].Harkins AL, Yuan G, London SD, o JW, An oleate-stimulated, phosphatidylinositol 4,5-bisphosphate-independent phospholipase D in Schizosaccharomyces pombe, FEMS Yeast Res. 10 (6) (2010) 717–726. [DOI] [PubMed] [Google Scholar]
- [220].Munnik T, Arisz SA, De Vrije T, Musgrave A, G protein activation stimulates phospholipase D signaling in plants, Plant Cell 7 (12) (1995) 2197–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Peters NT, Logan KO, Miller AC, Kropf DL, Phospholipase D signaling regulates microtubule organization in the fucoid alga Silvetia compressa, Plant Cell Physiol. 48 (12) (2007) 1764–1774. [DOI] [PubMed] [Google Scholar]
- [222].Peters NT, Pol SU, Kropf DL, Phospholipid signaling during stramenopile development, Plant Signal. Behav 3 (6) (2008) 398–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Monovich L, Mugrage B, Quadros E, Toscano K, Tommasi R, LaVoie S, et al. , Optimization of halopemide for phospholipase D2 inhibition, Bioorg. Med. Chem. Lett 17 (8) (2007) 2310–2311. [DOI] [PubMed] [Google Scholar]
- [224].Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, et al. , Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness, Nat. Chem. Biol 5 (2) (2009) 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].van Rooij HH, Waterman RL, Kraak JC, Dynamic cation-exchange systems for the separation of drugs derived from butyrophenone and diphenylpiperidine by high-performance liquid chromatography and applied in the determination of halopemide in plasma, J. Chromatogr 164 (2) (1979) 177–185. [DOI] [PubMed] [Google Scholar]
- [226].Lavieri R, Scott SA, Lewis JA, Selvy PE, Armstrong MD, Alex Brown H, et al. , Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part II. Identification of the 1,3,8-triazaspiro[4,5]decan-4-one privileged structure that engenders PLD2 selectivity, Bioorg. Med. Chem. Lett 19 (8) (2009) 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Lewis JA, Scott SA, Lavieri R, Buck JR, Selvy PE, Stoops SL, et al. , Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity, Bioorg. Med. Chem. Lett 19 (7) (2009) 1916–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Lavieri RR, Scott SA, Selvy PE, Kim K, Jadhav S, Morrison RD, et al. , Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8- triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor, J. Med. Chem 53 (18) (2010) 6706–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Ganesan R, Mahankali M, Alter G, Gomez-Cambronero J, Two sites of action for PLD2 inhibitors: the enzyme catalytic center and an allosteric, phosphoinositide biding pocket, Biochim. Biophys. Acta 1851 (3) (2015) 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Song HI, Yoon MS, PLD1 regulates adipogénie differentiation through mTOR - IRS- 1 phosphorylation at serine 636/639, Sci. Rep 6 (2016) 36968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Yin H, Gui Y, Du G, Frohman MA, Zheng XL, Dependence of phospholipase D1 multi-monoubiquitination on its enzymatic activity and palmitoylation, J. Biol. Chem 285 (18) (2010) 13580–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Henkels KM, Muppani NR, Gomez-Cambronero J, PLD-specific small-molecule inhibitors decrease tumor-associated macrophages and neutrophils infiltration in breast tumors and lung and liver metastases, PLoS One 11 (11) (2016) e0166553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Burkhardt U, Beyer S, Klein J, Role of phospholipases D1 and 2 in astroglial proliferation: effects of specific inhibitors and genetic deletion, Eur. J. Pharmacol 761 (2015) 398–404. [DOI] [PubMed] [Google Scholar]
- [234].Scott SA, Xiang Y, Mathews TP, Cho HP, Myers DS, Armstrong MD, et al. , Regulation of phospholipase D activity and phosphatidic acid production after purinergic (P2Y6) receptor stimulation, J. Biol. Chem 288 (28) (2013) 20477–20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Bruntz RC, Taylor HE, Lindsley CW, Brown HA, Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells, J. Biol. Chem 289 (2) (2014) 600–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Rudge SA, Wakelam MJ, Inter-regulatory dynamics of phospholipase D and the actin cytoskeleton, Biochim. Biophys. Acta 1791 (9) (2009) 856–861. [DOI] [PubMed] [Google Scholar]
- [237].Du G, Altshuller YM, Kim Y, Han JM, Ryu SH, Morris AJ, et al. , Dual requirement for rho and protein kinase C in direct activation of phospholipase D1 through G protein-coupled receptor signaling, Mol. Biol. Cell 11 (12) (2000) 4359–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Liscovitch M, Chalifa V, Danin M, Eli Y, Inhibition of neural phospholipase D activity by aminoglycoside antibiotics, Biochem. J 279 (Pt 1) (1991) 319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Liscovitch M, Chalifa-Caspi V, Enzymology of mammalian phospholipases D: in vitro studies, Chem. Phys. Upids 80 (1–2) (1996) 37–44. [DOI] [PubMed] [Google Scholar]
- [240].Mohn H, Chalifa V, Liscovitch M, Substrate specificity of neutral phospholipase D from rat brain studied by selective labeling of endogenous synaptic membrane phospholipids in vitro, J. Biol. Chem 267 (16) (1992) 11131–11136. [PubMed] [Google Scholar]
- [241].Ohguchi K, Banno Y, Nakashima S, Nozawa Y, Regulation of membrane-bound phospholipase D by protein kinase C in HL60 cells. Synergistic action of small GTP-binding protein RhoA, J. Biol. Chem 271 (8) (1996) 4366–4372. [DOI] [PubMed] [Google Scholar]
- [242].Kodaki T, Yamashita S, Cloning, expression, and characterization of a novel phospholipase D complementary DNA from rat brain, J. Biol. Chem 272 (17) (1995) 11408–11413. [DOI] [PubMed] [Google Scholar]
- [243].Divecha N, Roefs M, Halstead JR, D’Andrea S, Fernandez-Borga M, Oomen L, et al. , Interaction of the type Ialpha PIPkinase with phospholipase D: a role for the local generation of phosphatidylinositol 4, 5-bisphosphate in the regulation of PLD2 activity, EMBO J. 19 (20) (2000) 5440–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Chung JK, Sekiya F, Kang HS, Lee C, Han JS, Kim SR, et al. , Synaptojanin inhibition of phospholipase D activity by hydrolysis of phosphatidylinositol 4,5-bi-sphosphate, J. Biol. Chem 272 (25) (1997) 15980–15985. [DOI] [PubMed] [Google Scholar]
- [245].McPherson PS, Garcia EP, Slepnev VI, David C, Zhang X, Grabs D, et al. , A presynaptic inositol-5-phosphatase, Nature 379 (6563) (1996) 353–357. [DOI] [PubMed] [Google Scholar]
- [246].Banno Y, Fujita H, Ono Y, Nakashima S, Ito Y, Kuzumaki N, et al. , Differential phospholipase D activation by bradykinin and sphingosine 1-phosphate in NIH 3T3 fibroblasts overexpressing gelsolin, J. Biol. Chem 274 (39) (1999) 27385–27391. [DOI] [PubMed] [Google Scholar]
- [247].Powner DJ, Wakelam MJ, The regulation of phospholipase D by inositol phospholipids and small GTPases, FEBS Lett. 531 (1) (2002) 62–64. [DOI] [PubMed] [Google Scholar]
- [248].Cross MJ, Stewart A, Hodgkin MN, Kerr DJ, Wakelam MJ, Wortmannin and its structural analogue demethoxyviridin inhibit stimulated phospholipase A2 activity in Swiss 3T3 cells. Wortmannin is not a specific inhibitor of phosphatidy-linositol 3-kinase, J. Biol. Chem 270 (43) (1995) 25352–25355. [DOI] [PubMed] [Google Scholar]
- [249].Macia E, Chabre M, Franco M, Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6, J. Biol. Chem 276 (27) (2001) 24925–24930. [DOI] [PubMed] [Google Scholar]
- [250].Stain JC, Sander EE, Michiels F, van Leeuwen FN, Kain HE, van der Kammen RA, et al. , Targeting of Tiaml to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain, J. Biol. Chem 272 (45) (1997) 28447–28454. [DOI] [PubMed] [Google Scholar]
- [251].Zhang W, Wang C, Qin C, Wood T, Olafisdottir G, Welti R, et al. , The oleate-stimulated phospholipase D, PLDdelta, and phosphatidic acid decrease H202-induced cell death in Arabidopsis, Plant Cell 15 (10) (2003) 2285–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Rohwedder A, Zhang Q, Rudge SA, Wakelam MJ, Lipid droplet formation in response to oleic acid in Huh-7 cells is mediated by the fatty acid receptor FFAR4, J. Cell Sci 127 (Pt 14) (2014) 3104–3115. [DOI] [PubMed] [Google Scholar]
- [253].Kim JH, Kim Y, Lee SD, Lopez I, Arnold RS, Lambeth JD, et al. , Selective activation of phospholipase D2 by unsaturated fatty acid, FEBS Lett. 454 (1–2) (1999) 42–46. [DOI] [PubMed] [Google Scholar]
- [254].Xie Z, Ho WT, Exton JH, Functional implications of post-translational modifications of phospholipases D1 and D2, Biochim. Biophys. Acta 1580 (1) (2002) 9–21. [DOI] [PubMed] [Google Scholar]
- [255].Sugars JM, Cellek S, Manifava M, Coadwell J, Ktistakis NT, Hierarchy of membrane-targeting signals of phospholipase D1 involving lipid modification of a pleckstrin homology domain, J. Biol. Chem 277 (32) (2002) 29152–29161. [DOI] [PubMed] [Google Scholar]
- [256].Xie Z, Ho WT, Exton JH, Requirements and effects of palmitoylation of rat PLD1, J. Biol. Chem 276 (12) (2001) 9383–9391. [DOI] [PubMed] [Google Scholar]
- [257].Lopez I, Burns DJ, Lambeth JD, Regulation of phospholipase D by protein kinase C in human neutrophils. Conventional isoforms of protein kinase C phosphorylate a phospholipase D-related component in the plasma membrane, J. Biol. Chem 270 (33) (1995) 19465–19472. [DOI] [PubMed] [Google Scholar]
- [258].Chen JS, Exton JH, Sites on phospholipase D2 phosphorylated by PKCalpha, Biochem. Biophys. Res. Commun 333 (4) (2005) 1322–1326. [DOI] [PubMed] [Google Scholar]
- [259].Farquhar MJ, Powner DJ, Levine BA, Wright MH, Ladds G, Hodgkin MN, Interaction of PLD1b with actin in antigen-stimulated mast cells, Cell. Signal 19 (2007) 349–358. [DOI] [PubMed] [Google Scholar]
- [260].Ganley IG, Walker SJ, Manifava M, Li D, Brown HA, Ktistakis NT, Interaction of phospholipase D1 with a casein-kinase-2-like serine kinase, Biochem. J 354 (Pt 2) 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Ahn BH, Min G, Bae YS, Bae YS, Min DS, Phospholipase D is activated and phosphorylated by casein kinase-II in human IJ87 astroglioma cells, Exp. Mol. Med 38 (1) (2006) 55–62. [DOI] [PubMed] [Google Scholar]
- [262].Kim JH, Park JM, Yea K, Kim HW, Suh PG, Ryu SH, Phospholipase D1 mediates AMP-activated protein kinase signaling for glucose uptake, PLoS One 5 (2010) e9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Lee HY, Yea K, Kim J, Lee BD, Chae YC, Kim HS, et al. , Epidermal growth factor increases insulin secretion and lowers blood glucose in diabetic mice, J. Cell. Mol. Med 12 (5A) (2008) 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Zeniou-Meyer M, Liu Y, Begle A, Olanich ME, Hanauer A, Becherer U, et al. , The coffin-Lowry syndrome-associated protein RSK2 is implicated in calcium-regulated exocytosis through the regulation of PLD1, Proc. Natl. Acad. Sci. U. S. A 105 (24) (2008) 8434–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Watanabe H, Kanaho Y, Inhibition of phosphatidylinositol 4,5-bisphosphate-stimulated phospholipase D2 activity by Ser/Thr phosphorylation, Biochim. Biophys. Acta 1495 (2) (2000) 121–124. [DOI] [PubMed] [Google Scholar]
- [266].Bourgoin S, Grinstein S, Peroxides of vanadate induce activation of phospholipase D in HL-60 cells. Role of tyrosine phosphorylation, J. Biol. Chem 267 (17) (1992) 11908–11916. [PubMed] [Google Scholar]
- [267].Gomez-Cambronero J, Immunoprecipitation of a phospholipase D activity with antiphosphotyrosine antibodies, J. Interf. Cytokine Res 15 (10) (1995) 877–885. [DOI] [PubMed] [Google Scholar]
- [268].Horn J, Lopez I, Miller MW, Gomez-Cambronero J, The uncovering of a novel regulatory mechanism for PLD2: formation of a ternary complex with protein tyrosine phosphatase PTP1B and growth factor receptor-bound protein GRB2, Biochem. Biophys. Res. Commun 332 (1) (2005) 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Henkels KM, Short S, Peng HJ, Di Fulvio M, Gomez-Cambronero J, PLD2 has both enzymatic and cell proliferation-inducing capabilities, that are differentially regulated by phosphorylation and dephosphorylation, Biochem. Biophys. Res. Commun 389 (2) (2009) 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Henkels KM, Peng HJ, Frondorf K, Gomez-Cambronero J, A comprehensive model that explains the regulation of phospholipase D2 activity by phosphorylation-dephosphorylation, Mol. Cell. Biol 30 (9) (2010) 2251–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Min DS, Ahn BH, Jo YH, Differential tyrosine phosphorylation of phospholipase D isozymes by hydrogen peroxide and the epidermal growth factor in A431 epidermoid carcinoma cells, Mol. Cells 11 (3) (2001) 369–378. [PubMed] [Google Scholar]
- [272].Slaaby R, Jensen T, Hansen HS, Frohman MA, Seedorf K, PLD2 complexes with the EGF receptor and undergoes tyrosine phosphorylation at a single site upon agonist stimulation, J. Biol. Chem 273 (50) (1998) 33722–33727. [DOI] [PubMed] [Google Scholar]
- [273].Di Fulvio M, Lehman N, Lin X, Lopez I, Gomez-Cambronero J, The elucidation of novel SH2 binding sites on PLD2, Oncogene 25 (21) (2006) 3032–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Di Fulvio M, Frondorf K, Henkels KM, Lehman N, Gomez-Cambronero J, The Grb2/PLD2 interaction is essential for lipase activity, intracellular localization and signaling in response to EGF, J. Mol. Biol 367 (3) (2007) 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Di Fulvio M, Frondorf K, Gomez-Cambronero J, Mutation of Y179 on phospholipase D2 (PLD2) upregulates DNA synthesis in a PI3K-and Akt-dependent manner, Cell. Signal 20 (1) (2008) 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Wu F, Wang P, Zhang J, Young LC, Lai R, Li L, Studies of phosphoproteomic changes induced by nucleophosmin-anaplastic lymphoma kinase (ALK) highlight deregulation of tumor necrosis factor (TOT)/Fas/TNF-related apoptosis-induced ligand signaling pathway in ALK-positive anaplastic large cell lymphoma, Mol. Cell. Proteomics 9 (7) (2010) 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Conricode KM, Brewer KA, Exton JH, Activation of phospholipase D by protein kinase C. Evidence for a phosphorylation-independent mechanism, J. Biol. Chem 267 (11) (1992) 7199–7202. [PubMed] [Google Scholar]
- [278].Exton JH, Regulation of phospholipase D, Biochim. Biophys. Acta 1439 (2) (1999) 121–133. [DOI] [PubMed] [Google Scholar]
- [279].Hodgkin MN, Clark JM, Rose S, Saqib K, Wakelam MJ, Characterization of the regulation of phospholipase D activity in the detergent-insoluble fraction of HL60 cells by protein kinase C and small G-proteins, Biochem. J 339 (Pt 1) (1999) 87–93. [PMC free article] [PubMed] [Google Scholar]
- [280].Kang DW, Park MH, Lee YJ, Kim HS, Kwon TK, Park WS, et al. , Phorbol ester upregulates phospholipase D1 but not phospholipase D2 expression through a PKC/Ras/ERK/NFkappaB-dependent pathway and enhances matrix metalloproteinase-9 secretion in colon cancer cells, J Biol Chem 283 (7) (2008) 4094–4104. [DOI] [PubMed] [Google Scholar]
- [281].Han JM, Kim JH, Lee BD, Lee SD, Kim Y, Jung YW, et al. , Phosphorylation-dependent regulation of phospholipase D2 by protein kinase C delta in rat Pheochromocytoma PC12 cells, J. Biol. Chem 277 (10) (2002) 8290–8297. [DOI] [PubMed] [Google Scholar]
- [282].Chen JS, Exton JH, Regulation of phospholipase D2 activity by protein kinase C alpha, J. Biol. Chem 279 (21) (2004) 22076–22083. [DOI] [PubMed] [Google Scholar]
- [283].Siddiqi AR, Srajer GE, Leslie CC, Regulation of human PLD1 and PLD2 by calcium and protein kinase C, Biochim. Biophys. Acta 1497 (1) (2000) 103–114. [DOI] [PubMed] [Google Scholar]
- [284].Lee TG, Park JB, Lee SD, Hong S, Kim JH, Kim Y, et al. , Phorbol myristate acetate-dependent association of protein kinase C alpha with phospholipase D1 in intact cells, Biochim. Biophys. Acta 1347 (2–3) (1997) 199–204. [DOI] [PubMed] [Google Scholar]
- [285].Hu T, Exton JH, Mechanisms of regulation of phospholipase D1 by protein kinase Calpha, J. Biol. Chem 278 (4) (2003) 2348–2355. [DOI] [PubMed] [Google Scholar]
- [286].Singer WD, Brown HA, Jiang X, Sternweis PC, Regulation of phospholipase D by protein kinase C is synergistic with ADP-ribosylation factor and independent of protein kinase activity, J. Biol. Chem 271 (8) (1996) 4504–4510. [DOI] [PubMed] [Google Scholar]
- [287].Luo B, Prescott SM, Topham MK, Protein kinase C alpha phosphorylates and negatively regulates diacylglycerol kinase zeta, J. Biol. Chem 278 (41) (2003) 39542–39547. [DOI] [PubMed] [Google Scholar]
- [288].Luo B, Prescott SM, Topham MK, Association of diacylglycerol kinase zeta with protein kinase C alpha: spatial regulation of diacylglycerol signaling, J. Cell Biol 160 (6) (2003) 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Han JM, Kim Y, Lee JS, Lee CS, Lee BD, Ohba M, et al. , Localization of phospholipase DI to caveolin-enriched membrane via palmitoylation: implications for epidermal growth factor signaling, Mol. Biol. Cell 13 (11) (2002) 3976–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Cook SJ, Briscoe CP, Wakelam MJ, The regulation of phospholipase D activity and its role in sn-1,2-diradylglycerol formation in bombesin- and phorbol 12-myristate 13-acetate-stimulated Swiss 3T3 cells, Biochem. J 280 (Pt 2) (1991) 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Shome K, Nie Y, Romero G, ADP-ribosylation factor proteins mediate agonist-induced activation of phospholipase D, J. Biol. Chem 273 (46) (1998) 30836–30841. [DOI] [PubMed] [Google Scholar]
- [292].Henage LG, Exton JH, Brown HA, Kinetic analysis of a mammalian phospholipase D: allosteric modulation by monomeric GTPases, protein kinase C, and polyphosphoinositides, J. Biol. Chem 281 (6) (2006) 3408–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Terui T, Kahn RA, Randazzo PA, Effects of acid phospholipids on nucleotide exchange properties of ADP-ribosylation factor 1. Evidence for specific interaction with phosphatidylinositol 4,5-bisphosphate, J. Biol. Chem 269 (45) (1994) 28130–28135. [PubMed] [Google Scholar]
- [294].Randazzo PA, Kahn RA, GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acid phospholipids, J. Biol. Chem 269 (14) (1994) 10758–10763. [PubMed] [Google Scholar]
- [295].Ge M, Cohen JS, Brown HA, Freed JH, ADP ribosylation factor 6 binding to phosphatidylinositol 4,5-bisphosphate-containing vesicles creates defects in the bilayer structure: an electron spin resonance study, Biophys. J 81 (2) (2001) 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Vinggaard AM, Jensen T, Morgan CP, Cockcroft S, Hansen HS, Didecanoyl phosphatidylcholine is a superior substrate for assaying mammalian phospholipase D, Biochem. J 319 (Pt 3) (1996) 861–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Jiang X, Gutowski S, Singer WD, Sternweis PC, Assays and characterization of mammalian phosphatidylinositol 4,5-bisphosphate-sensitive phospholipase D, Methods Enzymol. 345 (2002) 328–334. [DOI] [PubMed] [Google Scholar]
- [298].Baillie GS, Huston E, Scotland G, Hodgkin M, Gall I, Peden AH, et al. , TAPAS-1, a novel microdomain within the unique N-terminal region of the PDE4A1 cAMP-specific phosphodiesterase that allows rapid, Ca2+ − triggered membrane association with selectivity for interaction with phosphatidic acid, J. Biol. Chem 277 (31) (2002) 28298–28309. [DOI] [PubMed] [Google Scholar]
- [299].Kim JH, Lee SD, Han JM, Lee TG, Kim Y, Park JB, et al. , Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA, FEBS Lett. 430 (3) (1998) 231–235. [DOI] [PubMed] [Google Scholar]
- [300].Zhang GF, Patton WA, Lee FJ, Liyanage M, Han JS, Rhee SG, et al. , Different ARF domains are required for the activation of cholera toxin and phospholipase D, J. Biol. Chem 270 (1) (1995) 21–24. [DOI] [PubMed] [Google Scholar]
- [301].Jones DH, Bax B, Fensome A, Cockcroft S, ADP ribosylation factor 1 mutants identify a phospholipase D effector region and reveal that phospholipase D participates in lysosomal secretion but is not sufficient for recruitment of coatomer I, Biochem. J 341 (Pt 1) (1999) 185–192. [PMC free article] [PubMed] [Google Scholar]
- [302].Fensome A, Cunningham E, Troung O, Cockcroft S, ARF1(2–17) does not specifically interact with ARF1-dependent pathways. Inhibition by peptide of phospholipases C beta, D and exocytosis in HL60 cells, FEBS Lett. 349 (1) (1994) 34–38. [DOI] [PubMed] [Google Scholar]
- [303].Fensome A, Whatmore J, Morgan C, Jones D, Cockcroft S, ADP-ribosylation factor and Rho proteins mediate fMLP-dependent activation of phospholipase D in human neutrophils, J. Biol. Chem 273 (21) (1998) 13157–13164. [DOI] [PubMed] [Google Scholar]
- [304].Malcolm KC, Elliott CM, Exton JH, Evidence for Rho-mediated agonist stimulation of phospholipase D in rati fibroblasts. Effects of Clostridium botulinum C3 exoenzyme, J. Biol. Chem 271 (22) (1996) 13135–13139. [DOI] [PubMed] [Google Scholar]
- [305].Singer WD, Brown HA, Bokoch GM, Stemweis PC, Resolved phospholipase D activity is modulated by cytosolic factors other than Arf, J. Biol. Chem 270 (25) (1995) 14944–14950. [DOI] [PubMed] [Google Scholar]
- [306].Brown HA, Sternweis PC, Stimulation of phospholipase D by ADP-ribosylation factor, Methods Enzymol. 257 (1995) 313–324. [DOI] [PubMed] [Google Scholar]
- [307].Malcolm KC, Ross AH, Qiu RG, Symons M, Exton JH, Activation of rat liver phospholipase D by the small GTP-binding protein RhoA, J. Biol. Chem 269 (42) (1994) 25951–25954. [PubMed] [Google Scholar]
- [308].Alberts AS, Bouquin N, Johnston LH, Treisman R, Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7, J. Biol. Chem 273 (15) (1996) 8616–8622. [DOI] [PubMed] [Google Scholar]
- [309].Cai S, Exton JH, Determination of interaction sites of phospholipase D1 for RhoA, Biochem. J 355 (Pt 3) (2001) 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Yamazaki M, Zhang Y, Watanabe H, Yokozeki T, Ohno S, Kaibuchi K, et al. , Interaction of the small G protein RhoA with the C terminus of human phospholipase D1, J. Biol. Chem 274 (10) (1999) 6035–6038. [DOI] [PubMed] [Google Scholar]
- [311].Peng HJ, Henkels KM, Mahankali M, Marchai C, Bubulya P, Dinauer MC, et al. , The dual effect of Rac2 on phospholipase D2 regulation that explains both the onset and termination of chemotaxis, Mol. Cell. Biol 31 (11) (2011) 2227–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Peng HJ, Henkels KM, Mahankali M, Dinauer MC, Gomez-Cambronero J, Evidence for two CRIB domains in phospholipase D2 (PLD2) that the enzyme uses to specifically bind to the small GTPase Rac2, J. Biol. Chem 286 (18) (2011) 16308–16320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Mahankali M, Peng HJ, Henkels KM, Dinauer MC, Gomez-Cambronero J, Phospholipase D2 (PLD2) is a guanine nucleotide exchange factor (GEF) for the GTPase Rac2, Proc. Natl. Acad. Sci. U. S. A 108 (49) (2011) 19617–19622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Abramovici H, Mojtabaie P, Parks RJ, Zhong XP, Koretzky GA, Topham MK, et al. , Diacylglycerol kinase zeta regulates actin cytoskeleton reorganization through dissociation of Rac1 from RhoGDI, Mol. Biol. Cell 20 (7) (2009) 2049–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Nishikimi A, Fukuhara H, Su W, Hongu T, Takasuga S, Mihara H, et al. , Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemo taxis, Science 324 (5925) (2009) 384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Schlam D, Canton J, Every day I’m rufflin’: calcium sensing and actin dynamics in the growth factor-independent membrane ruffling of professional phagocytes, Small GTPases 8 (2) (2017) 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D, Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos, Nat. Cell Biol 9 (6) (2007) 706–712. [DOI] [PubMed] [Google Scholar]
- [318].Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, et al. , Phospholipase D1 is an effector of Rheb in the mTOR pathway, Proc. Natl. Acad. Sci. U. S. A 105 (24) (2008) 8286–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Luo JQ, Liu X, Hammond SM, Colley WC, Feig LA, Frohman MA, et al. , RalA interacts directly with the Arf-responsive, PIP2-dependent phospholipase D1, Biochem. Biophys. Res. Commun 235 (3) (1997) 854–859. [DOI] [PubMed] [Google Scholar]
- [320].Jang YH, Do SM, Intermolecular association between caspase-mediated cleavage fragments of phospholipase D1 protects against apoptosis, Int. J. Biochem. Cell Biol 44 (2) (2012) 358–365. [DOI] [PubMed] [Google Scholar]
- [321].Voelker DR, Organelle biogenesis and intracellular lipid transport in eukaryotes, Microbiol. Rev 55 (4) (1991) 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Athenstaedt K, Daum G, Phosphatidic acid, a key intermediate in lipid metabolism, Eur. J. Biochem 266 (1) (1999) 1–16. [DOI] [PubMed] [Google Scholar]
- [323].Athenstaedt K, Weys S, Paltauf F, Daum G, Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae, J. Bacteriol 181 (5) (1999) 1458–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Gibellini F, Smith TK, The Kennedy pathway-De novo synthesis of phosphatidy-lethanolamine and phosphatidylcholine, IUBMB Life 62 (6) (2010) 414–428. [DOI] [PubMed] [Google Scholar]
- [325].Kennedy EP, The biosynthesis of phospholipids, Am. J. Clin. Nutr 6 (3) (1958) 216–220. [DOI] [PubMed] [Google Scholar]
- [326].Ammar MR, Kassas N, Chasserot-Golaz S, Bader MF, Vitale N, Lipids in regulated exocytosis: what are they doing? Front. Endocrinol. (Lausanne) 4 (2013) 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J, Phosphatidic acid-mediated mitogenic activation of mTOR signaling, Science 294 (5548) (2001) 1942–1945. [DOI] [PubMed] [Google Scholar]
- [328].Ghosh S, Moore S, Bell RM, Dush M, Functional analysis of a phosphatidic acid binding domain in human Raf-1 kinase: mutations in the phosphatidate binding domain lead to tail and trunk abnormalities in developing zebrafish embryos, J. Biol. Chem 278 (46) (2003) 45690–45696. [DOI] [PubMed] [Google Scholar]
- [329].Ghosh S, Strum JC, Sciorra VA, Daniel L, Bell RM, Raf-1 kinase possesses distinct binding domains for phosphatidylserine and phosphatidic acid. Phosphatidic acid regulates the translocation of Raf-1 in 12–0-tetradecanoylphorbol-13-acetate-stimulated Madin-Darby canine kidney cells, J. Biol. Chem 271 (14) (1996) 8472–8480. [DOI] [PubMed] [Google Scholar]
- [330].Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman, et al. , Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway, J. Biol. Chem 274 (2) (1999) 1131–1139. [DOI] [PubMed] [Google Scholar]
- [331].Rizzo MA, Shome K, Watkins SC, Romero G, The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras, J. Biol. Chem 275 (31) (2000) 23911–23918. [DOI] [PubMed] [Google Scholar]
- [332].Nemoz G, Sette C, Conti M, Selective activation of rolipram-sensitive, cAMP-specific phosphodiesterase isoforms by phosphatidic acid, Mol. Pharmacol 51 (2) (1997) 242–249. [DOI] [PubMed] [Google Scholar]
- [333].Grange M, Sette C, Cuomo M, Conti M, Lagarde M, Prigent AF, et al. , The cAMP-62 specific phosphodiesterase PDE4D3 is regulated by phosphatidic acid binding. Consequences for cAMP signaling pathway and characterization of a phosphatidic acid binding site, J. Biol. Chem 275 (43) (2000) 33379–33387. [DOI] [PubMed] [Google Scholar]
- [334].Gillooly DJ, Melendez AJ, Hockaday AR, Harnett MM, Allen JM, Endoeytosis and vesicular trafficking of immune complexes and activation of phospholipase D by the human high-affinity IgG receptor requires distinct phosphoinositide 3-kinase activities, Biochem. J 344 (Pt 2) (1999) 605–611. [PMC free article] [PubMed] [Google Scholar]
- [335].Melendez A, Floto RA, Gillooly DJ, Harnett MM, Allen JM, FcgammaRI coupling to phospholipase D initiates sphingosine kinase-mediated calcium mobilization and vesicular trafficking, J. Biol. Chem 273 (16) (1998) 9393–9402. [DOI] [PubMed] [Google Scholar]
- [336].Humeau Y, Vitale N, Chasserot-Golaz S, Dupont JL, Du G, Frohman MA, et al. , A role for phospholipase D1 in neurotransmitter release, Proc. Natl. Acad. Sci. U. S. A 98 (26) (2001) 15300–15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Kaldi K, Szeberenyi J, Rada BK, Kovacs P, Geiszt M, Mocsai A, et al. , Contribution of phopholipase D and a brefeldin A-sensitive ARF to chemoattractant-induced superoxide production and secretion of human neutrophils, J. Leukoc. Biol 71 (4) (2002) 695–700. [PubMed] [Google Scholar]
- [338].Oh KJ, Lee SC, Choi HJ, Oh DY, Kim SC, Do SM, et al. , Role of phospholipase D2 in anti-apoptotic signaling through increased expressions of Bcl-2 and Bel-xL, J. Cell Biochem 101 (6) (2007) 1409–1422. [DOI] [PubMed] [Google Scholar]
- [339].Padron D, Tall RD, Roth MG, Phospholipase D2 is required for efficient endocytic recycling of transferrin receptors, Mol. Biol. Cell 17 (2) (2006) 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Koch T, Wu DF, Yang LQ, Brandenburg LO, Hollt V, Role of phospholipase D2 in the agonist-induced and constitutive endoeytosis of G-protein coupled receptors, J. Neurochem 97 (2) (2006) 365–372. [DOI] [PubMed] [Google Scholar]
- [341].Koch T, Brandenburg LO, Schulz S, Liang Y, Klein J, Hollt V, ADP-ribosylation factor-dependent phospholipase D2 activation is required for agonist-induced muopioid receptor endoeytosis, J. Biol. Chem 278 (11) (2003) 9979–9985. [DOI] [PubMed] [Google Scholar]
- [342].Banno Y, Nemoto S, Murakami M, Kimura M, Ueno Y, Ohguchi K, et al. , Depolarization-induced differentiation of PCI 2 cells is mediated by phospholipase D2 through the transcription factor CREB pathway, J. Neurochem 104 (5) (2008) 1372–1386. [DOI] [PubMed] [Google Scholar]
- [343].Watanabe H, Yamazaki M, Miyazaki H, Arikawa C, Itoh K, Sasaki T, et al. , Phospholipase D2 functions as a downstream signaling molecule of MAP kinase pathway in LI-stimulated neurite outgrowth of cerebellar granule neurons, J. Neurochem 89 (1) (2004) 142–151. [DOI] [PubMed] [Google Scholar]
- [344].Fisher GJ, Henderson PA, Voorhees JJ, Baldassare JJ, Epidermal growth factor- induced hydrolysis of phosphatidylcholine by phospholipase D and phospholipase C in human dermal fibroblasts, J. Cell. Physiol 146 (2) (1991) 309–317. [DOI] [PubMed] [Google Scholar]
- [345].Shome K, Rizzo MA, Vasudevan C, Andresen B, Romero G, The activation of phospholipase D by endothelin-1, angiotensin II, and platelet-derived growth factor in vascular smooth muscle A10 cells is mediated by small G proteins of the ADP-ribosylation factor family, Endocrinology 141 (6) (2000) 2200–2208. [DOI] [PubMed] [Google Scholar]
- [346].Wu DF, Yang LQ, Goschke A, Stumm R, Brandenburg LO, Liang YJ, et al. , Role of receptor internalization in the agonist-induced desensitization of cannabinoid type 1 receptors, J. Neurochem 104 (4) (2008) 1132–1143. [DOI] [PubMed] [Google Scholar]
- [347].Senogles SE, The D2s dopamine receptor stimulates phospholipase D activity: a novel signaling pathway for dopamine, Mol. Pharmacol 58 (2) (2000) 455–462. [DOI] [PubMed] [Google Scholar]
- [348].Ralevic Y, Burnstock G, Receptors for purines and pyrimidines, Pharmacol. Rev 50 (3) (1998) 413–492. [PubMed] [Google Scholar]
- [349].Neary JT, Kang Y, Bu Y, Yu E, Akong K, Peters CM, Mitogenic signaling by ATP/P2Y purinergic receptors in astrocytes: involvement of a calcium-independent protein kinase C, extracellular signal-regulated protein kinase pathway distinct from the phosphatidylinositol-specific phospholipase C/calcium pathway, J. Neurosci 19 (11) (1999) 4211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Berg KA, Evans KL, Cropper JD, Clarke WP, Temporal regulation of agonist efficacy at 5-hydroxytryptamine (5-HT)1A and 5-HT 1B receptors, J. Pharmacol. Exp. Ther 304 (1) (2003) 200–205. [DOI] [PubMed] [Google Scholar]
- [351].Antonescu CN, Danuser G, Schmid SL, Phosphatidic acid plays a regulatory role in clathrin-mediated endoeytosis, Mol. Biol. Cell 21 (16) (2010) 2944–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Jones D, Morgan C, Cockcroft S, Phospholipase D and membrane traffic. Potential roles in regulated exocytosis, membrane delivery and vesicle budding, Biochim. Biophys. Acta 1439 (2) (1999) 229–244. [DOI] [PubMed] [Google Scholar]
- [353].Shen Y, Xu L, Foster DA, Role for phospholipase D in receptor-mediated endocytosis, Mol. Cell. Biol 21 (2) (2001) 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [354].Koch T, Brandenburg LO, Liang Y, Schulz S, Beyer A, Schroder H, et al. , Phospholipase D2 modulates agonist-induced mu-opioid receptor desensitization and resensitization, J. Neurochem 88 (3) (2004) 680–688. [DOI] [PubMed] [Google Scholar]
- [355].Bhattacharya M, Babwah AV, Godin C, Anborgh PH, Dale LB, Poulter MO, et al. , Ral and phospholipase D2-dependent pathway for constitutive metabotropic glutamate receptor endoeytosis, J. Neurosci 24 (40) (2004) 8752–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [356].Lee JS, Kim IS, Kim JH, Cho W, Suh PG, Ryu SH, Determination of EGFR endoeytosis kinetic by auto-regulatory association of PLD1 with mu2, PLoS One 4 (9) (2009) e7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [357].Park MH, Choi KY, Min Do S, The pleckstrin homology domain of phospholipase D1 accelerates EGFR endoeytosis by increasing the expression of the Rab5 effector, rabaptin-5, Exp. Mol. Med 47 (2015) e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Shen Y, Zheng Y, Foster DA, Phospholipase D2 stimulates cell protrusion in v-Src- transformed cells, Biochem. Biophys. Res. Commun 293 (1) (2002) 201–206. [DOI] [PubMed] [Google Scholar]
- [359].Hatton N, Lintz E, Mahankali M, Henkels KM, Gomez-Cambronero J, Phosphatidic acid increases epidermal growth factor receptor expression by stabilizing mRNA decay and by inhibiting Lysosomal and proteasomal degradation of the internalized receptor, Mol. Cell. Biol 35 (18) (2015) 3131–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [360].Preininger AM, Henage LG, Oldham WM, Yoon EJ, Hamm HE, Brown HA, Direct modulation of phospholipase D activity by Gbetagamma, Mol. Pharmacol 70 (1) (2006) 311–318. [DOI] [PubMed] [Google Scholar]
- [361].Porat-Shliom N, Kloog Y, Donaldson JG, A unique platform for Η-Ras signaling involving clathrin-independent endoeytosis, Mol. Biol. Cell 19 (3) (2008) 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Scarselli M, Donaldson JG, Constitutive internalization of G protein-coupled receptors and G proteins via clathrin-independent endoeytosis, J. Biol. Chem 284 (6) (2009) 3577–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].Brandenburg LO, Pufe T, Koch T, Role of phospholipase d in g-protein coupled receptor function, Membranes (Basel) 4 (3) (2014) 302–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [364].Yang L, Seifert A, Wu D, Wang X, Rankovie V, Schroder H, et al. , Role of phospholipase D2/phosphatidic acid signal transduction in micro- and delta-opioid receptor endoeytosis, Mol. Pharmacol 78 (1) (2010) 105–113. [DOI] [PubMed] [Google Scholar]
- [365].Donaldson JG, Phospholipase D in endoeytosis and endosomal recycling pathways, Biochim. Biophys. Acta 1791 (9) (2009) 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].H Radhakrishna, Donaldson JG, ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway, J. Cell Biol 139 (1) (1997) 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Powelka AM, Sun J, Li J, Gao M, Shaw LM, Sonnenberg A, et al. , Stimulation-dependent recycling of integrin betal regulated by ARF6 and Rabll, Traffic 5 (1) (2004) 20–36. [DOI] [PubMed] [Google Scholar]
- [368].Brown FD, Rozelle AL, Yin HL, Balia T, Donaldson JG, Phosphatidylinositol 4,5- bisphosphate and Arf6-regulated membrane traffic, J. Cell Biol 154 (5) (2001) 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [369].Robertson SE, Setty SR, Sitaram A, Marks MS, Lewis RE, Chou MM, Extracellular signal-regulated kinase regulates clathrin-independent endosomal trafficking, Mol. Biol. Cell 17 (2) (2006) 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Vitale N, Chasserot-Golaz S, Bader MF, Regulated secretion in chromaffin cells: an essential role for ARF6-regulated phospholipase D in the late stages of exocytosis, Ann. N. Y. Acad. Sci 971 (2002) 193–200. [DOI] [PubMed] [Google Scholar]
- [371].Jovanovic OA, Brown FD, Donaldson JG, An effector domain mutant of Arf6 implicates phospholipase D in endosomal membrane recycling, Mol. Biol. Cell 17 (1) (2006) 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, et al. , EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization, EMBO J. 18 (6) (1999) 1480–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Chen YG, Siddhanta A, Austin CD, Hammond SM, Sung TC, Frohman MA, et al. , Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network, J. Cell Biol 138 (3) (1997) 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [374].Stutchfield J, Cockcroft S, Correlation between secretion and phospholipase D activation in differentiated HL60 cells, Biochem. J 293 (Pt 3) (1993) 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [375].Morgan CP, Sengelov H, Whatmore J, Borregaard N, Cockcroft S, ADP-ribosylation-factor-regulated phospholipase D activity localizes to secretory vesicles and mobilizes to the plasma membrane following N-formylmethionyl-leucyl-phenyla-lanine stimulation of human neutrophils, Biochem. J 325 (Pt 3) (1997) 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [376].Hitomi T, Zhang J, Nicoletti LM, Grodzki AC, Jamur MC, Oliver C, et al. , Phospholipase D1 regulates high-affinity IgE receptor-induced mast cell degranulation, Blood 104 (13) (2004) 4122–4128. [DOI] [PubMed] [Google Scholar]
- [377].Mead KI, Zheng Y, Manzotti CN, Perry LC, Liu MK, Burke F, et al. , Exocytosis of CTLA-4 is dependent on phospholipase D and ADP ribosylation factor-1 and stimulated during activation of regulatory T cells, J. Immunol 174 (8) (2005) 4803–4811. [DOI] [PubMed] [Google Scholar]
- [378].Cadwallader KA, Uddin M, Condliffe AM, Cowburn AS, White JF, Skepper JN, et al. , Effect of priming on activation and localization of phospholipase D-1 in human neutrophils, Eur. J. Biochem 271 (13) (2004) 2755–2764. [DOI] [PubMed] [Google Scholar]
- [379].Xie MS, Jacobs LS, Dubyak GR, Regulation of phospholipase D and primary granule secretion by P2-purinergic- and chemotactie peptide-receptor agonists is induced during granulocytic differentiation of HL-60 cells, J. Clin. Invest 88 (1) (1991) 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [380].Gruchalla RS, Dinh TT, Kennerly DA, An indirect pathway of receptor-mediated 1,2-diacyIglycerol formation in mast cells. I. IgE receptor-mediated activation of phospholipase D, J. Immunol 144 (6) (1990) 2334–2342. [PubMed] [Google Scholar]
- [381].Disse J, Vitale N, Bader MF, Gerke V, Phospholipase D1 is specifically required for regulated secretion of von Willebrand factor from endothelial cells, Blood 113 (4) (2009) 973–980. [DOI] [PubMed] [Google Scholar]
- [382].Liu L, Liao H, Castle, Zhang J, Casanova J, Szabo G, et al. , SCAMP2 interacts with Arf6 and phospholipase D1 and links their function to exocytotic fusion pore formation in PCI 2 cells, Mol. Biol. Cell 16 (10) (2005) 4463–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [383].Zeniou-Meyer M, Zabari N, Ashery U, Chasserot-Golaz S, Haeberle AM, Demais V, et al. , Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage, J. Biol. Chem 282 (30) (2007) 21746–21757. [DOI] [PubMed] [Google Scholar]
- [384].Marchini-Alves CM, Nicoletti LM, Mazucato VM, de Souza LB, Hitomi, Alves Cde P, et al. , Phospholipase D2: a pivotal player modulating RBL-2H3 mast cell structure, J. Histochem. Cytochem 60 (5) (2012) 386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [385].Lukowski S, Lecomte MC, Mira JP, Marin P, Gautero H, Russo-Marie F, et al. , Inhibition of phospholipase D activity by fodrin. An active role for the cytoskeleton, J. Biol. Chem 271 (39) (1996) 24164–24171. [DOI] [PubMed] [Google Scholar]
- [386].Gomez-Cambronero J, Di Fulvio M, Knapek K, Understanding phospholipase D (PLD) using leukocytes: PLD involvement in cell adhesion and chemotaxis, J. Leukoc. Biol 82 (2) (2007) 272–281. [DOI] [PubMed] [Google Scholar]
- [387].Lehman N, Di Fulvio M, McCray N, Campos I, Tabatabaian F, Gomez-Cambronero J, Phagocyte cell migration is mediated by phospholipases PLD1 and PLD2, Blood 108 (10) (2006) 3564–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [388].Gomez-Cambronero J, The exquisite regulation of PLD2 by a wealth of interacting proteins: S6K, Grb2, Sos, WASp and Rac2 (and a surprise discovery: PLD2 is a GEF), Cell. Signal 23 (12) (2011) 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [389].Gomez-Cambronero J, Phospholipase D in cell signaling: from a myriad of cell functions to cancer growth and metastasis, J. Biol. Chem 289 (33) (2014) 22557–22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].Ha KS, Exton JH, Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts, J. Cell Biol 123 (6 Pt 2) (1993) 1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Kam Y, Exton JH, Phospholipase D activity is required for actin stress fiber fomation in fibroblasts, Mol. Cell. Biol 21 (12) (2001) 4055–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [392].Powner DJ, Payne RM, Pettitt TR, Giudici ML, Irvine RF, Wakelam MJ, Phospholipase D2 stimulates integrin-mediated adhesion via phosphatidylinositol 4-phosphate 5-kinase Igamma b, J. Cell Sci 118 (Pt 13) (2005) 2975–2986. [DOI] [PubMed] [Google Scholar]
- [393].Speranza F, Mahankali M, Henkels KM, Gomez-Cambronero J, The molecular basis of leukocyte adhesion involving phosphatidic acid and phospholipase D, J. Biol. Chem 289 (42) (2014) 28885–28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [394].Iyer SS, Agrawal RS, Thompson CR, Thompson S, Barton JA, Kusner DJ, Phospholipase D1 regulates phagocyte adhesion, J. Immunol 176 (6) (2006) 3686–3696. [DOI] [PubMed] [Google Scholar]
- [395].Chae YC, Kim KL, Ha SH, Kim J, Suh PG, Ryu SH, Protein kinase Cdelta-mediated phosphorylation of phospholipase D controls integrin-mediated cell spreading, Mol. Cell. Biol 30 (21) (2010) 5086–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [396].Mikami K, Li L, Takahashi M, Saga N, Photosynthesis-dependent Ca2 + influx and functional diversity between phospholipases in the formation of cell polarity in migrating cells of red algae, Plant Signal. Behav 4 (9) (2009) 911–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Li L, Saga N, Mikami K, Ca2 + influx and phosphoinositide signalling are essential for the establishment and maintenance of cell polarity in monospores from the red alga Porphyra yezoensis, J. Exp. Bot 60 (12) (2009) 3477–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Zouwail S, Pettitt TR, Dove SK, Chibalina MV, Powner DJ, Haynes L, et al. , Phospholipase D activity is essential for actin localization and actin-based motility in Dictyostelium, Biochem. J 389 (Pt 1) (2005) 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [399].Santy LC, Casanova JE, Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D, J. Cell Biol 154 (3) (2001) 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [400].Chae YC, Kim JH, Kim KL, Kim HW, Lee HY, Heo WD, et al. , Phospholipase D activity regulates integrin-mediated cell spreading and migration by inducing GTP-Rac translocation to the plasma membrane, Mol. Biol. Cell 19 (7) (2008) 3111–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Kantonen S, Hatton N, Mahankali M, Henkels KM, Park H, Cox D, et al. , A novel phospholipase D2-Grb2-WASp heterotrimer regulates leukocyte phagocytosis in a two-step mechanism, Mol. Cell. Biol 31 (22) (2011) 4524–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [402].Arregui CO, Gonzalez A, Burdisso JE, Gonzalez Wusener AE, Protein tyrosine phosphatase PTP1B in cell adhesion and migration, Cell Adhes. Migr 7 (5) (2013) 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [403].Frondorf K, Henkels KM, Frohman MA, Gomez-Cambronero J, Phosphatidic acid is a leukocyte chemoattractant that acts through S6 kinase signaling, J. Biol. Chem 285 (21) (2010) 15837–15847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Lehman N, Ledford B, Di Fulvio M, Frondorf K, McPhail LC, Gomez-Cambronero J, Phospholipase D2-derived phosphatidic acid binds to and activates ribosomal p70 S6 kinase independently of mTOR, FASEB J. 21 (4) (2007) 1075–1087. [DOI] [PubMed] [Google Scholar]
- [405].Ling K, Doughman RL, Firestone AJ, Bunee MW, Anderson RA, Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions, Nature 420 (6911) (2002) 89–93. [DOI] [PubMed] [Google Scholar]
- [406].Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, et al. , Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of Talin, Nature 420 (6911) (2002) 85–89. [DOI] [PubMed] [Google Scholar]
- [407].Kusner DJ, Barton JA, Wen KK, Wang X, Rubenstein PA, Iyer SS, Regulation of phospholipase D activity by actin. Actin exerts bidirectional modulation of Mammalian phospholipase D activity in a polymerization-dependent, isoform-specific manner, J. Biol. Chem 277 (52) (2002) 50683–50692. [DOI] [PubMed] [Google Scholar]
- [408].Babior BM, Lambeth JD, Nauseef W, The neutrophil NADPH oxidase, Arch. Biochem. Biophys 397 (2) (2002) 342–344. [DOI] [PubMed] [Google Scholar]
- [409].Cummings R, Parinandi N, Wang L, Usatyuk P, Natarajan V, Phospholipase D/phosphatidic acid signal transduction: role and physiological significance in lung, Mol. Cell. Biochem 234–235 (1–2) (2002) 99–109. [PubMed] [Google Scholar]
- [410].Rossi F, Grzeskowiak M, Della Bianca V, Calzetti F, Gandini G, Phosphatidic acid and not diacylglycerol generated by phospholipase D is functionally linked to the activation of the NADPH oxidase by FMLP in human neutrophils, Biochem. Biophys. Res. Commun 168 (1) (1990) 320–327. [DOI] [PubMed] [Google Scholar]
- [411].Hall MN, mTOR-what does it do? Transplant. Proc 40 (10 Suppl) (2008) S5–S8. [DOI] [PubMed] [Google Scholar]
- [412].Ballou LM, Jiang YP, Du G, Frohman MA, Lin RZ, Ca(2 +)- and phospholipase D-dependent and -independent pathways activate mTOR signaling, FEBS Lett. 550 (1–3) (2003) 51–56. [DOI] [PubMed] [Google Scholar]
- [413].Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S, The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle, Proc. Natl. Acad. Sci. U. S. A 103 (12) (2006) 4741–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [414].O’Neil TK, Duffy LR, Frey JW, Hornberger TA, The role of phosphoinositide 3- kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions, J. Physiol 587 (Pt 14) (2009) 3691–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [415].You JS, Lincoln HC, Kim CR, Frey JW, Goodman CA, Zhong XP, et al. , The role of diacylglycerol kinase zeta and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy, J. Biol. Chem 289 (3) (2014) 1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [416].Chen J, Zheng XF, Brown EJ, Schreiber SL, Identification of an 11-kDa FKBP12- rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue, Proc. Natl. Acad. Sci. U. S. A 92 (11) (1995) 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [417].Yip CK, Murata K, Walz T, Sabatini DM, Kang SA, Structure of the human mTOR complex I and its implications for rapamycin inhibition, Mol. Cell 38 (5) (2010) 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [418].Laplante M, Sabatini DM, mTOR signaling at a glance, J. Cell Sci 122 (Pt 20) (2009) 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [419].Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA, Regulation of mTQRC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin, Mol. Cell. Biol 29 (6) (2009) 1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [420].Hornberger TA, Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle, Int. J. Biochem. Cell Biol 43 (9) (2011) 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Gangloff Y-G, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz J-F, et al. , Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development, Mol. Cell. Biol 24 (21) (2004) 9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [422].Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, et al. , Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1, Dev. Cell 11 (6) (2006) 859–871. [DOI] [PubMed] [Google Scholar]
- [423].Foster DA, Salloum D, Menon D, Frias MA, Phospholipase D and the maintenance of phosphatidic acid levels for regulation of mammalian target of rapamycin (mTOR), J. Biol. Chem 289 (33) (2014) 22583–22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [424].Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. , The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1, Science (New York, NY) 320 (5882) (2008) 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].Lagace TA, Ridgway ND, The role of phospholipids in the biological activity and structure of the endoplasmic reticulum, Biochim. Biophys. Acta 1833 (11) (2013) 2499–2510. [DOI] [PubMed] [Google Scholar]
- [426].Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM, Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids, Cell 141 (2) (2010) 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [427].Ward PS, Thompson CB, Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate, Cancer Cell 21 (3) (2012) 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [428].Sengupta S, Peterson TR, Sabatini DM, Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress, Mol. Cell 40 (2) (2010) 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [429].Foster DA, Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells, Biochim. Biophys. Acta 1791 (9) (2009) 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [430].Shi M, Zheng Y, Garcia A, Xu L, Foster DA, Phospholipase D provides a survival signal in human cancer cells with activated Η-Ras or Κ-Ras, Cancer Lett. 258 (2) 268–275, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [431].Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. , Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids, Proc. Natl. Acad. Sci. U. S. A 110 (22) (2013) 8882–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [432].Salloum D, Mukhopadhyay S, Tung K, Polonetskaya A, Foster DA, Mutant ras elevates dependence on serum lipids and creates a synthetic lethality for rapamycin, Mol. Cancer Ther 13 (3) (2014) 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [433].Zheng Y, Rodrik V, Toschi A, Shi M, Hui L, Shen Y, et al. , Phospholipase D couples survival and migration signals in stress response of human cancer cells, J. Biol. Chem 281 (23) (2006) 15862–15868. [DOI] [PubMed] [Google Scholar]
- [434].Marchesan D, Rutberg M, Andersson L, Asp L, Larsson T, Boren J, et al. , A phospholipase D-dependent process forms lipid droplets containing caveolin, adipocyte differentiation-related protein, and vimentin in a cell-free system, J. Biol. Chem 278 (29) (2003) 27293–27300. [DOI] [PubMed] [Google Scholar]
- [435].Asp L, Claesson C, Boren, Olofcson SO, ADP-ribosylation factor 1 and its activation of phospholipase D are important for the assembly of very low density lipoproteins, J. Biol. Chem 275 (34) (2000) 26285–26292. [DOI] [PubMed] [Google Scholar]
- [436].Olofsson SO, Asp L, Boren J, The assembly and secretion of apolipoprotein B-containing lipoproteins, Curr. Opin. Lipidol 10 (4) (1999) 341–346. [DOI] [PubMed] [Google Scholar]
- [437].Alexander CA, Hamilton RL, Havel RJ, Subcellular localization of B apoprotein of plasma lipoproteins in rat liver, J. Cell Biol 69 (2) (1976) 241–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [438].Andersson L, Bostrom P, Ericson J, Rutberg M, Magnusson B, Marchesan D, et al. , PLD1 and ERK2 regulate cytosolic lipid droplet formation, J. Cell Sci 119 (Pt 11) (2006) 2246–2257. [DOI] [PubMed] [Google Scholar]
- [439].Nakamura N, Banno Y, Tamiya-Koizumi K, Arf1-dependent PLD1 is localized to oleic acid-induced lipid droplets in NIH3T3 cells, Biochem. Biophys. Res. Commun 335 (1) (2005) 117–123. [DOI] [PubMed] [Google Scholar]
- [440].Andersson L, Bostrom P, Ericson J, Rutberg M, Magnusson B, Marchesan D, et al. , PLD1 and ERK2 regulate cytosolic lipid droplet formation, J. Cell Sci 119 (Pt 11) (2006) 2246–2257. [DOI] [PubMed] [Google Scholar]
- [441].Browning JD, Horton JD, Molecular mediators of hepatic steatosis and liver injury, J. Clin. Invest 114 (2) (2004) 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [442].den Boer M, Voshol PJ, Kuipers F, Havekes LM, Romijn JA, Hepatic steatosis: a mediator of the metabolic syndrome. Lessons from animal models, Arterioscler. Thromb. Vase. Biol 24 (4) (2004) 644–649. [DOI] [PubMed] [Google Scholar]
- [443].Plakkal Ayyappan J, Paul A, Goo YH, Lipid droplet-associated proteins in ather-osclerosis (review), Mol. Med. Rep 13 (6) (2016) 4527–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [444].Chen JS, Greenberg AS, Tseng YZ, Wang SM, Possible involvement of protein kinase C in the induction of adipose differentiation-related protein by Sterol ester in RAW 264.7 macrophages, J. Cell Biochem 83 (2) (2001) 187–199. [DOI] [PubMed] [Google Scholar]
- [445].Schonberger T, Jurgens T, Muller J, Armbruster N, Niermann C, Gorressen S, et al. , Pivotal role of phospholipase D1 in tumor necrosis factor-alpha-mediated inflammation and scar formation after myocardial ischemia and reperfusion in mice, Am. J. Pathol 184 (9) (2014) 2450–2464. [DOI] [PubMed] [Google Scholar]
- [446].Dhingra S, Rodriguez ME, Shen, Duan X, Stanton ML, Chen L, et al. , Constitutive activation with overexpression of the mTORC2-phospholipase D1 pathway in uterine leiomyosarcoma and STUMP: morphoproteomic analysis with therapeutic implications, Int. J. Clin. Exp. Pathol 4 (2) (2010) 134–146. [PMC free article] [PubMed] [Google Scholar]
- [447].Shen Q, Stanton ML, Feng W, Rodriguez ME, Ramondetta L, Chen L, et al. , Morphoproteomic analysis reveals an overexpressed and constitutively activated phospholipase D1-mTORC2 pathway in endometrial carcinoma, Int. J. Clin. Exp. Pathol 4 (1) (2010) 13–21. [PMC free article] [PubMed] [Google Scholar]
- [448].Vorland M, Holmsen H, Phospholipase D in human platelets: presence of isoenzymes and participation of autocrine stimulation during thrombin activation, Platelets 19 (3) (2008) 211–224. [DOI] [PubMed] [Google Scholar]
- [449].Sadler JE, Biochemistry and genetics of von Willebrand factor, Annu. Rev. Biochem 67 (1998) 395–424. [DOI] [PubMed] [Google Scholar]
- [450].Qin H, Frohman MA, Bollag WB, Phospholipase D2 mediates acute aldosterone secretion in response to angiotensin II in adrenal glomerulosa cells, Endocrinology 151 (5) (2010) 2162–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [451].Tsai YY, Rainey WE, Pan ZQ, Frohman MA, Choudhary V, Bollag WB, Phospholipase D activity underlies very-low-density lipoprotein (VLDL)-induced aldosterone production in adrenal glomerulosa cells, Endocrinology 155 (9) (2014) 3550–3560. [DOI] [PubMed] [Google Scholar]
- [452].Stegner D, Thielmann I, Kraft P, Frohman MA, Stoll G, Nieswandt B, Pharmacological inhibition of phospholipase D protects mice from occlusive thrombus formation and ischemic stroke-brief report, Arterioscler. Thromb. Vase. Biol 33 (9) (2013) 2212–2217. [DOI] [PubMed] [Google Scholar]
- [453].Wuescher LM, Takashima A, Worth RG, A novel conditional platelet depletion mouse model reveals the importance of platelets in protection against Staphylococcus aureus bacteremia, J. Thromb. Haemost 13 (2) (2015) 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [454].Ganesan R, Henkels KM, Wrenshall LE, Kanaho Y, Di Paolo G, Frohman MA, et al. , Oxidized LDL phagocytosis during foam cell formation in atherosclerotic plaques relies on a PLD2-CD36 functional interdependence, J. Leukoc. Biol 103 (5) (2018) 867–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [455].Taylor HE, Simmons GE Jr, Mathews TP, Khatua AK, Popik W, Lindsley CW, et al. , Phospholipase D1 couples CD4 + T cell activation to c-Myc-dependent deoxyribonucleotide Pool expansion and HIV-1 replication, PLoS Pathog. 11 (5) (2015) e1004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [456].Rouzer, Targeting Phospholipase D to Suppress HIV Infection, https://www.vanderbilt.edu/vicb/discovery_archives/targeting_phosD_suppress_HIV_infection.html, (2015) Accessed May 29, 2015.
- [457].Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, et al. , Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication, Nature 463 (7282) (2010) 818–822. [DOI] [PubMed] [Google Scholar]
- [458].Yamada Y, Hamajima N, Kato T, Iwata H, Yamamura Y, Shinoda M, et al. , Association of a polymorphism of the phospholipase D2 gene with the prevalence of colorectal cancer, J. Mol. Med. (Berl) 81 (2) (2003) 126–131. [DOI] [PubMed] [Google Scholar]
- [459].Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. , The genomic landscapes of human breast and colorectal cancers, Science 318 (5853) (2007) 1108–1113. [DOI] [PubMed] [Google Scholar]
- [460].Uchida N, Okamura S, Nagamachi Y, Yamashita S, Increased phospholipase D activity in human breast cancer, J. Cancer Res. Clin. Oncol 123 (5) (1997) 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [461].Uchida N, Okamura S, Kuwano H, Phospholipase D activity in human gastric carcinoma, Anticancer Res. 19 (1B) (1999) 671–675. [PubMed] [Google Scholar]
- [462].Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, et al. , Overexpression of phospholipase D1 in human breast cancer tissues, Cancer Lett. 161 (2) (2000) 207–214. [DOI] [PubMed] [Google Scholar]
- [463].Buchanan FG, McReynolds M, Couvillon A, Kam Y, Holla VR, Dubois RN, et al. , Requirement of phospholipase D1 activity in H-RasV12-induced transformation, Proc. Natl. Acad. Sci. U. S. A 102 (5) (2005) 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [464].Saito M, Iwadate M, Higashimoto M, Ono K, Takebayashi Y, Takenoshita S, Expression of phospholipase D2 in human colorectal carcinoma, Oncol. Rep 18 (5) (2007) 1329–1334. [PubMed] [Google Scholar]
- [465].Song JG, Pfeffer LM, Foster DA, v-Src increases diacylglycerol levels via a type D phospholipase-mediated hydrolysis of phosphatidylcholine, Mol, Cell. Biol 11 (1991) 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [466].Frankel P, Ramos M, Flom J, Bychenok S, Joseph T, Kerkhoff E, et al. , Ral and Rho-dependent activation of phospholipase D in v-Raf-transformed cells, Biochem. Biophys. Res. Commun 255 (2) (1999) 502–507. [DOI] [PubMed] [Google Scholar]
- [467].Jiang H, Lu Z, Luo JQ, Wolfman A, Foster DA, Ras mediates the activation of phospholipase D by v-Src, J. Biol. Chem 270 (11) (1995) 6006–6009. [DOI] [PubMed] [Google Scholar]
- [468].Jiang YW, Song J, Zang Q, Foster DA, Phosphatidylcholine-specific phospholipase D activity is elevated in v-Fps-transformed cells, Biochem. Biophys. Res. Commun 203 (2) (1994) 1195–2003. [DOI] [PubMed] [Google Scholar]
- [469].Park MH, Ahn BH, Hong YK, Do SM, Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-kappaB/Sp1-mediated signaling pathways, Carcinogenesis 30 (2) (2009) 356–365. [DOI] [PubMed] [Google Scholar]
- [470].Kang DW, Park MH, Lee YJ, Kim HS, Lindsley CW, Alex Brown H, et al. Autoregulation of phospholipase D activity is coupled to selective induction of phospholipase D1 expression to promote invasion of breast cancer cells, Int. J. Cancer 128 (4) (2011) 805–816. [DOI] [PubMed] [Google Scholar]
- [471].Williger BT, Ho WT, Exton JH, Phospholipase D mediates matrix metalloproteinase-9 secretion in phorbol ester-stimulated human fibrosarcoma cells, J. Biol. Chem 274 (2) (1999) 735–738. [DOI] [PubMed] [Google Scholar]
- [472].Moolenaar WH, Kruijer W, Tilly BC, Verlaan I, Bierman AJ, de Laat SW, Growth factor-like action of phosphatidic acid, Nature 323 (6084) (1986) 171–173. [DOI] [PubMed] [Google Scholar]
- [473].Rodrik Y, Zheng Y, Harrow F, Chen Y, Foster DA, Survival signals generated by estrogen and phospholipase D in MCF-7 breast cancer cells are dependent on Myc, Mol. Cell. Biol 25 (17) (2005) 7917–7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [474].Hui L, Rodrik V, Pielak RM, Knirr S, Zheng Y, Foster DA, mTOR-dependent suppression of protein phosphatase 2A is critical for phospholipase D survival signals in human breast cancer cells, J. Biol. Chem 280 (43) (2005) 35829–35835. [DOI] [PubMed] [Google Scholar]
- [475].Hui L, Zheng Y, Yan Y, Bargonetti J, Foster DA, Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D, Oncogene 25 (55) (2006) 7305–7310. [DOI] [PubMed] [Google Scholar]
- [476].Chen Y, Rodrik V, Foster DA, Alternative phospholipase D/mTOR survival signal in human breast cancer cells, Oncogene 24 (4) (2005) 672–679. [DOI] [PubMed] [Google Scholar]
- [477].Chen Y, Zheng Y, Foster DA, Phospholipase D confers rapamycin resistance in human breast cancer cells, Oncogene 22 (25) (2003) 3937–3942. [DOI] [PubMed] [Google Scholar]
- [478].Bacac M, Stamenkovic I, Metastatic cancer cell, Annu. Rev. Pathol 3 (2008) 221–247. [DOI] [PubMed] [Google Scholar]
- [479].Zhang Y, Frohman MA, Cellular and physiological roles for phospholipase D1 in cancer, J. Biol. Chem 289 (33) (2014) 22567–22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [480].Wang XX, Liao Y, Hong L, Zeng Z, Yuan TB, Xia X, et al. , Tissue microarray staining reveals PLD1 and Sp1 have a collaborative, pro-tumoral effect in patients with osteosarcomas, Oncotarget 8 (43) (2017) 74340–74347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [481].Yoshida M, Okamura S, Kodaki T, Mori M, Yamashita S, Enhanced levels of oleate-dependent and Arf-dependent phospholipase D isoforms in experimental colon cancer, Oncol. Res 10 (8) (1998) 399–406. [PubMed] [Google Scholar]
- [482].Su W, Chen Q, Frohman MA, Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis, Future Oncol. 5 (9) (2009) 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [483].Labelle M, Begum S, Hynes RO, Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis, Cancer Cell 20 (5) (2011) 576–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [484].Nieswandt B, Hafner M, Echtenacher B, Mannel DN, Lysis of tumor cells by natural killer cells in mice is impeded by platelets, Cancer Res. 59 (6) (1999) 1295–1300. [PubMed] [Google Scholar]
- [485].Im JH, Fu W, Wang H, Bhatia SK, Hammer DA, Kowalska MA, et al. , Coagulation facilitates tumor cell spreading in the pulmonary vasculature during early metastatic colony formation, Cancer Res. 64 (23) (2004) 8613–8619. [DOI] [PubMed] [Google Scholar]
- [486].Camerer E, Qazi AA, Duong DN, Cornelissen L, Advincula R, Coughlin SR, Platelets, protease-activated receptors, and fibrinogen in hematogenous metastasis, Blood 104 (2) (2004) 397–401. [DOI] [PubMed] [Google Scholar]
- [487].Zhou BH, Chen JS, Chai MQ, Zhao S, Liang J, Chen HH, et al. , Activation of phospholipase D activity in transforming growth factor-beta-induced cell growth inhibition, Cell Res. 10 (2) (2000) 139–149. [DOI] [PubMed] [Google Scholar]
- [488].Zeng XX, Zheng X, Xiang Y, Cho HP, Jessen JR, Zhong TP, et al. , Phospholipase D1 is required for angiogenesis of intersegmental blood vessels in zebrafish, Dev. Biol 328 (2) (2009) 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [489].Oshimoto H, Okamura S, Yoshida M, Mori M, Increased activity and expression of phospholipase D2 in human colorectal cancer, Oncol. Res 14 (1) (2003) 31–37. [DOI] [PubMed] [Google Scholar]
- [490].Knoepp SM, Chahal MS, Xie Y, Zhang Z, Brauner DJ, Hallman MA, et al. , Effects of active and inactive phospholipase D2 on signal transduction, adhesion, migration, invasion, and metastasis in EL4 lymphoma cells, Mol. Pharmacol 74 (3) (2008) 574–584. [DOI] [PubMed] [Google Scholar]
- [491].Henkels KM, Boivin GP, Dudley ES, Berberich SJ, Gomez-Cambronero J, Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model, Oncogene 32 (49) (2013) 5551–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [492].Ganesan R, Mallets E, Gomez-Cambronero J, The transcription factors Slug (SNAI2) and Snail (SNAI1) regulate phospholipase D (PLD) promoter in opposite ways towards cancer cell invasion, Mol. Oncol 10 (5) (2016) 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [493].Fite K, Gomez-Cambronero J, Down-regulation of MicroRNAs (MiRs) 203, 887, 3619 and 182 prevents Vimentin-triggered, phospholipase D (PLD)-mediated cancer cell invasion, J. Biol. Chem 291 (2) (2016) 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [494].Lee CS, Ghim J, Song P, Suh PG, Ryu SH, Loss of phospholipase D2 impairs VEGF-induced angiogenesis, BMB Rep. 49 (3) (2016) 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [495].Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L, VEGF receptor signalling - in control of vascular function, Nat. Rev. Mol. Cell Biol 7 (5) (2006) 359–371. [DOI] [PubMed] [Google Scholar]
- [496].Ferrara N, Frantz G, LeCouter J, Dillard-Telm L, Pham T, Draksharapu A, et al. , Differential expression of the angiogenic factor genes vascular endothelial growth factor (VEGF) and endocrine gland-derived VEGF in normal and polycystic human ovaries, Am. J. Pathol 162 (6) (2003) 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [497].Ferrara N, Gerber HP, LeCouter J, The biology of VEGF and its receptors, Nat. Med 9 (6) (2003) 669–676. [DOI] [PubMed] [Google Scholar]
- [498].Carmeliet P, Jain RK, Molecular mechanisms and clinical applications of angiogenesis, Nature 473 (7347) (2011) 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [499].Behl T, Kotwani A, Exploring the various aspects of the pathological role of vascular endothelial growth factor (VEGF) in diabetic retinopathy, Pharmacol. Res 99 (2015) 137–148. [DOI] [PubMed] [Google Scholar]
- [500].Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA, Vascular endothelial growth factor and angiogenesis, Pharmacol. Rev 56 (4) (2004) 549–580. [DOI] [PubMed] [Google Scholar]
- [501].Ghim J, Moon JS, Lee CS, Lee J, Song P, Lee A, et al. , Endothelial deletion of phospholipase D2 reduces hypoxic response and pathological angiogenesis, Arterioscler. Thromb. Vase. Biol 34 (8) (2014) 1697–1703. [DOI] [PubMed] [Google Scholar]
- [502].Kandori S, Kojima T, Matsuoka T, Yoshino T, Sugiyama A, Nakamura E, et al. , Phospholipase D2 promotes disease progression of renal cell carcinoma through the induction of angiogenin, Cancer Sci. 109 (6) (2018) 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [503].Ye Q, Kantonen S, Henkels KM, Gomez-Cambronero J, A new signaling pathway (JAK-Fes-phospholipase D) that is enhanced in highly proliferative breast cancer cells, J. Biol. Chem 288 (14) (2013) 9881–9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [504].Nagasaki A, Uyeda TQ, Screening of genes involved in cell migration in Dictyostelium, Exp. CeU Res 314 (5) (2008) 1136–1146. [DOI] [PubMed] [Google Scholar]
- [505].Carrigan SO, Pink DB, Stadnyk AW, Neutrophil transepithelial migration in response to the chemoattractant fMLP but not C5a is phospholipase D-dependent and related to the use of CD11b/CD18, J. Leukoc. Biol 82 (6) (2007) 1575–1584. [DOI] [PubMed] [Google Scholar]
- [506].Mazie AR, Spix JK, Block ER, Achebe HB, Klarlund JK, Epithelial cell motility is triggered by activation of the EGF receptor through phosphatidic acid signaling, J. Cell Sci 119 (Pt 8) (2006) 1645–1654. [DOI] [PubMed] [Google Scholar]
- [507].Block ER, Klarlund JK, Wounding sheets of epithelial cells activates the epidermal growth factor receptor through distinct short- and long-range mechanisms, Mol. Biol. Cell 19 (11) (2008) 4909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [508].Pilquil C, Dewald J, Cherney A, Gorshkova I, Tigyi G, English D, et al. , Lipid phosphate phosphatase-1 regulates lysophosphatidate-induced fibroblast migration by controlling phospholipase D2-dependent phosphatidate generation, J. Biol. Chem 281 (50) (2006) 38418–38429. [DOI] [PubMed] [Google Scholar]
- [509].Knapek K, Frondorf K, Post J, Short S, Cox D, Gomez-Cambronero J, The molecular basis of phospholipase D2-induced chemotaxis: elucidation of differential pathways in macrophages and fibroblasts, Mol. Cell. Biol 30 (18) (2010) 4492–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [510].Ngo Thai Bich V, Hongu T, Miura Y, Katagiri N, Ohbayashi N, Yamashita-Kanemaru Y, et al. , Physiological function of phospholipase D2 in anti-tumor immunity: regulation of CD8(+) T lymphocyte proliferation, Sci. Rep 8 (1) (2018) 6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [511].Saito M, Kanfer J, Solubilization and properties of a membrane-bound enzyme from rat brain catalyzing a base-exchange reaction, Biochem. Biophys. Res. Commun 53 (2) (1973) 391–398. [DOI] [PubMed] [Google Scholar]
- [512].Kobayashi M, Kanfer JN, Phosphatidylethanol formation via transphosphatidylation by rat brain synaptosomal phospholipase D, J. Neurochem 48 (5) (1987) 1597–1603. [DOI] [PubMed] [Google Scholar]
- [513].Kotter K, Klein J, Adrenergic modulation of astroglial phospholipase D activity and cell proliferation, Brain Res. 830 (1) (1999) 138–145. [DOI] [PubMed] [Google Scholar]
- [514].Guizzetti M, Costa P, Peters J, Costa LG, Acetylcholine as a mitogen: muscarinic receptor-mediated proliferation of rat astrocytes and human astrocytoma cells, Eur. J. Pharmacol 297 (3) (1996) 265–273. [DOI] [PubMed] [Google Scholar]
- [515].Klein J, Functions and pathophysiological roles of phospholipase D in the brain, J. Neurochem 94 (6) (2005) 1473–1487. [DOI] [PubMed] [Google Scholar]
- [516].Kotter K, Klein J, Ethanol inhibits astroglial cell proliferation by disruption of phospholipase D-mediated signaling, J. Neurochem 73 (6) (1999) 2517–2523. [DOI] [PubMed] [Google Scholar]
- [517].Kanaho Y, Funakoshi Y, Hasegawa H, Phospholipase D signalling and its involvement in neunte outgrowth, Biochim. Biophys. Acta 1791 (9) (2009) 898–904. [DOI] [PubMed] [Google Scholar]
- [518].Watanabe H, Yokozeki T, Yamazaki M, Miyazaki H, Sasaki T, Maehama, et al. , Essential role for phospholipase D2 activation downstream of ERK MAP kinase in nerve growth factor-stimulated neurite outgrowth from PCI2 cells, J. Biol. Chem 279 (36) (2004) 37870–37877. [DOI] [PubMed] [Google Scholar]
- [519].Guizzetti M, Moore NH, Giordano G, Costa LG, Modulation of neuritogenesis by astrocyte muscarinic receptors, J. Biol. Chem 283 (46) (2008) 31884–31897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [520].Saito S, Sakagami H, Kondo H, Localization of mRNAs for phospholipase D (PLD) type 1 and 2 in the brain of developing and mature rat, Brain Res. Dev. Brain Res 120 (1) (2000) 41–47. [DOI] [PubMed] [Google Scholar]
- [521].Duan S, Huang RS, Zhang W, Bleibel WK, Roe CA, Clark TA, et al. , Genetic architecture of transcript-level variation in humans, Am. J. Hum. Genet 82 (5) 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [522].Armey R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, et al. , A genome-wide scan for common alleles affecting risk for autism, Hum. Mol. Genet 19 (20) (2010) 4072–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [523].Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, et al. , Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans, Proc. Natl. Acad. Sci. U. S. A 106 (39) (2008) 16746–16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [524].Debette S, Bis JC, Fomage M, Schmidt H, Ikram MA, Sigurdsson S, et al. , Genome-wide association studies of MRI-defined brain infarcts: meta-analysis from the CHARGE Consortium, Stroke 41 (2) (2010) 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [525].Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. , Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis, Hum. Mol. Genet 18 (4) (2009) 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [526].Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, et al. , Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD, Sci. Transi. Med 3 (95) (2011) 95ra75. [DOI] [PubMed] [Google Scholar]
- [527].Curran S, Bolton P, Rozsnyai K, Chiocchetti A, Klauck SM, Duketis E, et al. , No association between a common single nucleotide polymorphism, rs4141463, in the MACROD2 gene and autism spectrum disorder, Am. J. Med. Genet. B Neuropsychiatr. Genet 156B (6) (2011) 633–639. [DOI] [PubMed] [Google Scholar]
- [528].Jin JK, Ahn BH, Na YJ, Kim JI, Kim YS, Choi EK, et al. , Phospholipase D1 is associated with amyloid precursor protein in Alzheimer’s disease, Neurobiol. Aging 28 (7) (2007) 1015–1027. [DOI] [PubMed] [Google Scholar]
- [529].Haass C, Selkoe DJ, Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide, Nat. Rev. Mol. Cell. Biol 8 (2) (2007) 101–112. [DOI] [PubMed] [Google Scholar]
- [530].Vassar R, Kovacs DM, Yan R, Wong PC, The beta-secretase enzyme BACE in health and Alzheimer’s disease: regulation, cell biology, function, and therapeutic potential, J. Neurosci 29 (41) (2009) 12787–12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [531].De Strooper B, Vassar R, Golde T, The secretases: enzymes with therapeutic potential in Alzheimer disease, Nat. Rev. Neurol 6 (2) (2010) 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [532].Small SA, Gandy S, Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis, Neuron 52 (1) (2006) 15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [533].Cox DA, Cohen ML, Amyloid beta-induced neurotoxicity is associated with phospholipase D activation in cultured rat hippocampal cells, Neurosci. Lett 229 (1) (1997) 37–40. [DOI] [PubMed] [Google Scholar]
- [534].Lee MJ, Oh JY, Park HT, Uhlinger DJ, Kwak JY, Enhancement of phospholipase D activity by overexpression of amyloid precursor protein in PI 9 mouse embryonic carcinoma cells, Neurosci. Lett 315 (3) (2001) 159–163. [DOI] [PubMed] [Google Scholar]
- [535].Kanfer JN, Hattori H, Orihel D, Reduced phospholipase D activity in brain tissue samples from Alzheimer’s disease patients, Ann. Neurol 20 (2) (1986) 265–267. [DOI] [PubMed] [Google Scholar]
- [536].Kanfer JN, Sorrentino G, Sitar DS, Phospholipases as mediators of amyloid beta peptide neurotoxicity: an early event contributing to neurodegeneration characteristic of Alzheimer’s disease, Neurosci. Lett 257 (2) (1998) 93–96. [DOI] [PubMed] [Google Scholar]
- [537].Singh IN, Sato K, Takashima A, Kanfer JN, Activation of LA-N-2 cell phospholipases by single alanine substitution analogs of amyloid beta peptide (25–35), FEBS Lett 405 (1) (1997) 65–67. [DOI] [PubMed] [Google Scholar]
- [538].Singh IN, Sorrentino G, Kanfer JN, Activation of LA-N-2 cell phospholipase D by amyloid beta protein (25–35), Neurochem. Res 23 (10) (1998) 1225–1232. [DOI] [PubMed] [Google Scholar]
- [539].Singh IN, Sorrentino G, Sitar DS, Kanfer JN, Indomethacin and nordihydroguaiaretic acid inhibition of amyloid beta protein (25–35) activation of phospholipases A2 and D of LA-N-2 cells, Neurosci. Lett 222 (1) (1997) 5–8. [DOI] [PubMed] [Google Scholar]
- [540].Raghu P, Coessens E, Manifava M, Georgiev P, Pettitt T, Wood E, et al. , Rhabdomere biogenesis in Drosophila photoreceptors is acutely sensitive to phosphatidic acid levels, J. Cell Biol 185 (1) (2009) 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [541].Leong SL, Cappai R, Barnham KJ, Pham CL, Modulation of alpha-synuclein aggregation by dopamine: a review, Neurochem. Res 34 (10) (2009) 1838–1846. [DOI] [PubMed] [Google Scholar]
- [542].R Kruger, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, et al. , Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease, Nat. Genet 18 (2) (1998) 106–108. [DOI] [PubMed] [Google Scholar]
- [543].Polymeropoulos MH, Autosomal dominant Parkinson’s disease and alpha-synuclein, Ann. Neurol 44 (3 Suppl 1) (1998) S63–S64. [DOI] [PubMed] [Google Scholar]
- [544].Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. , Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease, Science 276 (5321) (1997) 2045–2047. [DOI] [PubMed] [Google Scholar]
- [545].Payton JE, Perrin RJ, Woods WS, George JM, Structural determinants of PLD2 inhibition by alpha-synuclein, J. Mol. Biol 337 (4) (2004) 1001–1009. [DOI] [PubMed] [Google Scholar]
- [546].Ahn BH, Rhim H, Kim SY, Sung YM, Lee MY, Choi JY, et al. , alpha-Synuclein interacts with phospholipase D isozymes and inhibits pervanadate-induced phospholipase D activation in human embryonic kidney-293 cells, J. Biol. Chem 277 (2002) 12334–12342. [DOI] [PubMed] [Google Scholar]
- [547].Jenco JM, Rawlingson A, Daniels B Morris AJ, Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by alpha- and beta-synucleins, Biochemistry 37 (14) (1998) 4901–4909. [DOI] [PubMed] [Google Scholar]
- [548].Leng Y, Chase TN, Bennett MC, Muscarinic receptor stimulation induces translocation of an alpha-synuclein oligomer from plasma membrane to a light vesicle fraction in cytoplasm, J. Biol. Chem 276 (30) (2001) 28212–28218. [DOI] [PubMed] [Google Scholar]
- [549].Rappley I, Gitler AD, Selvy PE, LaVoie MJ, Levy BD, Brown HA, et al. , Evidence that alpha-synuclein does not inhibit phospholipase D, Biochemistry 48 (5) (2009) 1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [550].Bukhardt U, Klein J, Phospholipase D: A central switch in the development of Fetal alcohol Spectrum disorder, Neuropathy of Drug Addictions and Substance Misuse, Academic Press, 2016, pp. 488–499. [Google Scholar]
- [551].Clarren SK, Sampson PD, Larsen J, Donnell DJ, Barr HM, Bookstein FL, et al. , Facial effects of fetal alcohol exposure: assessment by photographs and morphometric analysis, Am. J. Med. Genet 26 (3) (1987) 651–666. [DOI] [PubMed] [Google Scholar]
- [552].Lundqvist C, Rodriguez FD, Simonsson P, Ailing C, Gustavsson L, Phosphatidylethanol affects inositol 1,4,5-trisphosphate levels in NG108–15 neuroblastoma X glioma hybrid cells, J. Neurochem 60 (2) (1993) 738–744. [DOI] [PubMed] [Google Scholar]
- [553].Rodriguez FD, Lundqvist C, Ailing C, Gustavsson L, Ethanol and phosphatidylethanol reduce the binding of [3H]inositol 1,4,5-trisphosphate to rat cerebellar membranes, Alcohol Alcohol. 31 (5) (1996) 453–461. [DOI] [PubMed] [Google Scholar]
- [554].Standaert ML, Bandyopadhyay G, Zhou X, Galloway L, Farese RV, Insulin stimulates phospholipase D-dependent phosphatidylcholine hydrolysis, Rho translocation, de novo phospholipid synthesis, and diacylglycerol/protein kinase G signaling in L6 myotubes, Endocrinology 137 (7) (1996) 3014–3020. [DOI] [PubMed] [Google Scholar]
- [555].Babenko NA, Kharchenko VS, Modulation of insulin sensitivity of hepatocytes by the pharmacological downregulation of phospholipase D, Int. J. Endocrinol 2015 (2015) 794838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [556].Slaaby R, Du G, Altshuller YM, Frohman MA, Seedorf K, Insulin-induced phospholipase D1 and phospholipase D2 activity in human embryonic kidney-293 cells mediated by the phospholipase C gamma and protein kinase C alpha signaling cascade, Biochem. J 351 (Pt 3) (2000) 613–619. [PMC free article] [PubMed] [Google Scholar]
- [557].Zhang C, Wendel AA, Keogh MR, Harris TE, Chen J, Coleman RA, Glycerolipid signals alter mTQR complex 2 (mTORC2) to diminish insulin signaling, Proe. Natl. Acad. Sci. U. S. A 109 (5) (2012) 1667–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [558].Doti N, Cassese A, Marasco D, Paturzo F, Sabatella M, Viparelli F, et al. , Residues 762–801 of PLD1 mediate the interaction with PED/PEA15, Mol. BioSyst 6 (10) (2010) 2039–2048. [DOI] [PubMed] [Google Scholar]
- [559].Greig FH, Nixon GF, Phosphoprotein enriched in astrocytes (PEA)-15: a potential therapeutic target in multiple disease states, Pharmacol. Ther 143 (3) (2014) 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [560].Viparelli F, Cassese A, Doti N, Paturzo F, Marasco D, Dathan NA, et al. , Targeting of PED/PEA-15 molecular interaction with phospholipase D1 enhances insulin sensitivity in skeletal muscle cells, J. Biol. Chem 283 (31) (2008) 21769–21778. [DOI] [PubMed] [Google Scholar]
- [561].Zhang Y, Redina O, Altshuller YM, Yamazaki M, Ramos J, Chneiweiss H, et al. , Regulation of expression of phospholipase D1 and D2 by PEA-15, a novel protein that interacts with them, J. Biol. Chem 275 (45) (2000) 35224–35232. [DOI] [PubMed] [Google Scholar]
- [562].Sulzmaier FJ, Valmiki MK, Nelson DA, Caliva MJ, Geerts D, Matter ML, et al. , PEA-15 potentiates H-Ras-mediated epithelial cell transformation through phospholipase D, Oncogene 31 (30) (2012) 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [563].Thorens B, Facilitated glucose transporters in epithelial cells, Annu. Rev. Physiol 55 (1993) 591–608. [DOI] [PubMed] [Google Scholar]
- [564].Burant CF, Sivitz WI, Fukumoto H, Kayano T, Nagamatsu S, Seino S, et al. , Mammalian glucose transporters: structure and molecular regulation, Recent Prog. Horm. Res 47 (1991) 349–387 [discussion 387–8]. [DOI] [PubMed] [Google Scholar]
- [565].Condorelli G, Vigliotta G, Iavarone C, Caruso M, Tocchetti CG, Andreozzi F, et al. , PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus, EMBO J. 17 (14) (1998) 3858–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [566].Sajan MP, Bandyopadhyay G, Kanoh Y, Standaert ML, Quon MJ, Reed BC, et al. , Sorbitol activates atypical protein kinase C and GLUT4 glucose transporter translocation/glucose transport through proline-rich tyrosine kinase-2, the extra-cellular signal-regulated kinase pathway and phospholipase D, Biochem. J 362 (Pt 3) (2002) 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [567].Chen HC, Bandyopadhyay G, Sajan MP, Kanoh Y, Standaert M, Farese RV Jr et al. , Activation of the ERK pathway and atypical protein kinase C isoforms in exercise- and aminoimidazole-4-carboxamide-1-beta-D-riboside (AICAR)-stimulated glucose transport, J. Biol. Chem 277 (26) (2002) 23554–23562. [DOI] [PubMed] [Google Scholar]
- [568].Emoto M, Klarlund JK, Waters SB, Hu V, Buxton JM, Chawla A, et al. , A role for phospholipase D in GLUT4 glucose transporter translocation, J. Biol. Chem 275 (2000) 7144–7151. [DOI] [PubMed] [Google Scholar]
- [569].Condorelli G, Vigliotta G, Trencia A, Maitan MA, Caruso M, Miele C, et al. , Protein kinase C (PKC)-alpha activation inhibits PKC-zeta and mediates the action of PED/PEA-15 on glucose transport in the L6 skeletal muscle cells, Diabetes 50 (6) (2001) 1244–1252. [DOI] [PubMed] [Google Scholar]
- [570].Condorelli G, Vigliotta G, Trencia A, Maitan MA, Caruso M, Miele C, et al. , Expression of concern. Protein kinase C (PKC)-alpha activation inhibits PKC-zeta and mediates the action of PED/PEA-15 on glucose transport in the L6 skeletal muscle cells, Diabetes 50 (2001) 1244–1252, 10.2337/diabetes.50.6.1244. [DOI] [PubMed] [Google Scholar]
- [571].Fiory F, Formisano P, Perruolo G, Beguinot F, Frontiers: PED/PEA-15, a multi-functional protein controlling cell survival and glucose metabolism, Am. J. Physiol. Endocrinol. Metab 297 (3) (2009) E592–E601. [DOI] [PubMed] [Google Scholar]
- [572].Cassese A, Raciti GA, Fiory F, Nigro C, Ulianich L, Castano I, et al. , Adenoviral gene transfer of PLD1-D4 enhances insulin sensitivity in mice by disrupting phospholipase D1 interaction with PED/PEA-15, PLoS One 8 (4) (2013) e60555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [573].Oriente F, lovino S, Cassese A, Romano C, Miele C, Troncone G, et al. , Overproduction of phosphoprotein enriched in diabetes (PED) induces mesangial expansion and upregulates protein kinase C-beta activity and TGF-betal expression, Diabetologia 52 (12) (2009) 2642–2652. [DOI] [PubMed] [Google Scholar]
- [574].lovino S, Oriente F, Botta G, Cabaro S, Iovane V, Paciello O, et al. , PED/PEA-15 induces autophagy and mediates TGF-betal effect on muscle cell differentiation, Cell Death Differ. 19 (7) (2012) 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [575].Lamb CA, Dooley HC, Tooze SA, Endocytosis and autophagy: shared machinery for degradation, Bioessays 35 (1) (2013) 34–45. [DOI] [PubMed] [Google Scholar]
- [576].Seglen PO, Bohley P, Autophagy and other vacuolar protein degradation mechanisms, Experientia 48 (2) (1992) 158–172. [DOI] [PubMed] [Google Scholar]
- [577].Barth JM, Kohler K, How to take autophagy and endocytosis up a notch, Biomed. Res. Int 2014 (2014) 960803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [578].Cai M, He J, Xiong J, Tay LW, Wang Z, Rog C, et al. , Phospholipase D1-regulated autophagy supplies free fatty acids to counter nutrient stress in cancer cells, Cell Death Dis. 7 (11) (2016) e2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [579].Yang J-S, Lu C-C, Kuo S-C, Hsu Y-M, Tsai S-C, Chen S-Y, et al. , Autophagy and its link to type II diabetes mellitus, Biomedicine (Taipei) 7 (2) (2017), 10.1051/bmdcn/2017070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [580].Bourdon DM, Wing MR, Edwards EB, Sondek J, Harden TK, Quantification of iozyme-specific activation of phospholipase C-beta2 by Rac GTPases and phospholipase C-epsilon by Rho GTPases in an intact cell assay system, Methods Enzymol 406 (2006) 489–499. [DOI] [PubMed] [Google Scholar]
- [581].Nelson RK, Ya-Ping J, Gadbery J, Abedeen D, Sampson N, Lin RZ, et al. , Phospholipase D2 loss results in increased blood pressure via inhibition of the endothelial nitric oxide synthase pathway, Scientific reports 7 (2017) 9112,, 10.1038/s41598-017-09852-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


