Abstract
Background
COVID-19 and pneumonia of other etiology share similar CT characteristics, contributing to the challenges in differentiating them with high accuracy.
Purpose
To establish and evaluate an artificial intelligence (AI) system in differentiating COVID-19 and other pneumonia on chest CT and assess radiologist performance without and with AI assistance.
Methods
521 patients with positive RT-PCR for COVID-19 and abnormal chest CT findings were retrospectively identified from ten hospitals from January 2020 to April 2020. 665 patients with non-COVID-19 pneumonia and definite evidence of pneumonia on chest CT were retrospectively selected from three hospitals between 2017 and 2019. To classify COVID-19 versus other pneumonia for each patient, abnormal CT slices were input into the EfficientNet B4 deep neural network architecture after lung segmentation, followed by two-layer fully-connected neural network to pool slices together. Our final cohort of 1,186 patients (132,583 CT slices) was divided into training, validation and test sets in a 7:2:1 and equal ratio. Independent testing was performed by evaluating model performance on separate hospitals. Studies were blindly reviewed by six radiologists without and then with AI assistance.
Results
Our final model achieved a test accuracy of 96% (95% CI: 90-98%), sensitivity 95% (95% CI: 83-100%) and specificity of 96% (95% CI: 88-99%) with Receiver Operating Characteristic (ROC) AUC of 0.95 and Precision-Recall (PR) AUC of 0.90. On independent testing, our model achieved an accuracy of 87% (95% CI: 82-90%), sensitivity of 89% (95% CI: 81-94%) and specificity of 86% (95% CI: 80-90%) with ROC AUC of 0.90 and PR AUC of 0.87. Assisted by the models’ probabilities, the radiologists achieved a higher average test accuracy (90% vs. 85%, Δ=5, p<0.001), sensitivity (88% vs. 79%, Δ=9, p<0.001) and specificity (91% vs. 88%, Δ=3, p=0.001).
Conclusion
AI assistance improved radiologists' performance in distinguishing COVID-19 from non-COVID-19 pneumonia on chest CT.
Summary
AI assistance improved radiologists’ performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT.
Key Results:
■ An AI model had higher test accuracy (96% vs 85%, p<0.001), sensitivity (95% vs 79%, p<0.001), and specificity (96% vs 88%, p=0.002) than radiologists.
■ In an independent test set, our AI model achieved an accuracy of 87%, sensitivity of 89% and specificity of 86%.
■ With AI assistance, the radiologists achieved a higher average accuracy (90% vs 85%, p<0.001), sensitivity (88% vs 79%, p<0.001) and specificity (91% vs 88%, p=0.001).
Introduction
It has been hypothesized that COVID-19 infection is difficult to contain because of its potential transmission from asymptomatic carriers (1, 2). Common symptoms include fever, cough, and dyspnea while the disease has potential to cause a host of severe and potentially fatal cardiorespiratory complications in vulnerable populations—particularly the elderly with comorbid conditions (3, 4). While distinguishing COVID-19 from normal lung or other lung pathologies such as cancer on chest CT may be straightforward, a major hurdle in controlling the current pandemic is making out subtle radiological differences between COVID-19 and pneumonia of other etiology. For example, manual radiologist interpretation of chest CT is a specific modality for recognizing COVID-19 by its characteristic patterns including peripheral ground-glass opacities (GGO), but unfortunately this measure often has low specificity in distinguishing COVID-19 from other pneumonia (5, 6). The exception to this is in screening populations with high disease prevalence, such as in Wuhan at the beginning of the outbreak and in Italy presently. In these cases, the sensitivity of chest CT for COVID-19 is high while specificity is low due to an abundance of false positives (7, 8).
Current literature has revealed that it is possible for artificial intelligence (AI) to identify COVID-19 from other pneumonia with good accuracy (9). However, published studies have limitations such as small sample size, lack of external validation, no comparison with radiologist performance, no gold standard for the “other pneumonia” diagnosis, etc. (10-13).
To capture and properly manage all cases of COVID-19, it is essential to develop testing methods that accurately recognize the disease as distinct from other causes of pneumonia on chest CT. The purpose of this study was to establish and evaluate an AI system in differentiating COVID-19 and other pneumonia on chest CT and assess radiologist performance without and with AI assistance.
Material and Methods
Patient Cohorts
The institutional review board of all nine hospitals in Hunan Providence, China and Rhode Island Hospital (RIH) in Providence, RI and Hospital of the University of Pennsylvania (HUP) in Pennsylvania, PA from the U.S. approved this retrospective study, and written informed consent was waived. A total of 521 patients with confirmed positive COVID-19 by RT-PCR and chest CT imaging were retrospectively identified from RIH and 9 hospitals in Hunan Providence, China from January 6 to April 1, 2020. The RT-PCR results were extracted from the patients’ electronic medical records in the hospital information system. The RT-PCR assays were performed by using TaqMan One-Step RT-PCR Kits from Shanghai Huirui Biotechnology Co., Ltd or Shanghai BioGerm Medical Biotechnology Co., Ltd, both of which have approved their use by the China Food and Drug Administration for the Chinese cohorts and the COVID-19 RT-PCR test (Laboratory Corporation of America) for US cohorts. For patients with multiple RT-PCR assays, positive result on the last performed test was adopted as a confirmation of diagnosis.
The radiology search engine MONTAGE (Nuance Communications, Burlington, MA) at RIH and HUP was used to identify cases that contain the word “pneumonia” in the impression section of the radiology CT reports from January 1, 2017 to December 30, 2019. The impression sections of these CT reports were initially reviewed by a research assistant (BH) followed by verification by a radiologist (HXB) board-certified in general diagnostic radiology and interventional radiology with one year of practice experience to identify cases with final CT impression being “consistent with” or “highly suspicious for” pneumonia. Then, the images were further reviewed by a radiologist (HXB) to ensure agreement with the original report. A Chinese radiology search engine was used to identify a similar non-COVID-19 pneumonia cohort from Xiangya Hospital in Hunan Providence, China from 2017 to 2019, followed by verification by a radiologist (DW). The identified CT scans were directly downloaded from the hospital Picture Archiving and Communications Systems (PACS), and non-chest CTs were excluded.
Data on the respiratory pathogen were collected from Respiratory Pathogen Panel (RPP) results for the RIH cohort, as described in a previous study (5). The tests of ePlex Respiratory Pathogen panel (GenMark Diagnostics, Carlsbad, CA) were performed in the Microbiology Lab of Rhode Island Hospital Pathology Department according to its manufacture protocol (14).
Our final cohort consisted of 665 non-COVID pneumonia patients. A diagram illustrating patient inclusion and exclusion is shown in Figure 1. The number of cases included from each hospital is shown in Supplementary Table E1. The chest CT protocols from all 11 hospitals are shown in Supplementary Table E2. 214 patients from the COVID-19 group and 202 patients from the non-COVID pneumonia group (RIH cohort) overlapped with a previous study (5).
Figure 1.
Diagram illustrating patient inclusion and exclusion. Abbreviations: RIH, Rhode Island Hospital; HUP, Hospital of the University of Pennsylvania; AI, artificial intelligence; RT-PCR, reverse transcriptase polymerase chain reaction.
Lung Segmentation
To exclude non-pulmonary regions of the CT, the lungs were first segmented on the basis of Hounsfield Unit (HU) with -320 HU used as the thresholding value. Manual editing of the lung segmentation was performed by radiologists using the manual active contour segmentation method with 3D Slicer software (v4.6) when auto-segmentation was insufficient.
Image Preprocessing
The whole dataset was preprocessed by setting the CT window width and level to the lung window (WL: -400; WW: 1500). The slices with lesions (COVID-19 or pneumonia) were manually labeled by radiologists (HXB and DW) in consensus and used as gold standard for training the deep neural network for identifying slices with abnormal lung findings. Images were padded, if necessary, to equal height and width and rescaled to 224x224 pixels. Lung windowing was applied to the Hounsfield units to generate an 8-bit image for each individual 2D axial slice in a CT scan. Images were preprocessed by first normalizing pixel values from the range [0, 255] to [0, 1], then standardizing using the normalized ImageNet mean and standard deviation.
Development of the deep learning model
A classification model was trained to distinguish between slices with and without pneumonia-like findings (both COVID and non-COVID). The EfficientNet architecture (15), which consists of mobile inverted bottleneck MBConv blocks (16) was employed for the classification task. It possessed a smaller number of model parameters and improved the accuracy and efficiency over the existing convolutional networks. Pretrained on ImageNet, an EfficientNet-B3 convolutional neural network with a single fully connected 2-class classification layer was used. Dropout with probability 0.5 was applied to the fully connected layer. Data augmentation was performed dynamically during training and included flips, scaling, rotations, random brightness and contrast manipulations, random noise, and blurring. Training was performed for 20 epochs, where each epoch was defined as 16,000 slices. The AdamW optimizer was used with default parameters. A one cycle policy was used for the learning rate schedule, with an initial learning rate of 4.0 x 10^-6 to a max of 1.0 x 10^-4. Validation was carried out on a separate validation set every 2 epochs. The area under the curve (AUC) was used to track model performance, and the checkpoint with the highest validation AUC was selected as the final model. The choice of compound scaling metrics was made empirically based on validation set performance. Specifically, a larger network was used when it resulted in high performance on the validation set. If increasing the network size did not result in higher performance on the validation set, a smaller network was used to maintain computational efficiency.
Pneumonia Classification
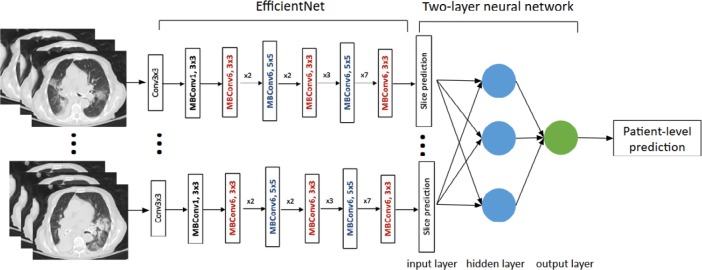
The EfficientNet B4 architecture was employed for the pneumonia classification task. Each slice was stacked to three channels as the input of EfficientNet to use the pre-trained weights on ImageNet. EfficientNets with dense top fully-connected layers were used. The configuration of dense top layers was as follows: four fully-connected layers of (256, 128, 64, 32) combined with 0.5 dropout using rectified linear unit (ReLU) activations with batch normalization layers replacing the top fully-connected layers of EfficientNet. A fully-connected layer with 16 neurons with batch normalization and a classification layer (one neuron) with sigmoid activation were at the end of EfficientNet to make predictions of COVID-19 vs. non-COVID pneumonia slices. Then, the slices were pooled using a 2 layer fully-connected neural network to make prediction at the patient level. Stochastic gradient descent optimizer with 0.0001 learning rate was utilized. Batch size was a set to 64. Figures 2-3 illustrate our deep learning workflow. A heatmap for important image regions that lead our model to classify a case as COVID-19 or non-COVID-19 was generated using Gradient-weighted Class Activation Mapping (Grad-CAM) (17).
Figure 2.
Flow diagram illustrating our AI model for distinguishing COVID-19 from non-COVID-19 pneumonia. Abbreviations: ROC AUC: Receiver Operator Characteristics Area Under the Curve; PR AUC: Precision Recall area under curve.
Figure 3:
COVID-19 Classification Neural Network Model.
Radiologist Interpretation
Six radiologists with 10, 10, 20, 20 (XZ), 20, and 10 years of chest CT experience reviewed the test set consisting of 119 chest CT images and scored each case as COVID-19 or pneumonia of other etiology. All identifying information was removed from the CT studies, which were shuffled and uploaded to 3D slicer for interpretation. All radiologists were given information on patient age when reviewing images. All radiologists then reviewed the test set again, with prediction from the AI. The studies were shuffled between the two evaluation sessions. The two sessions were separated by at least one day. The radiologists were not given feedback on their performance after the first session.
Statistical Analysis
Demographic and clinicopathologic characteristics were compared between COVID-19 and non-COVID-19 pneumonia groups by means of chi-square test for categorical variables and Student t test for continuous variables. CT slice thickness was compared between the COVID-19 and non-COVID-19 pneumonia groups using the Mann-Whitney U test and among training, validation and test sets using the Kruskal-Wallis H test. Accuracy, sensitivity, specificity, area under Receiver Operating Characteristic curve (ROC AUC), and area under Precision-Recall curve (PR AUC) were calculated for classification model. 95% confidence intervals on accuracy, sensitivity, and specificity were determined using the adjusted Wald method (18). Model performance was compared to average radiologist performance. Radiologist performance without AI assistance was compared with that with AI assistance. The p-values were calculated using the permutation method. All analyses were performed with the use of the R statistical computing language (R version 3.4.2, The R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org).
Code Availability
The implementation of the deep learning models was based on the Keras package (version 2.2.5) with the Tensorflow library (version 1.12.3) on the backend. The models were trained on a computer with two NVIDIA V100 GPUs. To allow other researchers to develop their models, the code is publicly available on Github at http://github.com/robinwang08/COVID19.
Results
Patient Characteristics
Our final cohort consisted of 1,186 patients, of which 521 were patients with COVID-19 and 665 were patients with non-COVID pneumonia. The average age of patients with COVID-19 was lower than that of patients with non-COVID-19 pneumonia (48 vs. 62 years, p<0.001). Patients with COVID-19 were less likely to have an elevated white blood cell count than patients with non-COVID-19 illness (2% vs. 51%, p<0.001) or reduced lymphocyte (36% vs. 55%, p<0.001). The clinical characteristics of the COVID-19 and non-COVID-19 pneumonia patients including co-morbidities are shown in Table 1. A diagram illustrating the breakdown of the viral pathogen species for the RIH cohort is shown in Supplementary Figure E1. The average number of days between chest CT and COVID-19 diagnosis was 2.8±4.0. A diagram illustrating the breakdown of days between symptom onset and CT scan is shown in Supplementary Figure E2. There was no significant difference in median CT slice thickness COVID-19 and non-COVID-19 cases (COVID-19: 1.25 mm, non-COVID-19: 1.00 mm, p=0.869). Supplementary Table E3 shows CT slice thickness among training, validation and test sets for the different splits.
Table 1.
Clinical Characteristics of COVID-19 and non-COVID-19 pneumonia patient cohorts
Slice Identification
The classifier to distinguish between slices with and without pneumonia-like findings (both COVID and non-COVID) achieved a final test AUC of 0.83. A naïve classification threshold of 0.5 was used to binarize predictions. Additional metrics included average mean precision (0.675), F1 score (0.675), and positive predictive value (0.795).
Pneumonia Classification
The CT images of the 1,186 patients (132,583 slices) were divided into training, validation, and test sets in a 7:2:1 ratio (i.e., 830, 237 and 119 patients, respectively). The number of patients and slices in training, validation and testing sets is shown in Supplementary Table E4. Our final model achieved a test accuracy of 96% (95% CI: 90-98%), sensitivity of 95% (95% CI: 83-100%) and specificity of 96% (95% CI: 88-99%) with ROC AUC of 0.95 and PR AUC of 0.90. Compared to average radiologist, our model had higher test accuracy (96% vs. 85%, p<0.001), sensitivity (95% vs. 79%, p<0.001), and specificity (96% vs. 88%, p=0.002) (Table 2). The ROC curve comparing model with radiologist performance is shown in Figure 4. A model trained on random equal split of training, validation and test sets (i.e., 396, 395 and 395 patients, respectively) achieved a test accuracy of 91% (95% CI: 87-93%), sensitivity of 94% (95% CI: 90-97%) and specificity of 87% (95% CI: 82-91%) with ROC AUC of 0.95 and PR AUC of 0.92. The number of patients and slices in training, validation and testing sets is shown in Supplementary Table E4. Model performance on training, validation and test sets for the different splits is shown in Supplementary Table E5.
Table 2.
The results of an artificial intelligence (AI) model and six radiologists without AI assistance on the test set (n=119) in differentiating between COVID-19 pneumonia and non-COVID-19 pneumonia
Figure 4.

ROC curve of deep neural network on the test set compared to radiologist performance. ROC = receiver operating curve.
Independent testing was performed by leaving out cohorts from one US hospital (HUP) and three Chinese hospitals (Yongzhou Central Hospital, Zhuzhou Central Hospital, and Yiyang No.4 Hospital). Our model achieved a test accuracy of 87% (95% CI: 82-90%), sensitivity of 89% (95% CI: 81-94%) and specificity of 86% (95% CI: 80-90%) with ROC AUC of 0.90 and PR AUC of 0.87.
Grad-CAM on representative CT slices from test set demonstrates that the model focused on the area of abnormality (Figure 5). Figure 6 shows five cases (Figure 6a-e) in the test set where the deep learning model was correct but most of the radiologists were incorrect (at least 4/6) as well as one case (Figure 6f) where both AI and most of the radiologists were incorrect. The three COVID-19 cases (Figure 6a-c) demonstrate atypical findings (e.g., focal abnormality) that could have been mistaken for non-COVID-19 pneumonia, while the three non-COVID-19 pneumonia cases (Figure 6d-f) demonstrate GGOs that mimic COVID-19 cases.
Figure 5.
Representative slices corresponding to Grad-CAM images on the test set.
Figure 6.
Representative cases that the majority of radiologists misclassified. A-C (top row, left to right): COVID-19 pneumonia. Our model correctly classified all three cases. A. 4/6 radiologists (radiologists 3-6) said it was non-COVID-19. With AI assistance, 2/6 radiologists (radiologists 5 and 6) continued to say it was non-COVID-19. B. 4/6 radiologists (radiologists 3-6) said it was non-COVID-19. With AI assistance, 3/6 radiologists (radiologists 3-5) continued to say it was non-COVID-19. C. 4/6 radiologists (radiologists 2 and 4-6) said it was non-COVID-19. With AI assistance, 1/6 radiologist (radiologist 2) continued to say it was non-COVID-19. D-F (bottom row, left to right): Non-COVID-19 pneumonia. Our model correctly classified D and E. D. 5/6 radiologists (radiologists 1-5) said it was COVID-19. With AI assistance, all 5/6 radiologists continued to say it was COVID-19. E. 4/6 radiologists (radiologists 1, 2, 4, and 6) said it was COVID-19. With AI assistance, 3/6 radiologists (radiologists 1, 2, and 4) continued to say it was COVID-19. F. 4/6 radiologists (radiologists 1-3 and 6) said it was COVID-19. With AI assistance, 5/6 radiologists (radiologists 1-4 and 6) said it was COVID-19.
Radiologist Performance
For blind review on the test set without AI prediction, six radiologists had an average accuracy of 85% (95% CI: 77-90%), sensitivity of 79% (95% CI: 64-89%) and specificity of 88% (95% CI: 78-94%). Assisted by the model’s probabilities, the radiologists achieved a higher average accuracy (90% vs. 85%, Δ=5, p<0.001), sensitivity (88% vs. 79%, Δ=9, p<0.001) and specificity (91% vs. 88%, Δ=3, p=0.001). Table 3 summarizes the comparison of radiologist performances without and with AI assistance.
Table 3.
Comparison of six radiologists without and with assistance of an artificial intelligence (AI) model in differentiating between COVID-19 pneumonia and non-COVID-19 pneumonia
Discussion
COVID-19 can be difficult to distinguish from other types of pneumonia on chest CT . It has been revealed that the gold standard diagnostic test, real time reverse-transcriptase polymerase chain reaction (RT-PCR), frequently produces false-negatives or fluctuating results that make it difficult to diagnose and contain active COVID-19 infections with confidence (19). Therefore, chest CT is often relied on as a supplementary diagnostic measure that helps physicians build a more complete patient assessment. AI has shown efficacy in differentiating COVID-19 from pneumonia of other etiology on chest CT, yet the practical application of AI augmentation to radiologists’ COVID-19 diagnostic workflow has not been explored in the literature (9). Our study revealed that when compared to a radiologist only approach, AI augmentation significantly improved radiologists’ performance in distinguishing COVID-19 from pneumonia of other etiology yielding higher measures of accuracy, sensitivity, and specificity.
The diagnostic accuracy produced by manual interpretation of COVID-19 chest CT is good but needs to be improved to make resource allocation and disease management during the current pandemic less strenuous on healthcare systems and economies worldwide. Current clinical algorithms for the management of COVID-19 patients are contingent on the amount of resources available and require definitive imaging results (20). While distinguishing COVID-19 from normal lung or other lung pathologies such as cancer on chest CT may be straightforward, differentiation between COVID-19 and other pneumonia can be particularly troublesome for physicians because of the radiographic similarities (21). Inaccurate imaging interpretation makes it harder for patient management strategies to work efficiently.
Our study is relevant and novel for demonstrating the effect of AI augmentation on radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT. The results that we present suggest that integrating AI into radiologists’ routine workflow has potential to improve diagnostic outcomes related to COVID-19. In addition, external validation was used in our study while other recent AI studies either lacked external validation completely or had poor outcomes associated with external validation. The slight decrease in performance on external validation is secondary to some lack of generalization, which is expected across institutions due to differences in patient population and imaging acquisition (22, 23). This research makes progress on the practical use of AI in COVID-19 diagnosis, and a future study will explore the prospective use of AI in real-time to assist physician diagnosis.
Our study has several limitations. First, there could be bias as a product of the radiologists in this study evaluating the same cases twice, first without and then with AI assistance. However, this limitation cannot be overcome without a prospective design. Second, our COVID-19 cohort was heterogenous in the distribution of time between symptom onset and CT. Although this reflected a spectrum of chest CT presentations that likely represented the real-world scenario, the most difficult distinction between COVID-19 and pneumonia of other etiology remains during the early disease stage. The limited sample size of early stage COVID-19 CT prevented us from performing a subgroup analysis focusing on this cohort. Third, the composition of other pneumonia cases is heterogeneous and not all the patients in the non-COVID-19 pneumonia cohort underwent RPP testing or had the test results available. For those without RPP testing, the cases were selected by searching the impression section of the original report and further review of the images by a second radiologist, which could have introduced selection bias. Furthermore, there is a possibility of pneumonia of other etiology (e.g., viral pneumonia by influenza) superimposed on COVID-19. Although we did our best to standardize CT images, a possibility remains for AI or radiologists to notice subtle differences between scans from different countries, institutions, or CT instruments. Lastly, there was significant difference in baseline characteristics between COVID-19 and non-COVID-19 pneumonia patients which could have introduced bias. For example, patients in the non-COVID-19 pneumonia cohort were predominately from the United States, and significantly older with more comorbid conditions than those in our COVID-19 cohort, which by contrast mainly contained patients from China. Any of these factors could have complicated the appearance of non-COVID-19 chest CTs and influenced performance measures in our study. Lastly, although we had a multi-national, multi-institutional cohort, our model training could have benefited from a larger cohort size.
AI assistance improved radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT. Future study will investigate integration of these algorithms into routine clinical workflow to assist radiologists in accurately diagnosing COVID-19.
H.X.B. and R.W. contributed equally to this article.
Abbreviations:
- COVID-19
- Coronavirus Disease 2019
- AI
- Artificial Intelligence
- ROC
- Receiver Operating Characteristic
- PR
- Precision-Recall
- RT-PCR
- Reverse Transcription Polymerase Chain Reaction
- RPP
- Respiratory Pathogen Panel
- RIH
- Rhode Island Hospital
- HUP
- Hospital of the University of Pennsylvania
- GGO
- Ground-Glass Opacities
- Grad-CAM
- Gradient-weighted Class Activation Mapping
References
- 1. Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, et al . Presumed Asymptomatic Carrier Transmission of COVID-19 . JAMA . 2020. . Epub 2020/02/23. doi: 10.1001/jama.2020.2565. PubMed PMID: 32083643; PubMed Central PMCID: PMCPMC7042844 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. . Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study . The Lancet Infectious Diseases . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China . Lancet . 2020. ; 395 ( 10223 ): 497 - 506 . Epub 2020/01/28. doi: 10.1016/S0140-6736(20)30183-5. PubMed PMID: 31986264 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al . Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China . Jama . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, et al . Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT . Radiology . 2020. : 200823 -. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi H, Qi X, Yoon SH, Park SJ, Lee KH, Kim JY, et al . Extension of Coronavirus Disease 2019 (COVID-19) on Chest CT and Implications for Chest Radiograph Interpretation . Radiology: Cardiothoracic Imaging . 2020. ; 2 ( 2 ): e200107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al . Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases . Radiology . 2020 : 200642 . Epub 2020/02/27. doi: 10.1148/radiol.2020200642. PubMed PMID: 32101510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caruso D, Zerunian M, Polici M, Pucciarelli F, Polidori T, Rucci C, et al . Chest CT Features of COVID-19 in Rome, Italy . Radiology . 2020. : 201237 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al . Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia on Chest CT . Radiology . 0 ( 0 ): 200905. . doi: 10.1148/radiol.2020200905. PubMed PMID: 32191588 . [Google Scholar]
- 10. Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, et al . A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) . medRxiv; . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu X, Jiang X, Ma C, Du P, Li X, Lv S, et al . Deep Learning System to Screen Coronavirus Disease 2019 Pneumonia . arXiv preprint arXiv : 200209334 . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, et al . Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study . medRxiv . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi W, Peng X, Liu T, Cheng Z, Lu H, Yang S, et al . Deep Learning-Based Quantitative Computed Tomography Model in Predicting the Severity of COVID-19: A Retrospective Study in 196 Patients . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. GenMark Diagnostics I. ePlex Pipeline: GenMarkDx; [cited 2020 March 9]. Available from: https://genmarkdx.com/solutions/panels/eplex-panels/eplex-pipeline/ .
- 15. Tan M, Le QV. . EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks . arXiv e-prints [Internet] . 2019. May 01, 2019:[arXiv:1905.11946 p.]. Available from: https://ui.adsabs.harvard.edu/abs/2019arXiv190511946T . [Google Scholar]
- 16. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. . MobileNetV2: Inverted Residuals and Linear Bottlenecks . arXiv e-prints [Internet] . 2018. January 01, 2018:[arXiv:1801.04381 p.]. Available from: https://ui.adsabs.harvard.edu/abs/2018arXiv180104381S . [Google Scholar]
- 17. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. , editors. Grad-cam: Visual explanations from deep networks via gradient-based localization . Proceedings of the IEEE international conference on computer vision ; 2017. . [Google Scholar]
- 18. Agresti A, Coull BA. . Approximate is Better than “Exact” for Interval Estimation of Binomial Proportions . The American Statistician . 1998. ; 52 ( 2 ): 119 - 26 . doi: 10.1080/00031305.1998.10480550 . [Google Scholar]
- 19. Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, et al . Stability Issues of RT-PCR Testing of SARS-CoV-2 for Hospitalized Patients Clinically Diagnosed with COVID-19 . Journal of Medical Virology . 2020. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, et al . The Role of Chest Imaging in Patient Management during the COVID-19 Pandemic: A Multinational Consensus Statement from the Fleischner Society . Radiology . 0 ( 0 ): 201365. . doi: 10.1148/radiol.2020201365. PubMed PMID: 32255413 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, et al . Coronavirus Disease 2019 (COVID-19): A Perspective from China . Radiology . 0 ( 0 ): 200490 . doi: 10.1148/radiol.2020200490. PubMed PMID: 32083985 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. . Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study . PLoS medicine . 2018. ; 15 ( 11 ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. AlBadawy EA, Saha A, Mazurowski MA. . Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing . Medical physics . 2018. ; 45 ( 3 ): 1150 - 8 . [DOI] [PubMed] [Google Scholar]
- 24. Du Q. . Clinical classification. Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment ( 7th edition ) [Internet]. 2020 April 6, 2020 April 8, 2020]. Available from: http://kjfy.meetingchina.org/msite/news/show/cn/3337.html . [Google Scholar]