Abstract
MYB-related transcription factors play important roles in plant development and response to various environmental stresses. In the present study, a novel MYB gene, designated as BnMYB2 (GenBank accession number: MF741319.1), was isolated from Boehmeria nivea using rapid amplification of cDNA ends (RACE) and RT-PCR on a sequence fragment from a ramie transcriptome. BnMYB2 has a 945 bp open reading frame encoding a 314 amino acid protein that contains a DNA-binding domain and shares high sequence identity with MYB proteins from other plant species. The BnMYB2 promoter contains several putative cis-acting elements involved in stress or phytohormone responses. A translational fusion of BnMYB2 with enhanced green fluorescent protein (eGFP) showed nuclear and cytosolic subcellular localization. Real-time PCR results indicated that BnMYB2 expression was induced by Cadmium (Cd) stress. Overexpression of BnMYB2 in Arabidopsis thaliana resulted in a significant increase of Cd tolerance and accumulation. Thus, BnMYB2 positively regulated Cd tolerance and accumulation in Arabidopsis, and could be used to enhance the efficiency of Cd removal with plants.
Introduction
With the rapid development of industry and mining, excessive emission of industrial waste water, gas and residue containing heavy metals (Cu, Pb, Cd, As, Zn and so on) has resulted in serious pollution of farmland soil [1, 2]. Cadmium (Cd) is a highly toxic metal element, with high mobility, accumulation and non-degradability, that is considered non-essential for living organisms. Once exogenous Cd gets into the soil, it progressively accumulates and causes long-term harm to farmland and crops [3, 4]. More importantly, Cd may jeopardize food security and human health through food chain bioaccumulation [5]. Research on remediation techniques for Cd polluted farmlands has increased since Cd pollution was reported to have caused "Itai-itai" disease in Japan in 1961 [6].
Phytoextraction has been promoted as an environmentally-friendly, low-input alternative for remediation of soil contaminated with heavy metals [7–10]. Recently, plant species used for phytoremediation have been extended from hyperacuumulators to non-hyperaccumulators such as Brassica napus [11], Solanum nigrum [12], Boehmeria nivea [13] and Helianthus annuus [14], which have significantly higher growth rates and biomass than hyperaccumulators. However, the use of these species is often limited by low levels of heavy metal accumulation related to their lower tolerance to heavy metals. Use of genetic engineering technology to increase heavy metal tolerance, uptake and accumulation in plants has expanded new methods in phytoremediation.
Among several mechanisms of toxicity, Cd is known to markedly increase oxygen free radicals and reactive oxygen species (ROS) in plants; to induce the oxidation of protein, DNA, lipid and carbohydrate; and to inhibit many biological and biochemical processes in organelles through its high affinity for sulfhydryl groups of proteins [15–19]. Transcriptional regulation contributes to plant response to environment stresses [20]. Transcription factors (TFs) regulate diverse physiological and biochemical responses to different abiotic stresses by activating a variety of stress-related genes [21].
The MYB (v-myb myeloblastosis viral oncogene homolog (avian)) gene family exists widely in many eukaryotes and is the largest transcription factor family in plants. All MYB proteins contain MYB domains that are highly conserved in animals, plants and yeast [22]. The MYB domain is comprised of 51–52 amino acid residues. In accordance with the number of MYB domain repeats, plant MYB proteins have been classified into 4 subfamilies: 1R-MYB/MYB-related, R2R3-MYB, R1R2R3-MYB and 4R-MYB [23, 24]. MYB genes play important roles in abiotic stress response in plants [25, 26]. Up-regulation of OsMYB48-1, an Oryza sativa MYB transcription factor, promotes drought tolerance in rice by influencing ABA synthesis [27]. Transgenic Arabidopsis (Arabidopsis thaliana) overexpressing OsMYB3R-2 exhibit enhanced cold stress tolerance [28]. AtMYB15 overexpression in transgenic Arabidopsis, which negatively regulated the expression of CBF genes, improved tolerance to cold [29] and drought stress [30]. Excessive copper ions (Cu2+) in rice significantly influenced the expression of several kinds of transcription factors, including MYB [31]. The R2R3-MYB transcription factor OsARM1 was implicated in arsenic (As) stress response in rice [32]. About 20 percent of all MYB family genes in Arabidopsis thaliana are differentially expressed in response to Cd stress [33]. Overexpression of the Cd-induced MYB49 gene in Arabidopsis resulted in a significant improvement in Cd accumulation, whereas MYB49 knockout plants and plants expressing chimeric repressors of MYB49: ERF exhibited reduced Cd accumulation [34]. In ramie, the transcript levels of fourteen MYB transcription factor genes were found to be affected by Cd stress. The expression of seven MYB genes were up-regulated and seven genes were down-regulated, respectively [35].
Ramie (Boehmeria nivea L.), also known as “China grass”, is a perennial herb from the Urticaceae family with a large shoot biomass, fast growth rate and strong root system [36]. Ramie is one of the most important natural fiber crops and can usually be harvested three times per year. It is widely cultivated in China, India, and other Southeast Asian and Pacific Rim countries. In southern China, ramie has been cultivated for at least five thousand years, yielding fiber production that is second only to cotton. Previous studies have indicated that ramie has strong tolerance to and ability to accumulate certain heavy metals, such as Cd, Pb and As, from soil [36–38]. More importantly, ramie can accumulate a relatively large amount of heavy metals in its aboveground parts. The mean concentration (mg·kg-1) of Cd, Pb and As in leaves is 2.7, 27.2 and 55.6, respectively, under field conditions [39]. In addition, it is a good candidate for phytoextraction of heavy metal polluted soils because its fiber is not in the food chain and is easy to harvest. In our previous studies on the transcriptome profiling of cadmium response genes in ramie, unigene35788 was found to be significantly up-regulated in cadmium treated groups, which was annotated as MYB transcription factors using NR and KEGG database [40]. MYB transcription factors play critical roles in plant resistance to environment stress. However, scant research on the function of MYB genes in ramie has been reported. This study focused on the identification of a putative MYB transcription factor (BnMYB2) from ramie and analysis of its promoter, expression profile, subcellular localization and transgenic overexpression in order to investigate the function of this gene in Cd stress response in ramie.
Materials and methods
Plant materials and stress treatments
Boehmeria nivea cultivar Zhongzhu-1 used as plant material in this research was grown in the experimental farm at Yichun University, Yichun, China. The 20-day-old ramie seedlings were grown in perlite pots and irrigated with half strength Hoagland solution. For tissue specific expression analysis, roots, stems and leaves from 20-day-old ramie seedlings were collected separately and stored at -80°C until used. To assess the effects of various Cd exposure times, seedlings were irrigated with Cd solution (100 μM) for 0, 3, 6, 12, 24 or 48 hours, and then leaves were sampled and stored at -80°C until RNA extraction. For treatments with varying Cd concentration, seedlings were irrigated with solutions with different Cd concentrations (0, 10, 30, 50,100 and 150 μM) for 24 hours, and then the leaves were sampled, frozen in liquid nitrogen and stored in -80°C until RNA extraction.
Isolation of full length BnMYB2 gene
From transcriptome screening, the unigene35788 was identified with BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against National Center for Biotechnology Information (NCBI) nucleotide database based on its homology with known MYB transcription factors. Sequence analysis revealed that both the 3’ and 5’ ends of unigene35788 were incomplete. Therefore, full-length BnMYB2 cDNA was obtained by performing 3’ RACE using primer MYB2-3FO/3FI and 5’ RACE using primer MYB2-5RO/5RI (S1 Table) with UAP primer and instructions provided by the SMARTer RACE 5’/3’ Kit (Clontech, USA). PCR products were run on a 1.5% agarose gel and then purified with the MiniBEST Agarose Gel DNA Extraction Kit (TaKaRa, China). Next, purified PCR product was inserted into pMD19-T plasmid (TaKaRa, China) and sequenced at the Shanghai Shenggong Company (Shanghai, China). In order to amplify full-length BnMYB2 cDNA, a pair of specific primers MYB2-F/MYB2-R (S1 Table) was designed on the basis of full-length sequence obtained from an alignment of the partial sequence, 3’RACE product and 5’RACE product. The PCR amplification procedure was 5 min at 94°C, 32 cycles (30 s at 94°C, 30 s at 52°C and 90 s at 72°C) and final extension for 7 min at 72°C. The PCR product was run on a 1.5% agarose gel, purified with a MiniBEST Agarose Gel DNA Extraction Kit (TaKaRa, China) and sequenced at the Shanghai Shenggong Company (Shanghai, China). The BnMYB2 nucleotide sequence was submitted to GenBank (accession number: MF741319.1).
Sequence analysis
ORF Finder in the NCBI website (https://www.ncbi.nlm.nih.gov/orffinder/) was used to search the BnMYB2 open reading frame. The basic physical and chemical properties of the deduced amino acid were analyzed with ProtParam tool in the ExPASy website (https://web.expasy.org/protparam). Multiple sequence alignment was performed with DNAMAN software (version 6.0) set to default parameters. The conserved domains of the BnMYB2 protein were predicted using the Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The phylogenetic tree analysis was performed with MEGA software (version 5.0) using the neighbor-joining (NJ) method (1000 replication of bootstrap) according to Kimura 2-parameter distance with BnMYB2 and other species provided in Genbank of NCBI. Protein subcellular location was predicted using WoLF PSORT (https://www.genscript.com/wolf-psort.html).
Isolation of the BnMYB2 promoter
Genomic DNA was extracted from the fresh ramie leaves. Specific primers MYB2-PF/MYB2-PR (S1 Table) were designed to clone the 1947 bp upstream of the BnMYB2 initiation codon. PCR cycling conditions were 5 min at 94°C, 32 cycles (30 s at 94°C, 30 s at 59°C and 2 min at 72°C) and final extension for 7 min at 72°C. PCR product was run on a 1.5% agarose gel, purified with a MiniBEST Agarose Gel DNA Extraction Kit (TaKaRa, China) and sequenced at the Shanghai Shenggong Company (Shanghai, China). Putative cis-acting elements in the BnMYB2 promoter were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html).
Subcellular localization of BnMYB2 protein
The complete coding sequence of BnMYB2 was amplified using MYB2-XbaI-F and MYB2-BamHI-R primers with a Xba I site and a BamH I site (S1 Table). Then, the PCR products were digested with Xba I/BamH I and inserted into pAN580 vector containing a 35S promoter-driven GFP reporter gene. The 35S::OsGhd7-CFP plasmid was used to produce the nuclear marker while the empty vector 35S::GFP was used as control [41]. The constructs was sequenced and transformed into Arabidopsis protoplasts. Protoplast isolation and transfection procedures were performed according to the method described by Yoo et al. [42]. Briefly, mesophyll protoplasts were isolated from rosette leaves collected from 3 to 4-weeks-old Col wild type Arabidopsis thaliana plants. Then, the nuclear marker construct 35S::OsGhd7-CFP was co-transformed with 35S::BnMYB2-GFP construct or 35S::GFP construct into Arabidopsis protoplasts. Finally, the GFP and CFP fluorescences were observed using Olympus FV1200 confocal microscopy imaging system after the protoplasts were incubated at room temperature for 20–22 h under darkness.
Tissue-specific and Cd-induced BnMYB2 expression
To analyze the tissue-specific expression pattern of BnMYB2, total RNA was extracted from ramie roots, stems and leaves using TRIzol reagent. The PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, China) was used for cDNA synthesis by following the manufacturer’s instructions. For gene expression analysis under Cd treatment, total RNA was extracted from leaves harvested from Cd-treated ramie seedlings. Then total RNA was reverse-transcribed into first strand cDNA in accordance with the user’s manual. Quantitative real time PCR (qRT-PCR) was performed in triplicate using TB Green Premix Ex Taq II kit (TaKaRa, China) in the StepOnePlus Real-Time PCR System (Applied biosystems) by following the manufacturer’s instructions. The primers used were MYB2-qF/MYB2-qR and BnActin-F/BnActin-R (S1 Table). The relative quantification method (ΔΔ-CT) was used to quantify the relative expression normalized to the reference gene (β-actin) [43]. The qRT-PCR assays were conducted with three biological replicates and three independent technical replicates for each sample.
Construction of plant expression vector and Arabidopsis thaliana transformation
The BnMYB2 ORF was cloned with the specific primers MYB2-BamHI-F and MYB2-SacI-R (S1 Table) and subcloned into pBI121, a plant expression vector carrying the CaMV35S promoter, to construct the fusion plasmid pBI121-BnMYB2. Then, recombinant plasmid was transformed into Arabidopsis thaliana cultivar Columbia-0 using Agrobacterium tumefaciens strain EHA105 in the floral dipping method. Transformants (T1 generation) were selected by planting seeds on MS plates containing 30 mg/L kanamycin. Positive transformants were confirmed with genomic PCR using the MYB2-35SF/MYB2-SPR primers (S1 Table). For this, genomic DNA was isolated from independent BnMYB2 overexpression and wild-type plants. The PCR cycling conditions were 5 min at 94°C, 32 cycles (30 s at 94°C, 30 s at 64°C and 30 s at 72°C) and final extension for 7 min at 72°C. To further confirm positive transformants, RNA extracted from leaves of transformants (T2 generation) was reverse-transcribed into cDNA, and then RT-PCR was performed as described above.
Cd stress assay
Phenotypic observation
To test the effect of Cd on the growth of transgenic Arabidopsis seedlings, seeds of transgenic plants (T3 generation) and wild-type plants were sterilized with 70 percent ethanol and sodium hypochlorite, plated on MS (Murashige and Skoog) solid medium containing Cd (0, 100 or 150 μM) and grown for 14 days. Phenotypes were photographed, and then the fresh weight and root length were measured. Wild-type Arabidopsis served as a control. There were three experimental replicates.
Detection of Cd content in plant
Wild-type and transgenic Arabidopsis were planted in plastics pots with unpolluted soil and irrigated with half-strength Hoagland nutrient solution for 30 days. Then, CdCl2 salt was added to the half-strength Hoagland nutrient solution to the final concentration of 0 and 50 μM, and the plants were watered with the Cd solution one time for dose-response experiment. After 7 days of cultivation, the plants were harvested, washed with tap water and rinsed with deionized water three times. Then, the roots and shoots were separated and dried at 75°C for 3 days. After that, Cd in the samples was measured by flame AAS with HNO3-HClO4 digestion. There were three experimental replicates.
Statistical analysis
Statistical analysis was conducted using SPSS version 17.0 and Microsoft excel 2013 software. All data were expressed as the mean of three biological replicates ± standard deviation (SD). Comparisons between different groups were tested by one-way ANOVA, followed by Student’s t-test. A p-value less than 0.05 was considered to be significantly different.
Results
Isolation and characterization of BnMYB2
Cd is a major environmental pollutant that affects human health and plant growth. Ramie could tolerate and accumulate Cd when grown on polluted soils [38, 39, 44]. Searching for novel genes involved in Cd tolerance and accumulation has become a priority in ramie breeding [45]. In our previous studies on the transcriptome profiling of cadmium response genes in ramie, unigene35788 was found to be significantly up-regulated in cadmium treated groups, which was annotated as MYB transcription factors using NR and KEGG database [40]. In the present study, a 1 733 bp full-length cDNA for this gene (named as BnMYB2) was isolated using 3’-RACE, 5’-RACE and RT-PCR methods. Sequence analysis revealed that the cDNA contains a 945 bp open reading frame encoding a 314 amino acid protein (S1 Fig). The physicochemical properties of the predicted BnMYB2 protein were analyzed in ExPASy database. The results indicated that the BnMYB2 protein has a molecular mass of 33.91 kDa and an isoelectric point value of 8.87. The secondary structure of the BnMYB2 protein was predicted using NPS@ server. The result indicated that BnMYB2 protein consists of α-helices (21.66%), extended strands (5.73%) and random coils (72.61%).
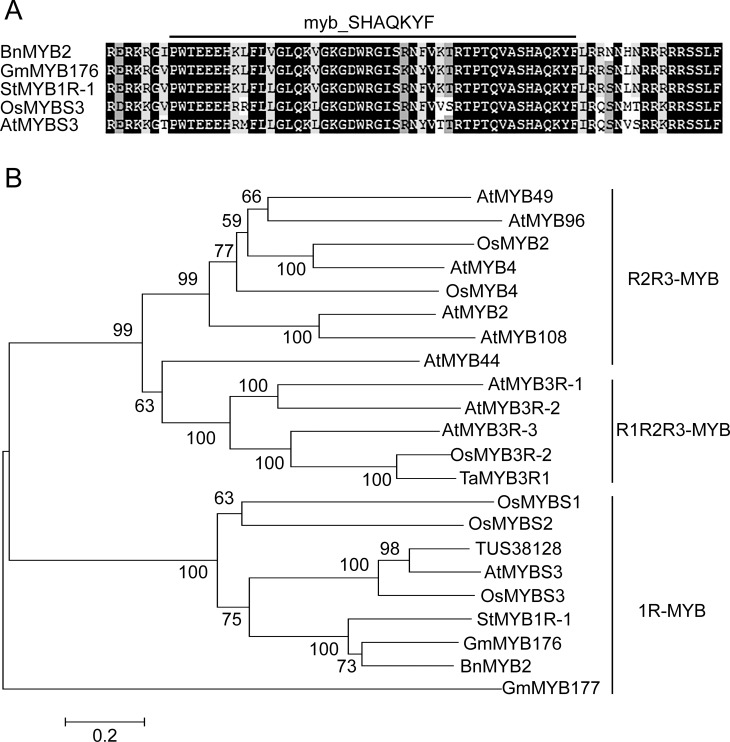
Conserved domain analysis showed that BnMYB2 contains a highly conserved MYB DNA-binding domain (myb_SHAQKYF) within residues 101–146, which indicated BnMYB2 belongs to 1R-MYB subfamily. BLAST analysis showed that BnMYB2 matches with 1R-MYB proteins reported from Solanum tuberosum (NP_001236048.2), Glycine max (NP_001236048.2), Oryza sativa (AAN63154.1) and Arabidopsis thaliana (At5g47390) with approximately 65% identity at the amino acid level. Multiple alignment demonstrated that the MYB-DNA binding domain of 1R-MYBs from different plants are conserved (Fig 1A). Thus, this gene was designated as BnMYB2 (GenBank accession number: MF741319.1), which was the second MYB gene identified in ramie.
Fig 1. Multiple alignment and phylogenetic analysis of BnMYB2 and MYB proteins related to abiotic stress from other plants.
A, The alignment of BnMYB2 and its ortholog proteins of different plants constructed using DNAMAN 8.0 software. Black shadow represents the conserved residues. The line indicates a highly conserved MYB DNA-binding domain (myb_SHAQKYF). B, Neighbor-joining phylogenetic tree of BnMYB2 from Boehmeria nivea and other MYBs constructed using MEGA 5.0 software. The scale bar represents 0.05 amino acid substitutions per site. The statistical reliability of individual nodes of the tree is assessed by bootstrap analyses with 1 000 replications. GenBank accession numbers of the proteins are as follows: AtMYB49 (At5g54230), AtMYB96 (At5g62470), AtMYB4 (At4g38620), AtMYB2 (At2g47190), AtMYB108 (At3g06490), AtMYB44 (At5g67300), AtMYB3R-1 (At4G32730), AtMYB3R-2 (At4g00540), AtMYB3R-3 (At3g09370) and AtMYBS3 (At5g47390) from Arabidopsis thaliana; OsMYB2 (BAA23338), OsMYB4 (BAA23340), OsMYB3R-2 (BAD81765.1), OsMYBS1 (AAN63152.1), OsMYBS2 (AAN63153.1) and OsMYBS3 (AAN63154.1) from Oryza sativa; TaMYB3R1 (ADO32617.1) from Triticum aestivum; TUS38128 (XP_004508939.1) from Cicer arietinum; StMYB1R-1 (ABB86258.1) from Solanum tuberosum; GmMYB176 (NP_001236048.2) and GmMYB177 (ABH02866.1) from Glycine max.
BnMYB2 protein and twenty-one abiotic-stress-related MYB proteins from Arabidopsis thaliana, Oryza sativa, Triticum aestivum, Cicer arietinum, Solanum tuberosum and Glycine max downloaded from GenBank were used to construct a phylogenetic tree by the neighbor-joining (NJ) method with MEGA 5.0 software (Fig 1B). The results suggested that MYB proteins were divided into three groups (1R-MYB, R2R3MYB and R1R2R3-MYB). The BnMYB2 protein is more similar to TUS38128, AtMYBS3, OsMYBS3, StMYB1R-1 and GmMYB176 which belong to 1R-MYB transcription factor subfamily.
Isolation and analysis of BnMYB2 promoter sequences
A 1947 bp BnMYB2 promoter was obtained from Boehmeria nivea by PCR amplification with a pair of specific primers (S2 Fig). The promoter sequence was analyzed using PlantCARE software (Table 1). The results indicated that the BnMYB2 promoter contained the CAAT-box and TATA-box elements, which are typical core eukaryotic promoter regions. In the BnMYB2 promoter sequence, two abscisic acid response elements (ABRE) were found at +1120 and +1767 bp, one anaerobic regulatory element (ARE) was found at -866 bp, one MeJA response element (CGTCA-motif) was found at +756 bp, one cis-acting element associated with circadian rhythm was found at +24 bp, one ethylene response element (ERE) was found at -957 bp, two gibberellin response elements (P-box) were found at +232 and -1732 bp, one stress response element (STRE) was found at -1822 bp and four wound response elements (WUN-motif) were found at +1162, +1395, +1227 and +1548 bp. The above analysis suggested that BnMYB2 could respond to different plant hormones and multiple environmental stresses.
Table 1. Putative cis-acting regulatory elements identified in the BnMYB2 promoter sequence using the PlantCARE database.
| Cis element | Position | Sequence | Function of site |
|---|---|---|---|
| ABRE | +1120,+1767 | ACGTG | cis-acting element involved in the abscisic acid responsiveness |
| ARE | -866 | AAACCA | cis-acting regulatory element essential for the anaerobic induction |
| AT-rich element | +29 | ATAGAAATCAA | binding site of AT-rich DNA binding protein (ATBP-1) |
| ATCT-motif | -867 | AATCTAATCC | part of a conserved DNA module involved in light responsiveness |
| Box 4 | +39, +760 | ATTAAT | part of a conserved DNA module involved in light responsiveness |
| CAAT-box | +67,-271,+130,+202,+311,+318,+326,-489,-500,+884,-894,-908,-928,-950,+982,-1001,-1007,+1025,+1053,+1115,-1147,+1265,+1394,+1430,-1433,+1614,+1683 | CAAT, CAAAT, CCAAT | common cis-acting element in promoter and enhancer regions |
| CGTCA-motif | +756 | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| circadian | +24 | CAAAGATATC | circadian |
| ERE | -957 | ATTTTAAA | ethylene-responsive element |
| G-Box | -1766,+1119 | CACGTG, TACGTG | cis-acting regulatory element involved in light responsiveness |
| I-box | +1307 | GATAAGGGT | part of a light responsive element |
| P-box | +232,-1732 | CCTTTTG | gibberellin-responsive element |
| STRE | -1822 | AGGGG | stress response elements |
| TATA-box | -45,+47,+103, +124, -125, +126, +172,+265, -302, +303,-445, +447, +464,-555,+556, 577, -609, +610,-739,-819, +822,-964,967,-1064, -1156,-1201,-1442, +1511,-1542,-1644, +1812 | TATACA,ATTATA, TATAA,ATATAA, ATATAT,TATATAA,TATATA,TATAAAA,TATAAA,TAAAGATT, | core promoter element around -30 of transcription start |
| TCT-motif | -493 | TCTTAC | part of a light responsive element |
| WUN-motif | +1162, +1395, +1227,+1548 | AAATTTCTT | wound-responsive element |
Subcellular localization of BnMYB2 protein
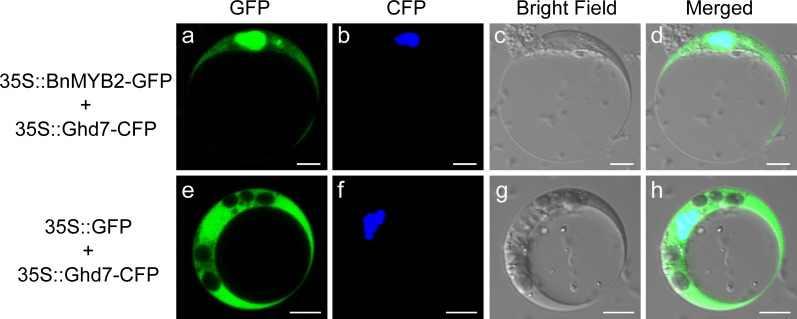
Using WoLF PSORT, BnMYB2 was predicted to be subcellularly localized to the nucleus. In order to verify the prediction, a transient expression vector pAN-BnMYB2 (BnMYB2 protein fused with GFP) was constructed and transformed into isolated Arabidopsis protoplasts. Microscopic observation showed that the green fluorescent signal of BnMYB2-GFP fusion protein was detected in the nucleus along with the cyan fluorescent signal of nuclear marker OsGhd7-CFP. In addition, a weak signal for BnMYB2-GFP was also observed in the cytoplasm. In contrast, green fluorescence was observed in the nucleus, cytoplasm and cytomembrane of cells expressing the GFP gene alone (Fig 2). The prediction and experimental observations indicated that the nucleus and cytoplasm are the main locations for BnMYB2 protein.
Fig 2. Subcellular localization of BnMYB2.
Constructs 35S::BnMYB2-GFP and empty vector 35S::GFP were separately co-transformed into Arabidopsis protoplasts with nuclear marker 35S::OsGhd7-CFP. GFP and CFP fluorescences were observed using a laser confocal microscope. a, 35S::BnMYB2-GFP; b, 35S::OsGhd7-CFP; c, Bright field; d, Overlap images of (a), (b) and (c); e, 35S::GFP; f, 35S::OsGhd7-CFP; g, Bright field; h, Overlap images of (e), (f) and (g). The bar indicates 5 μm.
BnMYB2 expression patterns under Cd stress
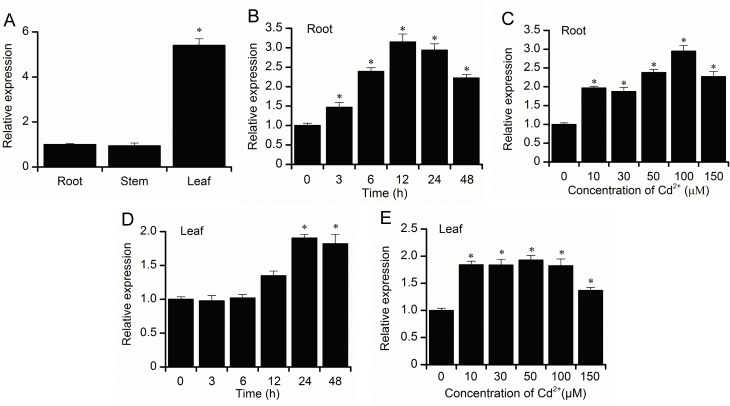
Real-time PCR experiments were carried out to study BnMYB2 expression patterns in different Boehmeria nivea tissues and under Cd stress. The tissue specificity analysis showed that BnMYB2 expression can be detected in the root, stem and leaf of ramie. BnMYB2 expression was significantly higher in leaves than in roots and stems, and the expression level in roots and stems were similar (Fig 3A). Different BnMYB2 expression patterns were detected when ramie seedlings were treated with Cd. The results indicated that BnMYB2 expression in the roots and leaves were up-regulated significantly compared to the control (0 h or 0 μM Cd) after treatment with Cd. In time-course experiment, the greatest changes in expression were observed at 12 h in roots and 24 h in leaves, which resulted in a 3.2-fold increase (Fig 3B) and a 1.9-fold increase (Fig 3D), respectively. In dose-response experiment, the greatest changes in expression were observed at 100 μM Cd treatment in roots and 50 μM Cd treatment in leaves, which resulted in a 2.9-fold increase (Fig 3C) and a 1.9-fold increase (Fig 3E), respectively.
Fig 3. Expression pattern of BnMYB2.
A, qRT-PCR analysis of BnMYB2 transcript levels in roots, stems and leaves of 20-day-old ramie seedlings. B, BnMYB2 transcript levels in the roots of ramie seedlings treated with 100 μM Cd for 0, 3, 6, 12, 24 and 48 h. C, BnMYB2 transcript levels in the roots of ramie seedlings treated with 0, 10, 30, 50, 100 and 150 μM Cd for 24 h. D, BnMYB2 transcript levels in the leaves of ramie seedlings treated with 100 μM Cd for 0, 3, 6, 12, 24 and 48 h. E, BnMYB2 transcript levels in the leaves of ramie seedlings treated with 0, 10, 30, 50, 100 and 150 μM Cd for 24 h. Data are presented as the means of three biological replicates with SE shown by vertical bars. Asterisk indicates significantly difference (p<0.05) from the untreated group.
Overexpression of BnMYB2 improved the tolerance and accumulation of Cd in transgenic plants
In order to identify the function of BnMYB2 in regulating plant response to Cd stress, the open reading frame of BnMYB2 was inserted into a pBI121 vector with a 35S promoter to construct pBI121-BnMYB2, which was then transformed into Col-0 ecotype Arabidopsis. The transgenic Arabidopsis seedlings were screened using MS medium containing 30 mg·L-1 kanamycin (S3 Fig). To confirm expression of the exogenous BnMYB2 in Arabidopsis plants, genomic DNA and total RNA were extracted from Arabidopsis plants (T1 generation) and analyzed by PCR and RT-PCR (S4 and S5 Figs). PCR results confirmed that the BnMYB2 gene was integrated into the genome of transformed plants. RT-PCR results indicated that BnMYB2 expression was highest in 35S::BnMYB2 carrying transgenic lines L3 and L6, which were selected for additional analysis.
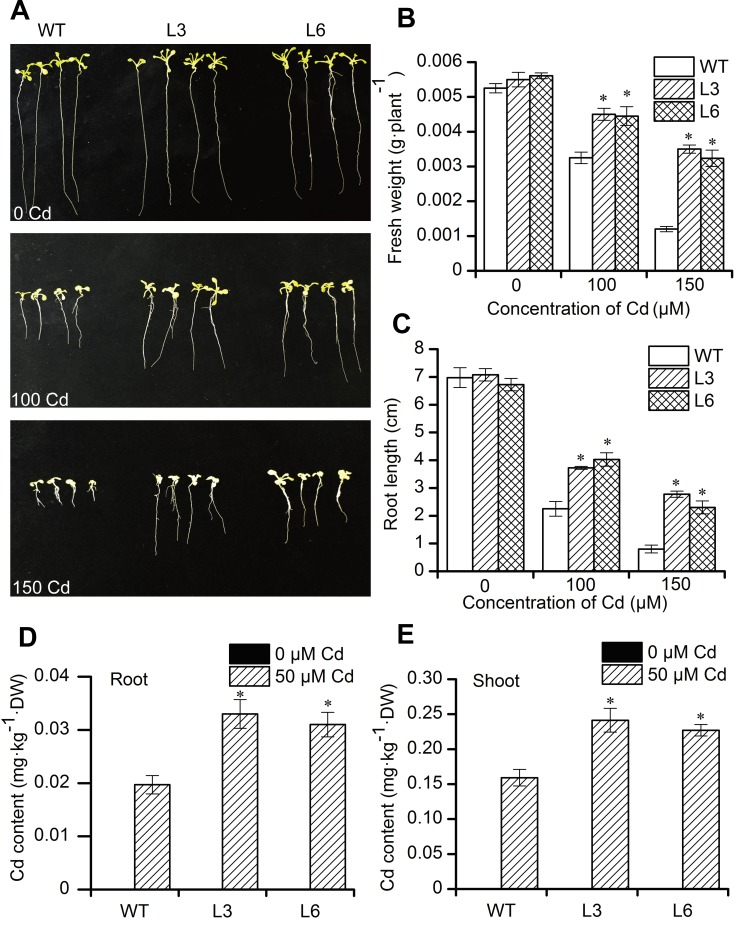
Then, T3 seeds of WT (wild-type), L3 and L6 Arabidopsis were germinated and grown on MS solid medium containing Cd (0, 100 or 150 μM) for 14 days. In normal MS medium, no significant morphological differences were observed between WT, L3 and L6 lines (Fig 4A). When the seedlings were exposed to 100 or 150 μM Cd, the growth of WT, L3 and L6 lines were all significantly inhibited. However, the L3 and L6 transgenic lines both showed significantly better growth than WT. Under Cd exposure, the fresh weight of L3 and L6 transgenic plants root were 1.4–2.9 fold heavier than that of WT (Fig 4B). Similarly, the primary root elongation of L3 and L6 transgenic seedlings were 1.7–3.5 fold longer than that of WT when seedlings were exposed to Cd (Fig 4C). To examine the effect of BnMYB2 gene expression on Cd accumulation in Arabidopsis, 30-day-old WT and transgenic Arabidopsis grown in soil were irrigated with half-strength Hoagland nutrient solution containing 0 μM or 50 μM Cd and allowed to grow for 7 days. Then the Cd content in roots and shoots were measured. In the presence of 0 μM Cd, the content of Cd in WT and transgenic Arabidopsis plants was undetectable because the soil and half-strength Hoagland nutrient solution were not contaminated by Cd. In the presence of 50 μM Cd, the two transgenic lines overexpressing BnMYB2 showed significantly higher levels of Cd than WT plants in both roots and shoots. Cd content in the roots and shoots of transgenic lines was 1.6–1.7 and 1.4–1.5 fold higher than that of WT, respectively (Fig 4D and 4E). These results indicated that BnMYB2 was able to enhance both Cd tolerance and accumulation in Arabidopsis.
Fig 4. Analysis of the phenotype and Cd content of wild-type (WT) and BnMYB2 transgenic Arabidopsis plants (L3 and L6) treated with Cd.
A, Phenotype of Arabidopsis seedlings grown on MS medium with 0, 100 or 150 μM Cd for 14 d. B, Fresh weight of BnMYB2 transgenic Arabidopsis seedlings grown on MS medium with 0, 100 or 150 μM Cd for 14 d. C, Root length of BnMYB2 transgenic Arabidopsis seedlings grown on MS medium with 0, 100 or 150 μM Cd for 14 d. D and E, Concentration of Cd in roots and shoots of WT and transgenic Arabidopsis plants grown on soil irrigated with 0 or 50 μM Cd for 7 d. Data are presented as the means of three biological replicates with SE shown by vertical bars. Asterisk indicates significantly difference (p<0.05) from wild-type (WT).
Discussion
As one of the largest plant transcription factor families, MYB transcription factors play important roles in the transcriptional regulation of plant growth, development and response to environment stress by employing their conserved DNA-binding domain to specifically bind the promoters of target genes [24, 46]. In the present work, we isolated a MYB transcription factor gene (named BnMYB2) in ramie using RACE and RT-PCR methods. The conserved domain analysis indicated that BnMYB2 protein contains one conserved MYB DNA-binding domain. Sequence alignment and phylogenetic tree analysis revealed that BnMYB2 shares high homology with known stress resistance-related 1R-MYB proteins from other plant species, such as TUS38128 [47], AtMYBS3 [48], OsMYBS3 [49], StMYB1R-1 [50] and GmMYB176 [51]. Among these proteins, AtMYBS3, a homolog of rice OsMYBS3, was reported to be activated by ABA and CdCl2. Thus, BnMYB2 from ramie was predicted belonging to the 1R-MYB subfamily, and may play an important role in ramie adaption to adversity stress including Cd stress.
Furthermore, we obtained a 1947 bp promoter sequence of BnMYB2 by PCR and analyzed this promoter sequence as detailed in additional file: S2 Fig. Sequence analysis showed that the promoter contains several stress-related hormone response elements, such as ABRE, CGTCA-motif, and P-box elements. Usually, plant tolerance to environmental stresses can be enhanced through the regulation of hormone metabolism and signal transduction to change morphology and physiological activities. For example, abscisic acid (ABA) plays an important role in plant response to diverse environmental abiotic stresses. Seo et al. [52, 53] reported that AtMYB60 and AtMYB96 act through the ABA signaling cascade to regulate drought stress and disease resistance. Abe et al. [54] confirmed that ABA could induce the expression of MYB gene AtMYB2 in Arabidopsis, which was also induced by dehydration or salt stress. In plants, MeJA was shown to act as a signaling molecule in response to biotic and abiotic stresses [55]. Anjum et al. [56] found that the exogenous MeJA can improve soybean drought resistance by inducing alterations in lipid peroxidation and the antioxidative defense system. ABRE and CGTCA-motif elements found in the promoter indicated that BnMYB2 expression may be induced by ABA and MeJA and involved in abiotic stress tolerance.
Initiating signaling cascades that regulate the expression of defense genes is an important way to manage plant's response to Cd [57]. Previous studies have shown that the expression of transcription factors belonging to MYB families is highly responsive to Cd stress [58]. About 20 percent of all MYB family genes in Arabidopsis thaliana are differentially expressed in response to Cd stress [33]. She et al. [40] studied the expression profile of Cd response genes in ramie roots using high throughput sequencing technology, finding that the expression of some MYB genes was significantly induced by Cd. Similarly, Liu et al. [59] identified ramie genes involved in Cd stress response using Illumina pair-end sequencing on two Cd-stressed plants grown on soil contaminated with 100 mg·kg-1 Cd, finding that a MYB3 transcription factor was up-regulated to 2.29-fold. The transcript level of AIM1, a MYB transcription factor in tomato, was induced about 1.5-fold under Cd stress (100 μM). In the present study, the transcript level of BnMYB2 in ramie roots and leaves were significantly increased under Cd stress; the change in the expression of BnMYB2 in roots was more obvious than that in leaves. The result indicated that BnMYB2 has an important role in mediating Cd stress response in ramie, especially in roots.
The analysis of the subcellular localization of a protein can provide insight to its function. In the present study, BnMYB2 fused to GFP showed a strong signal in the nucleus and a weak signal in the cytoplasm when expressed in Arabidopsis protoplasts. Similar results also have been observed in other plant species. In Salvia miltiorrhiza, a R2R3 MYB transcription factor gene SmMYB87 was found localized in the nucleus and cytomembrane of onion epidermal cells [60]. Several R3 MYB transcription factors such as ETC3, TCL1 and CPC related to trichome development in Arabidopsis thaliana, were found localized in the nucleus as well as in the cytoplasm when expressed in Arabidopsis protoplasts [61–63]. Isoflavonoids are natural compounds in plant which play important roles in plant growth, development and abiotic stress response. A 1R-MYB transcription factor GmMYB176 which regulates the biosynthesis of isoflavonoids in soybean, was found localized mainly in the nucleus and partially in the cytoplasm when expressed in tobacco leaf epidermal cell [64]. Meanwhile, GmMYB176 shares the nearest phylogenetic relationship with BnMYB2 (Fig 1B). In our study, BnMYB2 was localized both in nucleus and cytoplasm. The location of BnMYB2 in the nucleus implied that BnMYB2 is compatible with a role in transcriptional regulation of (as yet unknown) downstream target genes. However, the location of BnMYB2 in the cytoplasm is not yet understood. It could be due to an artifactual mislocalization due to the use of a 35S promoter to drive the expression of BnMYB2-GFP.
The functions of MYB proteins have been investigated in numerous plant species, such as Arabidopis, Oryza sativa, Malus domestica and maize, using genetic and molecular analysis. To illustrate that BnMYB2 was able to regulate plant response to Cd stress, we investigated the effect of BnMYB2 overexpression on Cd tolerance and accumulation in Arabidopsis. The 35S::BnMYB2 seedlings exhibited improved fresh weight, root elongation and higher Cd content compared to those of WT seedlings. These results above indicated that BnMYB2 can mediate the Cd response in Arabidopsis. Some evidence have implicated MYB transcription factors in the accumulation of heavy metals in plants. Overexpression of MYB49, a Cd-induced MYB transcription factor in Arabidopsis, significantly increased Cd accumulation by directly regulating HIPP22, HIPP44, bHLH38 and bHLH101. Whereas, the accumulation of Cd in Arabidopsis was significantly reduced when MYB49 was knocked out [34]. OsARM1, an arsenite-responsive MYB transcription factor from rice, was able to regulate the As tolerance and uptake in rice by regulating the expression levels of As-associated transporters genes like OsABCC1 by binding to their promoters [32]. Overexpression of a Raphanus sativus transcription factor gene RsMYB1, strongly enhance the tolerance of petunia to multi-heavy metals stress (K2Cr2O7, MnSO4, ZnSO4 and CuSO4) by enhancing the expression level of antioxidant genes (CAT and SOD) and abiotic stress-tolerant genes (PCs and GSH) [65]. Our results suggest that BnMYB2 from ramie plays an important function in mediating Cd tolerance and accumulation in plants and provide new clues to explain the mechanisms of Cd tolerance and accumulation in plants.
Conclusion
In this study, we isolated the BnMYB2 gene, which belongs to the 1R-MYB transcription factor subfamily, from ramie. The BnMYB2 promoter contains several cis-acting elements involved in phytohormone signaling and a variety of stress responses. It has a close phylogenetic relationship with other 1R-MYB transcription factors that contribute to different environment stress response. Analysis indicated that BnMYB2 is localized to the nucleus and cytoplasm. BnMYB2 transcription was significantly up-regulated by Cd stress. Heterologous overexpression of BnMYB2 in Arabidopsis enhanced Cd tolerance and accumulation in transgenic plants.
Supporting information
Nucleotides were numbered on the left. The deduced amino acid residues were showed under the corresponding codons. Asterisk indicates the stop codon.
(DOC)
The length of BnMYB2 gene promoter sequence was 1 947 bp. The A of the ATG initiation codon is defined as +1. The CAAT-box, TATA-box, ABRE, ARE, CGTCA-motif, ERE, P-box, STRE, WUN-motif and other important cis-regulatory elements are boxed and labeled.
(DOCX)
T0 seeds of transgenic Arabidopsis thaliana seedlings were germinated on MS medium supplemented with 30 mg⋅L-1 kanamycin for screening. Transgenic lines remained green, while the WT type turned yellow.
(DOCX)
M: Trans2K Plus II DNA Marker; P: positive plasmid control; CK: no template negative control; WT: wild type Arabidopsis thaliana seedling; L1-L12: BnMYB2 transgenic seedlings. The pBI121-BnMYB2 vectors were transferred by Agrobacterium tumefaciens-mediated genetic transformation into Arabidopsis thaliana. All overexpressing 35S::BnMYB2 transgenic lines (T1 generation) were verified by PCR using MYB2-35SF and MYB2-SPR primer.
(DOCX)
WT: wild type Arabidopsis thaliana seedling; L2-L12: BnMYB2 transgenic seedlings. Total RNA were extracted from leaves of overexpressing 35S:BnMYB2 transgenic lines (T2 generation) for qRT-PCR. BnMYB2 transcript levels were significantly high in several transgenic lines, the overexpression effect is excellent in L3 and L6 lines. Data are presented as the means of three biological replicates with SE shown by vertical bars.
(DOCX)
(DOCX)
Abbreviations
- AAS
Atomic absorption spectroscopy
- ABA
Abscisic acid
- ABRE
Abscisic acid response elements
- ARE
Anaerobic regulatory element
- As
Arsenic
- CAT
Catalase
- CBF
C-repeat binding factors
- Cd
Cadmium
- CFP
Cyan fluorescent protein
- Cu
Copper
- ERE
Ethylene response element
- GFP
Green fluorescent protein
- GSH
glutathione
- MeJA
Methyl jasmonate
- MS
murashige and skoog
- MYB
V-myb avian myeloblastosis viral oncogene homolog
- ORF
open reading frame
- Pb
Lead
- PCR
Polymerase chain reaction
- PCS
phytochelatin synthase
- RACE
Rapid amplification of cDNA end
- ROS
Reactive oxygen species
- RT-PCR
Reverse transcription polymerase chain reaction
- SANT
Switching-defective protein 3 [Swi3], adaptor 2 [Ada2], nuclear receptor co-repressor [N-CoR], transcription factor [TFIIB]
- SOD
Superoxide dismutase
- TFs
Transcription factors
- UAP
Universal Amplification Primer
- WT
wild-type
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by National Natural Science Foundation of China (No. 31371704, No. 31872877). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Duruibe J, Ogwuegbu MOC, Egwurugwu J. Heavy metal pollution and human biotoxic effects. Int J Phys Sci. 2007;2(5):112–118. [Google Scholar]
- 2.Zeng F, Ali S, Zhang H, Ouyang Y, Qiu B, Wu F, et al. The influence of pH and organic matter content in paddy soil on heavy metal availability and their uptake by rice plants. Environ Pollut. 2010;159(1):84–91. 10.1016/j.envpol.2010.09.019 [DOI] [PubMed] [Google Scholar]
- 3.Metwally A, Safronova VI, Belimov AA, Dietz KJ. Genotypic variation of the response to cadmium toxicity in Pisum sativum L. J Exp Bot. 2005;56(409):167–178. 10.1093/jxb/eri017 . [DOI] [PubMed] [Google Scholar]
- 4.Farinati S, DalCorso G, Varotto S, Furini A. The Brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants. New Phytol. 2010;185(4):964–978. 10.1111/j.1469-8137.2009.03132.x . [DOI] [PubMed] [Google Scholar]
- 5.Clemens S, Aarts MG, Thomine S, Verbruggen N. Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci. 2013;18(2):92–99. 10.1016/j.tplants.2012.08.003 . [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Hu Y, Liu Y. Heavy metal accumulation in iron plaque and growth of rice plants upon exposure to single and combined contamination by copper, cadmium and lead. Acta Ecolo Sin. 2009;29(6):320–326. 10.1016/j.chnaes.2009.09.011 [DOI] [Google Scholar]
- 7.Raskin I, Kumar PBAN, Dushenkov S, Salt DE. Bioconcentration of heavy metals by plants. Curr Opin Biotech. 1994;5(3):285–290. 10.1016/0958-1669(94)90030-2 [DOI] [Google Scholar]
- 8.Dahmani-Muller H, van Oort F, Gelie B, Balabane M. Strategies of heavy metal uptake by three plant species growing near a metal smelter. Environ Pollut. 2000;109(2):231–238. 10.1016/s0269-7491(99)00262-6 . [DOI] [PubMed] [Google Scholar]
- 9.Nascimento CWAd, Xing B. Phytoextraction: a review on enhanced metal availability and plant accumulation. Sci Agr. 2006;63(3):299–311. 10.1590/S0103-90162006000300014 [DOI] [Google Scholar]
- 10.Baldwin PR, Butcher DJ. Phytoremediation of arsenic by two hyperaccumulators in a hydroponic environment. Microchem J. 2007;85(2):297–300. 10.1016/j.microc.2006.07.005 [DOI] [Google Scholar]
- 11.Cojocaru P, Gusiatin Z, Cretescu I. Phytoextraction of Cd and Zn as single or mixed pollutants from soil by rape (Brassica napus). Environ Sci Pollut Res Int. 2016;23(11):10693–10701. 10.1007/s11356-016-6176-5 [DOI] [PubMed] [Google Scholar]
- 12.Xu P, Wang Z. A comparison study in cadmium tolerance and accumulation in two cool-season turfgrasses and Solanum nigrum L. Water Air Soil Poll. 2014;225 10.1007/s11270-014-1938-5 [DOI] [Google Scholar]
- 13.Peng X, Su S, Xia M, Lou K, Yang F, Peng S, et al. Fabrication of carboxymethyl-functionalized porous ramie microspheres as effective adsorbents for the removal of cadmium ions. Cellulose. 2018;25:1921–1938. 10.1007/s10570-018-1656-z [DOI] [Google Scholar]
- 14.Adesodun J, Atayese M, Agbaje T, Osadiaye B, Mafe O, Soretire A. Phytoremediation potentials of sunflowers (Tithonia diversifolia and Helianthus annuus) for metals in soils contaminated with zinc and lead nitrates. Water Air Soil Poll. 2010;207:195–201. 10.1007/s11270-009-0128-3 [DOI] [Google Scholar]
- 15.Maroco J, Rodrigues M, Lopes C, Chaves M. Limitations to leaf photosynthesis in field-grown grapevine under drought—metabolic and modelling approaches. Funct Plant Biol. 2002;29:451–459. 10.1071/PP01040 [DOI] [PubMed] [Google Scholar]
- 16.Balestrasse K, Gardey L, Gallego S, Tomaro M. Response of antioxidant defence system in soybean nodules and roots subjected to cadmium stress. Funct Plant Biol. 2001;28:497–504. 10.1071/PP00158 [DOI] [Google Scholar]
- 17.Sanita' di Toppi L, Gabbrielli R. Response to cadmium in higher plants. Environ Exp Bot. 1999;41(2):105–130. 10.1016/S0098-8472(98)00058-6 [DOI] [Google Scholar]
- 18.Fagioni M, D'Amici G, Timperio A, Zolla L. Proteomic analysis of multiprotein complexes in the thylakoid membrane upon cadmium treatment. J Proteome Res. 2008;8(1):310–326. 10.1021/pr800507x [DOI] [PubMed] [Google Scholar]
- 19.Qadir S, Qureshi MI, Javed S, Abdin M. Genotypic variation in phytoremediation potential of Brassica juncea cultivars exposed to Cd stress. Plant Sci. 2004;167:1171–1181. 10.1016/j.plantsci.2004.06.018 [DOI] [Google Scholar]
- 20.Lenka S, Katiyar A, Chinnusamy V, Bansal K. Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol J. 2011;9(3):315–327. 10.1111/j.1467-7652.2010.00560.x [DOI] [PubMed] [Google Scholar]
- 21.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58(2):221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- 22.Lipsick J. One billion years of MYB. Oncogene. 1996;13(2):223–235. [PubMed] [Google Scholar]
- 23.Du H, Wang YB, Xie Y, Liang Z, Jiang SJ, Zhang SS, et al. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013;20(5):437–448. 10.1093/dnares/dst021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 25.Supriya A, Sharma P, Yadav N, Yadav R. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Pla. 2013;19(3):307–321. 10.1007/s12298-013-0179-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Ng C, Fan L-M. MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot. 2014;114:80–91. 10.1016/j.envexpbot.2014.06.014 [DOI] [Google Scholar]
- 27.Xiong H, Li J, Pengli L, Duan J, Zhao Y, Guo X, et al. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS One. 2014;9(3):e92913 10.1371/journal.pone.0092913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feller A, Machemer K, Braun E, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 2011;66(1):94–116. 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- 29.Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281(49):37636–37645. 10.1074/jbc.M605895200 [DOI] [PubMed] [Google Scholar]
- 30.Ding Z, Li S, An X, Liu X, Huanju Q, Wang D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics. 2009;36(1):17–29. 10.1016/S1673-8527(09)60003-5 [DOI] [PubMed] [Google Scholar]
- 31.Lin CY, Nam TN, Fu SF, Hsiung YC, Chia LC, Lin CW, et al. Comparison of early transcriptome responses to copper and cadmium in rice roots. Plant Mol Biol. 2013;81(4–5):507–522. 10.1007/s11103-013-0020-9 [DOI] [PubMed] [Google Scholar]
- 32.Wang FZ, Chen MX, Yu LJ, Xie LJ, Yuan LB, Qi H, et al. OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front Plant Sci. 2017;8:1868 10.3389/fpls.2017.01868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Yang X, Kun H, Liu M, Li J, Gao Z, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- 34.Zhang P, Wang R, Ju Q, Li W, Tran LP, Xu J. The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiol. 2019;180(1):529–542. 10.1104/pp.18.01380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng X, Zhu S, Tang S, Liu T. Identification of drought, cadmium and root-lesion nematode infection stress-responsive transcription factors in ramie. Open Life Sci. 2016;11(1):191–199. 10.1515/biol-2016-0025 [DOI] [Google Scholar]
- 36.Yang B, Zhou M, Shu WS, Lan CY, Ye ZH, Qiu RL, et al. Constitutional tolerance to heavy metals of a fiber crop, ramie (Boehmeria nivea), and its potential usage. Environ Pollut. 2010;158(2):551–558. 10.1016/j.envpol.2009.08.043 [DOI] [PubMed] [Google Scholar]
- 37.Lei M, Yue QL, Chen TB, Huang ZC, Liao X, Liu YR, et al. Heavy metal concentrations in soils and plants around Shizuyuan mining area of Hunan Province. Acta Ecolo Sin. 2005;25(5):1146–1151. [Google Scholar]
- 38.She W, Jie Y, Xing H, Lu Y, Kang W, Wang D. Heavy metal concentrations and bioaccumulation of ramie (Boehmeria nivea) growing on 3 mining areas in shimen, lengshuijiang and Liuyang of Hunan province. Acta Ecolo Sin. 2011;31:874–881. [Google Scholar]
- 39.She W, Jie Y, Xing H, Luo Z, Kang W, Huang M, et al. Absorption and accumulation of cadmium by ramie (Boehmeria nivea) cultivars: a field study. Acta Agr Scand B-S P. 2011;61:641–647. 10.1080/09064710.2010.537678 [DOI] [Google Scholar]
- 40.She W, Zhu S, Jie Y, Xing H, Cui G. Expression profiling of cadmium response genes in ramie (Boehmeria nivea L.) root. Bull Environ Contam Toxicol. 2015;94(4):453–459. 10.1007/s00128-015-1502-z [DOI] [PubMed] [Google Scholar]
- 41.Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- 42.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 44.Corguinha A, Amaral de Souza G, Gonçalves V, Carvalho C, Lima W, Martins FAD, et al. Assessing arsenic, cadmium, and lead contents in major crops in Brazil for food safety purposes. J Food Compos Anal. 2014;37:143–150. 10.1016/j.jfca.2014.08.004 [DOI] [Google Scholar]
- 45.Viehweger K. How plants cope with heavy metals. Bot Stud. 2014;55:35 10.1186/1999-3110-55-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: Their role in drought response mechanisms. Int J Mol Sci. 2015;16(7):15811–15851. 10.3390/ijms160715811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramalingam A, Kudapa H, Pazhamala LT, Garg V, Varshney RK. Gene expression and yeast two-hybrid studies of 1R-MYB transcription factor mediating drought stress response in Chickpea (Cicer arietinum L.). Front Plant Sci. 2015;6:1117 10.3389/fpls.2015.01117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol. 2006;60(1):107–124. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- 49.Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant physiol. 2010;153(1):145–158. 10.1104/pp.110.153015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin D, Moon SJ, Han S, Kim BG, Park SR, Lee SK, et al. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011;155(1):421–432. 10.1104/pp.110.163634 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS, et al. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008;18(10):1047–1060. 10.1038/cr.2008.280 . [DOI] [PubMed] [Google Scholar]
- 52.Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, et al. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant physiol. 2009;151(1):275–289. 10.1104/pp.109.144220 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15(13):1196–1200. 10.1016/j.cub.2005.05.048 . [DOI] [PubMed] [Google Scholar]
- 54.Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9(10):1859–1868. 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nazarli H, Ahmadi A, Hadian J. Salicylic acid and methyl jasmonate enhance drought tolerance in chamomile plants. J Herbmed Pharmacol. 2014;3(2):87–92. [Google Scholar]
- 56.Anjum S, Wang L, Farooq M, Khan I, Xue L. Methyl Jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J Agron Crop Sci. 2011;197(4):296–301. 10.1111/j.1439-037X.2011.00468.x [DOI] [Google Scholar]
- 57.Maksymiec W. Signaling responses in plants to heavy metal stress. Acta Physiol Plant. 2007;29(3):177–187. 10.1007/s11738-007-0036-3 [DOI] [Google Scholar]
- 58.Weber M, Trampczynska A, Clemens S. Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd(2+)-hypertolerant facultative metallophyte Arabidopsis halleri. Plant Cell Environ. 2006;29(5):950–963. 10.1111/j.1365-3040.2005.01479.x [DOI] [PubMed] [Google Scholar]
- 59.Liu T, Zhu S, Tang Q, Tang S. Genome-wide transcriptomic profiling of ramie (Boehmeria nivea L. Gaud) in response to cadmium stress. Gene. 2015;558(1):131–137. 10.1016/j.gene.2014.12.057 [DOI] [PubMed] [Google Scholar]
- 60.Zhang SC, Feng SN, Gu W, Feng LG, Wang YP, Liang ZS. Cloning, subcellular localization and expression analysis of a transcription factor gene SmMYB87 in Salvia miltiorrhiza. Chinese Traditional and Herbal Drugs. 2017;48:3597–3604. 10.7501/j.issn.0253-2670.2017.17.023 (in Chinese, English summary). [DOI] [Google Scholar]
- 61.Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hulskamp M. Functional diversity of R3 single-repeat genes in trichome development. Development. 2009;136(9):1487–1496. 10.1242/dev.021733 . [DOI] [PubMed] [Google Scholar]
- 62.Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, et al. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development. 2007;134(21):3873–3882. 10.1242/dev.009597 . [DOI] [PubMed] [Google Scholar]
- 63.Serna L. CAPRICE positively regulates stomatal formation in the Arabidopsis hypocotyl. Plant Signal Behav. 2008;3(12):1077–1082. 10.4161/psb.3.12.6254 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X, Chen L, Dhaubhadel S. 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 2012;71(2):239–250. 10.1111/j.1365-313X.2012.04986.x . [DOI] [PubMed] [Google Scholar]
- 65.Ai TN, Naing AH, Yun BW, Lim SH, Kim CK. Overexpression of RsMYB1 enhances anthocyanin accumulation and heavy metal stress tolerance in transgenic petunia. Front Plant Sci. 2018;9:1388–1388. 10.3389/fpls.2018.01388 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotides were numbered on the left. The deduced amino acid residues were showed under the corresponding codons. Asterisk indicates the stop codon.
(DOC)
The length of BnMYB2 gene promoter sequence was 1 947 bp. The A of the ATG initiation codon is defined as +1. The CAAT-box, TATA-box, ABRE, ARE, CGTCA-motif, ERE, P-box, STRE, WUN-motif and other important cis-regulatory elements are boxed and labeled.
(DOCX)
T0 seeds of transgenic Arabidopsis thaliana seedlings were germinated on MS medium supplemented with 30 mg⋅L-1 kanamycin for screening. Transgenic lines remained green, while the WT type turned yellow.
(DOCX)
M: Trans2K Plus II DNA Marker; P: positive plasmid control; CK: no template negative control; WT: wild type Arabidopsis thaliana seedling; L1-L12: BnMYB2 transgenic seedlings. The pBI121-BnMYB2 vectors were transferred by Agrobacterium tumefaciens-mediated genetic transformation into Arabidopsis thaliana. All overexpressing 35S::BnMYB2 transgenic lines (T1 generation) were verified by PCR using MYB2-35SF and MYB2-SPR primer.
(DOCX)
WT: wild type Arabidopsis thaliana seedling; L2-L12: BnMYB2 transgenic seedlings. Total RNA were extracted from leaves of overexpressing 35S:BnMYB2 transgenic lines (T2 generation) for qRT-PCR. BnMYB2 transcript levels were significantly high in several transgenic lines, the overexpression effect is excellent in L3 and L6 lines. Data are presented as the means of three biological replicates with SE shown by vertical bars.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.