Abstract
This is a narrative review on the potential of rapid and point-of-care microbiological testing in pneumonia patients, focusing particularly on hospital-acquired and ventilator-associated pneumonia, which have substantial mortality and diverse microbiology. This work is written from a United Kingdom perspective, but much of it is generalizable internationally. In a world where antimicrobial resistance is a major international threat, the use of rapid molecular diagnostics has great potential to improve both the management of pneumonia patients and the stewardship of antibiotics. Rapid tests potentially can distinguish patients with bacterial versus viral infection and can swiftly identify bacterial pathogens and their resistances. We seek to answer the question: “Can such tests be used as an antibiotic guardian?” Their availability at the bedside rather than in the laboratory should best ensure that results are swiftly used to optimize patient management but will raise new challenges, not the least with respect to maintaining quality control and microbiology/infection control input. A further challenge lies in assessing the degree of trust that treating clinicians will place in these molecular diagnostic tests, particularly when early de-escalation of antibiotic therapy is indicated.
Keywords: point-of-care testing (POCT), antimicrobial resistance, pneumonia, rapid molecular diagnostics, biomarkers, next-generation sequencing
Introduction
Antimicrobial resistance (AMR) is a major public health issue, increasing morbidity and mortality, as illustrated by population-level modeling across Europe.1 Numerous governmental and agency reports assert that the best routes to managing this growing challenge lie in (i) better infection control, minimizing the need to use antibiotics, (ii) reinvigorating antibiotic discovery and development, and (iii) better “stewardship” of available antibiotics, meaning swift administration of the “right antibiotic at the right dose for the right duration” for those patients with a significant bacterial infection, while ensuring that antibiotics are not given to patients with viral infections who will not benefit from them. Unfortunately, stewardship is complicated by the fact that most initial antibiotic use is empiric, with the physician not knowing the identity or antibiotic resistances of the pathogen, if one is grown at all, until 2–3 days into therapy. In countries with moderate rates of resistance, including the United States and much of Europe, this leads to precautionary overtreatment, with broad-spectrum antibiotics given to patients who transpire either to have not-very-resistant pathogens or not to have bacterial infections. In countries with high rates of resistance and particularly where carbapenemase-producing Gram-negative bacteria are prevalent (as in India), the delay in pathogen identification leads to forced empiric use of less effective and potentially toxic agents, notably colistin combinations. In some cases, even these fail to cover the pathogen(s) ultimately grown.
It is widely agreed (and reflected in the United Kingdom (UK) Government’s 5 year action plan on AMR)2 that this undesirable situation could be improved by deploying rapid diagnostics to better discriminate between bacterial and viral infection and/or to deliver accelerated profiling of bacterial pathogens and their resistances. Such information should drive much better antibiotic stewardship, promoting early use of narrow-spectrum agents targeted against the particular pathogen(s) present, instead of the current model, where guidelines favor broad-spectrum agents chosen to cover all pathogens likely to be present until microbiology results become available, based on epidemiological surveillance data. Rapid results should also prompt termination of antibacterial therapy in patients found to have viral infections. Ideally, a “rapid” diagnostic should give an immediate result; in practical terms, a result within one dosage interval (typically 8 h) for an initially given empiric antibiotic seems a good target, allowing early refinement of therapy.
This review focuses on the potential utility of rapid molecular and point-of-care testing (POCT) diagnostics in patients with respiratory infection, particularly those admitted to hospital with community-acquired pneumonia (CAP) and those who, during their stay, develop hospital-acquired or ventilator-associated pneumonia (HAP and VAP, respectively). This work is written from a UK perspective, but the content is internationally relevant, since the pathogens of pneumonia are largely identical worldwide, though resistance rates and healthcare delivery systems vary by country.
Severe pneumonia has high mortality and is a common cause of admission to an intensive care unit (ICU), with the current situation complicated by the COVID-19 pandemic. Empirical broad-spectrum antibiotics are ordinarily used to treat pneumonia and are often continued for prolonged periods even when no bacterial pathogen is cultured.3
In normal (i.e., pre-COVID-19) times, it was estimated that 0.5–1% of UK adults would develop CAP in a given year;4 moreover, pneumonia is diagnosed in 5–12% of adults who present to general practitioners (GPs) with symptoms of lower respiratory tract infection (LRTI). Around 22–42% of these CAP patients are admitted to hospital, where their mortality rate is between 5 and 14%.4 Between 1.2 and 10% of adults admitted to hospital with CAP are managed in intensive care, and among these, mortality exceeds 30%.4 In addition, debilitated patients commonly develop pneumonia while in hospital, and this risk increases with mechanical ventilation, where tubing and liquid traps become colonized by bacteria that then reach the lungs. HAP (even excluding VAP) is estimated by the UK’s National Institute of Clinical Excellence (NICE) to increase a hospital stay by 8 days on average and has a reported mortality between 30 and 70%, “with variations in clinical management and outcomes across the UK”.4 In our experience, such rates are for severe pneumonia and not for the many HAP patients “who have a few crackles”.
Current practice for a patient with clinically suspected pneumonia (regardless of type) is to take a sputum or endotracheal aspirate sample (ETA), send it to the microbiology laboratory for culture, and to start empirical antibiotics in line with the hospital’s local antibiotic prescribing policy, which generally reflects national guidelines (Figure 1). These sample types are easy to take but are prone to contamination with upper respiratory tract flora, leading to uncertainty about whether the organisms detected are pathogens or colonists.
Figure 1.
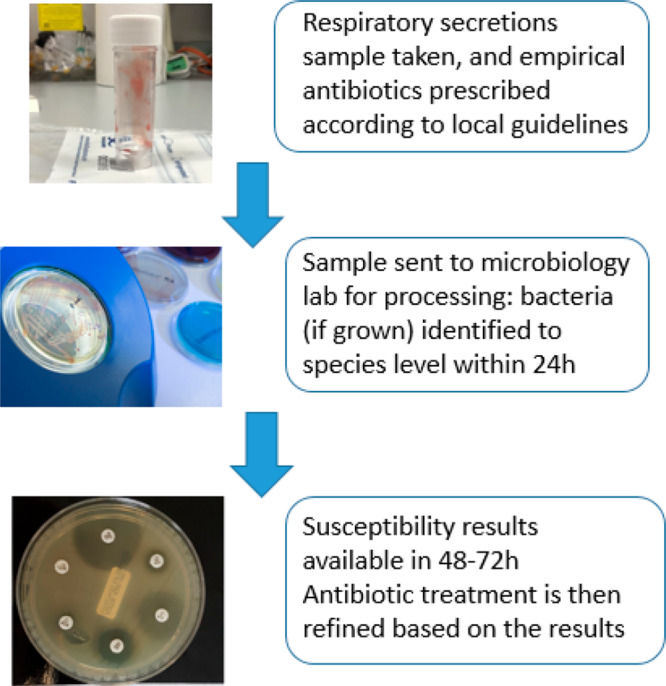
Classical microbiology processing.
Bronchoalveolar lavage (BAL) delivers a deep-lung specimen and is widely performed in some countries such as France and the United States (US), but it is rarely done in the UK, being seen as invasive and carrying some risk.5 Imaging findings, oxygen requirements, and inflammatory markers all contribute to the clinical differential diagnosis. Cultures with susceptibility tests on relevant pathogens take 2–3 days, and the processing, interpretation, and reporting of these results varies among laboratories. Adopting the model long established in Germany and increasingly seen in the UK, whereby a few centralized laboratories serve multiple hospitals, enhances test standardization and quality control, but this may delay sample processing for remote sites. Moreover, even in clinical trials, only around 30–40% of CAP patients have a pathogen grown; in routine practice, the proportion with a clear microbiological diagnosis was only 28.5%.6 Major factors here are (i) that common CAP pathogens, notably, Streptococcus pneumoniae and Haemophilus influenzae, are difficult to culture, leading to low recovery rates and (ii) that when scanty or mixed opportunist organisms are grown from HAP and VAP patients as common with sputa and ETA samples the microbiologist must make a subjective judgment of their significance. Furthermore, HAP and VAP are difficult to diagnose clinically in patients with other multiple pathologies.7 These multiple diagnostic uncertainties are reflected (i) in variations in treatment duration, (ii) in patients receiving repeated courses of antibiotics for vague ongoing symptoms thought to be due to a pneumonia, and (iii) in UK hospital guidelines advocating the antibiotic doxycycline in “mild HAP” despite an inappropriate spectrum and a lack of national or international guideline support.8 For community respiratory infections, patient expectation plays a big role in inappropriate antimicrobial prescribing, as do the constraints put on GPs, including their 10 min consultations in the UK.9
Etiology of Pneumonia
Bacterial CAP is usually attributable to a narrow range of pathogens, principally S. pneumoniae, H. influenzae, Moraxella catarrhalis, Legionella pneumophila, and the “atypicals” collectively including Chlamydophila pneumoniae and Mycoplasma pneumoniae.
The opportunistically pathogenic agents of HAP and VAP are more diverse: Enterobacterales, Pseudomonas aeruginosa, and Staphylococcus aureus are all prominent globally, each accounting for around a quarter to a third of cases.10 The importance of other pathogens in HAP/VAP varies geographically. This is particularly the case for Acinetobacter baumannii, which is widely listed among the most prominent ICU-VAP pathogens in East and South Asia as well as in Latin America11,12 but is uncommon in the UK. It is unclear whether this variation reflects (i) the effectiveness of infection control, (ii) climatic factors, with A. baumannii thriving in warmer and moister conditions, (iii) antibiotic pressure, or (iv) differing patient populations, particularly with respect to admission of terminally ill (and very-Acinetobacter-vulnerable) patients to the ICU. Although single Gram-negative and -positive bacteria strains are often responsible for HAP and VAP, Girard et al.13 described 276/946 (29.2%) of VAPs as polymicrobial.
Gram-negative bacteria are increasingly multi-drug-resistant in many countries, driving the sort of polypharmacy indicated in the Infectious Disease Society of America (IDSA) guidelines.14 These advocate that if a unit has a MRSA rate of >10–20% and >10% resistance to anti-Gram-negative agents (conditions that apply in virtually all ICUs across much of Asia or Latin America as well as many in Europe and the US), then empiric therapy for HAP/VAP should comprise two anti-pseudomonal agents plus an anti-staphylococcal agent active against MRSA. While this approach covers likely pathogens, it can hardly be seen as “stewarding” antibiotics or minimizing undesirable side effects, including disruption of the gut flora, which may allow proliferation of resistant organisms and/or Clostridium difficile. In settings with extreme rates of resistance, even colistin comes to be considered a component of empirical treatment for pneumonia, despite significant toxicity and questionable pharmacodynamics in the lung.15
Important resistance mechanisms among the bacterial pathogens of pneumonia are summarized in Table 1. This table also highlights the diversity of these mechanisms within species groups and whether resistance either largely involves acquired genes carried by plasmids or transposons or largely is attributable to chromosomal mutations. These aspects become critically important when seeking to predict resistance phenotypes from molecular data generated in the types of tests outlined below. It is far easier to use a molecular method to predict oxacillin resistance in staphylococci, almost always attributable to acquired mecA, than it is to predict oxyimino-cephalosporin resistance in Klebsiella pneumoniae, where the possible determinants include a wide range of different β-lactamase genes, some of them point mutants of genes that determine β-lactamases unable to attack these cephalosporins.
Table 1. Frequency and Genetic Diversity of Critical Modes of Resistance: Implications for the Development of Rapid Genetic Tests.
| Gram-negative bacteria | ampicillin | oxyimino-cephalosporins | carbapenems | aminoglycosides | fluoroquinolones | trimethoprim | colistin |
|---|---|---|---|---|---|---|---|
| Enterobacterales | endogenous (i.e., chromosomally mediated) β-lactamases in Klebsiella, indole-positive Proteeae Enterobacter, Citrobacter, Serratia; otherwise great diversity of plasmid-mediated β-lactamases, most often CTX-M types but mutant TEM and SHV types also frequenta | up-regulation of endogenous AmpC β-lactamases in indole-positive Enterobacter, Citrobacter, Proteeae, Serratia; otherwise acquired ESBLs of the CTX-M, TEM, and SHV familiesa | acquired OXA-48, KPC, IMP, VIM or NDM carbapenemases or (especially ertapenem) combinations of AmpC or ESBL activity together with mutational porin lossb | endogenous modifying enzymes in Serratia. Otherwise great diversity of acquired enzymes that acetylate, nucleotidylate or phosphorylate aminoglycosides or which methylate their target rRNAa | mostly via multiple mutations affecting gyrase and topoisomerase targets; sometimes augmented by acquired enzymes that cause efflux or modification of fluoroquinolonesc | mostly diverse acquired dihydrofolate reductase enzymesa | inherent in Proteeae and Serratia; otherwise mostly regulatory mutations that lead, circuitously, to the modification of lipopolysaccharides, reducing their binding of colistin; rarely via acquisition of plasmid mediated enzymes that similarly modify LPSa |
| P. aeruginosa | inherent resistance to ampicillin; piperacillin resistance largely via up-regulation of multiple endogenous efflux mechanisms or derepression of chromosomal AmpC β-lactamasesa | largely via up-regulation of multiple endogenous efflux mechanisms or derepression of chromosomal AmpC β-lactamasesa | largely by loss of porin OprD (together with efflux in the case of Meropenem); rarely via acquisition of carbapenemasesa | mostly via upregulation of endogenous efflux or reduction of permeability; less commonly via acquired enzymes that acetylate, nucleotidylate, or phosphorylate aminoglycosides or which methylate their target rRNAa | multiple mutations affecting gyrase and topoisomerase targetsc | inherent resistancef | regulatory mutations that lead, circuitously, to the modification of lipopolysaccharides, reducing their binding of colistind |
| A. baumannii | acquired plasmid-mediated β-lactamases nearly universala | up-regulation of chromosomal AmpC β-lactamasesa | mostly acquired OXA-23/40/58 β-lactamases; occasionally up-regulation of chromosomal OXA-51-like β-lactamasesb | great diversity of acquired enzymes that acetylate, nucleotidylate or phosphorylate aminoglycosides or which methylate their target rRNAa | mutations affecting gyrase and topoisomerase targetsc | inherent resistancef | regulatory mutations that lead, circuitously, to the modification of lipopolysaccharides, reducing their binding of colistind |
| H. influenzae | acquired TEM β-lactamase or mutation and mosaic formation in genes encoding penicillin-binding proteinsa | extremely rare: mutation and mosaic formation in genes encoding penicillin-binding proteinsf | extremely rare: mutation and mosaic formation in genes encoding penicillin-binding proteinsf | unknown: aminoglycosides not ordinarily used against H. influenzae infectionsd | multiple mutations affecting gyrase and topoisomerase targetsc | mutations affecting chromosomal dihydrofolate reductase enzymesf | extremely rare: drug not ordinarily used against speciesf |
| M. catarrhalis | acquired BRO-1 and BRO-2 β-lactamasese | extremely raref | extremely raref | unknown: aminoglycosides not ordinarily used against M. catarrhalis infections | extremely rare: mutations affecting gyrase and topoisomerase targetsc | inherentf | inherentf |
| Gram-positive bacteria | ampicillin | oxyimino-cephalosporins | carbapenems | aminoglycosides | fluoroquinolones | macrolides | tetracyclines |
|---|---|---|---|---|---|---|---|
| S. aureus | acquired β-lactamases to some penicillin; otherwise, resistance to β-lactamase-stable penicillins, anti-staphylococcal cephalosporins, and carbapenems almost always due to acquired mecA gene, encoding a PBP bypass mechanisme | diversity of acquired enzymes that acetylate, nucleotidylate or phosphorylate aminoglycosides or which methylate their target rRNAa | mutations affecting gyrase and topoisomerase targetsc | mostly owing to acquired enzymes that methylate rRNA preventing interaction with macrolidesb | acquired efflux pumps or ribosome modifying enzymesb | ||
| S. pneumoniae | diverse mutations and transformations leading to mosaic penicillin binding protein genesa | inherently resistantf | mutations affecting gyrase and topoisomerase targetsc | acquired enzymes that methylate rRNA preventing interaction with macrolides, or acquired efflux pumpsa | mostly acquired (TetM) ribosome-modifying enzymesb | ||
Resistance mechanisms highly diverse. This may because many different acquired enzymes can be involved, as with the multiplicity of different penicillin- and cephalosporin-hydrolyzing β-lactamases that occur in Enterobacterales16 or because, although a biochemical or biophysical mechanism is well conserved, its genetic underpinnings are variable. Thus, for example, many different mutations can disrupt porin OprD in P. aeruginosa, all leading to impermeability to carbapenems.17 PCR based prediction of such a resistance is difficult or impossible, though both sequencing and rapid phenotypic tests remain viable approaches.
Resistance is almost always due to one of 6 or fewer acquired genes: good scope to seek by PCR tests.
Resistance mechanisms are well-conserved, but mutational, making them difficult to seek without sequencing.
Diversity of mechanisms (at the genetic level) is uncertain.
Resistance is almost always due to 1 or 2 acquired genes which could be easily sought by molecular tests.
Inherent resistance, predictable for the species/genus identification.
The prevalence of resistance varies hugely among countries. In the UK, around 13% of K. pneumoniae isolated from LRTIs have extended-spectrum β-lactamases (ESBLs), conferring resistance to oxyimino-cephalosporins and around 0.9% have carbapenemases (according to BSAC surveillance data).71 In India, these proportions rise to 86.9 and 56.6%, respectively.18 The relative prevalence of particular mechanisms also varies geographically: KPC enzyme is the predominant carbapenemase of Enterobacterales in the Americas, China, Italy, Israel, Greece, and Portugal, but OXA-48 enzyme dominates elsewhere in Europe and in the Middle East. NDM is the major carbapenemase among Enterobacterales in the Indian subcontinent.19 As a second example, most resistance to β-lactams in P. aeruginosa is mutational in Europe and the US, whereas large proportions of resistant Pseudomonas in the Middle East have acquired ESBLs or carbapenemases.20,21
In summary, the slowness and poor pathogen-recovery rates of conventional microbiology impacts clinical decision-making and, in particular, delays the stopping or narrowing of antibiotic therapy for many patients with susceptible pathogens. Equally, it delays the initiation of active therapy for those patients with particularly resistant pathogens, delaying cures, increasing mortality, prolonging hospital stay, and adding to costs.22,23 Presently, there is no reliable tool to facilitate swift refinement of the patient’s empirical antibiotics. Such tests would play the part of an invaluable antibiotic guardian but face the challenge of having to seek multiple pathogens and, in some cases, a great diversity of resistance mechanisms.
Point-of-Care Testing: What’s the Point?
Rapid diagnostics potentially could improve both the care of the pneumonia patient and antibiotic stewardship.24,25 Potentially useful types of tests, in context, include those to (i) measure inflammatory markers to better distinguish bacterial versus viral pneumonia, (ii) seek specific respiratory viruses such as influenza, (iii) seek broad arrays of bacteria, viruses, and resistance genes, (iv) detect atypical bacteria, meaning those that cannot readily be grown in the laboratory, and (v) detect urinary antigens specific for Legionella and pneumococci.
To be most useful, a rapid method needs to be deployed as a Point of Care Test (POCT), providing a result at the hospital patient’s bedside or in the GP surgery. Bedside tests eliminate the transport delay to the laboratory, accelerating decisions about patient management, although they also introduce challenges of their own, discussed later in this article. Sexual Health Services have been using POCTs several years, and these are highly valued among their patients.26
POCT of Inflammatory Markers to Distinguish Viral and Bacterial Infection
C-reactive protein (CRP) is an acute phase protein expressed in response to infection or inflammation. CRP tests can be used for the differential diagnosis of bacterial and viral respiratory infections in the community, albeit mostly for conditions less serious than pneumonia. Such tests are widely employed by GPs in Scandinavia and The Netherlands, partly because of their direct value in identifying patients likely to benefit from antibiotics but also because a negative result provides objective justification to deny unnecessary antibiotics to a demanding patient.27
A randomized cluster study including 20 general practices in The Netherlands showed that deployment of CRP POCT significantly reduced antibiotic prescribing in LRTI, without compromising care.28 A large trial, involving 25 497 Spanish patients, demonstrated that CRP testing was useful in reducing prescriptions to GP patients who specifically asked for antibiotics.29 A UK study suggested a financial benefit to using CRP tests in the community, though these may be hard to achieve given the low acquisition cost of generic antibiotics.30 A review concluded that CRP tests could aid appropriate antibiotic prescriptions as well as being cost-effective.27 In the UK, NICE guidelines advocate the use of CRP tests in primary care to reduce antibiotic use, suggesting prescribing antibiotics only if the CRP is above 100 mg/L.4 This view may (or may not) be supported by the ongoing POCT to Target Antibiotics for Chronic Obstructive Pulmonary Disease Exacerbations (PACE) randomized control trial, using CRP tests to target antibiotic prescribing in community patients with acute exacerbations of chronic pulmonary disease.31
Procalcitonin, a peptide precursor of the hormone calcitonin, has been the subject of much debate as an indicator of bacterial infection, and there is evidence to suggest its use as an adjunctive tool for antimicrobial stewardship.32 Briefly, procalcitonin concentrations are raised in bacterial pneumonia but also can be elevated owing to chronic kidney disease, malignancies, burns, trauma, and by some immune-modulating drugs. One UK-based study used procalcitonin testing in 99 acute medical and 42 ICU patients who were thought to have an infection, denying antibiotics if concentrations were low. This approach led to a reduction in antimicrobial use, with no infection-related deaths.33 A Swiss trial used procalcitonin tests in patients admitted with CAP, randomizing them to a standard care arm or to one where antibiotic use was guided by procalcitonin concentrations.34 A total of 302 patients were recruited, and the duration of antibiotic therapy was reduced from a median of 12 days in the standard care arm to 5 days in the procalcitonin arm (55% reduction; p < 0.001).
On the basis of positive data such as these, a 1 day meeting between 19 experts from 12 countries (funded by ThermoFisher Scientific, as a test manufacturer)35 configured three algorithms (Table 2) to guide the use of procalcitonin with the aim of aiding antimicrobial stewardship, based on interventional trials. Clinical assessment, radiographic assessment, and microbiological workup were used to assess the probability of bacterial infection.
Table 2. Algorithms Developed for Procalcitonin Use35.
| bacterial infection uncertain and PCT below threshold | bacterial infection uncertain and PCT at or above threshold | bacterial infection highly suspected and PCT below threshold | bacterial infection highly suspected and PCT at or above threshold | |
|---|---|---|---|---|
| patient with mild illness outside ICU; PCT threshold 0.25 ng/mL | Bacterial infection unlikely. Withhold antibiotics (Abx); consider other diagnostic tests to establish diagnosis. | Bacterial infection likely. Use Abx based on clinical judgment. | Bacterial infection possible. Use empiric Abx based on clinical judgment; consider other diagnostic tests. | Bacterial infection highly likely. Use Abx based on clinical judgment. |
| patient with moderate illness outside ICU; PCT threshold 0.25 ng/mL | Bacterial infection unlikely. Use empiric Abx based on clinical judgment; consider other diagnostic tests. | Bacterial infection likely. Use Abx based on clinical judgment. | Bacterial infection possible. Use empiric Abx based on clinical judgment; consider other diagnostic tests. | Bacterial infection highly likely. Use Abx based on clinical judgment. |
| patient with severe illness in ICU; PCT threshold 0.5 ng/mL | Bacterial infection unlikely. Use empiric Abx based on clinical judgment; consider other diagnostic tests. | Bacterial infection likely. Use Abx based on clinical judgment. | Bacterial infection possible. Use empiric Abx based on clinical judgment; consider other diagnostic tests. | Bacterial infection highly likely. Use Abx based on clinical judgment. |
Other analyses are less positive. A meta-analysis, which included 12 studies on the use of procalcitonin testing in patients with CAP,36 concluded that it was an unreliable test when deciding whether or not to initiate antibiotics, finding a pooled sensitivity of 0.55 and a specificity of 0.76. Similarly, the UK’s NICE concluded that more evidence is needed with respect to using procalcitonin data to support stopping antibiotic treatment in ICU cases with confirmed or strongly suspected sepsis and or starting and stopping antibiotic treatment in people with suspected bacterial infection presenting to the emergency department.37
One possible refinement is to monitor changes in procalcitonin over time rather than obtaining a single “snapshot” reading. On this basis, the multicenter PRORATA randomized clinical trial (RCT),38 undertaken in France and Germany, monitored procalcitonin concentrations daily for patients on antibiotics and at each infectious episode, until day 28. It recruited 307 patients to the procalcitonin-guided group and 314 to a control group, given empirical antibiotics according to guidelines. Patients in the procalcitonin group had significantly more antibiotic-free days (out of the 28) than those in the control group (14.3 vs 11.6 days; 95% CI 1.4 to 4.1, p < 0.0001). However, an observational study on an ICU in Israel concluded that serial early procalcitonin measurements (at 0, 6, and 12 h) were of no more use than a single initial value.39
Other biomarkers may have potential to guide antimicrobial stewardship, besides CRP and procalcitonin, but have been studied less extensively. Hellyer et al. undertook the VAPrapid-2 study, where BAL samples were tested for IL-8 and IL-1β40 as markers for VAP, which was defined as growth of ≥104 colony forming units (CFU) per milliliter of BAL fluid. Initial observational work showed that the combined measurement of IL-1β and IL-8 could be configured to exclude VAP with a sensitivity of 100%, a specificity of 44.3%, and a negative predictive value (NPV) of 1, making it a good “rule out” test. A subsequent RCT, with 104 patients randomized to the biomarker-guided recommendation on antibiotics and 106 randomized to routine use of antibiotics, found no significant difference in days of antibiotic treatment in the week following BAL in the intention-to-treat analysis (p = 0·58). The authors attributed this failure to local prescribing culture/habits and to the fact that testing was laboratory-based, not as a POCT, extending turnaround time and precluding immediate impact on the treating physicians.40,41 Horonenko et al.42 assayed Soluble Triggering Receptor (sTREM-1, a receptor expressed on myeloid cell-1) in BAL fluid and exhaled ventilator condensate as a possible predictor of VAP, which they defined using a clinical pulmonary infection scoring system.43 The results were promising, although numbers are small, with sTREM-1 detected in the condensate from 11 of the 14 subjects with VAP, but from only 1 of 9 subjects without infection.42
Future diagnostic prospects, presently in their infancy, include using large batteries of host transcriptome biomarkers. One panel suggested to be of value in pneumonia, the HostDx Sepsis test, has been developed by Inflammatix in the US.44 This uses a blood sample from the patient to identify the presence, type (bacterial/viral), and severity of an acute infection in 30 min. It seeks 29 mRNAs produced by white cells in peripheral blood that may have their expression modulated by infection. Likelihood ratios are calculated using proprietary bioinformatics data at Inflammatix’ laboratory. Specific details of the gene set have not been released, and the test requires clinical evaluation. For a wider review of biomarkers in sepsis, rather than pneumonia, readers are referred to Teggert et al.45
Rapid Methods to Seek Respiratory Viruses and “Atypical” Bacteria
To date, both pre-COVID-19 and currently, rapid microbiological PCR-based diagnostics, mostly run in the laboratory rather than as POCT, have been used more to seek respiratory viruses rather than bacterial pathogens, typically among emergency admissions with respiratory symptomology. Their wide adoption in this role is partly because detection of a pathogenic virus, which should not be present in a healthy patient, gives a clear diagnosis, whereas detection of a low burden of, for example, Acinetobacter in a ventilated ICU patient does not. Moreover, classical virology, unlike classical bacteriology, is complex and costly. These systems also potentially allow for the rapid detection of high consequence infections in patients, including Middle East respiratory syndrome (MERS) coronavirus, essential for patient management/infection control purposes. If a virus is found, then it may prompt specific antiviral therapy, particularly in the case of influenza. Moreover, finding a pathogenic virus, including SARS-CoV2, should discourage the use of broad-spectrum antibacterials, though narrower spectrum coverage against likely secondary pathogens (e.g., S. aureus and S. pneumoniae following influenza) may be preferred.
Available systems and the pathogens they seek are tabulated in Table 3; it should be assumed that all manufacturers are adjusting panels to include SARS-CoV2. Some include difficult-to-grow bacteria as well as viruses, notably including the agents of atypical pneumonia. Some (e.g., the BioFire FilmArray) can also run different PCR panels, suited to seeking wider ranges of bacteria, including agents of HAP and VAP. There are also many specific POCT panels for influenza. These are beyond the scope of this article, and readers are directed to Egilmezer et al.46 for a detailed review.
Table 3. Respiratory Molecular Panels: Organisms and Resistance Genes Soughta.
| manufacturer | Curetis
Unyvero |
BioFire
FilmArray |
Nanosphere | Hologic | Seegene | |||
|---|---|---|---|---|---|---|---|---|
| panel | P50 | P55 | lower respiratory tract (pneumonia) | “respiratory” | “pneumonia” | Verigene RV+ | Gen-Probe Prodesse assays | respiratory panel |
| sample type | sputum, BAL | sputum, BAL | tracheal aspirate | naso-pharyngeal swab (NPS) | BAL, sputum, tracheal aspirate | NPS | NPS | NPS, NP aspirate, BAL, sputum |
| Viruses | ||||||||
| adenovirus | × | × | × | × | ||||
| coronavirus | × | × | × | |||||
| human metapneumovirus | × | × | × | × | ||||
| human rhinovirus/enterovirus | × | × | × | |||||
| influenza A | × | × | × | × | × | |||
| influenza B | × | × | × | × | × | |||
| parainfluenza virus | × | × | × | × | ||||
| respiratory syncytial virus | × | × | × | × | × | |||
| human bocavirus | × | |||||||
| MERS Co-V | ×b | |||||||
| Bacteria | ||||||||
| Acinetobacter calcoaceticus-baumannii complex | × | × | × | × | ||||
| Stenotrophomonas maltophilia | × | × | × | |||||
| Enterobacter cloacae complex | × | × | × | |||||
| Enterobacter spp. | × | |||||||
| Escherichia coli | × | × | × | × | ||||
| H. influenzae | × | × | × | × | × | |||
| Klebsiella aerogenes | × | × | ||||||
| Klebsiella oxytoca | × | × | × | × | ||||
| K. pneumoniae | × | × | × | × | ||||
| Klebsiella variicola | × | × | ||||||
| Moraxella catarrhalis | × | |||||||
| Proteus spp. | × | × | × | × | ||||
| Morganella morganii | × | × | × | |||||
| Citrobacter freundii | × | × | ||||||
| Moraxella catarrhalis | × | × | × | |||||
| P. aeruginosa | × | × | × | × | ||||
| Serratia marcescens | × | × | × | × | ||||
| S. aureus | × | × | × | × | ||||
| Streptococcus agalactiae | × | |||||||
| S. pneumoniae | × | × | × | × | ||||
| Streptococcus pyogenes | × | |||||||
| Streptococcus mitis group | × | |||||||
| C. pneumoniae | × | × | × | × | × | × | ||
| L. pneumophila | × | × | × | × | × | |||
| M. pneumoniae | × | × | × | × | × | |||
| Bordetella parapertussis | × | × | ||||||
| Bordetella pertussis | × | × | ||||||
| Fungi | ||||||||
| Pneumocystis jirovecii | × | × | × | |||||
| Resistance Genes | ||||||||
| ermA | × | |||||||
| ermB | × | × | ||||||
| ermC | × | |||||||
| msrA | × | |||||||
| mefA/E | × | |||||||
| mecA | × | × | × | × | ||||
| mecC | × | × | ||||||
| blaTEM | × | × | × | |||||
| blaSHV | × | × | ||||||
| blaCTX-M | × | × | × | × | ||||
| blaEBC | × | |||||||
| blaDHA | × | |||||||
| blaKPC | × | × | × | × | ||||
| blaIMP | × | × | ||||||
| blaNDM | × | × | × | |||||
| blaVIM | × | × | × | |||||
| blaOXA-23 | × | × | ||||||
| blaOXA24/40 | × | × | ||||||
| blaOXA-48 | × | × | × | |||||
| blaOXA-58 | × | × | ||||||
| blaOXA 51 like | × | |||||||
| int1 | × | |||||||
| sul1 | × | × | ||||||
| gyrA83 | × | × | ||||||
| gyrA87 | × | × | ||||||
| parC | × | |||||||
This table include the major tests which seek bacteria and viruses; other more specific tests are available, but are excluded.
Only on the Pneumonia Plus Panel.
A systematic review by Huang et al. in 201847 considered the performance of the FilmArray, Verigene RV+, and Prodesse for diagnosis of viral respiratory infections. The authors included 20 studies representing a total of 5510 upper and lower respiratory tract samples from children and adults and variously compared performance to virus culture, direct fluorescent antibody tests, and commercial and local “in-house” real-time PCR (RT-PCR). Viruses included were influenza A and B, respiratory syncytial virus, human metapneumovirus, and adenovirus; parainfluenza was excluded due to lack of data. The most promising results were for influenza A, where the platforms reviewed had a combined sensitivity of 0.940 (95% CI, 0.902–0.964) and high specificity 0.987 (95% CI, 0.979, 0.992). All platforms individually had an area under the receiver operating characteristic curve (AUROC) of 0.99. For influenza B, the FilmArray performed slightly worse (sensitivity 0.822 (0.689, 0.905), AUROC 0.98) compared with the performance of Prodesse (sensitivity 0.963, AUROC 0.99). Adenovirus was only sought by the FilmArray panel, which had the poorest diagnostic accuracy (AUROC 0.89). The review concluded that these systems helped early diagnosis of viral respiratory infections.
The BioFire FilmArray respiratory panel has been evaluated by multiple groups, including in RCTs against the standard of care; results for this panel, which has been the most evaluated, are summarized here and are likely to be generalizable across similar systems. One UK-based RCT of this panel, ResPOC (2017), enrolled 720 patients presenting in Accident and Emergency (A&E) or the acute medical unit of a large UK teaching hospital with acute respiratory illness and/or a fever of ≥37.5 °C.48,49 Patients were randomized to routine care or to testing using the FilmArray panel. In the latter case, research staff took a nose and throat swab and ran it immediately on the FilmArray. Strikingly, 91% of patients (52/57) with confirmed influenza in the FilmArray group received appropriate antivirals, compared with 65% (24/37) of those diagnosed with influenza in the control group (p = 0.0026). In reality, this differential was greater because in the control arm, only patients with clinically suspected infection were tested with the in-house laboratory PCR, as reflected in the low denominator for this group. The authors highlight that cases may well be missed with reliance upon clinical diagnosis based on a syndrome. Patients in the FilmArray arm had a reduced length of stay (5.7 days versus 6.8 days for the control group (95% CI, −2.2 to −0.3 days; p = 0·0443)) and more often had shorter or discontinued antibiotic courses (difference 7.8%, 95% CI, 2.5–13.1; p = 0·0047). Reduced hospital stay in the FilmArray arm was due to earlier discharge of patients confirmed to have respiratory viruses. However, the proportion of patients treated with intravenous antibiotics and their average duration of antibiotic treatment (in days) did not differ between the two arms; moreover, fully 301/360 (84%) patients in the POCT group received antibiotics during their admission compared with 294/354 (83%) in the control group.
In Japan, Kitano et al. introduced the FilmArray to the management of pediatric patients with respiratory infections in March 2018.50 Using nasopharyngeal swabs from 149 patients over the subsequent year to April 2019, performance was compared with the use of rapid antigen tests in the same demographic (1132 patients) over the preceding 6 years from March 2012 to March 2018. The average duration of antibiotic therapy fell from 12.82 to 8.56 days (p < 0.001), and the length of stay decreased from 8.18 to 6.83 days (p = 0.032). These results appear striking but are complicated by the historic control; it is unclear whether durations of hospitalization or antibiotic therapy were already falling during the years prior to implementation of the test or whether its introduction resulted in a step change.
An 800 patient RCT in China used the FilmArray with the respiratory panel for hospitalized cases with LRTI,51 compared with routine real-time PCR assays in the hospital laboratory. Courses of intravenous antibiotics were found to be shorter in the FilmArray group (7.0 days (interquartile range (IQR) 5.0–9.0) versus 8.0 days (IQR 6.0–11.0; p < 0.001)), as was the duration of the hospital stay (8.0 days (IQR 7.0–11.0) versus 9.0 days (IQR 7.0–12.0; p < 0.001), which directly related to the duration of IV antibiotics. Unlike Kitano’s study50 above, this was a comparison of PCR techniques (as opposed to a control arm using rapid antigen tests or culture); nonetheless, the median turnaround time (including processing, running, and reporting) in the FilmArray group was 1.6 h versus 29.0 h (p < 0.001) in the control group. Financial implications were analyzed, with a lower overall cost per patient when the FilmArray Panel was used ($1804.7 (IQR 1298.4–2633.8) versus $2042.5 (IQR 1427.4–2926.2); p = 0.002)). These savings may not translate to other healthcare systems with models different from that in China; however, this study suggests that commercial real-time PCR tests could play a part both in antimicrobial stewardship and in reducing hospitalization costs.
In contrast to these broadly positive results from the UK, Japan, and China, another UK hospital-based study, which used the FilmArray respiratory panel on 606 patients who presented with upper or lower respiratory tract infection or “influenza-like” symptoms from January 2015 to July 2015 found that although antivirals were given a 1.5 days quicker in the intervention arm (p < 0.001), the length of stay was not reduced compared with that of patients managed by routine laboratory respiratory PCR and serological testing.52 The authors describe their study as a “quasi-randomized trial”, meaning that patients were enrolled on odd-numbered days of the month into the control arm and on even-numbered days into the intervention arm. The machines were placed on two acute medical adult wards and a medical assessment unit. The authors account for their failure to reduce length of stay by remarking frequent delays in FilmArray testing, which often was performed only when the study investigators visited the wards. This suggests a flaw in the study design but underscores the point expanded upon below (“Practicalities of Introducing Molecular Methods” section) that any gain arising from rapid tests will only translate into a practical advantage if the technology is close to the patient and if tests are performed without delays arising from the need for specialist personnel or from transport issues.
Three further studies merit comment. First, a retrospective observational analysis by Li et al.53 reviewed patients presenting with a viral respiratory tract infection at three A&E departments in California between October 2016 and March 2017. In this study, 323/424 (76.2%) patients had a positive viral PCR result from a nasopharyngeal swab tested using the FilmArray respiratory panel available to the clinician before they were discharged from A&E; the remainder had results available postdischarge. Among the former 323 patients, only one-fifth were prescribed antibiotics, far fewer than would ordinarily be expected, underscoring the potential of this multiplex PCR as a stewardship tool. Patients diagnosed with influenza by PCR were particularly unlikely to receive antibiotics. Antiviral prescribing was not reviewed. Multivariate analysis identified factors influencing the antibiotic prescribing decision, many related to concerns over secondary bacterial infection. Decision-making is complex, and patients can have more than one pathology. Second, a 2017 study by Chen et al.54 in China used the FilmArray respiratory panel for 74 in-patient cases with CAP. In parallel, the authors performed (i) multiplex PCR for the same 14 viruses as sought by the FilmArray panel, (ii) bacterial/fungal cultures by Vitek, and (iii) IgM tests for C. pneumoniae and M. pneumoniae. Agreement between the FilmArray and multiplex PCR was complete for coronaviruses (the study predates COVID-19, which was not included), human metapneumovirus, influenza A and B, and parainfluenza viruses. However, the FilmArray panel has fewer bacterial targets, meaning that the 25 positive bacterial cultures identified by conventional culture could not be matched to its results. Last, Sails et al.55 compared the Luminex NxTAG respiratory pathogen panel to an in-house multiplex real-time PCR panel for the detection of respiratory viruses, using 314 samples (122 nasopharyngeal secretions, 53 throat swabs, 47 endotracheal secretions, 41 combined nose and throat swabs, 24 sputa, 17 BALs, and 10 others) from symptomatic respiratory tract infections. Agreement was 96.2% for enterovirus/rhinovirus and 100% for influenza A and B.
Broad-Spectrum Tests to Inform Antibiotic Use
The systems outlined above predominantly or solely seek viruses and atypical agents of pneumonia. Their potential to improve stewardship lies in discouraging unnecessary antibiotic use in patients found to have viral infections. No insight is gleaned as to which antibiotic should be used when a virus is not found and a bacterial infection is inferred. Moreover, such inference may or may not be correct: The clinical diagnosis of pneumonia is often uncertain, and patients are misdiagnosed, leading to the unnecessary use of broad-spectrum antibiotics.56
To address this aspect, diagnostics that seek the common bacterial agents of CAP, HAP, and VAP have been launched recently or are in advanced development. Relevant PCR-based systems, all using respiratory secretions without culture, include the BioFire FilmArray, used with the pneumonia panel rather than the virus-directed respiratory panel discussed above, Seegene’s respiratory test with 7 bacterial targets, and the Curetis Unyvero P50/P55 and lower respiratory tract panels (Table 3). The most comprehensively evaluated of these are the BioFire FilmArray, used with the pneumonia panel, and the Curetis Unyvero, used with any of the three panels detailed in Table 3. These have similar total numbers of targets and a near-total overlap with respect to the bacterial species sought, including the common agents of CAP, HAP, and VAP, but are different in that the BioFire FilmArray seeking a wide range of viruses whereas the Curetis Unyvero seeks more antibiotic-resistance genes, including blaTEM, blaSHV (penicillinase/ESBL gene), and erm (macrolide resistance) determinants (Table 3). The turnaround time for the Unyvero system is around 4–5 h; that for the FilmArray system is shorter at a little over 1 h. Both the manufacturers and independent researchers have carried out assessments of sensitivity and specificity in terms of pathogen detection (Table 4).
Table 4. Performance of PCR Tests for the Detection of Bacterial Pathogens in HAP/VAP Patients.
| country year | system | turnaround time | throughput in 24 h | footprint | number of samples | types of sample | sensitivity | specificity | authors/ref |
|---|---|---|---|---|---|---|---|---|---|
| USA 2018 | Curetis Unyvero lower respiratory tract panel | 5 h | 10 samples | 7.4 square feet | 788 | tracheal aspirate samples | 92.5% | 97.4% | Qi et al.59 |
| Germany 2018 | Curetis Unyvero P50 cartridge | 5 h | 10 samples | 7.4 square feet | 79 | 43 BAL fluids; 30 tracheal aspirates; 6 pleural fluids | 73.1% | 97.9% | Papan et al.60 |
| UK 2016 | Curetis Unyvero P50 cartridge | 90 | 55 sputa; 32 endotracheal tubes aspirates; 3 BAL | >95% | 33% | Personne et al.61 | |||
| UK 2017 | Curetis Unyvero P55 pneumonia cartridge | 5 h | 10 samples | 7.4 square feet | 85 | 52 sputa; 31 endotracheal tubes aspirate;1 BAL | 88.8% | 94.9% | Ozongwu et al.62 |
| UK 2019 | Curetis Unyvero P55 pneumonia cartridge | 74 | BAL | 56.9% | 58.5% | Gadsby et al.63 | |||
| France 2019 | BioFire FilmArray pneumonia panel | 1.25 h | 264 samples (FilmArray Torch) | 4.25 square feet (or less, depending on number of modules) | 63 | not specified | 93% positive agreement | 95% negative agreement | Gastli et al.64 |
| UK 2019 | BioFire FilmArray, pneumonia panel and Curetis Unyvero pneumonia panel | 634 | sputa, endotracheal tube exudates, BAL | FilmArray: 89.2–99.3% Unyvero: 88.6–97.0% | FilmArray: 93.8–99.9% Unyvero: 99.2–99.9% | Enne et al.65 | |||
| not quoted | BioFire FilmArray pneumonia panel | not quoted | sputa | 96.3% | 97.2% | data on file at BioFire Diagnostics (https://www.biofiredx.com/products/the-filmarray-panels/filmarray-pneumonia/) |
As an alternative approach, Accelerate’s Pneumonia Test,57 which is in development following the successful launch of a similar system testing bacteria recovered from blood,58 instead uses rapid phenotypic testing, employing automated microscopy to analyze the early growth of antibiotic-challenged bacteria recovered from the sample and thereby predicting their susceptibility. In parallel, it uses automated fluorescence in situ hybridization technology to identify the pathogens present. This gives a total turnaround of 8–12 h,57 which narrowly missed our target of “results within the first dosage interval of a typical thrice-daily antibiotic” but remains rapid compared with conventional methodology. Moreover, this approach also has some advantages over PCR. First, it seeks all bacteria that can grow, not just those represented on a PCR panel. Second, it delivers results as minimum inhibitory concentrations, which can be related to pharmacodynamics, guiding antibiotic dosage, rather than detecting the genetic signatures of organisms that may no longer be viable or mechanisms that may not be expressed. Last, it seeks all resistances, irrespective of the underlying mechanism, not just the few resistance determinants (Table 3) targeted by the PCR systems. This is potentially valuable in cases (see Table 1) where the molecular bases of resistance are diverse, multifactorial, or due to mutations that cannot readily be sought by simple PCR.
A review of Table 4 shows considerable trial-to-trial variation in the reported sensitivity and specificity for both the Curetis Unyvero and BioFire FilmArray PCR systems. This divergence may partly reflect the types of specimens used: BALs are more likely to contain a single organism from deep within the lung, whereas sputa are prone to more or less substantial contamination by colonizing bacteria from the upper airways. In large part, though, we believe that the major issue lies in variability in routine bacteriological culture, which is taken as the reference “gold standard” for the evaluations summarized in the table. Unfortunately, as noted already, routine culture is far from perfect. Many pneumonia patients do not have a pathogen grown, perhaps because an antibiotic was given before the respiratory specimen was taken, interfering with recovery in microbiological culture. This leads to “false positive” results when the PCR system is scored against culture, reducing the calculated specificity. Second, and more difficult to measure objectively, is the degree of subjectivity in the reading of conventional respiratory cultures. Essentially, the microbiologist makes a judgment on what is grown and may discount small numbers of potentially pathogenic organisms, especially if these are heavily mixed with normal upper-respiratory-tract flora. We believe that microbiologists’ reading of these plates is highly variable, basing this conclusion on results from the INHALE study66 presented at the European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) 2019 and now being prepared for publication.65 We made two critical observations. First, when Curetis Unyvero, BioFire FilmArray, and 16S rDNA analyses were all applied in parallel to the same respiratory samples, their findings tended to agree, with different molecular systems finding the same organism(s) even when these were not grown by the diagnostic laboratory. Evidently, it is implausible that different molecular systems would find the same organism unless it was actually present, meaning that it is unreasonable (and prejudicial) to call these “false positives”. Second, when we compared the results of centrally performed molecular test to locally performed culture results, from the 15 participating sites, we found that the ratio of PCR-positive/culture-positive varied from 29 to 87%. The likeliest explanation of such variation is site-to-site differences in the execution or interpretation of the routine microbiology. We consequently believe that these molecular systems are not only swifter than conventional microbiology but also deliver a standardization that is presently absent. However, it is arguable whether their finding more organisms than culture is advantageous or not: It can be countered that this will promote polypharmacy, some of it directed against organisms that were of little or no significance or those that were already dead but had left their genetic signatures in the respiratory secretions.
Several trials have explored the impact, or potential impact, of these PCR systems on patient outcomes. Gastli et al.67 performed a prospective cohort study on four wards, using the FilmArray pneumonia panel on endotracheal aspirates or induced sputa from 63 pneumonia episodes among 61 patients, concluding that the test results could have led to an early switch of antibiotics in 79% of these episodes. Similarly, Lejeune et al.68 analyzed 60 samples (30 BAL, 21 mini-BAL, 5 sputa, and 4 tracheal aspirates), again using the FilmArray pneumonia panel, and estimating that the approach could have led to an earlier change in antibiotics in 53% of patients. While these authors point to scope for better stewardship, neither demonstrated that the gain was realized in actuality. The INHALE study66 seeks to explore these aspects with a trial across 12 UK ICUs. HAP/VAP patients are randomized to receive antimicrobial chemotherapy guided either by standard policy (empirical treatment followed by culture-based refinement) or the FilmArray pneumonia plus panel result and an algorithm encourage use of the narrowest spectrum agents to cover the pathogens and resistances found (shown in simplified form in Figure 2).66 Final results are anticipated in 2021. In this context, we stress that such a prescribing algorithm is essential to translate the machine’s output of pathogens and resistance genes found into prescribing guidance. For INHALE, this algorithm was developed in liaison with microbiology and intensive care at participating sites, allowing some site-to-site variation (e.g., because pediatric sites are included and some favored agents lack pediatric licenses), retaining the core principle of preferring the narrowest spectrum agent to cover the pathogen(s) found. In the future, if rapid testing becomes the norm, then we envisage such algorithms replacing hospitals’ empirical treatment guidelines for clinical syndromes such as HAP and VAP.
Figure 2.
Simplified flow chart of the INHALE trial intervention arm.
A general limitation of PCR systems is that they can only detect targets for which there exist PCR primers. While the organisms represented on the Curetis lower respiratory tract (pneumonia) panel and BioFire FilmArray pneumonia panel (Table 3) cause around 90–95% of pneumonia cases,14 it would be difficult, owing to limitations on the number of primers that can be multiplexed, to expand this proportion to 99%. S. maltophilia is a notable omission from the FilmArray pneumonia panel, accounting for around 1–6% of VAP cases,69,70 while Citrobacter koseri and Raoultella spp. are absent from both the FilmArray and Curetis Unyvero systems. Issues of omission are far greater in respect of resistance genes. FilmArray seeks only the determinants of various carbapenemases (blaKPC, blaOXA-48-like, blaNDM, blaIMP, and blaVIM), along with mecA (conferring methicillin resistance in staphylococci) and blaCTX-M (encoding the principal family of extended-spectrum β-lactamases, ESBLs) (Table 3). The carbapenemases sought are significant both with respect to infection control and treatment choice, but these remain extremely rare in many countries, present in <1% of Enterobacterales from HAP and VAP in the UK.71 CTX-M ESBLs (encoded by blaCTX-M) and methicillin resistance in S. aureus (encoded by mecA) are more prevalent worldwide and have implications for treatment choice; however, only 50–70% of ESBLs in Klebsiella spp. are CTX-M types (most of the others are TEM and SHV variants), meaning that a negative result does not exclude the possibility that an isolate has an ESBL. The Curetis Unyvero system seeks more resistance genes (Table 3), including blaTEM and blaSHV, encoding TEM and SHV β-lactamases, respectively, but does not distinguish whether the genes detected encode ESBL or penicillinase variants within these families. Moreover, blaTEM and some of the other genes sought (e.g., erm, encoding macrolide, lincosamides streptogramin B resistance) are common in respiratory commensals,72 meaning that it is difficult to be sure that the determinant “belongs” to the pathogen found.
Given (i) the great diversity of acquired resistance genes that are important for many combinations of antibiotic and species (Table 1), (ii) the importance of diverse mutational resistances (which are inherently difficult to detect by PCR) in some species, notably P. aeruginosa (Table 1), and (iii) the fact that the resistance to β-lactam−β-lactamase inhibitor combinations can depend on the amount of β-lactamase, rather than the simple presence of a β-lactamase gene,73 it is difficult to see how these PCR systems will ever be able give a comprehensive prediction of the antibiogram(s) of the organism(s) found. Rather, as with PCR-based detection of gonococci from urine samples and genital swabs, it will remain necessary for culture to be performed in parallel, in the hope of growing the organism(s) detected and confirming its susceptibility. Local resistance surveillance programs will also be needed to inform treatment choices during the interval when the pathogen(s) have been identified but their susceptibility remains uncertain. These points should not, however, detract from the fact that swift detection of key carbapenemase genes in particular will likely prove very useful in countries where these are widespread. Moreover, swift determination of the particular carbapenemase type will guide choices among the various new β-lactamase inhibitor combinations now reaching the clinic.74
A brief mention should also be made of a few studies that have explored PCR to seek specific pathogens in respiratory secretions and of urinary antigen testing. Coppens et al. sought to detect S. aureus in 79 ETAs,75 using the GeneXpert MRSA/SA ETA assay (Cepheid), a PCR system designed for use with sputum samples. They found that the method was 100% sensitive and specific compared with routine culture, was easy to use, and had a turnaround time of approximately 1 h. Urinary antigen testing is often used for the rapid detection of pneumococcal infection and L. pneumophila. Notably, the IDSA recommends pneumococcal urinary antigen testing specifically to promote antimicrobial stewardship, since confirmation of pneumococcal infection often facilitates narrow-spectrum treatment. A recent American study emphasized this point and noted that the test was not being used as often as it could be.76 A retrospective Belgian multicenter study reviewed urinary antigen tests for 71 patients who had L. pneumophila detected by PCR of respiratory samples.77 Urinary antigen tests had been requested for 45 of these 71, and 44.4% (20/45) of these were negative.
Sequencing Directly from Respiratory Samples
The fundamental limitation of PCR methods (i.e., that they seek only a limited range of pathogens and a few resistance genes) can be addressed by metagenomic sequencing directly from respiratory samples. Unlike PCR, sequencing is comprehensive, with the potential to find all species, genes, and gene variants. Several workers have shown this approach to be practicable, including Yang et al.78 and Charalampous et al.,72 both using nanopore technology, and Langelier et al.,79 using metagenomic next-generation sequencing (NGS), combining both RNA and DNA sequencing. Even more than with PCR, sample quality is important, since metagemonic sequencing will detect any DNA (bacterial or human) in a sample. A “clean” sample such as BAL may be preferred to minimize the amount of human DNA; however, some workers prefer ETAs and sputa to BALs, as they have higher bacterial loads. If ETAs and sputa are used, then a timely and cost-effective host-DNA-depletion step becomes vital; moreover, such samples are likely to contain more oral and upper respiratory tract flora, complicating interpretation.
According to the technology used, DNA sequencing can produce long or short reads. Although long-read sequencing, as with nanopore technology, does not yet have the read accuracy of short-read technologies (e.g. Illumina), it has key advantages, providing analyzable sequence data in real-time, helping to establish which organisms are hosting resistance genes, and permitting easier genome assembly. These advantage are crucial for the development of rapid/POCT diagnostics.
In China, Li et al.80 used short-read metagenomic sequencing on 35 BAL samples from 32 ICU patients with respiratory failure. DNA was extracted using the TIANamp Micro DNA Kit, and libraries were sequenced on the BGISEQ-50 platform with the Burrows–Wheeler Alignment tool.81 Pathogen read numbers were low, partly reflecting the lack of a host-DNA-depletion step, and with most positive results were based on <50 reads/organism detected. Despite these limitations, the method achieved a sensitivity of 88.9% and a specificity of 74.1% compared with routine culture in terms of species identification. The authors commented that for approximately a third of their cohort the metagenomic NGS led to a change in treatment. The time to result is not stated, nor is the range of genome coverage. No information is given on the resistance genes found; it seems likely that the approach gave too few reads for reliable detection of resistance genes. Two other studies using Illumina sequencing deserve mention. In Japan, Takeuchi et al.,82 investigated 10 pediatric patients with respiratory failure, and in Spain, Lopez et al. investigated 55 chronic obstructive pulmonary disease (COPD) patients.83 Both concluded that metagenomic sequencing was useful for pathogen detection.
Nanopore sequencing, with the advantages of speed and long-reads, was first explored for respiratory samples by Charalampous et al.72 in the UK, with a novel saponin-based host-DNA-depletion method. They achieved a 6 h turnaround time, including the host-DNA-depletion step and found 96.6% sensitivity and 41.7% specificity compared with those of culture. Both these values increased to 100% following PCR verification of discordant results and posthoc specific gene analysis for all H. influenzae and/or S. pneumoniae positive samples. Resistance genes were found but often without an obvious host being recognized by sequencing or culture. In particular, the authors often found tet, erm, and blaTEM determinants when the only pathogen(s) grown remained susceptible to tetracyclines, macrolides, or β-lactams or were organisms inherently unlikely to host these genes. It was inferred that these genes originated in the commensal flora, not the pathogens, and the scope for uncertainty, if the method was deployed clinically, is evident. Bioinformatic analysis of the DNA flanking resistance genes might resolve this aspect when resistance genes are chromosomally located, but it is less likely to be useful when they are plasmid-borne, since the plasmid’s host would remain uncertain. A further issue remains when as in P. aeruginosa resistance is commonly effected by combinations of mechanisms (e.g., upregulation of efflux pumps and increased expression of AmpC β-lactamase) (Table 1), with these upregulations arising from diverse mutations in any of several regulatory genes. Almost all P. aeruginosa isolates have some functioning efflux via MexA–MexB–OprM: What matters is the extent to which this system is expressed as a result of regulatory mutations in mexR(84) or is augmented by upregulation of multiple further pumps.85 A possible solution is “genomic neighbor typing”, a method that matches genome sequences from the clinical sample to a database of sequences and susceptibility data from known variants, to find the closest match. Brinda et al.86 used this approach with nanopore sequence data for S. pneumoniae and Neisseria gonorrheae, both organisms where resistance to β-lactams depends on complex mixtures of mutation and gene recombination, achieving good prediction of susceptibility and resistance within 4 h of sample collection. Future work may allow this bioinformatic strategy to be expanded to other “difficult” organisms, including P. aeruginosa; nonetheless, these aspects underscore the complexity of the metagenomic pipelines that will be needed to translate sequence data into prescribing decisions. More generally, although the broad steps involved in sequencing are identical across methods, every metagenomic pipeline is different. Consequently, no uniform algorithm to interpret results in likely to be possible. Nevertheless, key themes will be universal, such as the threshold number of reads considered significant, and controlling for contamination.
In the US, Yang et al.78 used the Oxford MinION nanopore device with endotracheal aspirates from 9 patients with culture-positive pneumonia, 5 with culture-negative pneumonia, and 8 controls. Results were compared with both bacteriological cultures and 16S rRNA gene sequencing, and the team reported excellent concordance between the nanopore and both culture and 16S rRNA results. Nanopore sequencing and data analyses were not performed in real time because of a retrospective study design; however, a turnaround time of 6–8 h was achieved including host DNA depletion, with good pathogen genome coverage and detection of antimicrobial resistance genes. Notably, the authors detected organisms (S. aureus, H. influenzae, or S. pneumoniae) that can be either pathogens or commensals (“pathobionts”) in patients with no evidence of pneumonia, highlighting the need for caution in interpreting results.
Advances in rapid host DNA depletion and in bioinformatics make it likely that metagenomics will become the future for the rapid microbiological investigation of respiratory tract infections, with far greater comprehensiveness than PCR. As yet, however, these methods require substantial technical skill and are not yet ready for routine use. A further issue is cost, which is likely to be in the low hundreds of dollars per sample, which is expensive compared with culture though not when compared with an extra day stay in the ICU.
Practicalities of Introducing Molecular Methods
Our analysis, like those of others,87,88 indicates that molecular methods (particularly PCR and, in the future, metagenomic sequencing) have considerable potential for the better investigation of pneumonia, increasing diagnostic yield and providing early treatment guidance. Nonetheless, great challenges remain. Some are technical and have been discussed already; others relate to the practicalities and logistics of deployment.
First, there is the question of where tests will be done. If the answer is “In the laboratory, where else?”, then it is likely that transport to the laboratory, “booking in”, and possible batch processing will take longer than the tests themselves, partly negating the advantage of test speed. This issue will be exacerbated if the laboratory is physically separate from the hospital and does not work “24/7”. The potential of rapid tests run counter to the recent trend (predicated on objectives of standardization, quality control, and cost minimization) to centralize laboratories and to have them serve multiple hospitals. A recent review by Vandenberg et al.89 discusses this issue in greater detail. Retaining a small “hot” laboratory at every hospital for urgent tests would resolve this issue, but it would add significant cost, over and above that of the tests themselves.
If instead POCT systems are performed by the patient’s bedside, not in a laboratory, then ward staff will need to be trained to use them, meaning that the tests must not require specialist laboratory expertise and that their results and interpretation cannot be operator-dependent. Alternatively, communication systems must facilitate swift microbiology, infectious disease, and infection control advice on a 24/7 basis. Quality control also needs to be considered, as ward-based POCT would no longer be under the remit of laboratory accreditation. Further factors to consider are machine maintenance and interfacing with hospital computer systems. Crucial to test interpretation is the issue of distinguishing colonist and pathogens, as highlighted in a Swiss study that tested 35 diagnostic BAL specimens using a rapid broad-range PCR and microarray-based nucleic acid amplification test called Prove-it Sepsis Assay.90 The authors concluded that the clinical relevance of the results was uncertain and that colonizing organisms may be difficult to differentiate from pathogens. This underscores the point made earlier that PCR-based diagnostics tend to find more positives than culture, giving credence to the idea that DNA from small numbers of organisms that might have been discounted by a skeptical biomedical scientist reading culture plates. This issue is partly addressed on newer commercial platforms, including the Unyvero and FilmArray, where results are semiquantitative, giving an indication of the number of gene copies and bacterial load. If this is high, then infection is inferred to be more likely.
Second, linked to this first point, is the issue of where rapid tests best fit into hospital and community care pathways. Should they be used to decide when to stop/start antibiotics or when to admit patients presenting at A&E? Which settings would benefit most from POCTs: ICU, A&E, or community care? Answers may vary according to the hospital and the health system.
Third, there is the question of costs: Molecular tests are considerably more expensive than bacterial culture, costing anywhere from $60–400 versus ca. $12–25. A comprehensive health economic analysis is required to establish whether swifter refinement of patients’ treatment translates into cost savings. Moreover, unless they are comprehensive (which is unlikely, except for sequencing-based systems and rapid phenotypic systems), rapid tests for pathogens and resistances will be in addition to conventional testing, not a replacement. While gains in stewardship and patient management may recoup some additional costs, savings accrue remotely from the cost center for the test, complicating accounting. Moreover, efficiency gains are notoriously difficult to translate into cash savings in socialized healthcare, such as the UK National Health Service, operating at near-full capacity. A patient may be discharged earlier, giving a notional “saving”, but their bed is immediately filled by a new patient, whose new costs are likely to exceed those of an extra day’s stay by the original patient. Cost savings may be more obviously realizable in settings where the patient, or their insurer, pays directly.
Last, most subtle: Behavioral aspects are crucial and are apt to vary with place and human culture. ICU decisions relating to antibiotic prescribing in particular are multifactorial and complex, as outlined in systematic reviews by Warreman et al. and Krockow et al.91,92 Key factors include fear of adverse outcomes and the experience of clinician. Adding molecular tests that detect more organisms and resistance genes than culture may prompt more polypharmacy rather than better stewardship. It is unclear, as yet, how much clinicians will trust these tests and how this will change if the test machine sits in the ICU rather than remotely in the laboratory. The INHALE trial, outlined earlier, aims to explore these aspects: Behavioral psychologists will investigate decision making among ICU clinicians in relation to both the FilmArray and routine culture results, identifying key intervention points to optimize stewardship. Again, the likely solution will lie in different ways of working for microbiologists, who will need to provide urgent advice on test interpretation, rather than running tests in their “own” laboratories.
Conclusion: Taking Deployment Forward
This review has outlined the potentials, with respect to antimicrobial stewardship, of the various rapid diagnostics relevant for pneumonia, including (i) those that examine human biomarkers as predictors of infection type, (ii) those already widely used that seek respiratory viruses, and (iii) those now reaching the market that use PCR or rapid phenotypic testing to seek ranges of bacterial pathogens and antibiotic resistance genes. Sequencing-based tests are for the future, but they have potential to be far more comprehensive than PCR, particularly with respect to predicting resistance. It is likely that no single approach will be overwhelmingly successful but that collectively these approaches will facilitate a major shift in the management of respiratory infection.
Although POCT has the greatest potential as bedside tools, their use for pneumonia patients will need strong microbiology and/or infectious disease advice if their often-complex findings are to be best-translated into treatment advice and antimicrobial stewardship. Perhaps the biggest barriers to change and progress are people and tradition; deployment of these tests will demand significant changes to the ways of working both in the clinic and in the microbiology department. In closing, it should be added that COVID-19 (unknown when this review was first drafted, but pandemic and massively disruptive worldwide by the time of its publication) seems certain to drive major changes in hospital practice. In the short term, emphasis will be on detecting SARS-CoV2 specifically; in the longer term, depending on how the pandemic evolves, the virus may prover a driver to much wider changes in diagnostic practice.
Acknowledgments
We are grateful to all members of the INHALE Project Team for discussions, which have helped us to develop and refine the ideas and concepts outlined in this article. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Author Present Address
# Z.D.: Department for Clinical Microbiology, second Floor Royal Free Hospital, University College London, Rowland Hill Street, London NW3 2PF, United Kingdom.
This paper includes independent research funded by the National Institute for Health Research (NIHR) under its Programme Grants for Applied Research Programme (Reference Number RPPG-0514-20018).
The authors declare the following competing financial interest(s): Z.D.: Nothing to declare. V.I.E. has received research support, speaking honoraria, consultancy fees, and in-kind contributions from Curetis GmbH, BioMrieux, and Oxford Nanopore. J.O.G.: Has received research funding, financial support for attending conferences and free flowcells and reagents from Oxford Nanopore Technologies. Is a consultant for Simcere Diagnostics. V.G.: Has been paid to give presentations for Gilead, Biomrieux, Pfizer, and MSD. D.M.L.: Advisory Boards or ad-hoc consultancy Accelerate, Allecra, Antabio, Centauri, Entasis, Integra-Holdings, Meiji, Melinta, Menarini, Mutabilis, Nordic, ParaPharm, Pfizer, QPEX, Roche, Shionogi, T.A.Z., Tetraphase, VenatoRx, Wockhardt, Zambon. Paid lectures: Astellas, bioMerieux, Beckman Coulter, Cardiome, Cepheid, Merck/MSD, Menarini, Nordic, Pfizer, and Shionogi. Relevant shareholdings or options: Dechra, GSK, Merck, Perkin Elmer, Pfizer, T.A.Z, amounting to less than 10% of portfolio value.
References
- Cassini A.; Hogberg L. D.; Plachouras D.; Quattrocchi A.; Hoxha A.; Simonsen G. S.; Colomb-Cotinat M.; Kretzschmar M. E.; Devleesschauwer B.; Cecchini M.; et al. (2019) Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19 (1), 56–66. 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HM Government . (2019) Tackling antimicrobial resistance 2019–2024. https://www.gov.uk/government/publications/uk-5-year-action-plan-for-antimicrobial-resistance-2019-to-2024.
- Guitor A. K.; Wright G. D. (2018) Antimicrobial Resistance and Respiratory Infections. Chest 154 (5), 1202–1212. 10.1016/j.chest.2018.06.019. [DOI] [PubMed] [Google Scholar]
- NICE . (2014) Pneumonia in Adults. Diagnosis and management of community and hospital acquired pneumonia in adults. NICE Clinical Guidelines, Vol. 191, National Institute for Health and Care Excellence, London. [PubMed] [Google Scholar]
- Tai C.; Stoyanova R.; Brealey D. (2018) Novel diagnostics of respiratory infection in the intensive care unit. Annals of Research Hospitals 2, 9–9. 10.21037/arh.2018.08.01. [DOI] [Google Scholar]
- Blasi F.; Garau J.; Medina J.; Avila M.; McBride K.; Ostermann H. (2013) Current management of patients hospitalized with community-acquired pneumonia across Europe: outcomes from REACH. Respir. Res. 14, 44. 10.1186/1465-9921-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollef M. H.; Burnham C.-A. D. (2017) Ventilator-Associated Pneumonia: The Role of Emerging Diagnostic Technologies. Semin. Respir. Crit. Care Med. 38 (3), 253–263. 10.1055/s-0037-1599224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. H.; Horner C.; Livermore D. M.; MacGowan A. P. (2018) Doxycycline in UK guidelines for hospital-acquired pneumonia: where is the evidence base?. J. Antimicrob. Chemother. 73 (11), 3212–3215. 10.1093/jac/dky306. [DOI] [PubMed] [Google Scholar]
- McNulty C. A.; Nichols T.; French D. P.; Joshi P.; Butler C. C. (2013) Expectations for consultations and antibiotics for respiratory tract infection in primary care: the RTI clinical iceberg. Br. J. Gen. Pract. 63 (612), e429–36. 10.3399/bjgp13X669149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterton R. G.; Galloway A.; French G.; Street M.; Armstrong J.; Brown E.; Cleverley J.; Dilworth P.; Fry C.; Gascoigne A. D.; Knox A.; Nathwani D.; Spencer R.; Wilcox M. (2008) Guidelines for the management of hospital-acquired pneumonia in the UK: report of the working party on hospital-acquired pneumonia of the British Society for Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 62 (1), 5–34. 10.1093/jac/dkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. B.; Pham Thanh D.; Tran Do Hoan N.; Wick R. R.; Ingle D. J.; Hawkey J.; Edwards D. J.; Kenyon J. J.; Phu Huong Lan N.; Campbell J. I.; et al. (2016) Repeated local emergence of carbapenem-resistant Acinetobacter baumannii in a single hospital ward. Microb. Genom. 2 (3), e000050. 10.1099/mgen.0.000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio D.; Castagnino J.; Vazquez W.; Vergara G.; Curiale A. (2010) Tigecycline in the treatment of ventilator-associated pneumonia: experience from the Latin American Tigecycline Use Registry. Infez. Med. 18 (1), 27–34. [PubMed] [Google Scholar]
- Giard M.; Lepape A.; Allaouchiche B.; Guerin C.; Lehot J. J.; Robert M. O.; Fournier G.; Jacques D.; Chassard D.; Gueugniaud P. Y.; Artru F.; Petit P.; Robert D.; Mohammedi I.; Girard R.; Cetre J. C.; Nicolle M. C.; Grando J.; Fabry J.; Vanhems P. (2008) Early- and late-onset ventilator-associated pneumonia acquired in the intensive care unit: comparison of risk factors. J. Crit. Care 23 (1), 27–33. 10.1016/j.jcrc.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Kalil A.; Metersky M. L.; Klompas M. (2016) Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 63 (5), e61–e111. 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros J. M.; Rosso-Fernandez C. M.; Roca-Oporto C.; De Pascale G.; Jimenez-Jorge S.; Fernandez-Hinojosa E.; Matthaiou D. K.; Ramirez P.; Diaz-Miguel R. O.; Estella A.; et al. (2019) Colistin versus Meropenem in the empirical treatment of ventilator-associated pneumonia (Magic Bullet study): an investigator-driven, open-label, randomized, noninferiority controlled trial. Crit. Care 23 (1), 383. 10.1186/s13054-019-2627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. (2018) Past and Present Perspectives on beta-Lactamases. Antimicrob. Agents Chemother. 62 (10), e01076-18. 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier D.; Richardot C.; Muller E.; Robert-Nicoud M.; Llanes C.; Plesiat P.; Jeannot K. (2013) Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68 (8), 1772–80. 10.1093/jac/dkt098. [DOI] [PubMed] [Google Scholar]
- BSAC Resistance Surveillance Programme. http://www.bsacsurv.org/reports/respiratory/.
- Government of India . (2017) Scoping Report on Antimicrobial Resistance in India. https://cddep.org/wp-content/uploads/2017/11/AMR-INDIA-SCOPING-REPORT.pdf.
- Jaggi N.; Chatterjee N.; Singh V.; Giri S. K.; Dwivedi P.; Panwar R.; Sharma A. P. (2019) Carbapenem resistance in Escherichia coli and Klebsiella pneumoniae among Indian and international patients in North India. Acta Microbiol. Immunol. Hung. 66 (3), 367–376. 10.1556/030.66.2019.020. [DOI] [PubMed] [Google Scholar]
- Zowawi H. M.; Syrmis M. W.; Kidd T. J.; Balkhy H. H.; Walsh T. R.; Al Johani S. M.; Al Jindan R. Y.; Alfaresi M.; Ibrahim E.; Al-Jardani A.; Al Salman J.; Dashti A. A.; Sidjabat H. E.; Baz O.; Trembizki E.; Whiley D. M.; Paterson D. L. (2018) Identification of carbapenem-resistant Pseudomonas aeruginosa in selected hospitals of the Gulf Cooperation Council States: dominance of high-risk clones in the region. J. Med. Microbiol. 67 (6), 846–853. 10.1099/jmm.0.000730. [DOI] [PubMed] [Google Scholar]
- Ayoub Moubareck C.; Hammoudi Halat D.; Akkawi C.; Nabi A.; AlSharhan M. A.; AlDeesi Z. O.; Peters C. C.; Celiloglu H.; Karam Sarkis D. (2019) Role of outer membrane permeability, efflux mechanism, and carbapenemases in carbapenem-nonsusceptible Pseudomonas aeruginosa from Dubai hospitals: Results of the first cross-sectional survey. Int. J. Infect. Dis. 84, 143–150. 10.1016/j.ijid.2019.04.027. [DOI] [PubMed] [Google Scholar]
- Kollef K. E.; Schramm G. E.; Wills A. R.; Reichley R. M.; Micek S. T.; Kollef M. H. (2008) Predictors of 30-day mortality and hospital costs in patients with ventilator-associated pneumonia attributed to potentially antibiotic-resistant gram-negative bacteria. Chest 134 (2), 281–287. 10.1378/chest.08-1116. [DOI] [PubMed] [Google Scholar]
- Guillamet C. V.; Vazquez R.; Noe J.; Micek S. T.; Kollef M. H. (2016) A cohort study of bacteremic pneumonia: The importance of antibiotic resistance and appropriate initial therapy?. Medicine (Philadelphia, PA, U. S.) 95 (35), e4708. 10.1097/MD.0000000000004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M.; Wain J. (2013) Revolutionising bacteriology to improve treatment outcomes and antibiotic stewardship. Infect. Chemother. 45 (1), 1–10. 10.3947/ic.2013.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B. B.; Shankar P.; Jerris R. C.; Kotzbauer D.; Anderson E. J.; Watson J. R.; O’Brien L. A.; Uwindatwa F.; McNamara K.; Bost J. E. (2015) Impact of a rapid respiratory panel test on patient outcomes. Arch. Pathol. Lab. Med. 139 (5), 636–41. 10.5858/arpa.2014-0257-OA. [DOI] [PubMed] [Google Scholar]
- Hoyos J.; Belza M. J.; Fernandez-Balbuena S.; Rosales-Statkus M. E.; Pulido J.; de la Fuente L. (2013) Preferred HIV testing services and programme characteristics among clients of a rapid HIV testing programme. BMC Public Health 13, 791. 10.1186/1471-2458-13-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J.; Butler C.; Hopstaken R.; Dryden M. S.; McNulty C.; Hurding S.; Moore M.; Livermore D. M. (2015) Narrative review of primary care point-of-care testing (POCT) and antibacterial use in respiratory tract infection (RTI). BMJ. open respiratory research 2 (1), e000086. 10.1136/bmjresp-2015-000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cals J. W.; Butler C. C.; Hopstaken R. M.; Hood K.; Dinant G. J. (2009) Effect of point of care testing for C reactive protein and training in communication skills on antibiotic use in lower respiratory tract infections: cluster randomised trial. BMJ. 338, b1374. 10.1136/bmj.b1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llor C.; Bjerrum L.; Munck A.; Cots J. M.; Hernandez S.; Moragas A.; (2014) Access to point-of-care tests reduces the prescription of antibiotics among antibiotic-requesting subjects with respiratory tract infections. Respir. Care 59 (12), 1918–1923. 10.4187/respcare.03275. [DOI] [PubMed] [Google Scholar]
- Oppong R.; Jit M.; Smith R. D.; Butler C. C.; Melbye H.; Molstad S.; Coast J. (2013) Cost-effectiveness of point-of-care C-reactive protein testing to inform antibiotic prescribing decisions. Br. J. Gen. Pract. 63 (612), e465–71. 10.3399/bjgp13X669185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates J.; Francis N. A.; White P.; Gillespie D.; Thomas-Jones E.; Breen R.; Kirby N.; Hood K.; Gal M.; Phillips R.; et al. (2017) General practitioner use of a C-reactive protein point-of-care test to help target antibiotic prescribing in patients with acute exacerbations of chronic obstructive pulmonary disease (the PACE study): study protocol for a randomised controlled trial. Trials 18 (1), 442. 10.1186/s13063-017-2144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. L.; Snider R.; Nylen E. S. (2008) Procalcitonin assay in systemic inflammation, infection, and sepsis: clinical utility and limitations. Crit. Care Med. 36 (3), 941–52. 10.1097/CCM.0B013E318165BABB. [DOI] [PubMed] [Google Scholar]
- Saeed K.; Dryden M.; Bourne S.; Paget C.; Proud A. (2011) Reduction in antibiotic use through procalcitonin testing in patients in the medical admission unit or intensive care unit with suspicion of infection. J. Hosp. Infect. 78 (4), 289–92. 10.1016/j.jhin.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Christ-Crain M.; Stolz D.; Bingisser R.; Muller C.; Miedinger D.; Huber P. R.; Zimmerli W.; Harbarth S.; Tamm M.; Muller B. (2006) Procalcitonin guidance of antibiotic therapy in community-acquired pneumonia: a randomized trial. Am. J. Respir. Crit. Care Med. 174 (1), 84–93. 10.1164/rccm.200512-1922OC. [DOI] [PubMed] [Google Scholar]
- Schuetz P.; Beishuizen A.; Broyles M.; Ferrer R.; Gavazzi G.; Gluck E. H.; Gonzalez Del Castillo J.; Jensen J. U.; Kanizsai P. L.; Kwa A. L. H.; Krueger S.; Luyt C. E.; Oppert M.; Plebani M.; Shlyapnikov S. A.; Toccafondi G.; Townsend J.; Welte T.; Saeed K. (2019) Procalcitonin (PCT)-guided antibiotic stewardship: an international experts consensus on optimized clinical use. Clin. Chem. Lab. Med. 57 (9), 1308–1318. 10.1515/cclm-2018-1181. [DOI] [PubMed] [Google Scholar]
- Kamat I. S.; Ramachandran V.; Eswaran H.; Guffey D.; Musher D. M. (2019) Procalcitonin to Distinguish Viral From Bacterial Pneumonia: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 70 (3), 538–542. 10.1093/cid/ciz545. [DOI] [PubMed] [Google Scholar]
- NICE . (2015) Procalcitonin testing for diagnosing and monitoring sepsis. Diagnostics Guidance [DG18]; National Institute for Health and Care Excellence, London. [Google Scholar]
- Bouadma L.; Luyt C. E.; Tubach F.; Cracco C.; Alvarez A.; Schwebel C.; Schortgen F.; Lasocki S.; Veber B.; Dehoux M.; Bernard M.; Pasquet B.; Regnier B.; Brun-Buisson C.; Chastre J.; Wolff M. (2010) Use of procalcitonin to reduce patients’ exposure to antibiotics in intensive care units (PRORATA trial): a multicentre randomised controlled trial. Lancet 375 (9713), 463–74. 10.1016/S0140-6736(09)61879-1. [DOI] [PubMed] [Google Scholar]
- Levin P. D.; Cohen M. J.; Ohev-Zion E.; Tannus S.; Stohl S.; Avidan A.; Cohen-Poraduso R.; Moses A. E.; Sprung C. L.; Benenson S. (2019) The Effect of Repeated Versus Initial Procalcitonin Measurements on Diagnosis of Infection in the Intensive Care Setting: A Prospective Observational Study. Anesth. Analg. 129 (4), e114–e117. 10.1213/ANE.0000000000003313. [DOI] [PubMed] [Google Scholar]
- Hellyer T. P.; Anderson N. H.; Parker J.; Dark P.; Van Den Broeck T.; Singh S.; McMullan R.; Agus A. M.; Emerson L. M.; Blackwood B.; et al. (2016) Effectiveness of biomarker-based exclusion of ventilator-acquired pneumonia to reduce antibiotic use (VAPrapid-2): study protocol for a randomised controlled trial. Trials 17 (1), 318. 10.1186/s13063-016-1442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellyer T. P.; McAuley D. F.; Walsh T. S.; Anderson N.; Conway Morris A.; Singh S.; Dark P.; Roy A. I.; Perkins G. D.; McMullan R. (2020) Biomarker-guided antibiotic stewardship in suspected ventilator-associated pneumonia (VAPrapid2): a randomised controlled trial and process evaluation. Lancet Respir. Med. 8, 182. 10.1016/S2213-2600(19)30367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horonenko G.; Hoyt J. C.; Robbins R. A.; Singarajah C. U.; Umar A.; Pattengill J.; Hayden J. M. (2007) Soluble triggering receptor expressed on myeloid cell-1 is increased in patients with ventilator-associated pneumonia: a preliminary report. Chest 132 (1), 58–63. 10.1378/chest.06-2731. [DOI] [PubMed] [Google Scholar]
- Fartoukh M.; Maitre B.; Honore S.; Cerf C.; Zahar J. R.; Brun-Buisson C. (2003) Diagnosing pneumonia during mechanical ventilation: the clinical pulmonary infection score revisited. Am. J. Respir. Crit. Care Med. 168 (2), 173–9. 10.1164/rccm.200212-1449OC. [DOI] [PubMed] [Google Scholar]
- Sweeney T. E.; Perumal T. M.; Henao R.; Nichols M.; Howrylak J. A.; Choi A. M.; Bermejo-Martin J. F.; Almansa R.; Tamayo E.; Davenport E. E.; et al. (2018) A community approach to mortality prediction in sepsis via gene expression analysis. Nat. Commun. 9 (1), 694. 10.1038/s41467-018-03078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teggert A.; Datta H.; Ali Z. (2020) Biomarkers for Point-of-Care Diagnosis of Sepsis. Micromachines 11 (3), 286. 10.3390/mi11030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egilmezer E.; Walker G. J.; Bakthavathsalam P.; Peterson J. R.; Gooding J. J.; Rawlinson W.; Stelzer-Braid S. (2018) Systematic review of the impact of point-of-care testing for influenza on the outcomes of patients with acute respiratory tract infection. Rev. Med. Virol. 28 (5), e1995. 10.1002/rmv.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. S.; Tsai C. L.; Chang J.; Hsu T. C.; Lin S.; Lee C. C. (2018) Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin. Microbiol. Infect. 24 (10), 1055–1063. 10.1016/j.cmi.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendish N. J.; Malachira A. K.; Armstrong L.; Houghton R.; Aitken S.; Nyimbili E.; Ewings S.; Lillie P. J.; Clark T. W. (2017) Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): a pragmatic, open-label, randomised controlled trial. Lancet Respir. Med. 5 (5), 401–411. 10.1016/S2213-2600(17)30120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendish N. J.; Malachira A. K.; Clark T. W. (2017) Molecular point-of-care testing for respiratory viruses versus routine clinical care in adults with acute respiratory illness presenting to secondary care: a pragmatic randomised controlled trial protocol (ResPOC). BMC Infect. Dis. 17 (1), 128. 10.1186/s12879-017-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano T.; Nishikawa H.; Suzuki R.; Onaka M.; Nishiyama A.; Kitagawa D.; Oka M.; Masuo K.; Yoshida S. (2020) The impact analysis of a multiplex PCR respiratory panel for hospitalized pediatric respiratory infections in Japan. J. Infect. Chemother. 26, 82. 10.1016/j.jiac.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shengchen D.; Gu X.; Fan G.; Sun R.; Wang Y.; Yu D.; Li H.; Zhou F.; Xiong Z.; Lu B.; Zhu G.; Cao B. (2019) Evaluation of a molecular point-of-care testing for viral and atypical pathogens on intravenous antibiotic duration in hospitalized adults with lower respiratory tract infection: a randomized clinical trial. Clin. Microbiol. Infect. 25, 1415. 10.1016/j.cmi.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D.; Chetty Y.; Cooper B. S.; Virk M.; Glass S. K.; Letters A.; Kelly P. A.; Sudhanva M.; Jeyaratnam D. (2017) Multiplex PCR point of care testing versus routine, laboratory-based testing in the treatment of adults with respiratory tract infections: a quasi-randomised study assessing impact on length of stay and antimicrobial use. BMC Infect. Dis. 17 (1), 671. 10.1186/s12879-017-2784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Kang-Birken S. L.; Mathews S. K.; Kenner C. E.; Fitzgibbons L. N. (2019) Role of rapid diagnostics for viral respiratory infections in antibiotic prescribing decision in the emergency department. Infect. Control Hosp. Epidemiol. 40, 974. 10.1017/ice.2019.166. [DOI] [PubMed] [Google Scholar]
- Chen H.; Weng H.; Lin M.; He P.; Li Y.; Xie Q.; Ke C.; Jiao X. (2017) The Clinical Significance of FilmArray Respiratory Panel in Diagnosing Community-Acquired Pneumonia. BioMed Res. Int. 2017, 7320859. 10.1155/2017/7320859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sails A. D.; Eltringham G.; Valappil M.; Waugh S.; Saunders D. (2017) Comparison of the Luminex NxTAG respiratory pathogen panel and a multiplex in-house real-time PCR panel for the detection of respiratory viruses in symptomatic patients. J. Med. Microbiol. 66 (9), 1291–1296. 10.1099/jmm.0.000562. [DOI] [PubMed] [Google Scholar]
- Brendish N. J.; Malachira A. K.; Beard K. R.; Armstrong L.; Lillie P. J.; Clark T. W. (2019) Hospitalised adults with pneumonia are frequently misclassified as another diagnosis. Respir. Med. 150, 81–84. 10.1016/j.rmed.2019.02.013. [DOI] [PubMed] [Google Scholar]
- Millot G.; Voisin B.; Loiez C.; Wallet F.; Nseir S. (2017) The next generation of rapid point-of-care testing identification tools for ventilator-associated pneumonia. Annals of translational medicine 5 (22), 451. 10.21037/atm.2017.11.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnot-Katsikas A.; Tesic V.; Love N.; Hill B.; Bethel C.; Boonlayangoor S.; Beavis K. G. (2018) Use of the Accelerate Pheno System for Identification and Antimicrobial Susceptibility Testing of Pathogens in Positive Blood Cultures and Impact on Time to Results and Workflow. J. Clin. Microbiol. 56 (1), e01166-17. 10.1128/JCM.01166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C., Wunderink R. G., and Sims M. (2018) Multicentre clinical trial of the Unyvero Lower Respiratory Tract Infection Application, 28th ECCMID 2018, Abstract P0559.
- Papan C.; Meyer-Buehn M.; Laniado G.; Nicolai T.; Griese M.; Huebner J. (2018) Assessment of the multiplex PCR-based assay Unyvero pneumonia application for detection of bacterial pathogens and antibiotic resistance genes in children and neonates. Infection 46 (2), 189–196. 10.1007/s15010-017-1088-y. [DOI] [PubMed] [Google Scholar]
- Personne Y.; Ozongwu C.; Platt G.; Basurto-Lozada P.; Shamin M.; Gant V. A.; Zumla A.; Enne V. I. (2016) ’Sample-in, answer-out’? Evaluation and comprehensive analysis of the Unyvero P50 pneumonia assay. Diagn. Microbiol. Infect. Dis. 86 (1), 5–10. 10.1016/j.diagmicrobio.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Ozongwu C.; Personne Y.; Platt G.; Jeanes C.; Aydin S.; Kozato N.; Gant V.; O’Grady J.; Enne V. I. (2017) The Unyvero P55 ’sample-in, answer-out’ pneumonia assay: A performance evaluation. Biomol Detect Quantif 13, 1–6. 10.1016/j.bdq.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby N. J.; McHugh M. P.; Forbes C.; MacKenzie L.; Hamilton S. K. D.; Griffith D. M.; Templeton K. E. (2019) Comparison of Unyvero P55 Pneumonia Cartridge, in-house PCR and culture for the identification of respiratory pathogens and antibiotic resistance in bronchoalveolar lavage fluids in the critical care setting. Eur. J. Clin. Microbiol. Infect. Dis. 38 (6), 1171–1178. 10.1007/s10096-019-03526-x. [DOI] [PubMed] [Google Scholar]
- Gastli N. A. S., and Loubinoux J. (2019) Evaluation of the FILMARRAY Pneumonia Panel for rapid microbiological diagnosis of pneumonia. 29th ECCMID 2019, Abstract P1560.
- Enne V. I., Aydin A., Gant V., Livermore D., and O’Grady J. (2019) INHALE WP1: An observational study comparing the performance of two multiplex PCR platforms against routine microbiology for the detection of potential pathogens in patients with suspected hospital-acquired/ventilator associated pneumonia (HAP/VAP) across 15 UK intensive care units (ICUs). 29th ECCMID 2019, Abstract P1561.
- UCL INHALE project. https://www.ucl.ac.uk/inhale-project.
- Alviset S. G. N., Loubinoux J., and Kerneis S. (2019) Impact of the FILMARRAY pneumonia panel on antimicrobial stewardship in pneumonia patients. 29th ECCMID 2019, Abstract O0449.
- Lejeune M. V. E., and D’Humieres C. (2019) Performance of the FILMARRAY Pneumonia Panel and impact of antibiotic treatment in ICU patients with severe pneumonia. 29th ECCMID 2019, Abstract P1562.
- Chastre J.; Fagon J. Y. (2002) Ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 165 (7), 867–903. 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- Ibn Saied W.; Merceron S.; Schwebel C.; Le Monnier A.; Oziel J.; Garrouste-Orgeas M.; Marcotte G.; Ruckly S.; Souweine B.; Darmon M.; et al. (2020) Ventilator-associated pneumonia due to Stenotrophomonas maltophilia: Risk factors and outcome. J. Infect. 80, 279. 10.1016/j.jinf.2019.10.021. [DOI] [PubMed] [Google Scholar]
- Charalampous T.; Kay G. L.; Richardson H.; Aydin A.; Baldan R.; Jeanes C.; Rae D.; Grundy S.; Turner D. J.; Wain J.; Leggett R. M.; Livermore D. M.; O’Grady J. (2019) Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 37 (7), 783–792. 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- Livermore D. M. (1993) Determinants of the activity of beta-lactamase inhibitor combinations. J. Antimicrob. Chemother. 31, 9–21. 10.1093/jac/31.suppl_A.9. [DOI] [PubMed] [Google Scholar]
- Livermore D. M.; Nicolau D. P.; Hopkins K. L.; Meunier D. (2020) Carbapenem-Resistant Enterobacterales, Carbapenem Resistant Organisms, Carbapenemase-Producing Enterobacterales, and Carbapenemase-Producing Organisms: Terminology Past its “Sell-By Date” in an Era of New Antibiotics and Regional Carbapenemase Epidemiology. Clin. Infect. Dis. ciaa122. 10.1093/cid/ciaa122. [DOI] [PubMed] [Google Scholar]
- Coppens J.; Van Heirstraeten L.; Ruzin A.; Yu L.; Timbermont L.; Lammens C.; Matheeussen V.; McCarthy M.; Jorens P.; Ieven M.; et al. (2019) Comparison of GeneXpert MRSA/SA ETA assay with semi-quantitative and quantitative cultures and nuc gene-based qPCR for detection of Staphylococcus aureus in endotracheal aspirate samples. Antimicrobial resistance and infection control 8, 4. 10.1186/s13756-018-0460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J. J.; Haessler S.; Imrey P.; Lindenauer P. K.; Richter S. S.; Yu P. C.; Rothberg M. B. (2019) Pneumococcal urinary antigen testing in US hospitals: A missed opportunity for antimicrobial stewardship. Clin. Infect. Dis. ciz983. 10.1093/cid/ciz983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans A.; Descheemaeker P.; Boel A.; Desmet S.; Van Gasse N.; Reynders M. (2019) What is the risk of missing legionellosis relying on urinary antigen testing solely? A retrospective Belgian multicenter study. Eur. J. Clin. Microbiol. Infect. Dis. 39, 729–734. 10.1007/s10096-019-03785-8. [DOI] [PubMed] [Google Scholar]
- Yang L.; Haidar G.; Zia H.; Nettles R.; Qin S.; Wang X.; Shah F.; Rapport S. F.; Charalampous T.; Methe B.; et al. (2019) Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir. Res. 20 (1), 265. 10.1186/s12931-019-1218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier C.; Kalantar K. L.; Moazed F.; Wilson M. R.; Crawford E. D.; Deiss T.; Belzer A.; Bolourchi S.; Caldera S.; Fung M.; Jauregui A.; Malcolm K.; Lyden A.; Khan L.; Vessel K.; Quan J.; Zinter M.; Chiu C. Y.; Chow E. D.; Wilson J.; Miller S.; Matthay M. A.; Pollard K. S.; Christenson S.; Calfee C. S.; DeRisi J. L. (2018) Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. U. S. A. 115 (52), E12353–e12362. 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Sun B.; Tang X.; Liu Y. L.; He H. Y.; Li X. Y.; Wang R.; Guo F.; Tong Z. H. (2019) Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients. Eur. J. Clin. Microbiol. Infect. Dis. 39, 369–374. 10.1007/s10096-019-03734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 (14), 1754–60. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S.; Kawada J. I.; Horiba K.; Okuno Y.; Okumura T.; Suzuki T.; Torii Y.; Kawabe S.; Wada S.; Ikeyama T.; Ito Y. (2019) Metagenomic analysis using next-generation sequencing of pathogens in bronchoalveolar lavage fluid from pediatric patients with respiratory failure. Sci. Rep. 9 (1), 12909. 10.1038/s41598-019-49372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Caro J. C.; Santibanez M.; Garcia Rivero J. L.; Villanueva M.; Sainz J.; Gonzalez Astorqui P.; Hierro M.; Rodriguez Porres M.; Paras Bravo P.; Mira A.; Rodriguez J. C.; Galiana A. (2019) Sputum Microbiome Dynamics in Chronic Obstructive Pulmonary Disease Patients during an Exacerbation Event and Post-Stabilization. Respiration 98 (5), 447–454. 10.1159/000501988. [DOI] [PubMed] [Google Scholar]
- Andresen C.; Jalal S.; Aili D.; Wang Y.; Islam S.; Jarl A.; Liedberg B.; Wretlind B.; Martensson L. G.; Sunnerhagen M. (2010) Critical biophysical properties in the Pseudomonas aeruginosa efflux gene regulator MexR are targeted by mutations conferring multidrug resistance. Protein Sci. 19 (4), 680–92. 10.1002/pro.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3 (2), 255–264. [PubMed] [Google Scholar]
- Brinda K.; Callendrello A.; Ma K. C.; MacFadden D. R.; Charalampous T.; Lee R. S.; Cowley L.; Wadsworth C. B.; Grad Y. H.; Kucherov G.; et al. (2020) Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat. Microbiol 5, 455–464. 10.1038/s41564-019-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte F. S. (2008) Molecular diagnostics for detection of bacterial and viral pathogens in community-acquired pneumonia. Clin. Infect. Dis. 47, S123–S126. 10.1086/591392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterer G. W. (2017) Diagnosing Viral and Atypical Pathogens in the Setting of Community-Acquired Pneumonia. Clin. Chest Med. 38 (1), 21–28. 10.1016/j.ccm.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O.; Durand G.; Hallin M.; Diefenbach A.; Gant V.; Murray P.; Kozlakidis Z.; van Belkum A. (2020) Consolidation of Clinical Microbiology Laboratories and Introduction of Transformative Technologies. Clin. Microbiol. Rev. 33 (2), e00057-19. 10.1128/CMR.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K.; Kuisma M.; Maki M.; Grendelmeier P.; Hirsch H. H.; Tamm M.; Papakonstantinou E.; Stolz D. (2015) Molecular diagnostics for bacterial infections in bronchoalveolar lavage--a case-control, pilot study. Swiss Med. Wkly. 145, w14193. 10.4414/smw.2015.14193. [DOI] [PubMed] [Google Scholar]
- Warreman E. B.; Lambregts M. M. C.; Wouters R. H. P.; Visser L. G.; Staats H.; van Dijk E.; de Boer M. G. J. (2019) Determinants of in-hospital antibiotic prescription behaviour: a systematic review and formation of a comprehensive framework. Clin. Microbiol. Infect. 25 (5), 538–545. 10.1016/j.cmi.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Krockow E. M.; Colman A. M.; Chattoe-Brown E.; Jenkins D. R.; Perera N.; Mehtar S.; Tarrant C. (2019) Balancing the risks to individual and society: a systematic review and synthesis of qualitative research on antibiotic prescribing behaviour in hospitals. J. Hosp. Infect. 101 (4), 428–439. 10.1016/j.jhin.2018.08.007. [DOI] [PubMed] [Google Scholar]



