Abstract
The first crucial step in the developmental program occurs during pre-implantation, the time after the oocyte has been fertilized and before the embryo implants in the uterus. This period represents a vulnerable window as the epigenome undergoes dynamic changes in DNA methylation profiles. Alterations in the early embryonic reprogramming wave can impair DNA methylation patterns and induce permanent changes to the developmental program, leading to the onset of adverse health outcomes in offspring. Although there is an increasing body of evidence indicating that harmful exposures during pre-implantation embryo development can trigger lasting epigenetic alterations in offspring, the mechanisms are still not fully understood. Since physiological or pathological changes in DNA methylation can occur as a response to environmental cues, proper environmental milieu plays a critical role in the success of embryonic development. In this review, we depict the mechanisms behind the embryonic epigenetic reprogramming of DNA methylation and highlight how maternal environmental stressors (e.g., alcohol, heat stress, nutrient availability) during pre-implantation and assisted reproductive technology procedures affect development and DNA methylation marks.
Keywords: epigenetics, DNA methylation, pre-implantation embryos, prenatal exposures, developmental programming
Introduction
The rapidly emerging field of epigenetics studies genome modifications that regulate gene expression without altering the content of the genetic sequence. DNA and histones —the structural proteins of the chromatin— can possess a layer of reversible epigenetic modifications that contribute to how genes are expressed and how they interact within a cell. Epigenetic modifications are chemical tags, such as phosphate, methyl and acetyl groups, affixed to the histone proteins and DNA by a highly dynamic and synergic network of nuclear enzymes that modulate chromatin availability thereby regulating gene expression (Jenuwein and Allis, 2001; Gibney and Nolan, 2010). The epigenome is of utmost importance and comprehensively susceptible to environmental factors, (Marsit, 2015; Legault et al., 2018; Norouzitallab et al., 2019) particularly during the early stages of embryo development as its epigenetic regulation is concomitant with proper cell fate determination (Morey et al., 2015; Ohbo and Tomizawa, 2015; Vougiouklakis et al., 2017). The most notable epigenetic mechanism during mammalian pre-implantation is the epigenetic reprogramming of DNA methylation that triggers embryonic genome activation, a pivotal step for proper embryo development. While these processes are similar between species, they differ in regards to the rate and timing of events and sex-specific variations. Although many studies have shown the highly significant physiological roles of the epigenome in mammalian development, it is still considerably misunderstood and insufficiently studied during pre-implantation, partially due to technological limitations as a consequence of the very small number of cells and the short duration of this stage of development. This review will depict the mechanisms behind the embryonic epigenetic reprogramming of DNA methylation and will assess the epigenetic consequences of various assisted reproductive technology (ART) procedures as well as how environmental stressors during pre-implantation will affect short-term and long-term development, focussing specifically on the maternal environment.
DNA Methylation
DNA methylation is the most widely understood epigenetic modification as a mechanism for gene expression mediation and is involved in many key physiological processes such as genomic imprinting, transposable elements silencing, X-chromosome inactivation and aging (Bird, 2002; Smith and Meissner, 2013). In mammals, DNA methylation occurs mainly on the cytosines of cytosine-guanine dinucleotides known as CpG sites (CpGs) (Razin and Cedar, 1991; Weber and Schubeler, 2007), though non-CpG (i.e. CpA, CpT, CpC) methylation can also be found at specific stages of cellular development, primarily in stem cells and brain tissues (Lister et al., 2013; Patil et al., 2014). The enzymes directly responsible for the methylation of DNA are DNA methyltransferases (DNMTs). DNMT3A and DNMT3B add de novo methylation thus they have been identified to be involved in the establishment of methylation patterns required for cell lineage determination during development (Okano et al., 1999; Li, 2002). For optimal activity, DNMT3A and DNMT3B require the accessory protein DNMT3L (DNA methyltransferase-like), a protein similar to DNMTs but lacking methyltransferase activity (Suetake et al., 2004). In contrast, DNMT1 carries out heritable DNA methylation pattern maintenance during cellular division due to its preference for substrates with hemi-methylated CpGs, i.e. only one methylated DNA strand, which would naturally occur during semi-conservative DNA replication (Leonhardt et al., 1992; Lei et al., 1996; Pradhan et al., 1999). Although DNA methylation patterns are heritable from cell to cell, it remains strikingly dynamic in nature. Physiological or pathological changes in DNA methylation can occur as a response to environmental cues, therefore demethylation is conjointly a greatly relevant process. Contrary to enzyme-mediated methylation, demethylation can occur passively or actively (Kishikawa et al., 2003). Passive genome demethylation is replication-dependant, caused by DNMT1 reduced activity resulting in an unspecific progressive dilution of DNA methylation over multiple consecutive cell divisions (Kagiwada et al., 2013; Wu and Zhang, 2014). Conversely, active demethylation is specific and is catalytically directed by ten-eleven translocation enzymes (TET1, TET2, TET3) (Tahiliani et al., 2009; Ito et al., 2010). The addition and erasure of DNA methylation and other epigenetic marks are the driving forces behind embryo development, as they dictate how, when and at what level genes are expressed.
CpGs are present all across the mammalian genome but their methylation will bear different consequences depending upon their location (e.g. promoter regions, gene bodies, enhancers) and upon their level of enrichment. CpG methylation located in gene bodies has been shown to promote high levels of gene expression whereas when located in promoter regions, it is associated with transcriptional silencing, which coordinates cellular differentiation (Goll and Bestor, 2005; Jones, 2012). In mammalian genomes, promoters comprised of highly CpG dense sequences, known as CpG islands (CGIs), control approximately 60-80% of genes depending on the species (Antequera and Bird, 1993; Saxonov et al., 2006). Methylation of CGIs of a promoter accompanied with repressive histone modifications (H3K9me3, H3K27me3) induces nucleosome compaction and prevents transcription factor (TF) binding, causing the repression of gene transcription. On the other hand, the promoter regions of transcribed genes have CGIs devoid of methylation along with active histone modifications (H3K4me2/3, H3K9ac) (Barski et al., 2007; Koch et al., 2007; Henikoff and Shilatifard, 2011; Severin et al., 2011), thus ensuring the open chromatin configuration that allows for TF binding and gene activation. However, recent studies have started to refute this general rule suggesting that, in some cases, the loss of methylation can be a consequence of TF binding as opposed to the cause of action, leading some to believe that gene activation may not always be methylation driven (Zhu et al., 2016a; Pacis et al., 2019).
A particularly important role of DNA methylation in mammalian development is genomic imprinting. A small cohort of genes called imprinted genes possesses germline differentially methylated regions (gDMRs). gDMRs acquire monoallelic genomic methylation in a parent-of-origin manner causing only one allele to be expressed (Reik et al., 2001; Ferguson-Smith, 2011). A more specific type of gDMR is imprinting control regions (ICRs) that are directly implicated in the binding of TFs and regulate the expression of multiple imprinted genes at a time (e.g., H19 and Igf2; Insulin-Like Growth Factor 2) (Thorvaldsen et al., 1998; Fitzpatrick et al., 2002; Ideraabdullah et al., 2008). These genomic imprints are determined prior to fertilization in growing diplotene oocytes and in perinatal prospermatogonia and must be maintained throughout the entire lifespan of the new generation (Stoger et al., 1993; Kono et al., 1996; Davis et al., 2000; Ueda et al., 2000).
Embryonic Epigenetic Reprogramming
In early embryogenesis, the embryo undergoes a reprogramming wave of DNA methylation during which the global methylation profiles, with the exception of gDMRs, are remodeled. Shortly after fertilization, the zygotic genome remains separated into two distinct paternal and maternal pronuclei which must sustain extensive global demethylation to erase the germ cell-specific methylation profiles and implement totipotency prior to implantation of the embryo (Seisenberger et al., 2013). The demethylation mechanisms are known to be distinct for both pronuclei, but have not been fully characterized thus far. Another perplexing part of the reprogramming process is the maintenance of gDMR methylation patterns as it is a major requirement for normal mammalian development. In fact, the loss of genomic imprints during embryo development causes permanent damage to cellular functions since the embryo is unable to restore them (Howell et al., 1998; Howell et al., 2001; McGraw et al., 2013; McGraw et al., 2015). Since only one allele is inherently active, imprinted gene expression is hypersensitive to changes in regulation, which can cause dramatic effects on development as many imprinted genes have growth regulatory functions (Plasschaert and Bartolomei, 2014). Many studies in mice have demonstrated the prevalent involvement of DNMT1 variants (DNMT1o; DNMT1-oocyte, DNMT1s; somatic) in the maintenance of genomic imprints throughout embryonic epigenetic reprograming (Bostick et al., 2007; Arita et al., 2008; Avvakumov et al., 2008). How DNMT1 specifically recognizes and maintains gDMRs but does not maintain global methylation remains mostly unclear. However, Dnmt1-/- mice are embryonic lethal as the absence of Dnmt1 causes the exhaustive loss of genomic imprints and does not allow for de novo methylation to be properly maintained during remethylation (Li et al., 1992). Moreover, we observed that the loss of Dnmt1o caused sex-specific placental defects in female embryos as well as perturbed imprinted X-inactivation (McGraw et al., 2013). These data highlight how a brief perturbation in the DNA methylation maintenance process of early stage embryoscan influence development, and further emphasize the importance of studying the impact of the maternal environment and sex-specific alterations during this critical period.
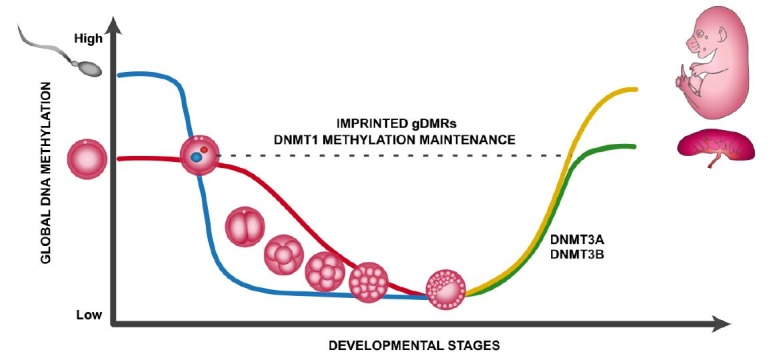
The loss of global DNA methylation during reprogramming initiates the embryonic genome activation, vital for proper development. The thoroughly timed expression of genes in embryonic genome activation is controlled by chromatin structural changes. A schematic representation of the mouse embryonic epigenetic reprogramming of DNA methylation is depicted in Figure 1, showing the dynamics of DNA methylation from the fertilization of the zygote to the maturation of embryonic and placental tissues. After global demethylation, the inner cell mass and the trophoblast gain de novo methylation catalyzed by DNMT3A and DNMT3B to implement the epigenetic patterns for the development of the embryo and the placenta (Red-Horse et al., 2004; Marikawa and Alarcon, 2009). Studies conducted in mice have indicated separate specific phenotypes in Dnmt3a-/- versus Dnmt3b-/-. Dnmt3a-/- mice make it to term, albeit severely runted and die shortly thereafter, whereas Dnmt3b-/- are embryonic lethal (Niakan et al., 2012). The altered expression of these key enzymes is unmistakably symptomatic of epigenetic developmental disturbances, but it is becoming more and more evident that the regulation of the epigenome is also staggeringly sensitive to embryonic environment, particularly throughout the pre-implantation period.
Figure 1. Global DNA demethylation and remethylation during the epigenetic reprogramming of early embryogenesis in mice. Soon after fertilization, the zygotic paternal and maternal pronuclei undergo global demethylation during the pre-implantation stages, except for gDMRs which are maintained via DNMT1 activity. The paternal genome (blue) is initially actively demethylated by the TET3 enzyme followed by passive demethylation, whereas the maternal genome (red) demethylation is solely passive due to DNMT1 inactivity, hence the sharper demethylation slope for the paternal curve. After implantation, the blastocyst acquires de novo methylation patterns catalyzed by DNMT3A and DNMT3B to establish the embryonic and placental programs imperative for development initiation.
Early embryonic environment and impact on epigenetic reprogramming events
A considerable amount of evidence has begun to show how epigenetic programming is susceptible to early embryonic environment, such as nutrient availability and toxin exposures, prior to implantation of the blastocyst. The dynamics of the embryonic wave of DNA methylation – proper erasure and de novo methylation or methylation maintenance – that is crucial to trigger the developmental program can become disturbed in response to these environmental cues leading to changes in gene expression and growth defects. Many have investigated the effects of the environment on embryo development using mainly in vitro models. However, the direct epigenetic impacts of the embryonic environment and the lasting effects on long-term development have been poorly studied, due to technological barriers and limited number of cells during pre-implantation stages. When studying early embryonic development exposures, one must be cautious when comparing in utero and in vitro models being that in vitro culture technologies have yet to accurately reproduce the maternal tract conditions. We have thus divided the following section describing the effects of environmental factors during early embryonic development in two parts: firstly, the influence of in vitro reproductive technologies of mammalian embryos, and secondly, the intrauterine exposures during pregnancy.
Assisted Reproductive Technology (ART)
ART is an umbrella term used to describe the assortment of medical procedures and approaches (e.g., superovulation, in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI), embryo in vitro culture (IVC)) that can be performed to achievepregnancy. Such procedures are now an integral part of human infertility treatment, as the number of children born by ART is estimated at more than 8 million worldwide (Weinerman, 2018). ART also plays a major role in animal reproduction to increase reproductive efficiency and genetic improvement in livestock, as well as to conserve endangered species. The outcomes of human pregnancies produced by ART have undergone intense scrutiny and while most children conceived using ART are healthy, these procedures have been associated with an increased risk of preeclampsia, intrauterine growth restriction, birth defects (Qin et al., 2016; Zhu et al., 2016b; Choux et al., 2018; Choufani et al., 2019) and imprinting disorders (Market-Velker et al., 2010; White et al., 2015). Although various studies have shown that ART may lead to epigenetic perturbations (El Hajj and Haaf, 2013; Urrego et al., 2014; Duranthon and Chavatte-Palmer, 2018), the etiology associated with ART and increased risk of perinatal complications is still poorly understood. However, the dynamic epigenome reprogramming during germ cell development and the pre-implantation period, especially of DNA methylation patterns, are processes that are prone to being affected by approaches used in ART and could provide biological plausibility.
ART procedures
Although it is unclear which ART procedure has the greatest influence, we know that the dramatic changes in embryo environment can induce long-term effects on the epigenome. In ovarian superstimulation, various studies suggest that the acquisition of imprinting patterns in the oocyte might be perturbed and lead to abnormal allelic expression in later embryo and placenta development (reviewed in Anckaert et al., 2013; McGraw and Trasler, 2013). However, studies show that superovulation treatments do not alter normal imprinted methylation acquisition in oocytes, but rather disrupt maternal-effect gene products that are required during pre-implantation for imprint maintenance (Denomme et al., 2011; Uysal et al., 2018). We also showed that some of these induced errors of imprinted gene expression (H19, Igf2) present in mid-gestation mouse placenta are no longer apparent at the end of the gestation (Fortier et al., 2008; Fortier et al., 2014). This suggests that even though superovulation produces abnormal oocytes that initiate altered expression of imprinted genes in embryos, compensatory mechanisms regulating imprinted gene networks are able to restore proper levels of gene expression during development. Although, as highlighted across the literature, the alterations in DNA methylation following ART procedures are not always striking and vary between studies (reviewed in Berntsen et al., 2019), in part because of distinctions in treatments used. It was recently reported that human placenta, but not cord blood, from IVF/ICSI showed decreased DNA methylation levels for imprinted loci H19/IGF2 and KCNQ1OT1, as well as for specific repetitive elements (Choux et al., 2018), whereas in another recent study, no obvious overall differences in genome-wide DNA methylation differences in placental tissues were associated with ART (Choufani et al., 2019). Yet, a subset of ART pregnancies associated with ICSI showed marked decrease in placental DNA methylation levels at imprinted loci (GNAS, SGCE, KCNQ1OT1 and NNAT). Not only do these studies reveal that ICSI generates distinct DNA methylation alterations in specific tissues compared to controls as opposed to less invasive ART procedures, they highlight the importance of carefully pairing and comparing equivalent ART procedures when designing epigenetic studies.
Another ART procedure that is routine practice in commercial and clinical settings is cryopreservation of oocytes and embryos. Flash-freezing cryopreservation protocols (i.e., vitrification) have been linked to epigenetic alterations. Selective loss of DNA methylation of imprinted loci was observed in blastocysts subsequent to fertilization of vitrified bovine and mouse oocytes (Chen, Zhang et al., 2016; Cheng et al., 2014), whereas others found no effect on DNA methylation levels at the H19/IGF2 ICR loci at embryonic day 3 in human ICSI blastocysts following vitrification (Derakhshan-Horeh et al., 2016). When vitrification of mouse embryo at E2.5 (8-cell stage) was paired with IVC, transferred embryos revealed increased levels of global DNA methylation in both E9.5 fetus and placenta compared to IVC, but interestingly were similar to naturally mated derived samples (Ma et al., 2019). The long-term effect of vitrification was further observed in the fetus with increased DNA methylation levels at the imprinted KvDMR1 loci and significant gene expression increase of Dnmt1 and Dnmt3b compared to the IVC and natural mating groups. Together, the body of work on vitrification suggests that such exposures could influence the epigenome and lead to abnormal expression of imprinted genes. However, it is difficult to make any definitive conclusions regarding the influence of vitrification as most of these studies only assessed a limited number of loci for DNA methylation analyses, which are mostly restricted to imprinted genes, and did not investigate the long-term impact on postnatal development.
ART culture environment
As previously mentioned, although the vast majority of ART-conceived offspring are healthy, they have a higher frequency of birth defects suggesting epigenetic costs. A large body of research now supports that the in vitro culture environment has both long-lasting and significant repercussions on DNA methylation reprogramming events and embryonic development, but the exact mechanisms remain unclear. In humans, an increased prevalence of Beckwith-Wiedemann syndrome has been associated with ART procedures. This overgrowth disorder has similar adverse phenotypes and epigenetic profiles (e.g., loss of imprinting) as the large offspring syndrome in ruminants, (Chen et al., 2015) for which the incidence has been linked to the presence of serum in the culture media. (Young et al., 1998; Chen et al., 2013). As such,the ART field has mostly limited the use of serum and has designed various serum-free and chemically defined media for livestock, mice, and humans.
Various commercial and custom culture systems exist but are not as complex and dynamic as the oviduct fluid. They may present a lack or excess of different key factors and metabolites when compared to the maternal reproductive environment (Morbeck et al., 2014a; Morbeck et al., 2014b; Morbeck et al., 2017). A number of different studies have investigated the impact of culture systems on epigenetic profiles in humans and other animals, but a direct correlation between results is challenging as additional associated parameters (e.g., culture conditions, protocols, ART-procedures) may introduce a range of confounding and unpredictable variables. To circumvent these effects, Market-Velker et al. (2010) undertook a direct side-by-side comparison between naturally mated mouse embryos cultured from the 2-cell stage to the blastocyst stage in commercial systems and in vivo-derived blastocyst. They uncovered that all commercial media compromised the early embryo’s proficiency in maintaining genomic imprinting profiles of H19, Peg3, and Snrpn to a variable extent. Although some media systems appeared to be more suitable for maintaining DNA methylation levels on these imprinted loci, we cannot know how the rest of the genome behaves under these conditions because of the narrow epigenetic analyses that were performed. Interestingly, a recent study tested the addition of natural reproductive fluids in the culture system to safeguard the embryo’s epigenome. They showed that by using natural reproductive fluids they could produce IVF-blastocysts with reduced morphological, epigenetic and transcriptomic anomalies when compared to porcine blastocysts produced from unsupplemented IVF protocols (Canovas et al., 2017). Furthermore, by using both whole-genome DNA methylation and RNA-seq approaches of single blastocysts, they were able to demonstrate that the addition of oviductal tract fluid compensated for the lack of specific factors in standard culture medium required for proper development. Since this strategy has been successful so far in improving the ART procedures in mice, humans and other livestock animals, it shows great potential for rescuing troubled early embryo development and future negative impacts in offspring.
Maternal and environmental influences
It is now well established that the maternal environment (e.g., nutrition, stress, toxicants) can create an adverse in utero milieu that affects the fetal developmental program and increase disease susceptibility in adulthood (aka. Developmental Origins of Health and Disease; DoHaD hypothesis). Since the all-or-none phenomenon once presumed that exposure that occurs on early stage embryos results in either death or in no adverse outcome, little research on the impact of harmful maternal environment on pre-implantation embryos was done in the past. However, this once pervasive tenet is now being revisited as several studies demonstrate that the direct contact of pre-implantation embryo with the cells of the mother’s reproductive tract can influence future development via interference with epigenetic mechanisms (Adam, 2012). Here, we will underline how adverse in uterine conditions triggered by the maternal environment (alcohol, heat stress) can have deleterious effects on the early embryonic epigenome.
Adverse stressors
Alcohol has teratogenic and neurotoxic effects on numerous potential mechanisms such as folate metabolism and DNMTs activity (Garro et al., 1991; Bielawski et al., 2002; Bonsch et al., 2006; Varela-Rey et al., 2013). We know that an exposure to alcohol during pregnancy can lead to abnormal brain development and cause fetal alcohol spectrum disorders (FASD), with symptoms ranging from craniofacial abnormalities to intellectual deficiency, behavioral difficulties and learning disabilities (Chudley et al., 2005; Cook et al., 2016; Legault et al., 2018). Although pioneer work demonstrated that early embryonic alcohol exposure can negatively influence development (Checiu and Sandor, 1986; Fazakas-Todea et al., 1986; Wiebold and Becker, 1987; Padmanabhan and Hameed, 1988), we still don’t fully understand how alcohol directly impacts the early embryo, especially its epigenome. A recent report shows that porcine zygotes exposed to alcohol in vitro have a lower rate of blastocyst formation, with blastocysts having increased mitochondrial dysfunctions and abnormal gene expression (Page-Lariviere et al., 2017). Haycock and Ramsay (2009) did show in a mouse model that alcohol exposure at E1.5 and E2.5 was associated with loss of H19 imprinted DNA methylation in the placenta at E10.5 and growth restriction (Haycock and Ramsay, 2009). In early stage embryos, ethanol exposure seems to have a lasting impact on Dnmt1 by reducing its expression, whereas Dnmt3a and Dnmt3b expression levels remained the same (Dasmahapatra and Khan, 2015). Since Dnmt1 is required for the maintenance of DNA methylation profiles, especially imprinted gene methylation during the early embryonic reprogramming wave (Hirasawa et al., 2008; McGraw et al., 2013), alcohol exposure might compromise proper Dnmt1 function and lead to altered epigenetic phenotypes. Although prenatal cigarette and recreational drugs (e.g., cocaine, cannabis) exposure have been linked to lasting behavioral and neurodevelopmental impairments, low birth weight, preterm birth, poor intrauterine growth and even infant death (Wehby et al., 2011), as well as alterations in DNA methylation and epigenetic profiles (Novikova et al., 2008; Toro et al., 2008; Breton et al., 2009; Guerrero-Preston et al., 2010; Suter et al., 2010; Toledo-Rodriguez et al., 2010; DiNieri et al., 2011), none of the prenatal expositions were done on pre-implantation embryos. As such, preclinical animal models of early embryonic exposure are needed to determine the deleterious consequence of cigarette smoking and recreational drugs on development, epigenome and gene expression. By being aware of the deleterious effects of cigarette and drug expositions onthe embryo during the first days of gestation days, women wanting to conceive might stop their consumption as preventive measures to protect their embryo.
Livestock does not have comparable environmental stressors, however, they are exposed to changing environmental conditions that affect their fertility. For example, livestock fertility, especially in dairy cows, is particularly vulnerable to higher temperature and humidity. Heat stress disrupts many metabolism processes (e.g., microtubules and microfilaments reorganization, reactive oxygen species production, DNA fragmentation and apoptosis) in embryos, leading to disrupted embryo development and increased embryonic mortality (Zhu et al., 2008; Koyama et al., 2012; de Barros and Paula-Lopes, 2018). Heat stress has a greater impact on pre-implantation embryos since heat resistance mechanisms are not fully developed at this stage. Embryos at 2-cell or 4-cell stage will be more affected since the acquisition of these processes overlaps with zygotic genome activation (ZGA) and early embryos do not respond to proapoptotic signals (de Barros and Paula-Lopes, 2018). One study suggests that the epigenetic changes seem to predominantly impact paternal imprinting genes, as the paternal genome is demethylated faster in the first days of embryo development compared to the maternal genome (Zhu et al., 2008). They reported that blastocysts resulting from mouse zygotes exposed to a 1 hour 40°C heat shock prior to IVC, showed loss of DNA methylation for paternally imprinted genes H19 and Igf-2r, but normal DNA methylation for maternally imprinted genes Peg1 and Peg3. However, since these embryos were only treated and cultured in vitro, it would be pertinent to retrieve oocytes, zygotes or embryos from livestock animals exposed to heat stress to define how genome-wide DNA methylation profiles are disturbed.
Conclusion
Epigenetic modifications, specifically DNA methylation, play a crucial role in embryo development and are vulnerable to prenatal environmental factors and exposures occurring during the pre-implantation period. So far, a handful of in vitro studies have explored the effects of assisted reproductive technologies as well as prenatal environmental conditions and exposures, such as alcohol consumption and heat stress, during pre-implantation looking at short-term effects of severe epigenetic disturbances causing early manifestation of serious developmental phenotypes. Though mild impacts during pre-implantation are hugely understudied and may cause latent long-term effects on postnatal development. Therefore, there is a dire need to study the impacts of early embryo in vitro exposures past the blastocyst-stage using embryo transfer experiments, as well as early embryo in vivo exposure models, while also taking into consideration the importance of sex-specific variations and timing of exposure. Moreover, very few studies have been able to establish the direct link between DNA methylation alterations and observed phenotypes, mainly because of the limitations in studying the methylome in the early stages of development. Cutting-edge adaptations of standard whole-genome and reduced bisulfite genome sequencing technologies are now rapidly emerging, permitting high-resolution low-input single-cell methylation analyses. Thanks to these technological and intellectual advancements, as well as the integrative analysis of multi-omics layers, a promising future lies ahead for the study of pre-implantation epigenetics.
Acknowledgments and Funding disclosure statement
MBL and SM are supported by Fonds de Recherche du Québec en Santé (FRQS). MBL is also supported by Réseau Québécois en Reproduction (RQR), and EE is supported by Fonds de Recherche du Québec en Nature et Technologies (FRQNT). We also thank Karine Doiron for figure design and editing.
References
- Adam MP. The all-or-none phenomenon revisited. Birth Defects Res A Clin Mol Teratol. 2012;94:664–669. doi: 10.1002/bdra.23029. [DOI] [PubMed] [Google Scholar]
- Anckaert E, De Rycke M, Smitz J. Culture of oocytes and risk of imprinting defects. Hum Reprod Update. 2013;19:52–66. doi: 10.1093/humupd/dms042. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berntsen S, Soderstrom-Anttila V, Wennerholm UB, Laivuori H, Loft A, Oldereid NB, Romundstad LB, Bergh C, Pinborg A. The health of children conceived by ART: 'the chicken or the egg?'. Hum Reprod Update. 2019;25:137–158. doi: 10.1093/humupd/dmz001. [DOI] [PubMed] [Google Scholar]
- Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin Exp Res. 2002;26:347–351. [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm (Vienna) 2006;113:1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas S, Ivanova E, Romar R, Garcia-Martinez S, Soriano-Ubeda C, Garcia-Vazquez FA, Saadeh H, Andrews S, Kelsey G, Coy P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. Elife. 2017:6. doi: 10.7554/eLife.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checiu M, Sandor S. The effect of ethanol upon early development in mice and rats. IX. Late effect of acute preimplantation intoxication in mice. Morphol Embryol (Bucur) 1986;32:5–11. [PubMed] [Google Scholar]
- Chen Z, Hagen DE, Elsik CG, Ji T, Morris CJ, Moon LE, Rivera RM. Characterization of global loss of imprinting in fetal overgrowth syndrome induced by assisted reproduction. Proc Natl Acad Sci U S A. 2015;112:4618–4623. doi: 10.1073/pnas.1422088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Robbins KM, Wells KD, Rivera RM. Large offspring syndrome: a bovine model for the human loss-of-imprinting overgrowth syndrome Beckwith-Wiedemann. Epigenetics. 2013;8:591–601. doi: 10.4161/epi.24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choufani S, Turinsky AL, Melamed N, Greenblatt E, Brudno M, Berard A, Fraser WD, Weksberg R, Trasler J, Monnier P. Impact of assisted reproduction, infertility, sex and paternal factors on the placental DNA methylome. Hum Mol Genet. 2019;28:372–385. doi: 10.1093/hmg/ddy321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choux C, Binquet C, Carmignac V, Bruno C, Chapusot C, Barberet J, Lamotte M, Sagot P, Bourc'his D, Fauque P. The epigenetic control of transposable elements and imprinted genes in newborns is affected by the mode of conception: ART versus spontaneous conception without underlying infertility. Hum Reprod. 2018;33:331–340. doi: 10.1093/humrep/dex366. [DOI] [PubMed] [Google Scholar]
- Chudley AE, Conry J, Cook JL, Loock C, Rosales T, LeBlanc N. Fetal alcohol spectrum disorder: Canadian guidelines for diagnosis. Cmaj. 2005;172:S1–s21. doi: 10.1503/cmaj.1040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JL, Green CR, Lilley CM, Anderson SM, Baldwin ME, Chudley AE, Conry JL, LeBlanc N, Loock CA, Lutke J, Mallon BF, McFarlane AA, Temple VK, Rosales T. Fetal alcohol spectrum disorder: a guideline for diagnosis across the lifespan. Cmaj. 2016;188:191–197. doi: 10.1503/cmaj.141593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra AK, Khan IA. DNA methyltransferase expressions in Japanese rice fish (Oryzias latipes) embryogenesis is developmentally regulated and modulated by ethanol and 5-azacytidine. Comp Biochem Physiol C Toxicol Pharmacol. 2015;176-177:1–9. doi: 10.1016/j.cbpc.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- de Barros FRO, Paula-Lopes FF. Cellular and epigenetic changes induced by heat stress in bovine preimplantation embryos. Mol Reprod Dev. 2018;85:810–820. doi: 10.1002/mrd.23040. [DOI] [PubMed] [Google Scholar]
- Denomme MM, Zhang L, Mann MR. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil Steril. 2011;96:734–738.e732. doi: 10.1016/j.fertnstert.2011.06.055. [DOI] [PubMed] [Google Scholar]
- Derakhshan-Horeh M, Abolhassani F, Jafarpour F, Moini A, Karbalaie K, Hosseini SM, Nasr-Esfahani MH. Vitrification at Day3 stage appears not to affect the methylation status of H19/IGF2 differentially methylated region of in vitro produced human blastocysts. Cryobiology. 2016;73:168–174. doi: 10.1016/j.cryobiol.2016.08.003. [DOI] [PubMed] [Google Scholar]
- DiNieri JA, Wang X, Szutorisz H, Spano SM, Kaur J, Casaccia P, Dow-Edwards D, Hurd YL. Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol Psychiatry. 2011;70:763–769. doi: 10.1016/j.biopsych.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranthon V, Chavatte-Palmer P. Long term effects of ART: What do animals tell us? Mol Reprod Dev. 2018;85:348–368. doi: 10.1002/mrd.22970. [DOI] [PubMed] [Google Scholar]
- El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil Steril. 2013;99:632–641. doi: 10.1016/j.fertnstert.2012.12.044. [DOI] [PubMed] [Google Scholar]
- Fazakas-Todea I, Sandor S, Hateganu M, Perta D, Checiu I, Stefanescu S. The effect of ethanol upon early development in mice and rats. X. The effect of acute intoxication with some alcoholic beverages upon preimplantation development in rats. Morphol Embryol (Bucur) 1986;32:159–164. [PubMed] [Google Scholar]
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–1665. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- Fortier AL, McGraw S, Lopes FL, Niles KM, Landry M, Trasler JM. Modulation of imprinted gene expression following superovulation. Mol Cell Endocrinol. 2014;388:51–57. doi: 10.1016/j.mce.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Guerrero-Preston R, Goldman LR, Brebi-Mieville P, Ili-Gangas C, Lebron C, Witter FR, Apelberg BJ, Hernandez-Roystacher M, Jaffe A, Halden RU, Sidransky D. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5:539–546. doi: 10.4161/epi.5.6.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biol Reprod. 2009;81:618–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Howell CY, Steptoe AL, Miller MW, Chaillet JR. cis-Acting signal for inheritance of imprinted DNA methylation patterns in the preimplantation mouse embryo. Mol Cell Biol. 1998;18:4149–4156. doi: 10.1128/mcb.18.7.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. Embo j. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa S, Murata T, Ugai H, Yamazaki T, Yokoyama KK. Control elements of Dnmt1 gene are regulated in cell-cycle dependent manner. Nucleic Acids Res. 2003;(Suppl):307–308. doi: 10.1093/nass/3.1.307. [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono T, Obata Y, Yoshimzu T, Nakahara T, Carroll J. Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat Genet. 1996;13:91–94. doi: 10.1038/ng0596-91. [DOI] [PubMed] [Google Scholar]
- Koyama H, Ikeda S, Sugimoto M, Kume S. Effects of Folic Acid on the Development and Oxidative Stress of Mouse Embryos Exposed to Heat Stress. Reproduction in Domestic Animals. 2012;47:921–927. doi: 10.1111/j.1439-0531.2012.01992.x. [DOI] [PubMed] [Google Scholar]
- Legault LM, Bertrand-Lehouillier V, McGraw S. Pre-implantation alcohol exposure and developmental programming of FASD: an epigenetic perspective. Biochem Cell Biol. 2018;96:117–130. doi: 10.1139/bcb-2017-0141. [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Ma Y, Wen L, Lei H, Chen S, Wang X. Changes in DNA methylation and imprinting disorders in E9.5 mouse fetuses and placentas derived from vitrified eight-cell embryos. Mol Reprod Dev. 2019;86:404–415. doi: 10.1002/mrd.23118. [DOI] [PubMed] [Google Scholar]
- Marikawa Y, Alarcon VB. Establishment of trophectoderm and inner cell mass lineages in the mouse embryo. Mol Reprod Dev. 2009;76:1019–1032. doi: 10.1002/mrd.21057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod. 2010;83:938–950. doi: 10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol. 2015;218:71–79. doi: 10.1242/jeb.106971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw S, Oakes CC, Martel J, Cirio MC, de Zeeuw P, Mak W, Plass C, Bartolomei MS, Chaillet JR, Trasler JM. Loss of DNMT1o disrupts imprinted X chromosome inactivation and accentuates placental defects in females. PLoS Genet. 2013;9:e1003873. doi: 10.1371/journal.pgen.1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw S, Trasler J. Biology & Pathology of the Oocyte. 2nd. Cambridge University Press; 2013. Oocyte epigenetics and the risks for imprinting disorders associated with assisted reproduction. [Google Scholar]
- McGraw S, Zhang JX, Farag M, Chan D, Caron M, Konermann C, Oakes CC, Mohan KN, Plass C, Pastinen T, Bourque G, Chaillet JR, Trasler JM. Transient DNMT1 suppression reveals hidden heritable marks in the genome. Nucleic Acids Res. 2015;43:1485–1497. doi: 10.1093/nar/gku1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbeck DE, Baumann NA, Oglesbee D. Composition of single-step media used for human embryo culture. Fertil Steril. 2017;107:1055–1060.e1051. doi: 10.1016/j.fertnstert.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Morbeck DE, Krisher RL, Herrick JR, Baumann NA, Matern D, Moyer T. Composition of commercial media used for human embryo culture. Fertil Steril. 2014;102:759–766.e759. doi: 10.1016/j.fertnstert.2014.05.043. a. [DOI] [PubMed] [Google Scholar]
- Morbeck DE, Paczkowski M, Fredrickson JR, Krisher RL, Hoff HS, Baumann NA, Moyer T, Matern D. Composition of protein supplements used for human embryo culture. J Assist Reprod Genet. 2014;31:1703–1711. doi: 10.1007/s10815-014-0349-2. b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Santanach A, Di Croce L. Pluripotency and Epigenetic Factors in Mouse Embryonic Stem Cell Fate Regulation. Mol Cell Biol. 2015;35:2716–2728. doi: 10.1128/MCB.00266-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Han J, Pedersen RA, Simon C, Pera RA. Human pre-implantation embryo development. Development. 2012;139:829–841. doi: 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzitallab P, Baruah K, Vanrompay D, Bossier P. Can epigenetics translate environmental cues into phenotypes? Sci Total Environ. 2019;647:1281–1293. doi: 10.1016/j.scitotenv.2018.08.063. [DOI] [PubMed] [Google Scholar]
- Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS, Undieh AS. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS One. 2008;3:e1919. doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbo K, Tomizawa S. Epigenetic regulation in stem cell development, cell fate conversion, and reprogramming. Biomol Concepts. 2015;6:1–9. doi: 10.1515/bmc-2014-0036. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pacis A, Mailhot-Leonard F, Tailleux L, Randolph HE, Yotova V, Dumaine A, Grenier JC, Barreiro LB. Gene activation precedes DNA demethylation in response to infection in human dendritic cells. Proc Natl Acad Sci U S A. 2019;116:6938–6943. doi: 10.1073/pnas.1814700116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R, Hameed MS. Effects of acute doses of ethanol administered at pre-implantation stages on fetal development in the mouse. Drug Alcohol Depend. 1988;22:91–100. doi: 10.1016/0376-8716(88)90042-7. [DOI] [PubMed] [Google Scholar]
- Page-Lariviere F, Campagna C, Sirard MA. Mechanisms Involved in Porcine Early Embryo Survival following Ethanol Exposure. Toxicol Sci. 2017;156:289–299. doi: 10.1093/toxsci/kfw256. [DOI] [PubMed] [Google Scholar]
- Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9:823–828. doi: 10.4161/epi.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert RN, Bartolomei MS. Genomic imprinting in development, growth, behavior and stem cells. Development. 2014;141:1805–1813. doi: 10.1242/dev.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- Qin J, Liu X, Sheng X, Wang H, Gao S. Assisted reproductive technology and the risk of pregnancy-related complications and adverse pregnancy outcomes in singleton pregnancies: a meta-analysis of cohort studies. Fertil Steril. 2016;105:73-85–e71-76. doi: 10.1016/j.fertnstert.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, Fisher SJ. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest. 2004;114:744–754. doi: 10.1172/JCI22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin PM, Zou X, Gaub HE, Schulten K. Cytosine methylation alters DNA mechanical properties. Nucleic Acids Res. 2011;39:8740–8751. doi: 10.1093/nar/gkr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stoger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, Barlow DP. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- Suter M, Abramovici A, Showalter L, Hu M, Shope CD, Varner M, Aagaard-Tillery K. In utero tobacco exposure epigenetically modifies placental CYP1A1 expression. Metabolism. 2010;59:1481–1490. doi: 10.1016/j.metabol.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, Lotfipour S, Leonard G, Perron M, Richer L, Veillette S, Pausova Z, Paus T. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am J Med Genet B Neuropsychiatr Genet. 2010;153b:1350–1354. doi: 10.1002/ajmg.b.31109. [DOI] [PubMed] [Google Scholar]
- Toro R, Leonard G, Lerner JV, Lerner RM, Perron M, Pike GB, Richer L, Veillette S, Pausova Z, Paus T. Prenatal exposure to maternal cigarette smoking and the adolescent cerebral cortex. Neuropsychopharmacology. 2008;33:1019–1027. doi: 10.1038/sj.npp.1301484. [DOI] [PubMed] [Google Scholar]
- Ueda T, Abe K, Miura A, Yuzuriha M, Zubair M, Noguchi M, Niwa K, Kawase Y, Kono T, Matsuda Y, Fujimoto H, Shibata H, Hayashizaki Y, Sasaki H. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells. 2000;5:649–659. doi: 10.1046/j.1365-2443.2000.00351.x. [DOI] [PubMed] [Google Scholar]
- Urrego R, Rodriguez-Osorio N, Niemann H. Epigenetic disorders and altered gene expression after use of Assisted Reproductive Technologies in domestic cattle. Epigenetics. 2014;9:803–815. doi: 10.4161/epi.28711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal F, Ozturk S, Akkoyunlu G. Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J Assist Reprod Genet. 2018;35:503–513. doi: 10.1007/s10815-017-1087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol Res. 2013;35:25–35. [PMC free article] [PubMed] [Google Scholar]
- Vougiouklakis T, Nakamura Y, Saloura V. Critical roles of protein methyltransferases and demethylases in the regulation of embryonic stem cell fate. Epigenetics. 2017;12:1015–1027. doi: 10.1080/15592294.2017.1391430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Wehby GL, Prater K, McCarthy AM, Castilla EE, Murray JC. The Impact of Maternal Smoking during Pregnancy on Early Child Neurodevelopment. J Hum Cap. 2011;5:207–254. doi: 10.1086/660885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinerman R. In vitro fertilization (IVF): Where are we now? Birth Defects Res. 2018;110:623–624. doi: 10.1002/bdr2.1227. [DOI] [PubMed] [Google Scholar]
- White CR, Denomme MM, Tekpetey FR, Feyles V, Power SG, Mann MR. High Frequency of Imprinted Methylation Errors in Human Preimplantation Embryos. Sci Rep. 2015;5:17311. doi: 10.1038/srep17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebold JL, Becker WC. In-vivo and in-vitro effects of ethanol on mouse preimplantation embryos. J Reprod Fertil. 1987;80:49–57. doi: 10.1530/jrf.0.0800049. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3:155–163. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17:551–565. doi: 10.1038/nrg.2016.83. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JQ, Liu JH, Liang XW, Xu BZ, Hou Y, Zhao XX, Sun QY. Heat stress causes aberrant DNA methylation of H19 and Igf-2r in mouse blastocysts. Mol Cells. 2008;25:211–215. [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Liu Y, Zhang R, Wu Y, Huang Y, Liu F, Li M, Sun S, Xing L, Zhu Y, Chen Y, Xu L, Zhou L, Huang H, Zhang D. Maternal and Live-birth Outcomes of Pregnancies following Assisted Reproductive Technology: A Retrospective Cohort Study. Sci Rep. 2016;6:35141. doi: 10.1038/srep35141. b. [DOI] [PMC free article] [PubMed] [Google Scholar]


