Abstract
Purpose
The aim of this study was to identify the subtype, characterize the antimicrobial resistance, determine the virulence gene distribution, and analyze the biofilm production of Staphylococcus aureus isolates from bovine mastitis milk samples in the Liaoning Province of China.
Materials and Methods
In total, 56 Staph. aureus isolates were collected and identified in this study; the isolates were divided into different spa types based on the sequence of the polymorphic X region of the spa gene. Additionally, antimicrobial susceptibility was investigated using the broth microdilution method, and 18 virulence genes were detected using PCR. Biofilm formation was measured by spectrophotometry with crystal violet staining and observed using confocal laser scanning microscopy.
Results
There were 12.12% (56/462) milk samples that were positive for Staph. aureus. These isolates were nonsusceptible to sulfamethoxazole (100%), penicillin (76.9%), daptomycin (76.79%), clindamycin (69.64%), and oxacillin (60.71%); however, the majority of the isolates (80.4%) were susceptible to amoxicillin/clavulanate. The predominant virulence genes encoded the cytotoxins, hla (94.64%) and hlb (89.29%), and the adhesion factors clfA (89.29%), clfB (89.29%), and fnbB (80.36%). Comparatively, virulence genes related to other adhesion factors such as cna (8.93%) and enterotoxins, such as seg (26.79%), sea (16.07%), seb (7.14%), and sec (7.14%) were detected at relatively lower rates. The following eight spa types were identified: t267 (35.84%), t730 (22.64%), t518 (15.09%), t1190 (11.32%), t1456 (9.43%), t224 (1.88%), t9129 (1.88%), and t177 (1.88%). The highest biofilm production was observed for t267. Staph. aureus exhibited various patterns of biofilm formation, with the biofilm often being associated with a tower-shaped structure or a thicker biofilm.
Conclusion
Our results indicated that Staph. aureus isolates from dairy cows with mastitis in the Liaoning Province of China were non-susceptible to sulfamethoxazole, penicillin, daptomycin, oxacillin, and clindamycin. Additionally, the most prevalent subtype was t267, which displayed resistance to multiple antimicrobial agents and harbored several virulence genes, including clfA, clfB, fnbB, hla, and hlb.
Keywords: Staph. aureus, antimicrobial resistance, virulence factors, biofilm formation, mastitic dairy
Introduction
Mastitis, defined as inflammation of the mammary gland, causes severe economic loss in the dairy industry due to decreased milk production, decreased reproductive performance, additional costs for treatments, and the increased risk of culling or death of the affected animals.1 Moreover, milk from dairy cows with mastitis represents a food issue and thus maybe a threat to human health. Pathogens, such as Streptococcus uberis, Escherichia coli, and coagulase-positive staphylococci (CPS), are implicated as the causative agents of mastitis.2,3
Staphylococcus aureus, a member of CPS, usually inhabits the skin and mucosal surfaces of humans and animals but can also be found in the environment. Staph. aureus leads to a variety of diseases in humans, is associated with nosocomial morbidity and mortality,4 and it has been regarded as a major pathogen in leading to bovine mastitis.5 It is important to study the clonal origin of Staph. aureus isolated from milk samples of dairy cows with mastitis because it may provide information about pathogen-associated factors.6 Notably, spa typing is a standardized method for distinguishing Staph. aureus isolates from different areas around the world. As previously reported, t529, t267 and t359 were three main spa types identified in Canada, India, the USA and Germany.7 However, the predominant Staph. aureus spa type in the Liaoning Province of China has rarely been reported.
Antimicrobial agents, such as β-lactams, tetracyclines, and some quinolones, are used to treat bovine mastitis in China;8 however, the β-lactams penicillin, oxacillin, and ceftiofur and the fluoroquinolone enrofloxacin are mainly used to treat bovine mastitis in the Liaoning Province of China (data unpublished). S. aureus isolates, however, have developed resistance to a variety of antimicrobial agents, particularly penicillin, ampicillin, erythromycin, and tetracycline.8
Several studies have been conducted to investigate the prevalence of antibiotic resistance in Staph. aureus isolates from dairy cows with mastitis in China.9,10 Unfortunately, very few studies have been performed to investigate the antimicrobial resistance of Staph. aureus in the Liaoning Province of China. The study could be improved by comparing the antibiotic resistance of Staph. aureus isolates from the Liaoning Province to those from other provinces in China.
Staph. aureus infections that lead to mastitis in dairy cows are associated with the expression of a variety of virulence factors.7 These virulence factors allow Staph. aureus to adhere to the mucosal surface, avoid elimination by the host’s immune system, and ultimately lead to infection of the host. For example, the virulence gene hlb allows organisms evade the host’s immune system by encoding for hemolysin beta; several virulence genes such as clfA, clfB, fnbA and fnbB are involved in the process of cell surface adhesion, which is a crucial step in establishing infection; many Staph. aureus isolates also possess genes that code for superantigens, such as toxic syndrome toxin-1 related gene (tsst-1) and heat-stable enterotoxins related genes (sea, seb, sec, sed, and seg).11 Therefore, the objective of this study was to determine the antimicrobial resistance of Staph. aureus isolates from dairy cows with mastitis in the Liaoning Province of China, and to investigate their virulence gene distribution and biofilm production.
Materials and Methods
Sample Collection
All the milk samples were collected between March 2016 and October 2016 using an aseptic technique as previously reported.12 Briefly, the teats were cleaned using towels and disinfected with 75% ethanol; immediately following disinfection, approximately 3 mL of milk was collected from each quarter of the udder by veterinarians who previously received training on the procedures for sampling as well as how to diagnose clinical mastitis and subclinical mastitis. All of the samples were stored in a cooler box containing ice and were transported to the laboratory within 6 h of collection.
To obtain a representative sample of the herds in the Liaoning Province, we selected the farms for this study according to the following criteria: (i) the farm had a minimum of 105 lactating cows, (ii) the average milk production was above 5500 kg of milk per cow per year, (iii) the cows were milked twice daily at 4:00 a.m. and 4:00 p.m., (iv) the cows were primarily Holstein and Chinese Holstein (hybrid of Holstein and Yellow cattle); additionally, we selected at least two herds in the north, middle and south regions of the Liaoning Province to carry out our experiment. Milk samples from dairy cows with mastitis were obtained from seven herds in total from three districts (2 herds in Shenyang, which is located in the middle of the Liaoning Province, 2 herds at Fuxin, which is located in the northern part of the Liaoning Province; and 3 herds at Wafangdian, which is located in the southern part of the Liaoning Province), and the mean herd size was 181. All of the Staph. aureus isolates used in this study were collected from milk samples from dairy cows with clinical mastitis (n=186; which includes 55 milk samples from Shenyang; 51 milk samples from Fuxin; 80 milk samples from Wafangdian) or subclinical mastitis (n=276; which includes 84 milk samples from Shenyang; 77 milk samples from Fuxin; 115 milk samples from Wafangdian). Each farm’s veterinarian diagnoses clinical mastitis according to previous reports.13 Briefly, the veterinarian examined the cow for signs of systemic illness, measured its rectal temperature, and examined the milk for signs of clinical mastitis. Clinical mastitis was defined as at least one of the following symptoms, clots in the milk, elevated udder temperature, or swelling of the udder. For this study, the mastitic cows had received no systemic or local treatment by antimicrobial agents within the last 15 days. For subclinical mastitis, the California mastitis test was initially used as a diagnostic technique to determine whether the bovine was undergoing subclinical mastitis (scored as -, +, ++, and +++, corresponding to negative, weak positive, positive, and strong positive, respectively), and then a somatic cell count (SCC) was determined using a Fossomatic FC (Foss, Hilleroød, Denmark) to confirm the result. The bovine was considered to be undergoing subclinical mastitis if the SCC of the milk samples from a mammary quarter were higher than 200,000.14 The SCC was carried out in duplicated quarter milk samples to confirm whether the cow was undergoing a subclinical mastitis.
Staph. aureus Isolates
Staph. aureus was isolated from milk samples according to the National Mastitis Council guidelines.15 In short, 10 µl of each milk sample was streaked onto an Aureus ChromoSelect Agar Base (Sigma, Shanghai, China) that contained one vial of polymyxin B per 500 mL (Sigma, Shanghai, China). The plates were incubated at 37 ºC for 48 h, and then single colonies that were a brownish to blackish color were isolated and cultured.16 The suspected colonies were initially identified by appearance, Gram staining, catalase testing, and coagulase testing (rabbit plasma tube test, TransGen, Beijing China). Finally, the isolates were identified using the API Staph system (Mérieux, Shanghai, China). A pure culture of each isolate was stored at −80 ºC in cryogenic vials (Biologix, Shandong, China), that contained 1 mL of trypticase soy broth (AoBox, Beijing, China) and 30% glycerin (Solarbio, Beijing, China).
DNA Extraction
The stored cultures were streaked onto Mueller-Hinton agar (MH(A), AoBox, Beijing, China) containing 5% sterile defibrinated sheep blood (Land Bridge, Beijing, China) and were incubated at 37 ºC for 24 h. Then, a single colony was isolated, inoculated into 3 mL of sterile MH(B) (AoBox), and cultured at 37 ºC for 24 h in an incubator (Thermo Fisher, Shanghai, China). The overnight culture was centrifuged at 3000 × g (ThermoFisher, Shanghai, China), and the supernatant was discarded. The precipitate was resuspended in phosphate buffer saline (PBS, pH=7.4; Solarbio, Beijing, China) containing 20 mg/mL lysostaphin (Sigma, Shanghai, China). The suspension was incubated at 37 ºC for 30 min. DNA extraction was carried out using a bacterial DNA extraction kit (TaKaRa, Dalian, China) according to the manufacturer’s protocol. All of the DNA preparations were stored at −20 ºC until subsequent use.
Spa Typing
The primers spa-1113F and spa-1514R were used to amplify the polymorphic X region of the spa gene (Table 1). Briefly, the spa gene was amplified by a Bio-Rad S1000 system (Bio-Rad, CA, USA) using Ex Taq® DNA polymerase (TaKaRa, Dalian, China). Each reaction mixture (25 μL) contained 2.5 μL of 10 × Ex Taq buffer (containing 3 mM MgCl2), 100 μM dNTP mixture, 200 nM of each primer, 0.63 U of Ex Taq, and 10 ng of chromosomal DNA. The PCR products were resolved by electrophoresis in 1.0% agarose gels in a 0.5 × Tris-acetate-EDTA buffer (Solarbio, Beijing, China); the bands were visualized using the System GelDoc XR+ (Bio-Rad, CA, USA) after being stained with GoldView (Solarbio). The PCR amplicons were purified using the MiniBEST DNA Fragment Purification Kit (TaKaRa, Dalian, China) and then sequenced; the spa types were determined using the spa-server (http://spa.ridom.de/).17
Table 1.
Primer Sequences and PCR Conditions Used in This Study
| Primers | Primer Sequence (5ʹ→3ʹ) | Programa | Size of PCR Product (bp) | Reference |
|---|---|---|---|---|
| spa-1113F | TAAAGACGATCCTTCGGTGAG | 1 | Variable | Aires-de-Sousa et al 200640 |
| spa-1514R | CAGCAGTAGTGCCGTTTGCTT | |||
| seaF | TTGGAAACGGTTAAAACGAA | 2 | 120 | Park et al, 201142 |
| seaR | GAACCTTCCCATCAAAAACA | |||
| sebF | TCGCATCAAACTGACAAACG | 2 | 478 | Park et al, 201142 |
| sebR | GCAGGTACTCTATAAGTGCC | |||
| secF | GCATAAAAGCTAGGAATTT | 2 | 257 | Park et al, 201138 |
| secR | AAATCGGATTAACATTATCC | |||
| sedF | CTAGTTTGGTAATATCTCCT | 2 | 317 | Park et al, 201142 |
| sedR | TAATGCTATATCTTATAGGG | |||
| segF | AATTATGTGAATGCTCAACCCGATC | 2 | 642 | Park et al, 201142 |
| segR | AAACTTATATGGAACAAAAGGTACTAGTTC | |||
| tsst-1F | ATGGCAGCATCAGCTTGATA | 2 | 350 | Park et al, 201142 |
| tsst-1R | TTTCCAATAACCACCCGTTT | |||
| bbpF | AACTACATCTAGTACTCAACAACAG | 3 | 575 | Tristan et al, 200343 |
| bbpR | ATGTGCTTGAATAACACCATCATCT | |||
| cnaF | GTCAAGCAGTTATTAACACCAGAC | 3 | 423 | Tristan et al, 200343 |
| cnaR | AATCAGTAATTGCACTTTGTCCACTG | |||
| fnbAF | GTGAAGTTTTAGAAGGTGGAAAGATTAG | 4 | 643 | Tristan et al, 200343 |
| fnbAR | GCTCTTGTAAGACCATTTTTCTTCAC | |||
| fnbBF | GTAACAGCTAATGGTCGAATTGATACT | 4 | 524 | Tristan et al, 200343 |
| fnbBR | CAAGTTCGATAGGAGTACTATGTTC | |||
| clfAF | ATTGGCGTGGCTTCAGTGCT | 4 | 292 | Tristan et al, 200343 |
| clfAR | CGTTTCTTCCGTAGTTGCATTTG | |||
| clfBF | ACATCAGTAATAGTAGGGGGCAAC | 4 | 205 | Tristan et al, 200343 |
| clfBR | TTCGCACTGTTTGTGTTTGCAC | |||
| hlaF | CTGATTACTATCCAAGAAATTCGATTG | 5 | 209 | Jarraud et al, 200241 |
| hlaR | CTTTCCAGCCTACTTTTTTATCAGT | |||
| hlbF | GTGCACTTACTGACAATAGTGC | 5 | 309 | Jarraud et al, 200241 |
| hlbR | GTTGATGAGTAGCTACCTTCAGT | |||
| hldF | AAGAATTTTTATCTTAATTAAGGAAGGAGTG | 5 | 111 | Jarraud et al, 200241 |
| hldR | TTAGTGAATTTGTTCACTGTGTCGA | |||
| hlgAF | GTCAYAGAGTCCATAATGCATTTAA | 5 | 535 | Jarraud et al, 200241 |
| hlgAR | CACCAAATGTATAGCCTAAAGTG | |||
| lukAF | GAAGTATCTAATACTTCTTTAGCAGC | 6 | 625 | da Costa et al, 201434 |
| lukAR | TCATTTGACAATTCTACACTTCCAAC | |||
| lukBF | TGAAAAAGGTTCAAAGTTGATACGAG | 6 | 433 | da Costa et al, 201434 |
| lukBR | TGTATTCGATAGCAAAAGCAGTGCA |
Notes: aPCR program. 1: 1×(94ºC, 600s), 30×(94ºC, 20s, 62ºC, 20s, 72ºC, 30s), 1×(72ºC, 600s). 2: 1×(94ºC, 600s), 35×(94ºC, 60s, 55ºC, 60s, 72ºC, 60s), 1×(72ºC, 300s). 3: 1×(94ºC, 600s), 30×(94ºC, 60s, 58ºC, 120s, 72ºC, 60s), 1×(72ºC, 600s). 4: 1×(94ºC, 600s), 30×(94ºC, 60s, 52ºC, 60s, 72ºC, 60s), 1×(72ºC, 600s). 5: 1×(94ºC, 180s), 35×(94ºC, 60s, 60ºC, 60s, 72ºC, 180s), 1×(72ºC, 420s). 6: 1×(94ºC, 180s), 35×(94ºC, 60s, 57ºC, 60s, 72ºC, 180s), 1×(72ºC, 420s).
Antimicrobial Susceptibility Test
The antibiotic resistance phenotype was determined using the microdilution method in MH(B) broth (AoBox) according to the Clinical Laboratory and Standards Institute (CLSI) guidelines.18 For each isolate, five colonies from the agar medium were inoculated into the broth medium in order to reach adequate growth; the inoculums were then transferred into tubes containing sterile 0.85% saline (Solarbio, Beijing, China), and the inoculums were mixed with the saline. The turbidity was adjusted using sterile saline (Solarbio) to equal a 0.5 McFarland standard, by comparing turbidity under a light source to observe two tubes side by side. The trays were maintained in an incubator with normal atmosphere for 24 h at 37 ºC, and Staph. aureus ATCC 25923 was used as the reference strain. Breakpoints for different antimicrobial agents are shown in Table 2.
Table 2.
Dilution Range and Susceptibility Breakpoints of Antimicrobial Agents Used in This Study
| Antimicrobial Agent | Dilution Range (μg/mL) | Susceptibility Break Points (μg/mL) | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||
| Penicillina | 0.03–64 | ≤0.12 | ≥0.25 | |
| Oxacillinb | 0.03–64 | ≤2 | ≥4 | |
| Sulfamethoxazolea | 0.12–128 | ≤64 | ≥128 | |
| Tetracyclinea | 0.03–64 | ≤4 | 8 | ≥16 |
| Enrofloxacina | 0.03–16 | ≤0.25 | 0.5–1 | ≥2 |
| Amoxicillin/clavulanateb | 0.03–64 | ≤2 | 2/4 | ≥8 |
| Cefalexind | 0.03–64 | ≤2 | 4 | ≥8 |
| Ceftiofura | 0.03–64 | ≤24 | 4 | ≥8 |
| Clindamycinb | 0.03–64 | ≤2 | 1–2 | ≥4 |
| Daptomycinc | 0.03–64 | ≤1 | 1–2 | ≥2 |
Notes: aInterpretative breakpoint based on the CLSI breakpoints used in veterinary medicine; binterpretative breakpoint based on the CLSI breakpoints used in human medicine; cinterpretative break point based on EUCAST, 2015;44 dbreakpoint of cefalexin based on cephalothin.
The following antimicrobial agents were purchased from the Chinese Institute of Veterinary Drug Control (Beijing, China): amoxicillin/clavulanate (the ratio for amoxicillin:clavulanate is 4:1, the purity of amoxicillin and clavulanate is 98% and 94%, respectively), cefalexin (95% pure), cefquinome (80% pure), ceftiofur (98% pure), clindamycin (80% pure), daptomycin (78% pure), enrofloxacin (94.2% pure), oxacillin (93.7% pure), penicillin (84.6% pure), tetracycline (73.92% pure), sulfamethoxazole/trimethoprim (the ratio for sulfamethoxazole:trimethoprim is 20:1, the purity of sulfamethoxazole and trimethoprim is 93.2% and 81.36%, respectively), and gentamicin (61.57% pure).
Detection of Virulence Genes
Multiplex polymerase chain reaction (Multiplex PCR) was used to screen the distribution of the following virulence factors as previously reported: 6 cytotoxins (hla, hlb, hld, hlgA, lukA, and lukB), 6 adhesion factors (bbp, clfA, clfB, cna, fnbA, and fnbB), and 6 enterotoxins (sea, seb, sec, sed, seg, and tsst-1). The primers used are shown in Table 1.
Briefly, multiplex PCR was carried out by a Bio-Rad S1000 system (Bio-Rad) using Ex Taq® DNA polymerase (TaKaRa). Each reaction mixture (50 μL) contained 5 μL of 10 × Ex Taq buffer (containing 3 mM MgCl2), 100 μM dNTP mixture, 200 nM of each primer, 1.25 U of Ex Taq, and 10 ng of chromosomal DNA. The PCR products were resolved by electrophoresis in 2.0% agarose gels in a 0.5 × Tris-acetate-EDTA buffer (Solarbio), were stained with GoldView (Solarbio), and then were visualized using the System GelDoc XR+ (Bio-Rad).
Adhesion Testing
Biofilm formation was measured as previously reported using spectrophotometry with crystal violet staining.19 All Staph. aureus isolates were streaked onto tryptic soy agar plates (TSB; AoBox, Beijing, China), and incubated at 37 ºC for 18 h. Five colonies were inoculated into a brain heart infusion (BHI; AoBox, Beijing, China) containing 0.25% glucose (Solarbio, Beijing, China) to obtain a 0.5 McFarland standard (approximately 1.5 × 108 CFU/mL). One-hundred microliters was transferred into the wells of a 96-well microtiter plate (ThermoFisher, Shanghai, China), and 100 μL BHI medium (AoBox) containing 0.25% glucose (Solarbio) was added into each well. The plates were stored in an incubator (ThermoFisher) at 35 ºC for 24 h under aerobic conditions without agitation. The plates were slightly washed three times with 200 μL of phosphate buffer saline (PBS, pH=7.4; Solarbio) after discarding the supernatant. The plates were then dried and stained with 0.1% (w/v) crystal violet (Sigma, Beijing, China) for 30 min. Each well was washed three times with 200 μL of water and then dried again. Two hundred microliters of 95% ethanol (Anpel, Shanghai, China) was added to each well, and the plate was incubated at room temperature for 1 h with frequent agitation. A plate reader (BioRad, CA, USA) was used to measure the absorbance of each well at 560 nm. To avoid variations between different plates, the reference strain ATCC 29740 was used to normalize each plate.19 Each experiment was carried out at least three times. The biofilm formation for each isolate was calculated and normalized by dividing the average biofilm formation for each isolate by the biofilm production of the reference strain. Then, the normalized biofilm formation was calculated based on the three independent assays.
Confocal Laser Scanning Microscopy Observation
Biofilms were observed using an Olympus FV1000-IX81 inverted confocal microscope equipped with a 60 × oil immersion lens (Olympus, Tokyo, Japan) as previously reported.20 Briefly, 3 to 5 colonies were inoculated into TSB broth (AoBox) and incubated at 37ºC for 18 h. Two-milliliter aliquots of inocula, which were diluted to 105 and 106 CFU/mL, were transferred to 35-mm-diameter glass bottom petri dishes (ThermoFisher, Shanghai, China). After incubation for 18 h, the TSB broth (AoBox) was removed by washing with phosphate buffer saline (pH=7.4, Solarbio), and the biofilms were stained using the LIVE/DEAD BacLight Bacterial Viability kit (ThermoFisher, Shanghai, China) according to the kit instructions. Biofilms were imaged using an inverted confocal microscope (Olympus). The SYTO 9 dye was excited by a 488-nm argon laser, and emitted light was collected at 520 nm. The propidium iodide dye was excited by a 559-nm argon laser, and emitted light was collected at 619 nm.
Statistical Analysis
The values from adhesion testing by spa types were compared using an ANOVA test. These data were first analyzed using a repeated measure ANOVA, and then the Fisher protected least significant difference test was performed. For all analyses, P<0.05 was considered significant, and values are presented as the mean ± SEM.
Results
Prevalence of Staph. aureus
In this study, 12.12% (56/462) of milk samples obtained from dairy cows with mastitis were positive for Staph. aureus, including 13.97% (26/186) of milk samples from dairy cows with clinical mastitis and 10.86% (30/276) of milk samples from dairy cows with subclinical mastitis.
SpaTyping
The 53 Staph aureus isolates belonged to 8 spa types (Table 3), but three isolates were nontypable because PCR amplification was not achieved. The main types (n≥5) were t267 (n=19), t730 (n=12), t518 (n=8), t1190 (n=6), and t1456 (n=5).
Table 3.
Spa Type and Biofilm Production of the Staph. Aureus Isolates
| Spa Type | Number of Isolates | Biofilm Formation |
|---|---|---|
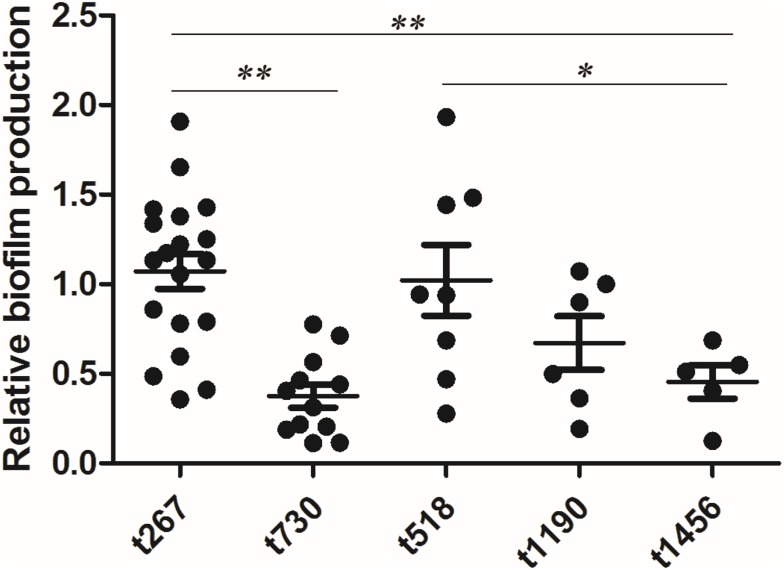
| t267 | 19 | 1.07±0.16 |
| t730 | 12 | 0.38±0.12 |
| t518 | 8 | 1.02±0.29 |
| t1190 | 6 | 0.67±0.17 |
| t1456 | 5 | 0.46±0.19 |
| t224 | 1 | 1.14±0.37 |
| t9129 | 1 | 0.42±0.15 |
| t177 | 1 | 0.64±0.24 |
Antimicrobial Resistance of Staph. aureus Isolates
The results of the antimicrobial susceptibility measurement of Staph. aureus isolates are shown in Table 4. All of the isolates were nonsusceptible to sulfamethoxazole, and forty-three (76.79%) of the isolates were nonsusceptible to penicillin and daptomycin. Additionally, most of the isolates were also nonsusceptible to oxacillin (34/56, 60.71%), clindamycin (39/56, 69.64%), and enrofloxacin (28/56, 50%). Only 11 (19.6%) isolates were nonsusceptible to amoxicillin/clavulanate. Notably, oxacillin showed a wide range of MIC distributions and exhibited a high antibiotic resistance rate. Moreover, 85.71% (48/56) of the isolates were nonsusceptible to at least three of the antimicrobial agents, and 23 out of the 56 isolates were nonsusceptible to five or more of the antimicrobial agents.
Table 4.
Minimum Inhibitory Concentrations (μg/mL) of the Staph. Aureus Isolates from the Milk of Dairy Cows with Mastitis (56 Isolates)
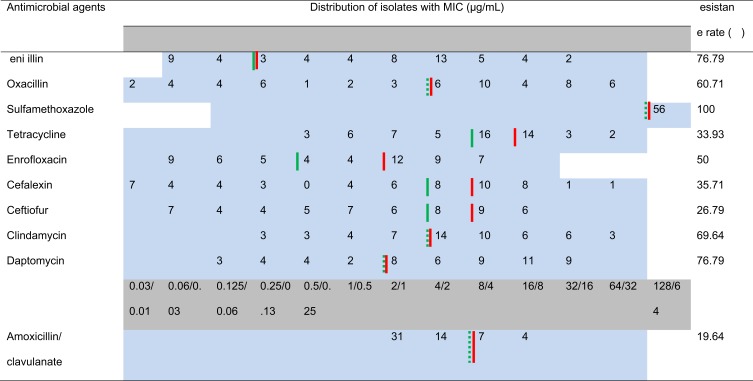 |
Notes: The dilution ranges tested are those contained within the light blue area, and the concentrations of the different antimicrobial agents are illustrated in the light gray area. The breakpoints were employed according to VET01S, 3rd ed, human, daptomycin was interpreted according to EUCAST, and the breakpoint of cefalexin was based on cephalothin. When available, susceptible and resistance breakpoints are indicated by the vertical green and red line, respectively. The dashed vertical green line represents the breakpoints from humans.
Prevalence and Distribution of the Virulence Genes
Among the Staph. aureus isolates, the detection rates for clfA, clfB, fnbB, and cna were 89.29% (50/56), 89.29% (50/56), 80.36% (45/56), and 8.93% (5/56), respectively. Conversely, genes bbp and fnbA were negative in all of the isolates. High detection rates were detected for hla (53/56, 94.64%) and hlb (50/56, 89.29%) among the Staph. aureus isolates; however genes encoding for other cytotoxins, including hld, hlgA, lukA, and lukB, were negative in all of the isolates. Of the 56 isolates, 23 isolates (41.07%) were found to have one or more genes that encode for enterotoxins. Isolates with seg (15/56, 26.79%) were the most prevalent followed by those with sea (9/56, 16.07%), seb (4/56, 7.14%), and sec (4/56, 7.14%). However, all of the isolates were negative for both sed and tsst-1 in this study.
Adhesion Testing
Biofilm formation was determined in vitro using the crystal violet staining method. A total of 53 out of the 56 isolates were compared (the nontypable strains were not included). The average biofilm formation for each spa type is shown in Table 3. The biofilm production of t267 was elevated in comparison with t730 (P<0.001), t1190 (P<0.001), t1456 (P<0.001), t9129 (P<0.001), and t177 (P=0.003), but t267 showed a similar biofilm production ability as t518 and t224. The distribution of biofilm production for each spa type is presented in Figure 1.
Figure 1.
Boxplots of the in vitro biofilm production of Staph. aureus strains of the major spa types (n≥5). Each dot represents one strain. Biofilm production was different among the main spa types, especially spa types t730 and t1456, which produced less biofilm than the other main spa type. Significance was determined by an ANOVA test (* P<0.05; ** P<0.01).
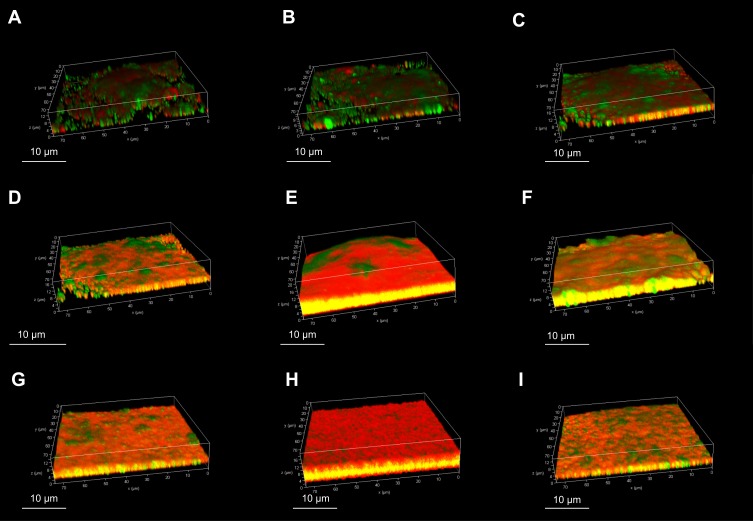
Confocal laser scanning microscopy was used to observe the biofilm production of the Staph. aureus isolates, and the results indicated that these isolates possessed a variety of biofilm production (Figure 2). The SA0020 (Figure 2D), SA0028 (Figure 2F), and SA0031 (Figure 2G) isolates grew biofilm with a tower-shaped structure, and the SA0023 (Figure 2E) and SA0047 (Figure 2H) isolates could produce thicker biofilm in comparison with the other isolates.
Figure 2.
Confocal laser scanning microscopy of in vitro biofilm production of Staph. aureus isolates. The biofilms were stained with LIVE/DEAD stain. Green represents live cells, red represents dead cells and eDNA, and yellow represents a mixture. (A) SA0007; (B) SA0012; (C) SA0019; (D) SA0020; (E) SA0023; (F) SA0028; (G) SA0031; (H) SA0047; (I) SA0052.
Discussion
Cow mastitis is a common infectious disease found all over the world, which leads to huge economic losses. A variety of factors can cause mastitis in dairy cows, including bacteria, viruses, fungi, and mycoplasma; however, Staph. aureus is one of the most frequently isolated etiological agents.21 In this study, we characterized 56 Staph. aureus isolates from milk samples of bovine mastitis in the Liaoning province of China. The Staph. aureus isolates were non-susceptible to a variety of antimicrobial agents, including sulfamethoxazole, penicillin, daptomycin, oxacillin, and clindamycin; however, the majority of the isolates were susceptible to amoxicillin/clavulanate. The virulence gene patterns did not differ systematically among the herds, eg hla, hlb, clfA, clfB,and fnbB were the predominant virulence genes found among all of the herds. The t267 spa type was the most spa type identified among all of the isolates and exhibited the highest biofilm production; whereas the t730 spa type exhibited the lowest biofilm formation; biofilm formation was also often associated with tower-shaped structures or thicker biofilm.
Staph. aureus is considered an important factor in causing mastitis in China.22–24 Our study indicated that 12.12% (56/462) of milk samples obtained from seven dairy farms in the Liaoning Province of China were positive for Staph. aureus. This prevalence is similar to previous reports from China and eastern Poland.9,24,25 Conversely, recent studies indicated that the rates of Staph. aureus in milk samples from bovine mastitis in Beijing and Jiangsu were 46.2% and 29.0%, respectively;23,24 41.7% of milk samples from dairy cows with clinical mastitis were positive for Staph. aureus in Northern Ethiopia;26 and Staph. aureus were identified among 21% of dairy cows in the Republic of Ireland.27 We speculate that better sanitation in herds, increased surveillance, and more control over Staph. aureus infections may contribute to the observed discrepancy, and thus, additional epidemiological investigation needs to be carried out to monitor the occurrence of bovine mastitis caused by Staph. aureus. An increased SCC was noticed in all isolates from both the clinical and subclinical mastitis samples. Clinical mastitis exhibited either a modest or pronounced increase in the SCC (with a mean SCC of 7.51×105 cells/mL); however, subclinical mastitis showed a weak increase (with an average SCC of 3.23×105 cells/mL). An elevated SCC is normally indicative of a severe infection within the udder and is usually associated with severe symptoms, including clots in the milk, elevated udder temperature, and/or swelling within the udder. However, no correlation has been observed between the spa-types and the SCC or clinical symptoms.
Antimicrobial susceptibility testing can provide information aid in choosing antibiotics for the treatment of bovine mastitis caused by Staph. aureus. In this study, high resistance to antimicrobial agents was observed in the Staph. aureus isolates; similarly, as previously reported, 47.15% of Staph. aureus isolates were nonsusceptible to 3 or more antimicrobial agents in north east of India.28 However, a much lower resistance rate to antimicrobial agents was observed in the United States and European countries.29 We speculate that the herd size may contribute to this discrepancy because farms with larger herd sizes normally have more dairy cows, there is often more residue in the environment within a herd due to the use of antimicrobial agents to treat bovine mastitis; thus, the pressure for pathogens to become resistant is increased. Recent reports indicated that Staph. aureus isolates from bovine mastitis have the highest resistance rate to penicillin (ranging from 10% to 80%),24 and similar results were achieved in our study (76.79%). In contrast, a considerably lower resistance rate to penicillin (33.33%) was reported by Rabello et al in Brazil.30 The reason for this phenomenon is that penicillin is normally used to treat bovine mastitis during the dry period, thus Staph. aureus is under extensive pressure to become resistant to penicillin. Moreover, the resistance patterns were different between the different dairy farms. For example, most of the Staph. aureus isolates from farm A were resistant to enrofloxacin, while those from farm B in Fuxin showed no resistance to enrofloxacin. This data indicated that the veterinarian in farm A may prefer to use enrofloxacin when treating infections, and thus, there was increased pressure for Staph. aureus to become resistant to enrofloxacin. Therefore, attention should be paid to the rational management and appropriate usage of antimicrobial agents in these herds, which has been reported to be important in the control and prevention of the transmission of antimicrobial resistance.31
Virulence genes are vital for bacterial pathogenicity. Staph. aureus isolates from cows with mastitis were reported to harbor different enterotoxin genes, indicating that these toxins are important in the development of mastitis.32 Consistent with previous reports, the adhesion genes clfA, clfB, and fnbB were the most prevalent in our study; however, Gogoi-Tiwari et al found fnbB only in 1.3% of Staph. aureus isolates,33 which was much lower than the rate found in our study (80.36%). Genes encoding for the adhesion factors clfA and clfB were more prevalent in comparison with other virulence genes; we share the same speculation with previous researchers that these adherence genes may be involved in the initial attachment of Staph. aureus to the epithelial cells of the teat canal and have become dominant in these herds.34 In the present study, none of the 56 Staph. aureus isolates from bovine mastitis were positive for the tsst gene; however, 30% of the clinical isolates contained the tsst gene in a previous report.23 The classic enterotoxin staphylococcus enterotoxin (SE) was also detected in this study, which is believed to cause food poisoning incidents. In our study, however, fewer isolates were positive for SEs genes in comparison with that from a previous report (46.42% vs 89.7%).35 The seg gene was the most prevalent gene found in our study, while Wang et al reported that the sec (65.6%) and the sea (60.4%) genes were the most frequently detected genes;24 moreover, another Chinese study reported that the seb gene was the most commonly detected.9 A previous study indicated that genotype B and genotype C were the most common genotypes found in bovine intramammary infections; genotype B was typically positive for sea, sed, and sej, but genotype C strains were positive for sec, seg, sei, and tst, but negative for sea, sed, and sej.36 We speculate that virulence gene patterns of Staph. aureus have variable distributions among herds from different regions because of local strain evolution.
Biofilm can protect bacteria from a variety of hazards, especially when bacteria are challenged with antibiotics. Biofilms decrease the amount of antibiotics that are able to reach the inner layer of the biofilm.37 Usually, the bacteria located within the inner layer are less susceptible to the antibiotics because they metabolize slowly.38 Our study analyzed the spa types in the isolates; t267 was the most prevalent spa type, but a recent study indicated that t267 was mainly located in cow nostrils.21 In this study, we mainly collected the milk samples in summer, so we speculate that flies may play an important role in the dissemination of these pathogens, leading to bovine mastitis. However, a previous report indicated that the t127 and t2279 spa types were the top two frequently distributed genotypes in milk samples.39 Therefore, we speculate that Staph. aureus isolates from different areas may belong to various spa types. The t267 spa type showed the highest ability to produce biofilm, which was also confirmed by Pichette-Jolette’s report.7 When observed under confocal laser scanning microscopy, the biofilm of the Staph. aureus isolates showed a tower-shaped structure, and some of the isolates produced a thicker biofilm. Similarly, Staph. aureus USA300 can produce biofilms with a more tower- and pillar-shaped structure when cultured in low-dose amoxicillin in comparison with their biofilm when grown in the absence of antibiotic.20
Conclusion
Our study provides an epidemiological survey on the prevalence of Staph. aureus in dairy cows with mastitis in the Liaoning Province of China. The Staph. aureus isolates were resistant to many β-lactams except ceftiofur and amoxicillin/clavulanate; speculate that this phenomenon may be caused by the priority of clinical applications when choosing antimicrobial agents. The predominant spa-type was t267, which exhibited severe antimicrobial resistance to penicillin, oxacillin, and enrofloxacin and harbored the virulence genes clfA, clfB, fnbB, hla, and hlb; this suggests that this spa-type is capable of adhesion to the teat leading to bovine mastitis. Our results suggest a need for continuous monitoring of the antimicrobial resistance, virulence phenotypes, and adhesion ability of Staph. aureus isolates from bovine mastitis. Additionally, data about antimicrobial agent management practices, which may influence antimicrobial resistance in bovine mastitis, should be collected in the Liaoning Province of China.
Acknowledgments
This research was supported by the National Key Research and Development Program of China (No. 2016YFD0501309) and National Natural Science Foundation of China (No. 31572564 and No. 31772795).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Francoz D, Wellemans V, Dupré JP, Roy JP, Labelle P, Dufour S. Invited review: a systematic review and qualitative analysis of treatments other than conventional antimicrobials for clinical mastitis in dairy cows. J Dairy Sci. 2017;100(10):7751–7770. [DOI] [PubMed] [Google Scholar]
- 2.Kateete DP, Kabugo U, Baluku H, et al. Prevalence and antimicrobial susceptibility patterns of bacteria from milkmen and cows with clinical mastitis in and around Kampala, Uganda. PLoS One. 2013;8(5):e63413. doi: 10.1371/journal.pone.0063413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boireau C, Cazeau G, Jarrige N, et al. Antimicrobial resistance in bacteria isolated from mastitis in dairy cattle in France, 2006–2016. J Dairy Sci. 2018;101(10):9451–9462. doi: 10.3168/jds.2018-14835 [DOI] [PubMed] [Google Scholar]
- 4.Zadoks R, van Leeuwen W, Barkema H, et al. Application of pulsed-field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J Clin Microbiol. 2000;38(5):1931–1939. doi: 10.1128/JCM.38.5.1931-1939.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergonier D, de Crémoux R, Rupp R, Lagriffoul G, Berthelot X. Mastitis of dairy small ruminants. Vet Res. 2003;34(5):689–716. doi: 10.1051/vetres:2003030 [DOI] [PubMed] [Google Scholar]
- 6.Feil EJ, Cooper JE, Grundmann H, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185(11):3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichette-Jolette S, Millette G, Demontier E, et al. Partial prediction of the duration and the clinical status of Staphylococcus aureus bovine intramammary infections based on the phenotypic and genotypic analysis of isolates. Vet Microbiol. 2019;228:188–195. doi: 10.1016/j.vetmic.2018.11.024 [DOI] [PubMed] [Google Scholar]
- 8.Cheng J, Qu W, Barkema HW, et al. Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J Dairy Sci. 2019;102(3):2416–2426. doi: 10.3168/jds.2018-15135 [DOI] [PubMed] [Google Scholar]
- 9.Gao J, Barkema HW, Zhang L, et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. 2017;100(6):4797–4806. doi: 10.3168/jds.2016-12334 [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Li S, Meng L, et al. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J Dairy Sci. 2017;100(11):8796–8803. doi: 10.3168/jds.2017-13370 [DOI] [PubMed] [Google Scholar]
- 11.Artursson K, Söderlund R, Liu L, Monecke S, Schelin J. Genotyping of Staphylococcus aureus in bovine mastitis and correlation to phenotypic characteristics. Vet Microbiol. 2016;193:156–161. doi: 10.1016/j.vetmic.2016.08.012 [DOI] [PubMed] [Google Scholar]
- 12.Gao J, Ferreri M, Yu F, et al. Molecular types and antibiotic resistance of Staphylococcus aureus, isolates from bovine mastitis in a single herd in China. Vet J. 2012;192(3):550–552. doi: 10.1016/j.tvjl.2011.08.030 [DOI] [PubMed] [Google Scholar]
- 13.Bryan M, Taylor K. Periparturient use of parenteral micronized procaine penicillin to reduce the risk of clinical mastitis in heifers after calving. Vet Microbiol. 2009;134(1–2):143–149. doi: 10.1016/j.vetmic.2008.09.021 [DOI] [PubMed] [Google Scholar]
- 14.Harmon RJ. Physiology of mastitis and factors affecting somatic cell counts. J Dairy Sci. 1994;77(7):2103–2112. doi: 10.3168/jds.S0022-0302(94)77153-8 [DOI] [PubMed] [Google Scholar]
- 15.Hogan JS, Gonzalez RN, Harmon R, et al. Laboratory Handbook on Bovine Mastitis. Verona, WI: National Mastitis Council Inc.; 1999. [Google Scholar]
- 16.Manafi M. New developments in chromogenic and fluorogenic culture media. Int J of Food Microbiol. 2000;60(2–3):205–218. doi: 10.1016/S0168-1605(00)00312-3 [DOI] [PubMed] [Google Scholar]
- 17.Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; CLSI Document M100-S27. Wayne, PA: Clinical Laboratory Standards Institute (CLSI); 2017. [Google Scholar]
- 19.Ster C, Lebeau V, Leclerc J, et al. In vitro antibiotic susceptibility and biofilm production of Staphylococcus aureus isolates recovered from bovine intramammary infections that persisted or not following extended therapies with cephapirin, pirlimycin or ceftiofur. Vet Res. 2017;48(1):56. doi: 10.1186/s13567-017-0463-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mlynek KD, Callahan MT, Shimkevitch AV, et al. Effects of low-dose amoxicillin on Staphylococcus aureus USA300 biofilms. Antimicrob Agents Chemother. 2016;60(5):2639–2651. doi: 10.1128/AAC.02070-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leuenberger A, Sartori C, Boss R, et al. Genotypes of Staphylococcus aureus: on-farm epidemiology and the consequences for prevention for intramammary infections. J Dairy Sci. 2019;102(4):3295–3309. doi: 10.3168/jds.2018-15181 [DOI] [PubMed] [Google Scholar]
- 22.Dan M, Yehui W, Qingling M, et al. Antimicrobial resistance, virulence gene profile and molecular typing of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, northwest China. J Glob Antimicrob Resist. 2019;16:98–104. doi: 10.1016/j.jgar.2018.08.024 [DOI] [PubMed] [Google Scholar]
- 23.Zhang LM, Gao J, Barkema HW, et al. Virulence gene profiles: alpha-hemolysin and clonal diversity in Staphylococcus aureus isolates from bovine clinical mastitis in China. BMC Vet Res. 2018;14(1):63. doi: 10.1186/s12917-018-1374-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Lin X, Jiang T, et al. Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China. Front Microbiol. 2018;9:1123. doi: 10.3389/fmib.2018.01123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagielski T, Puacz E, Lisowski A, et al. Antimicrobial susceptibility profiling and genotyping of Staphylococcus aureus isolates from bovine mastitis in Poland. J Dairy Sci. 2014;97(10):6122–6128. doi: 10.3168/jds.2014-8321 [DOI] [PubMed] [Google Scholar]
- 26.Haftu R, Taddele H, Gugsa G, Kalayou S. Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large-scale dairy farms of Northern Ethiopia. Trop Anim Health Prod. 2012;44(7):1765–1771. doi: 10.1007/s11250-012-0135-z [DOI] [PubMed] [Google Scholar]
- 27.Li L, Zhou L, Wang L, Xue H, Zhao X. Characterization of methicillin-resistant and -susceptible staphylococcal isolates from bovine milk in northwestern China. PLoS One. 2015;10(3):e0116699. doi: 10.1371/journal.pone.0116699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mistry H, Sharma P, Mahato S, Saravanan R, Kumar PA, Bhandari V. Prevalence and characterization of oxacillin susceptible mecA-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in India. PLoS One. 2016;11(9):e0162256. doi: 10.1371/journal.pone.0162256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver SP, Murinda SE. Antimicrobial resistance of mastitis pathogens. Vet Clin Food Anim. 2012;28(2):165–185. doi: 10.1016/j.cvfa.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 30.Rabello RF, Moreira BM, Lopes RM, Teixeira LM, Riley LW, Castro AC. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J Med Microbiol. 2007;56(11):1505–1511. doi: 10.1099/jmm.0.47357-0 [DOI] [PubMed] [Google Scholar]
- 31.Jessen LR, Sørensen TM, Lilja ZL, Kristensen M, Hald T, Damborg P. Cross-sectional survey on the use and impact of the Danish national antibiotic use guidelines form companion animal practice. Acta Vet Scan. 2017;59(1):81. doi: 10.1186/s13028-017-0350-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rall VL, Miranda ES, Castilho IG, et al. Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis. J Dairy Sci. 2014;97(2):829–837. doi: 10.3168/jds.2013-7226 [DOI] [PubMed] [Google Scholar]
- 33.Gogoi-Tiwari J, Waryah CB, Eto KY, et al. Relative distribution of virulence-associated factors among Australian bovine Staphylococcus aureus isolates: potential relevance to development of an effective bovine mastitis vaccine. Virulence. 2015;6(5):419–423. doi: 10.1080/21505594.2015.1043508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Da Costa LB, Rajala-Schultz PJ, Hoet A, Seo KS, Fogt K, Moon BS. Genetic relatedness and virulence factors of bovine Staphylococcus aureus isolated from teat skin and milk. J Dairy Sci. 2014;97(11):1–10. doi: 10.3168/jds.2014-7972 [DOI] [PubMed] [Google Scholar]
- 35.Song M, Bai Y, Xu J, Carter MQ, Shi C, Shi X. Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int J Food Microbiol. 2015;195 . [DOI] [PubMed] [Google Scholar]
- 36.Cosandey A, Boss R, Luini MK, et al. Staphylococcus aureus genotype B and other genotypes isolated from cow milk in European countries. J Dairy Sci. 2016;99(1):529–540. doi: 10.3168/jds.2015-9587 [DOI] [PubMed] [Google Scholar]
- 37.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2(2):114–122. doi: 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 38.Huang CT, Yu FP, McFeters GA, Stewart PS. Nonuniform spatial patterns of respiratory activity within biofilms during disinfection. Appl Environ Microbiol. 1995;61(6):2252–2256. doi: 10.1128/AEM.61.6.2252-2256.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao G, Bao G, Cao Y, et al. Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int J Food Microbiol. 2015;211:142–147. doi: 10.1016/j.ijfoodmicro.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 40.Aires-de-Sousa M, Boye K, de Lencastre H, et al. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J Clin Microbiol. 2006;44(2):619–621. doi: 10.1128/JCM.44.2.619-621.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarraud S, Mougel C, Thioulouse J, et al. Relationships between Staphylococcus aureus genetic background virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70(2):631–641. doi: 10.1128/IAI.70.2.631-641.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JY, Fox LK, Seo KS, et al. Detection of classical and newly described staphylococcal superantigen genes in coagulase-negative staphylococci isolated from bovine intramammary infections. Vet Microbiol. 2011;147(1–2):149–154. doi: 10.1016/j.vetmic.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tristan A, Ying L, Bes M, Etienne J, Vandenesch F, Lina G. Use of multiplex PCR to identify staphylococcus aureus adhesins involved in human hematogenous Infections. J Clin Microbiol. 2003;41(9):4465–4467. doi: 10.1128/JCM.41.9.4465-4467.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.EUCAST. The European committee on antimicrobial susceptibility testing. http://www.eucast.org/. Accessed September14, 2018
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- EUCAST. The European committee on antimicrobial susceptibility testing. http://www.eucast.org/. Accessed September14, 2018