Abstract
Historically, genetic engineering in livestock proved to be challenging. Without stable embryonic stem cell lines to utilize, somatic cell nuclear transfer (SCNT) had to be employed to produce many of the genetically engineered (GE) livestock models. Through the genetic engineering of somatic cells followed by SCNT, GE livestock models could be generated carrying site-specific modifications. Although successful, only a few GE livestock models were generated because of low efficiency and associated birth defects. Recently, there have been major strides in the development of genome editing tools: Zinc-Finger Nucleases (ZFNs), Transcription activator-like effector nucleases (TALENS), and Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated 9 (Cas9) system. These tools rely on the generation of a double strand DNA break, followed by one of two repair pathways: non-homologous end joining (NHEJ) or homology directed repair (HDR). Compared to the traditional approaches, these tools dramatically reduce time and effort needed to establish a GE animal. Another benefit of utilizing genome editing tools is the application of direct injection into developing embryos to induce targeted mutations, therefore, eliminating side effects associated with SCNT. Emerging technological advancements of genome editing systems have dramatically improved efficiency to generate GE livestock models for both biomedical and agricultural purposes. Although the efficiency of genome editing tools has revolutionized GE livestock production, improvements for safe and consistent application are desired. This review will provide an overview of genome editing techniques, as well as examples of GE livestock models for agricultural and biomedical purposes.
Keywords: GENETIC ENGINEERING, LIVESTOCK, GENOME EDITING
Introduction
The ability to engineer the genome has been limited in livestock species, in part due to the lack of stable embryonic stem (ES) cells that can contribute to the germline. In the mouse, targeted disruption of genes can be performed in ES cells, then the ES cells carrying desired genotypes can be introduced into recipient blastocysts to generate chimeric mice that can transmit the desired genotype to the germline [1–3]. This strategy has been effective, thus establishing the mouse as the main model in biomedicine to elucidate the function of target genes. Since true ES cells that can contribute to the germline have not been fully identified in livestock species, the development of somatic cell nuclear transfer (SCNT) technology [4] allowed researchers to generate genetically engineered (GE) livestock. Genetic modification of somatic cells followed by SCNT allowed for the generation of GE livestock with targeted modifications [5]. Although possible, efficiency to produce GE animals through the route is poor; therefore, only selective GE models have been produced [6, 7].
The recent development of genome editing systems [8, 9] dramatically changed the field of genetic engineering. The use of the genome editing technologies can dramatically increase the efficiency to introduce targeted modifications in livestock, thus increasing the number of available GE livestock models. Genetic engineering of the somatic genome is more feasible using genome editing system technology and application of SCNT.. These technological advancements have significantly reduced the time required to produce GE livestock, which should expand their use in biomedicine and agriculture.
Here, we provide a summary of available genome editing systems used in GE livestock production and discuss current possibilities and limitations of the technology.
Conventional approach to generate genetically engineered livestock
As noted above, the lack of embryonic stem cells hindered the production of GE livestock. Technologies such as pronuclear injection allowed generation of transgenic animals, including livestock [10]; however, targeted disruption was not possible using these approaches. In addition, pronuclear injection could not control the site of integration or the number of copies of the gene that integrate. The development of SCNT technology [4] allowed for production of livestock carrying targeted modifications. Specifically, genetic modifications are introduced to in vitro cultured cells, then SCNT is performed to produce GE animals. The genetic modifications introduced to the genome of somatic cells should be carried over to offspring via SCNT including targeted modifications. The strategy was first demonstrated in sheep [5, 11] and then in pigs by inserting an exogenous gene [12] and the targeted disruption of an endogenous gene [6], known to cause hyperacute rejection after organ transplantation. Using the approach, numerous other GE livestock models have been reported [7, 13, 14].
Although the production of GE animals with site-specific modifications is now achievable, only a limited number of GE animal models are reported due to the low efficiency. Targeted modifications in somatic cells rely on the frequency of endogenous homologous recombination; however, the efficiency is extremely low in somatic cells [6, 14]. The low efficiency results in modifying only one out of two alleles at a time. Since only heterozygous mutations could be introduced, breeding steps were necessary to produce GE animals with homozygous modifications. Some livestock species, especially large animals, have a prolonged gestation period, which adds days required for breeding steps to be completed. The low frequency of homologous recombination forced researchers to use a selectable marker such as Neomycin to select somatic cells carrying the proper genetic modification [6, 14]. Therefore, GE animals produced through the approach often carried a foreign gene that could potentially add antibiotic resistance to the animals. In addition, efficiency of SCNT is poor [15] and animals born through SCNT often present sudden death or health complications that are associated with the technology [16–18].
The lack of ES cells persists in poultry species and the conventional SCNT approach used to clone mammals cannot be applied in poultry. However, the presence of primordial germ cells (PGCs) in poultry species allows for the production of GE birds [19, 20]. Specifically, PGCs isolated from developing embryos can be genetically engineered, and injecting the GE PGCs into a host embryo allows for the production of a chimeric bird that can transmit the intended modifications through the germline. Although possible, success of the approach largely depends on the quality of PGCs, which could be variable. In fact, because of the low efficiency, the first knockout chicken was not reported until 2013 [21].
Development of genome editing technology.
The development of genome editing systems significantly increased the efficiency of generating GE animals. Although exceptions exist in certain genome editing systems that utilize single-strand breaks [8, 22], the basis of genome editing systems arises from the ability to recognize specific sequences on the genome and introduce site-specific double strand breaks (DSBs) [23]. The DNA breaks drive activation of the endogenous DNA repair process to restore the DSBs because the DNA breaks may result in cell death if not resolved. The introduction of site-specific genomic modification is facilitated by two main DNA repair pathways: non-homologous end joining (NHEJ) or homology-directed repair (HDR). The DSB-induced NHEJ process leads to random base insertions or deletions (indels), which result in gene knockout because it often causes a frameshift of the amino acid codons and formation of a premature stop codon. Alternatively, in the presence of donor DNA with homologous sequences to the target genome site, specific modifications at the nucleotide level can be introduced through the HDR pathway [24]. In general, lower frequency of HDR than NHEJ has been known in most cell types [25]. Because of their ability to recognize specific sequences on the genome and introduce site-specific DNA breaks, genome editing systems are also referred to as engineered endonucleases. The mechanistic action of genome editing systems is illustrated in Figure 1.
Figure 1.
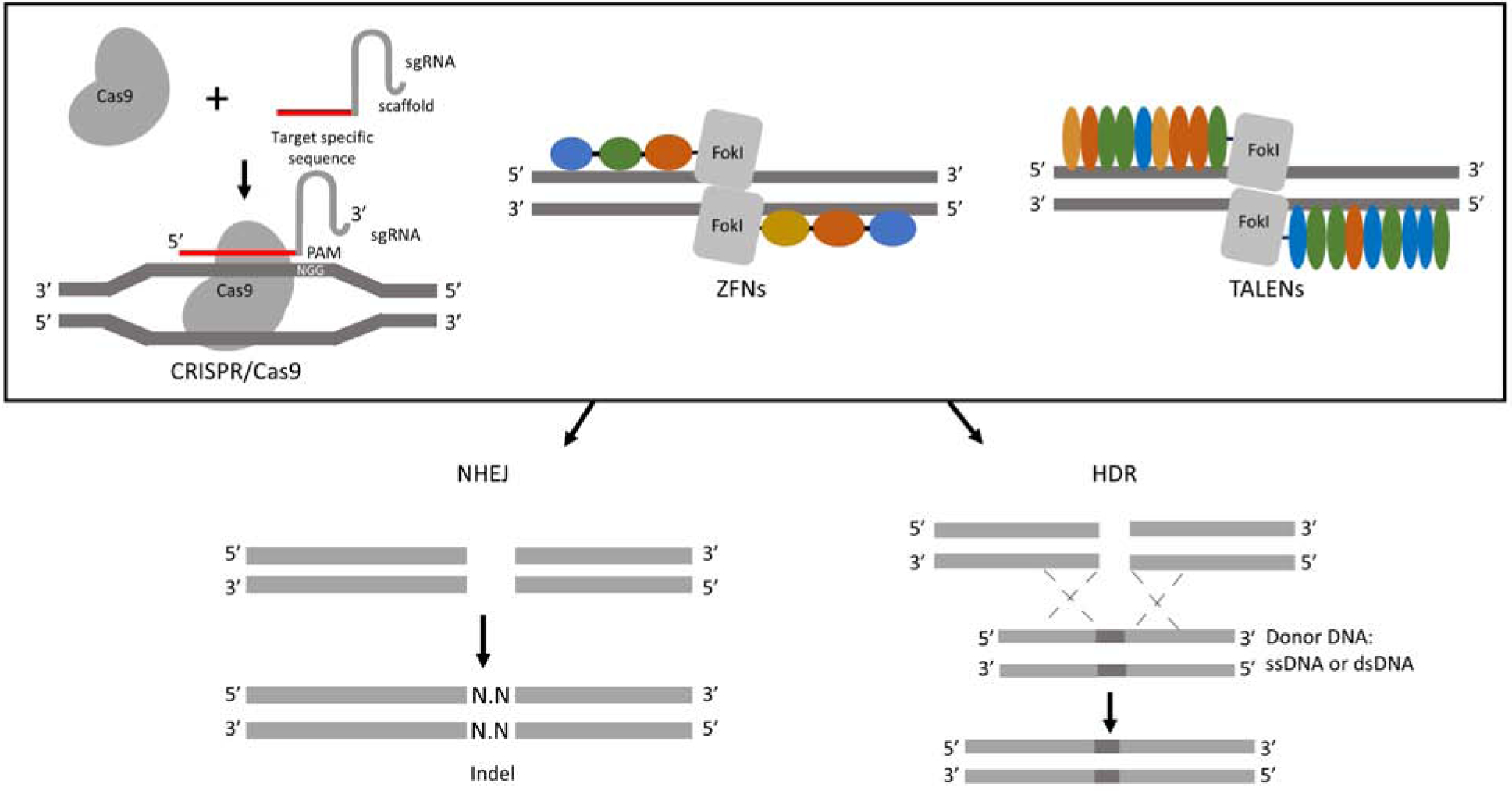
Basic mechanism of genome editing systems, demonstrated using CRISPR/Cas9 system, ZFNs, and TALENs. In the CRISPR/Cas9 system, after sgRNA binds to target site of genome, Cas9 cleaves the genomic DNA creating double strands breaks (DSBs). In ZFNs and TALENs, the non-specific endonuclease, FokI, binds and generates a DSB. Subsequently, random indel mutations or precise modification occurs by one of two DNA repair mechanisms: NHEJ or HDR. Both mechanisms are functional in somatic cells as well as in embryos. In general, frequency of NHEJ is more active compared to the HDR.
In recent years, three types of engineered endonucleases, Zinc-Finger Nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs), and Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system, have emerged that enable investigators to rapidly introduce modifications into the genomes of virtually all cell types and organisms.
The first genome editing system developed to modify the genome of mammals was the ZFNs. Zinc-finger motifs are known to recognize three specific nucleotide sequences [26]. By fusing engineered zinc-fingers to the non-specific endonuclease, FokI, the system can induce DSBs on a specific locus on the genome. The concept was first demonstrated using human cells; efficiency of gene targeting increased up to 1000 folds when comparing targeted disruption of exogenous GFP using ZFN instead of the conventional method [9]. The technology was quickly applied to GE livestock production. As a proof-of-concept, exogenous GFP gene was inactivated using ZFNs in somatic cells and GFP knockout pigs were produced using SCNT [27]. The enhanced genetic engineering efficacy of ZFNs allowed researchers to introduce targeted modifications on both alleles in livestock [28], which could dramatically reduce the time required to establish and expand GE livestock models.
The development of TALENs further expanded the use of genome editing systems in GE livestock production as the target recognition of TALENs is wider than ZFNs. TALENs are designed based on a plant pathogenic bacteria [29]. The engineered TALENs can identify specific sequences on the genome and introduce DSBs on the target locus. Then, the DNA repair system, activated by the DSBs, can introduce targeted modifications through NHEJ or HDR. The target sequence of TALENs appeared to be more flexible when compared to ZFNs because TALENs can effectively recognize AT-rich regions [30], whereas the activity of ZFNs is preferred in GC-rich target regions. This difference in recognition region stimulated the use of TALENs in the production of GE livestock; its first application to produce GE livestock was in 2012 [31].
Although the development of ZFNs and TALENs significantly increased the efficiency of producing GE livestock models, it was the development of CRISPR/Cas9 system that dramatically lowered the effort required to produce GE livestock models. The CRISPR/Cas9 system was originally characterized as an adaptive immune system of bacteria cells against exogenous DNA from an invading virus or phage [32, 33]. Because the system is designed to cleave exogenous DNA, engineering the CRISPR/Cas9 system allows researchers to induce site-specific DSB. Specifically, engineered complementary RNA to the target DNA combined with trans-activating CRISPR RNA (tracr-RNA), called single guide RNA (sgRNA), can bind to a target sequence, thus, locating Cas9 protein to the target site on the genome. Then, the Cas9 protein acts as an endonuclease to the target site if the protospacer adjacent motif sequence (PAM) is present [34]. Unlike ZFN or TALEN assemble, which requires a series of ligation reactions to form an array [35, 36], CRISPR/Cas9 system can be assembled by inserting a 20 bp target DNA sequence into a targeting vector [8]. In addition, several online based CRISPR design systems (CHOPCHOP or CRISPOR) are readily available; therefore, no specialized training is required to utilized the CRISPR/Cas9 technology. The user-friendly features of the CRISPR/Cas9 system made the tool become the leading genome editing system. Efficacy of the system was first demonstrated using human cells in 2013 [8], then quickly used for GE livestock production.
Application of genome editing technology in livestock.
The development of genome editing tools significantly modified the approaches used to generate GE livestock models. First, targeted modifications in somatic cells are more feasible with the use of genome editing tools. Unlike relying on endogenous homologous recombination, the DSB-induced DNA repair system can now effectively disrupt target genes through either NHEJ or HDR. The high efficiency allows us to perform genetic modifications in somatic cells without having to rely on the insertion of a selectable marker, i.e. antibiotic resistant gene. The modified cells are then used for SCNT to produce GE animals. In addition, now both alleles can be effectively modified using genome editing tools [28, 31, 37]. Because both alleles in cultured somatic cells can be modified using genome editing tools, the number of breeding steps required to establish a herd of GE animals became significantly lower.
Genome editing systems have also transformed GE livestock production by offering an alternative route that does not involve SCNT. Direct injection of engineered endonucleases into developing embryos resulted in targeted modifications. In livestock, the idea was first examined by injecting the RNA version of ZFN and TALEN into developing pig embryos [38]. The targeted modifications during embryogenesis demonstrated that SCNT is no longer required to introduce targeted modifications in livestock species. As expected, animals produced via this approach do not have any of the abnormalities that are commonly seen with cloned animals [37, 39]. The injection of engineered endonucleases, especially CRISPR/Cas9 system, consistently introduced targeted modifications at high efficiency as multiple publications indicate that all embryos/animals derived from the approach carry targeted modifications with no detectable wild-type allele [39–43]. The approach is simpler than SCNT-mediated genetic engineering, which requires specialized training (Figure 2). Similarly, a recent report in the quail suggests that genome editing tools can transform the conventional GE bird production system. Direct injection of CRISPR/Cas9 system into the quail blastoderm generated a chimeric bird that transmitted targeted modifications to the germline [44], indicating that the development of genome editing tools lead to a novel approach to produce GE poultry as well.
Figure 2.
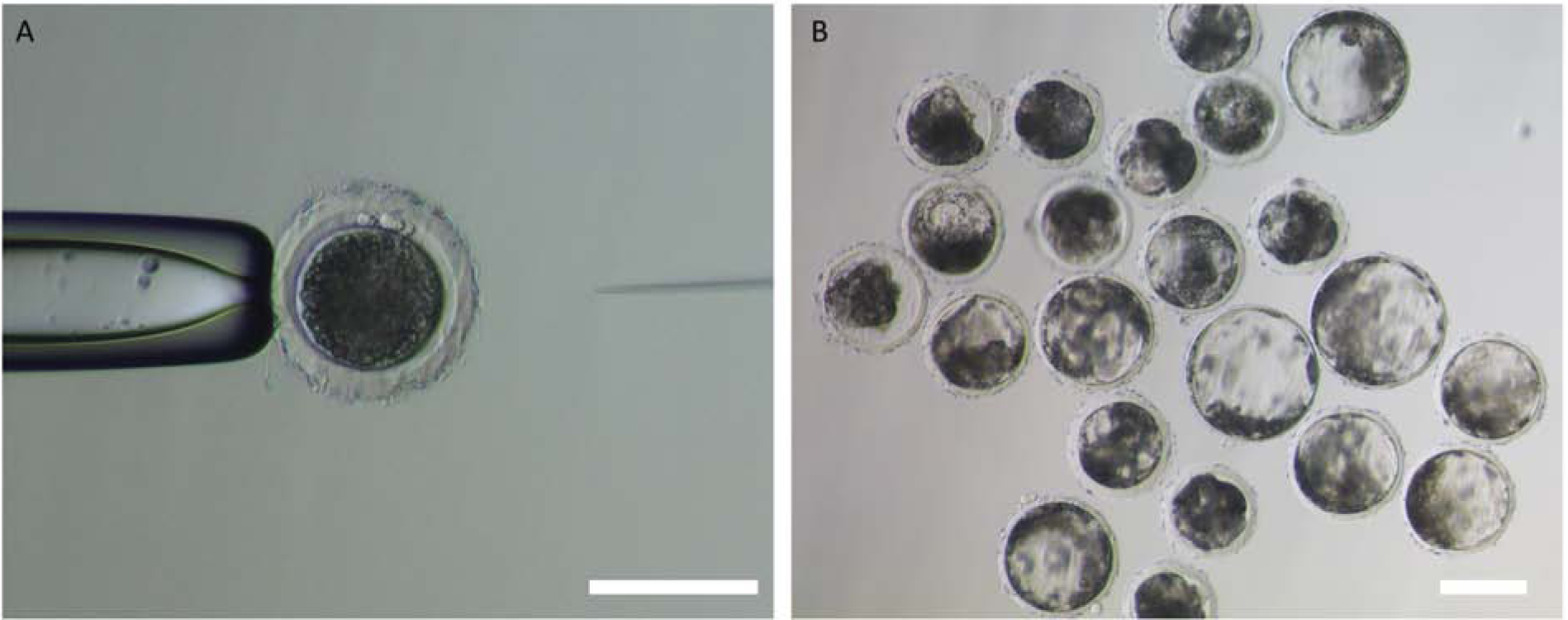
Image of direct injection of CRISPR/Cas9 system into presumable pig zygotes. (A) Microinjection of CRISPR/Cas9 system into developing embryo. The approach is simpler than somatic cell nuclear transfer. (B) CRISPR/Cas9 injected embryos at day 5 post fertilization, prior to embryo transfer. The scale bar indicates 100 μm.
As illustrated here, the use of genome editing technology has enhanced the efficiency to produce GE animals and offered alternative approaches that can overcome the shortcomings of current GE animal production routes, therefore sparking the production of GE livestock models for biomedical and agricultural applications.
Genetically engineered livestock produced using genome editing technology
The development of diverse genome editing tools increased access to GE livestock models. Because pigs are physiologically and anatomically similar to humans, pigs can accurately present symptoms of human diseases, making them a proper model in biomedicine [45]. Genome editing tools rapidly expanded pig models designed for xenotransplantation. Using ZFNs, GE pigs lacking both alleles of GGTA1 were produced without breeding steps [28]. Similarly, diverse GE pigs aimed to facilitate the use of pig organs for xenotransplantation have been generated [46–49]. Another line of GE pigs, produced using genome editing technology for biomedical application, are immunodeficient pigs. Animals lacking a functional immune system are important to study the mechanism of pathogenesis and immune response upon infection, vaccine development, and cell transplantation [50, 51]. Before the genome editing era, only one type of immunodeficient pig was established using the conventional approach [52] due to the low efficiency. Using TALENs, GE pigs lacking functional RAG2 were produced and the pigs could support proliferation and differentiation of transplanted induced pluripotent stem (iPS) cells [53]. To the best of our knowledge, this was the first report of a successful human stem cell transplantation into a large animal model and suggests that these immunodeficient pigs could be used to predict the outcomes of stem cell transplantation or progression of cancer cells. Furthermore, using CRISPR/Cas9 system, immunodeficient pigs lacking all major lymphocytes (T-B-NK-) were generated by disrupting RAG2 and IL2RG simultaneously during embryogenesis [39]. Phenotype of the pigs resemble the NOD-scid gamma (NSG) mouse model, the leading animal model used to perform xenograft experiments using human stem cells [54, 55]. Use of genome editing technology also assisted in the production of GE pigs having potential impact in agriculture. Disruption of CD163 using CRISPR/Cas9 system led to pigs resistant to porcine reproductive and respiratory syndrome virus (PRRSV) [37, 56]. Additionally, pigs carrying modified myostatin (MSTN) gene or uncoupling protein 1 (UCP1) were produced to improve meat productivity of pigs [57–59].
Sheep were the first species to be cloned through SCNT [4]. Development of genome editing tools has increased the number of GE small ruminant models, such as sheep and goat. To enhance meat production, myostatin (MSTN) gene was disrupted in sheep [60] and goat [61] using genome editing tools. Similarly, CRISPR/Cas9 system was used to remove the allergenic component in goat milk [62], which could potentially increase the safety of goat milk consumption. Small ruminants are applied in the biomedicine field as well with the utilization of genome editing tools. It was demonstrated that the disruption of PDX1 using CRISPR/Cas9 system led to apancreatic phenotype in the sheep [63], which can be used to grow a pancreas from human origin through a chimeric approach. Cystic fibrosis (CF) sheep models were produced using CRISPR/Cas9 system and recapitulated many of the human CF disease symptoms [64]; previously proper CF animal models reflecting human phenotype were only available in pigs [7].
The development of GE cow models has been slow, in part, due to prolonged gestation length (around 280 days) compared to other livestock species. The development of genome editing technology assisted in the utilization of GE cow models because rapid generation of GE cows is now possible. Using ZFNs, human lysozyme (hLYZ) gene was integrated into bovine β-casein locus to generate GE cows potentially resistant to mastitis [65]. TALENs were effectively used to modify the cow genome at high efficiency [31, 66] and produced dairy cattle lacking horns by editing the POLLED allele [67]. Others have utilized TALENs and CRISPR/Cas9 system to induce site-specific integration of an exogenous gene to enhance the value of cows [68, 69].
As mentioned above, genetic engineering in poultry is different from other livestock species because SCNT technology cannot be applied. The development of genome editing technology significantly improved GE bird production by improving the efficiency to introduce site-specific modifications into the genome of PGCs and inducing germ-line genetic modification at an in vivo level. Using TALENs, the genome of PGCs was successfully modified and used to generate knockout chickens [70]. CRISPR/Cas9 system was also successfully applied to produce GE birds carrying targeted disruption of endogenous genes [71, 72]. An interesting report recently came out where direct injection of adenovirus-based CRISPR/Cas9 system into the quail blastoderm could lead to the targeted disruption of an endogenous gene [44], indicating that the culture and transfection of PGCs are not absolutely necessary for the production of poultry knockout models during the genome editing era.
Future prospective and current limitations
The development of genome editing technology significantly lowered the barrier to produce GE livestock models. Figure 3 illustrates GE livestock models generated using various approaches. For biomedical application, animal models that resemble human diseases will significantly contribute to designing an optimal remedy for the diseases. Use of genome editing systems grant rapid production of GE pigs carrying multiple genetic modifications for xenotransplantation [73], which would take years of preparation through the conventional approaches. The development of animal models representing symptoms of cystic fibrosis in multiple species (pigs [7] and sheep [64]) will also contribute to novel cures through comparative medicine. In addition, the development of new immunodeficient large animal models [39, 53] offer possibilities to utilize the models for human cell transplantation. Transplantation of patient-specific cancer cells into the immunodeficient animals will allows us to monitor progression of the cancer cells and design patient-specific treatments. Large animal models carrying immunodeficiency can be used for stem cell transplantation to predict outcomes of stem cell therapy and organogenesis projects to grow cells/tissues that can be transplanted back into the patient without immune rejection. Development of genome editing systems facilitate production of these translational animal models that can complement findings from rodent data and bridge the findings to the clinic.
Figure 3.
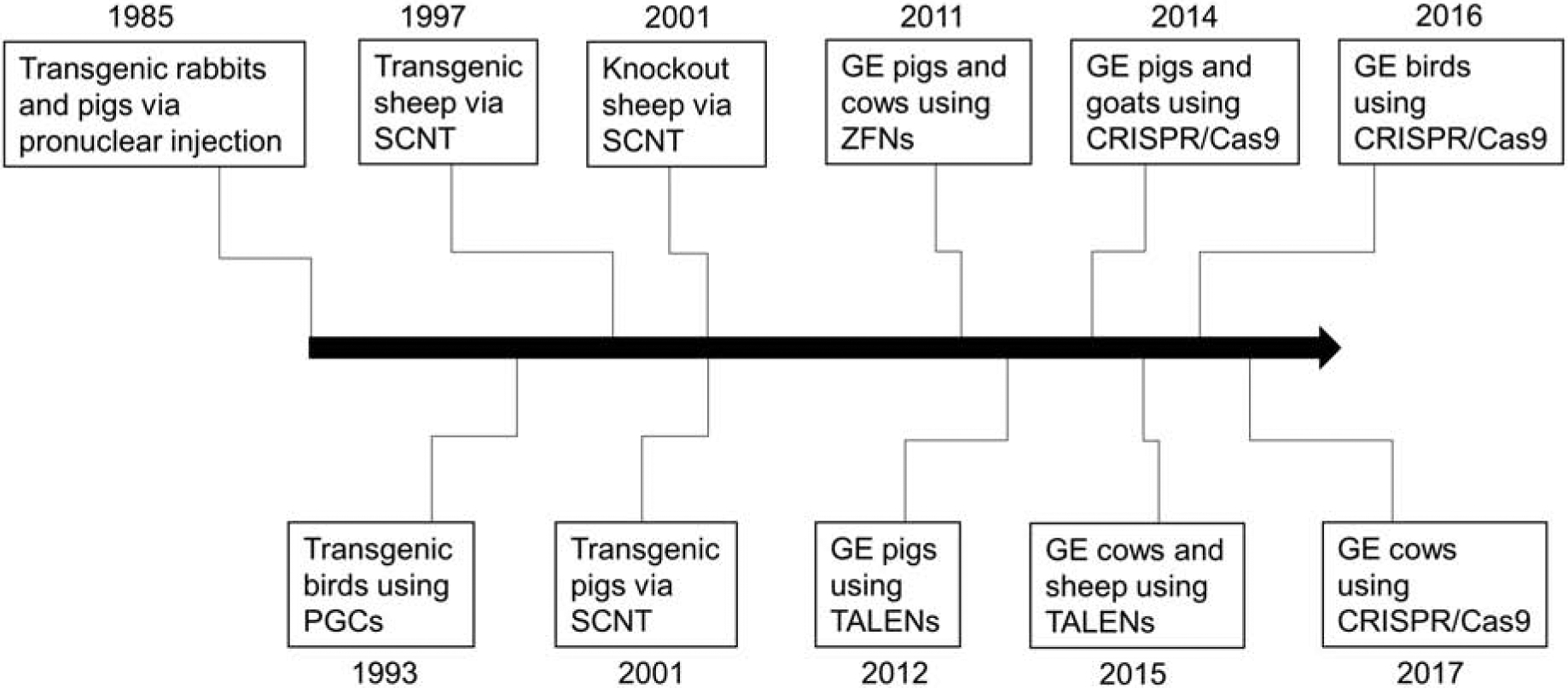
Milestones in the production of genetically engineered livestock animals. Development of genome editing systems significantly assisted the design and production of genetically engineered in livestock.
Use of genome editing systems in livestock can also improve genetic value of livestock for production. It has been demonstrated that the disruption of putative receptor of viruses that damage the swine industry leads to GE pigs resistant to the deadly viruses [56, 74]. Production of hornless cattle by the application of genome editing system [67] can lower the cost of producing cows, while also enhancing animal welfare. Genetic modifications introduced through HDR could specifically edit target genes at the nucleotide level without affecting the rest of the genome. The point mutations can be introduced as a form of precision genetic enhancement [66, 75]. The improvements through genome editing technology could require years of breeding to introduce a specific allele into a breeding line. It is anticipated that genome editing technology will be widely used to sustain productivity of livestock to supply a sufficient amount of meat for the growing population.
Although the potential applications stated above are promising, the application of genome editing systems requires attention, and refinement of the technologies are desired. Utilization of genome editing systems often follow the error prone nature of the NHEJ pathway. However, the system does not always lead to an expected phenotype. For instance, mutations introduced through NHEJ are random, therefore, insertions or deletions in triplets do not always lead to targeted disruption of the genes. The shortcomings have been reported in multiple studies [39, 76] and should be considered when introducing targeted modifications through NHEJ-mediated genome editing events. Targeted modifications through the HDR pathway could solve the uncertainties caused by NHEJ-mediated genetic modifications. However, the efficiency of HDR is, in general, lower compared to NHEJ because the HDR pathway is only active during the late S and G2 phases with the presence of sister chromatids serving as a repair template [77]. The use of NHEJ inhibitors has been employed to push genome editing induced DSBs repaired through HDR pathway [78–80]. Unfortunately, there is no standardized method that can control the efficiency of HDR. Ability to improve the frequency of HDR-mediated targeted modifications could assist in the utilization of genome editing systems.
As mentioned above, direct injection of genome editing systems into developing embryos has provided a novel route to generate GE livestock without having to incorporate SCNT. However, the approach comes with its unique shortcoming, the mosaicism. GE animals produced through the direct injection approach may carry more than two alleles in their genomic DNA if targeted modifications are introduced after the first cleavage. The mosaicism can be a hurdle in utilizing the approach because animals carrying mosaic genotype may not present a clear phenotype. Also, progenies derived from GE animals with mosaicism do not follow Mendelian ratio, which impedes designing predictable breeding scheme using the founder animals. In our previous reports, the frequency of mosaicism introduced due to the approach was 0–20% in pigs [39, 41, 42]; however, the frequency varies in other studies. Breeding of founder animals carrying mosaic genotype can solve the issues associated with the mosaicism. Unfortunately, considering gestation periods of farm animals, additional breeding scheme will further delay propagating GE livestock models. For instance, the gestation period of cattle is 282 days and typically only deliver one calf. Establishing homozygous GE animals in cattle from founder cows carrying mosaicism would require at least 2–3 breeding plans, which could take 4–5 years. Identifying genotype of each embryo by biopsy followed by genotyping can be performed to reduce GE animals carrying mosaicism; however, the process can be technically challenging. Therefore, a significant portion of genetic modifications in livestock is still introduced through somatic genome editing followed by SCNT.
Most of the currently available genome editing systems induce site-specific DSBs to trigger the DNA repair system. However, DSBs on unintended locations introduced by genome editing systems could result in undesirable mutations, i.e. off-targeting. Although their frequencies are variable, previous reports suggest that CRISPR/Cas9 system could introduce off-targeting events [81–83]. In our recent study, we identified one CRISPR/Cas9 system causing off-targeting events out of four CRISPR/Cas9 systems examined when injected into developing pig embryos [41]. A recent report also suggests that hornless cattle produced with the use of TALENs introduced unintended modifications to the founder genome [84]. The examples of off-targeting events raise concerns on applying the genome editing systems in human clinics. A recent report of genome edited babies became a controversial topic due to ethical concerns [85]. Because outcomes from genome editing systems may be unpredictable, the systems must be used responsibly and potential side effect should be fully analyzed.
Different approaches have been suggested to prevent off-targeting activity from the genome editing systems. For instance, the application of Cas9-nikase, a variant of Cas9 protein causing only a single strand break instead of DSBs, can be used to reduce off-targeting events and introduce targeted modifications through the HDR pathway [8, 86]. Furthermore, CRISPR/Cas9-guided use of base pair editors could introduce targeted modifications at the nucleotide level while minimizing off-targeting events [87]. These genome editing systems have been applied to generate GE livestock models [68, 78, 88–90]. Development of genome editing tools that can highly secure the integrity of the genome will alleviate risks associate with the use of genome editing systems in GE livestock production.
Conclusion
The development of genome editing systems has facilitated the production of GE livestock models, as the disadvantages of generating livestock can be relieved by using the newly available tools. GE livestock models will advance the field of biomedicine by supplying animal models that can more accurately present human disease symptoms and improve productivity of livestock for agriculture purposes. Although concerns about the genome editing systems remain, constant efforts to improve current systems will mitigate side effects associated with the systems. The era of genome editing systems will lift the existing limit on the use of GE livestock models.
Highlights.
Genetic engineering in livestock has been challenging due to the lack of stable embryonic stem cells
Development of genome editing technology significantly enhanced efficiency to produce genetically engineered livestock
Using genome editing technology, diverse genetically engineered livestock models have been produced for biomedical and agriculture purposes
More livestock models are expected to be produced at higher efficiency
Acknowledgement
This project was supported in part by the Virginia Agricultural Experiment Station through the NIFA, U.S. Department of Agriculture and NIH grant 1R21OD027062.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–12. [DOI] [PubMed] [Google Scholar]
- [2].Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R. Germ-line transmission of a disrupted beta 2-microglobulin gene produced by homologous recombination in embryonic stem cells. Nature. 1989;342:435–8. [DOI] [PubMed] [Google Scholar]
- [3].Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4–8+ cytolytic T cells. Nature. 1990;344:742–6. [DOI] [PubMed] [Google Scholar]
- [4].Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. [DOI] [PubMed] [Google Scholar]
- [5].Denning C, Burl S, Ainslie A, Bracken J, Dinnyes A, Fletcher J, et al. Deletion of the alpha(1,3)galactosyl transferase (GGTA1) gene and the prion protein (PrP) gene in sheep. Nature biotechnology. 2001;19:559–62. [DOI] [PubMed] [Google Scholar]
- [6].Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science (New York, NY). 2002;295:1089–92. [DOI] [PubMed] [Google Scholar]
- [7].Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science (New York, NY). 2008;321:1837–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY). 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. [DOI] [PubMed] [Google Scholar]
- [10].Hammer RE, Pursel VG, Rexroad CE Jr., Wall RJ, Bolt DJ, Ebert KM, et al. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315:680–3. [DOI] [PubMed] [Google Scholar]
- [11].Schnieke AE, Kind AJ, Ritchie WA, Mycock K, Scott AR, Ritchie M, et al. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278:2130–3. [DOI] [PubMed] [Google Scholar]
- [12].Park KW, Cheong HT, Lai L, Im GS, Kuhholzer B, Bonk A, et al. Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Animal biotechnology. 2001;12:173–81. [DOI] [PubMed] [Google Scholar]
- [13].Lai L, Kang JX, Li R, Wang J, Witt WT, Yong HY, et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nature biotechnology. 2006;24:435–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dai Y, Vaught TD, Boone J, Chen SH, Phelps CJ, Ball S, et al. Targeted disruption of the alpha1,3-galactosyltransferase gene in cloned pigs. Nature biotechnology. 2002;20:251–5. [DOI] [PubMed] [Google Scholar]
- [15].Liu T, Dou H, Xiang X, Li L, Li Y, Lin L, et al. Factors Determining the Efficiency of Porcine Somatic Cell Nuclear Transfer: Data Analysis with Over 200,000 Reconstructed Embryos. Cellular reprogramming. 2015;17:463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Carter DB, Lai L, Park KW, Samuel M, Lattimer JC, Jordan KR, et al. Phenotyping of transgenic cloned piglets. Cloning and stem cells. 2002;4:131–45. [DOI] [PubMed] [Google Scholar]
- [17].Park MR, Cho SK, Lee SY, Choi YJ, Park JY, Kwon DN, et al. A rare and often unrecognized cerebromeningitis and hemodynamic disorder: a major cause of sudden death in somatic cell cloned piglets. Proteomics. 2005;5:1928–39. [DOI] [PubMed] [Google Scholar]
- [18].Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Reviews of reproduction. 1998;3:155–63. [DOI] [PubMed] [Google Scholar]
- [19].Vick L, Li Y, Simkiss K. Transgenic birds from transformed primordial germ cells. Proceedings Biological sciences. 1993;251:179–82. [DOI] [PubMed] [Google Scholar]
- [20].van de Lavoir MC, Diamond JH, Leighton PA, Mather-Love C, Heyer BS, Bradshaw R, et al. Germline transmission of genetically modified primordial germ cells. Nature. 2006;441:766–9. [DOI] [PubMed] [Google Scholar]
- [21].Schusser B, Collarini EJ, Yi H, Izquierdo SM, Fesler J, Pedersen D, et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome research. 2012;22:1327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fernandez A, Josa S, Montoliu L. A history of genome editing in mammals. Mammalian genome : official journal of the International Mammalian Genome Society. 2017;28:237–46. [DOI] [PubMed] [Google Scholar]
- [24].Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA repair. 2006;5:1021–9. [DOI] [PubMed] [Google Scholar]
- [26].Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. The EMBO journal. 1985;4:1609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Whyte JJ, Zhao J, Wells KD, Samuel MS, Whitworth KM, Walters EM, et al. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Molecular reproduction and development. 2011;78:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12013–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gohre V, Robatzek S. Breaking the barriers: microbial effector molecules subvert plant immunity. Annual review of phytopathology. 2008;46:189–215. [DOI] [PubMed] [Google Scholar]
- [30].Gaj T, Gersbach CA, Barbas CF 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, et al. Efficient TALEN-mediated gene knockout in livestock. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. [DOI] [PubMed] [Google Scholar]
- [33].Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. [DOI] [PubMed] [Google Scholar]
- [34].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY). 2012;337:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ellis BL, Hirsch ML, Porter SN, Samulski RJ, Porteus MH. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther. 2013;20:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, et al. Use of the CRISPR/Cas9 System to Produce Genetically Engineered Pigs from In Vitro-Derived Oocytes and Embryos. Biology of reproduction. 2014;91:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C, et al. Live pigs produced from genome edited zygotes. Scientific reports. 2013;3:2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lei S, Ryu J, Wen K, Twitchell E, Bui T, Ramesh A, et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Scientific reports. 2016;6:25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kang JT, Cho B, Ryu J, Ray C, Lee EJ, Yun YJ, et al. Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reproductive biology and endocrinology : RB&E. 2016;14:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Carey K, Ryu J, Uh K, Lengi AJ, Clark-Deener S, Corl BA, et al. Frequency of off-targeting in genome edited pigs produced via direct injection of the CRISPR/Cas9 system into developing embryos. BMC biotechnology. 2019;19:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yugo DM, Heffron CL, Ryu J, Uh K, Subramaniam S, Matzinger SR, et al. Infection dynamics of hepatitis E virus in wild-type and immunoglobulin heavy chain knockout JH (−/−) gnotobiotic piglets. Journal of virology. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sheets TP, Park CH, Park KE, Powell A, Donovan DM, Telugu BP. Somatic Cell Nuclear Transfer Followed by CRIPSR/Cas9 Microinjection Results in Highly Efficient Genome Editing in Cloned Pigs. International journal of molecular sciences. 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee J, Ma J, Lee K. Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:13288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Whitelaw CB, Sheets TP, Lillico SG, Telugu BP. Engineering large animal models of human disease. The Journal of pathology. 2016;238:247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Li P, Estrada JL, Burlak C, Montgomery J, Butler JR, Santos RM, et al. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation. 2014. [DOI] [PubMed] [Google Scholar]
- [47].Kwon DN, Lee K, Kang MJ, Choi YJ, Park C, Whyte JJ, et al. Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Scientific reports. 2013;3:1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Beaton BP, Kwon DN, Choi YJ, Kim JH, Samuel MS, Benne JA, et al. Inclusion of homologous DNA in nuclease-mediated gene targeting facilitates a higher incidence of bi-allelically modified cells. Xenotransplantation. 2015;22:379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Burlak C, Bern M, Brito AE, Isailovic D, Wang ZY, Estrada JL, et al. N-linked glycan profiling of GGTA1/CMAH knockout pigs identifies new potential carbohydrate xenoantigens. Xenotransplantation. 2013;20:277–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang B, Duan Z, Zhao Y. Mouse models with human immunity and their application in biomedical research. Journal of cellular and molecular medicine. 2009;13:1043–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ito R, Takahashi T, Ito M. Humanized mouse models: Application to human diseases. Journal of cellular physiology. 2018;233:3723–8. [DOI] [PubMed] [Google Scholar]
- [52].Suzuki S, Iwamoto M, Saito Y, Fuchimoto D, Sembon S, Suzuki M, et al. Il2rg gene-targeted severe combined immunodeficiency pigs. Cell stem cell. 2012;10:753–8. [DOI] [PubMed] [Google Scholar]
- [53].Lee K, Kwon DN, Ezashi T, Choi YJ, Park C, Ericsson AC, et al. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. Journal of immunology (Baltimore, Md : 1950). 2005;174:6477–89. [DOI] [PubMed] [Google Scholar]
- [55].Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nature reviews Immunology. 2007;7:118–30. [DOI] [PubMed] [Google Scholar]
- [56].Whitworth KM, Rowland RR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nature biotechnology. 2016;34:20–2. [DOI] [PubMed] [Google Scholar]
- [57].Cyranoski D Super-muscly pigs created by small genetic tweak. Nature. 2015;523:13–4. [DOI] [PubMed] [Google Scholar]
- [58].Qian L, Tang M, Yang J, Wang Q, Cai C, Jiang S, et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Scientific reports. 2015;5:14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zheng Q, Lin J, Huang J, Zhang H, Zhang R, Zhang X, et al. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E9474–e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li H, Wang G, Hao Z, Zhang G, Qing Y, Liu S, et al. Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer. Scientific reports. 2016;6:33675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M, et al. Efficient gene knockout in goats using CRISPR/Cas9 system. PloS one. 2014;9:e106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhou W, Wan Y, Guo R, Deng M, Deng K, Wang Z, et al. Generation of beta-lactoglobulin knock-out goats using CRISPR/Cas9. PloS one. 2017;12:e0186056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Vilarino M, Rashid ST, Suchy FP, McNabb BR, van der Meulen T, Fine EJ, et al. CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Scientific reports. 2017;7:17472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fan Z, Perisse IV, Cotton CU, Regouski M, Meng Q, Domb C, et al. A sheep model of cystic fibrosis generated by CRISPR/Cas9 disruption of the CFTR gene. JCI insight. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Liu X, Wang Y, Tian Y, Yu Y, Gao M, Hu G, et al. Generation of mastitis resistance in cows by targeting human lysozyme gene to beta-casein locus using zinc-finger nucleases. Proceedings Biological sciences. 2014;281:20133368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Carlson DF, Lancto CA, Zang B, Kim ES, Walton M, Oldeschulte D, et al. Production of hornless dairy cattle from genome-edited cell lines. Nature biotechnology. 2016;34:479–81. [DOI] [PubMed] [Google Scholar]
- [68].Gao Y, Wu H, Wang Y, Liu X, Chen L, Li Q, et al. Single Cas9 nickase induced generation of NRAMP1 knockin cattle with reduced off-target effects. Genome biology. 2017;18:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Luo Y, Wang Y, Liu J, Cui C, Wu Y, Lan H, et al. Generation of TALE nickase-mediated gene-targeted cows expressing human serum albumin in mammary glands. Scientific reports. 2016;6:20657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Park TS, Lee HJ, Kim KH, Kim JS, Han JY. Targeted gene knockout in chickens mediated by TALENs. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12716–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Oishi I, Yoshii K, Miyahara D, Kagami H, Tagami T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Scientific reports. 2016;6:23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dimitrov L, Pedersen D, Ching KH, Yi H, Collarini EJ, Izquierdo S, et al. Germline Gene Editing in Chickens by Efficient CRISPR-Mediated Homologous Recombination in Primordial Germ Cells. PloS one. 2016;11:e0154303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Fischer K, Rieblinger B, Hein R, Sfriso R, Zuber J, Fischer A, et al. Viable pigs after simultaneous inactivation of porcine MHC class I and three xenoreactive antigen genes GGTA1, CMAH and B4GALNT2. Xenotransplantation. 2019:e12560. [DOI] [PubMed] [Google Scholar]
- [74].Whitworth KM, Rowland RRR, Petrovan V, Sheahan M, Cino-Ozuna AG, Fang Y, et al. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgenic research. 2019;28:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Niu Y, Zhao X, Zhou J, Li Y, Huang Y, Cai B, et al. Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9. Reproduction, fertility, and development. 2018;30:307–12. [DOI] [PubMed] [Google Scholar]
- [76].Park KE, Kaucher AV, Powell A, Waqas MS, Sandmaier SE, Oatley MJ, et al. Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Scientific reports. 2017;7:40176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Heyer W-D, Ehmsen KT, Liu J. Regulation of Homologous Recombination in Eukaryotes. Annual review of genetics. 2010;44:113–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Park KE, Powell A, Sandmaier SE, Kim CM, Mileham A, Donovan DM, et al. Targeted gene knock-in by CRISPR/Cas ribonucleoproteins in porcine zygotes. Scientific reports. 2017;7:42458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature biotechnology. 2015;33:538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature biotechnology. 2015;33:543–8. [DOI] [PubMed] [Google Scholar]
- [81].Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular therapy Nucleic acids. 2015;4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Li C, Zhou S, Li Y, Li G, Ding Y, Li L, et al. Trio-Based Deep Sequencing Reveals a Low Incidence of Off-Target Mutations in the Offspring of Genetically Edited Goats. Frontiers in genetics. 2018;9:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Young AE, Mansour TA, McNabb BR, Owen JR, Trott JF, Brown CT, et al. Genomic and phenotypic analyses of six offspring of a genome-edited hornless bull. Nature biotechnology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Cyranoski D, Ledford H. Genome-edited baby claim provokes international outcry. Nature. 2018;563:607–8. [DOI] [PubMed] [Google Scholar]
- [86].Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Komor AC, Badran AH, Liu DR. Editing the Genome Without Double-Stranded DNA Breaks. ACS chemical biology. 2018;13:383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Li Z, Duan X, An X, Feng T, Li P, Li L, et al. Efficient RNA-guided base editing for disease modeling in pigs. Cell discovery. 2018;4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ruan J, Li H, Xu K, Wu T, Wei J, Zhou R, et al. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific reports. 2015;5:14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Xie J, Ge W, Li N, Liu Q, Chen F, Yang X, et al. Efficient base editing for multiple genes and loci in pigs using base editors. Nature communications. 2019;10:2852. [DOI] [PMC free article] [PubMed] [Google Scholar]


