Abstract
Porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), and swine acute diarrhea syndrome-coronavirus (SADS-CoV) are three emerging and re-emerging coronaviruses (CoVs). Symptoms caused by these three viruses are extremely similar, including acute diarrhea, vomiting and even death in piglets. To date, strict biosecurity is still the most effective disease prevention and control measures, and the early detection of pathogens is the most important link. Here, we developed a microfluidic-RT-LAMP chip detection system for the first time, which could detected PEDV, PDCoV and SADS-CoV simultaneously, and had advantages of rapid, simple, sensitive, high-throughput, and accurate at point-of-care settings. The lowest detection limits of the microfluidic-RT-LAMP chip method are 101 copies/μL, 102 copies/μL and 102 copies/μL for PEDV, PDCoV and SADS-CoV, respectively. The whole detection procedure can be finished rapidly in 40 min without any cross-reaction with other common swine viruses. A total of 173 clinical swine fecal samples characterized with diarrheal symptoms were used to evaluate the performance of the newly developed system, which presented good stabilities (C.V.s<5%) and specificities (100%), and possessed sensitivities of 92.24%, 92.19% and 91.23% for PEDV, PDCoV and SADS-CoV respectively. In summary, the established microfluidic-RT-LAMP chip detection system could satisfy the demanding in field diagnoses, which was suitable for promotion in remote areas due to its fast, portable and cost-effective characters.
Keywords: Microfluidic chip, Loop mediated isothermal amplification (LAMP), POC, Emerging and re-emerging swine coronaviruses
Graphical abstract
Highlights
-
•
Microfluidic chip and LAMP were firstly integrated for high-throughput and multitarget detection of PEDV, PDCoV and SADS-CoV.
-
•
The microfluidic-RT-LAMP chip system established greatly facilitated the rapid diagnosis of the PEDV, PDCoV and SADS-CoV.
-
•
The microfluidic chip device provides excellent prospects in swine enteric coronaviruses surveillance for remote areas.
1. Introduction
Diarrhea disease is one of major threats that afflict the global pork industry in recent years, which has caused an estimated loss of multi-million dollar annually. Various pathogens that can cause diarrhea in swine have been identified, and swine enteric coronaviruses are the most destructive pathogens that are able to result vomiting, dehydration and lethal watery diarrhea in piglets [1]. To date, four swine enteric coronaviruses have been defined, including three Alphacoronaviruses [transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV)], and one porcine deltacoronavirus [porcine deltacoronavirus (PDCoV)] [2]. TGEV has been circulating in swine for decades since 1946 [3], whereas PEDV, PDCoV and SADS-CoV are regarded as three emerging and re-emerging coronaviruses [2]. PEDV was first discovered in England in feeder and fattening swine in early 1970s [4], and subsequently circulated in Asia, Europe and America [5]. In late 2010, a highly virulent PEDV caused epidemic diarrhea in all age groups of swine in China and US, with 80%–100% morbidity and 50%–90% mortality in suckling piglets which made the pork industry suffer destructive blow [[6], [7], [8]]. PDCoV is a relatively new coronavirus that was identified in 2009 in Hong Kong [9]. However, the first outbreak of PDCoV in Ohio in 2014 addressed its clinical significance [10]. Until now, the detection of PDCoV has been reported in Canada, South Korea, China, Thailand, Laos [[11], [12], [13], [14], [15]]. Among these viruses, SADS-CoV is the most newly discovered coronavirus, which has been first reported in 2017 in China and is considered to be an HKU2-related coronavirus with a bat-origin [[16], [17], [18]]. The fatal diarrhea caused by SADS-CoV has led to the death of 24,693 piglets in a short time in South China. Plentiful epidemiological investigations have shown that there is a co-infection between PEDV, PDCoV and SADS-CoV in swine, and their clinical symptoms and pathological characteristics are extremely similar [19,20], which makes it difficult to distinguish these coronaviruses in clinical infections.
Recently, various isothermal nucleic acid amplification methods have been developed to optimize conventional polymerase chain reaction (PCR) systems [21]. Representative techniques include loop-mediated isothermal amplification (LAMP) [22], recombinase polymerase amplification (RPA) [23], strand displacement amplification (SDA) [24], cross-priming amplification (CPA) [25] and so on. Among those, LAMP is an outstanding method which has been proved to be a reliable, simple, specific and rapid detection method for point-of-care (POC) diagnosis [26] and widely applied for detecting and identifying pathogens [[27], [28], [29], [30]].
Up to now, the predominant control measures of swine diarrhea disease are the rapid and accurate diagnoses of pathogens at the early stage to prevent further circulations in swine. Currently, the most commonly detection methods for swine enteric coronaviruses such as ELISA (enzyme-linked immunosorbent assay), PCR and IHC (immunohistochemistry), all depend on laboratory diagnosis and possessed the advantages of high sensitivity and accuracy, and have been widely used [[31], [32], [33], [34]]. However, most of these diagnostic assays are complex, time-consuming, and require a large of reagents and well-equipped laboratories, which limit their application in remote areas. Therefore, it is urgent to develop portable POC devices with characters of simple, timely, multi-target and strong pertinence, so as to detect pathogens early, prevent the spread of infectious disease and reduce mortality. Amongst the various biosensor-based methods that have been reported, centrifugal microfluidic lab-on-a-chip (LOC) is the most recent innovation tool in microfluidic field, which can make analytical assays perform in a multiple, high throughput and integrated format with high efficiency and speed [35,36]. Centrifugal microfluidic devices actually have the CD-like chip combining microfluidic unit operations, such as liquid mixing, metering, aliquot, switching, valving and storage, which are controlled by the centrifugal speed of the chip [37,38].
In the present study, we integrated the real-time reverse transcription loop-mediated isothermal amplification (RT-LAMP) into the CD-like microfluidic chip for rapid and convenient multiplex detection of PEDV, PDCoV and SADS-CoV. Our results showed that the developed microfluidic-RT-LAMP chip system was accurate, sensitive, specific and reproducible for point-of-care diagnosis, which was advantaged for the early diagnosis of diarrhea in swine farms, especially in remote areas, and adopting effective measures to control the spread of the diseases.
2. Experimental
2.1. Clinical samples, nucleic acid extraction and qRT-PCR assays
Clinical diarrheal samples including feces and intestinal contents were collected from swine in southern China in accordance with the recommendations of National Standards for Laboratory Animals of the People’s Republic of China (GB149258-2010). Samples were preserved at −80 °C from the time of original receipt until use.
Samples were homogenized in phosphate buffered saline (PBS) (20% w/v), frozen and thawed three times, then centrifuged for 10 min at 10,000 g. Viral nucleic acid was extracted following the manufacturer’s recommendations of Magnetic Viral DNA/RNA Kit (iGeneTech, Ningbo, China). The viral nucleic acid was stored at −80 °C until PCR was performed. The qRT-RCR detection of PEDV, PDCoV and SADS-CoV was carried out with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) according to the previously described methods [35,36].
2.2. Chips and microfluidic device
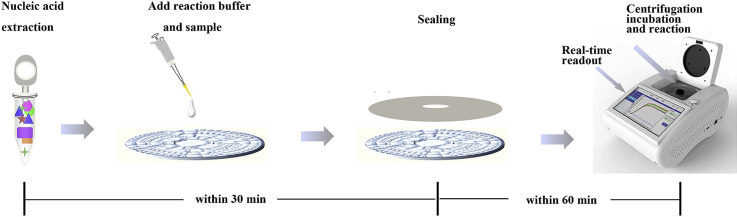
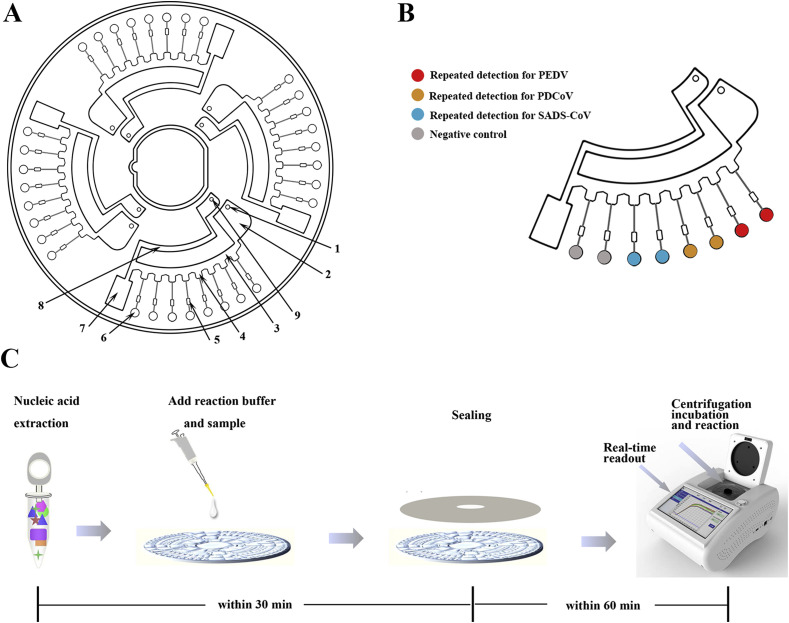
The microfluidic chip made of PMMA that was used in this study was a CD-shape with a diameter of 80 mm and a thickness of 2.5 mm, which was fabricated by the micro-injection molding technology. To form an integrated microfluidic chip, a kind of pressure-sensitive film was applied to seal the micro-channel of chip via suitable pressure. The microfluidic chip comprised four independent units, each of which was consisted of a sample well, a vent and eight micro reaction wells (Fig. 1 A). The ball valve played a significant role throughout the process ensuring that the reaction mixture diffused into the adjacent pipe and the reaction wells were filled with liquid. The volume of each micro reaction well was 5 μL. The supporting microfluidic detection equipment (dimensions: length, width, and height are 280 × 200 × 135 mm) was actually a device (Product Model: AJYGene TM MA2000) that was equipped with the functions of constant temperature control, high-speed centrifugation and fluorescence reading and analysis, and was provided by iGeneTech (Ningbo, China). Once microfluidic detection equipment starts running, the sample would be uniformly distributed in each reaction chambers at two step centrifugal forces (1600 RPM/min, 30 s and 4600 RPM/min, 10 s). While the reaction was performed at the given temperature, the microfluidic device gathered fluorescence signals of each micro chambered every minute and displayed in real time on the electronic screen.
Fig. 1.
Schematic of the microfluidic-RT-LAMP chip detection platform for the emerging and re-emerging swine enteric coronaviruses. A. The structure of the CD-like microfluidic chip. 1: Sample well; 2: liquid storage chamber; 3: the second arc passage; 4: reservoirs; 5: ball valve; 6: reaction chamber; 7: waste liquid tank; 8: the first arc passage; 9: vent hole; B. Detailed view of the chip with its sample adding area, channels, and reaction area; C. A complete set of procedures of microfluidic-RT-LAMP chip detection platform.
2.3. Primer design
The reference complete genomic sequences of PEDV, PDCoV and SADS-CoV were retrieved from Genbank with accession numbers of KT323979.1, KY078891, MG557844, respectively. Based on the highly conserved PEDV M, PDCoV N and SADS-CoV M gene nucleic acid fragments, the alternative primers were designed by PrimerExplorer V5 online software ((https://primerexplorer.jp/lampv5/index.html) and synthesized by BGI (Shenzhen, China).
2.4. Primer screen and integrated microfluidic-RT-LAMP system
First of all, the primers corresponding to different viruses were pre-immobilized in eight reaction chambers by heating drying. Six micro chambers were employed for detections of three viruses with one repetition, and the remaining two were used for negative controls (Fig. 1B). The related primer mixture was pre-loaded into the reaction holes. Secondly, the real-time RT-LAMP reaction solution was prepared in a volume of 10.2 μL comprising 50 μM SYBR Green (Invitrogen,USA), 800 U/μL of Bst DNA polymerase (New England BioLabs, USA), 5000 U/mL of AMV transcripitase (New England BioLabs, USA) and 40 U/μL RNase inhibitor (Vazyme, China). The premixed reaction solution containing 14.8 μL of nucleic acid was injected into sample wells of the microfluidic chip directly by pipette, and then sample wells and vent wells were sealed by the film. The preprocessed microfluidic chip was then placed in the provided microfluidic device to react at 63.5 °C for 60 min. To ensure the accuracy of the three LAMP systems, the amplification products were further determined by standard agarose gel electrophoresis.
Generally, the amplification result was regarded as positive only when experiments with one repetition shows a positive signal and the negative controls show negative signals in 60 min. A complete set of procedures was showed in Fig. 1C.
2.5. System evaluation and validation
2.5.1. Specificity
To investigate the specificity, 11 other swine viruses including porcine parvovirus (PPV), Japanese encephalitis virus (JEV), porcine circovirus type 2 (PCV2), classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV), pseudo rabies virus (PRV), swine influenza virus (SIV), foot and mouth disease virus (FMDV), seneca valley virus (SVA), rotavirus (RV) and transmissible gastroenteritis virus (TGEV) were employed for testing. RNase Free ddH2O was also contained in the run as negative control.
2.5.2. Sensitivity
To examine the detection limit of three RT-LAMP systems, in vitro-transcribed RNA standards of PEDV, PDCoV and SADS-CoV were diluted serially ten-fold with RNase free ddH2O (106-10° copies/μL) as templates and performed in three different times. The in vitro-transcribed RNA standards were prepared according to the previously reported method [37].
2.5.3. Stability and repeatability analysis
In order to confirm the stability and repeatability of three developed LAMP systems, ten-fold serial standards dilutions with high (106 copies/μL), medium (104 copies/μL) and low (detection limits of three coronavirus tested by microfluidic-RT-LAMP chip system) concentrations were used to evaluate coefficients of variation (C.V.) of the microfluidic-RT-LAMP chip. The assay for time to threshold values (Tt), defined as the reaction time required for a particular sample reach sufficiently positive signals above the baseline during the real-time amplification, were both included in repeated eight times. The threshold for choosing the Tt was based on the fluorescence value of the negative controls of each test. Three clinical samples were also conducted to determine the stability and reproducibility of the developed microfluidic-RT-LAMP chip system.
2.6. Microfluidic-RT-LAMP chip and qRT-PCR for the detection of clinical samples
A total of 173 swine clinical fecal samples characterized with diarrheal symptoms were collected and detected by the developed microfluidic-RT-LAMP chip system. Samples being amplified in 60 min and showing the typical amplification curves were considered to be positive. At the same time, the diagnostic accuracy of the microfluidic-RT-LAMP chip system was compared with the qRT-PCR assays, and was further evaluated by generating receiver operating characteristic curves (ROCs, using MedCalc statistical software, V19.1.4).
3. Results and discussion
3.1. Screening primers for three emerging and re-emerging swine coronaviruses
Several sets of LAMP primers were designed and screened for PEDV, PDCoV and SADS-CoV, and one for each pathogen was selected respectively to conduct the following experiments. The in vitro transcribed RNA of each coronavirus was used as template.
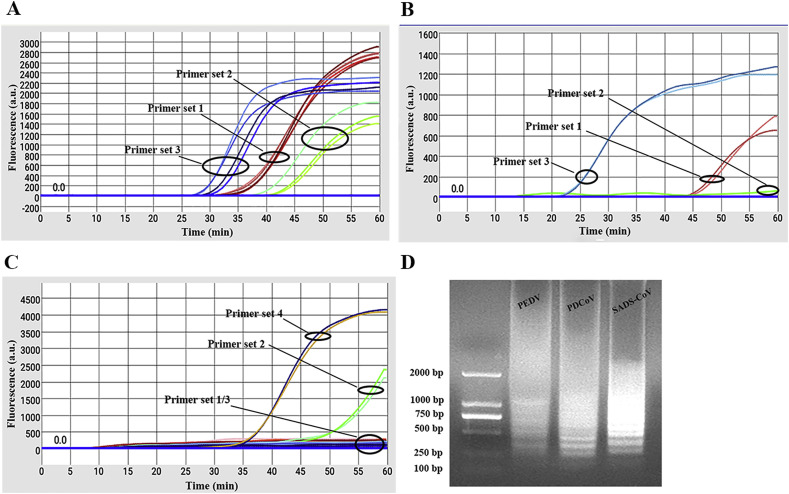
After the temperature reached to 63.5 °C for LAMP reaction, the fluorescence signals were recorded for each hole every minute. Obvious S-shaped amplification curves were observed on the display if successful experiments were processed. As shown in Fig. 2 (A-C), PEDV, PDCoV and SADS-CoV could be amplified by the candidate primers, and the primer set 3 (PEDV), 3 (PDCoV) and 4 (SADS-CoV) were selected, respectively. The primer sequences used in this study were listed in Table 1 . To determine the accuracy of the LAMP reaction, the target templates were successfully amplified from the target-contained sample and presented ladder-like bands with agarose gel electrophoresis (Fig. 2D).
Fig. 2.
Results of screening for primers. A-C. Primer screening of PEDV, PDCoV and SADS-CoV. D. Analysis of PEDV, PDCoV and SADS-CoV by RT-LAMP with agarose gel electrophoresis.
Table 1.
Primer sequences used in microfluidic-RT-LAMP chip system.
| Primer set | Primer ID | Sequence (5′-3′) | Target Gene |
|---|---|---|---|
| PEDV set 3 | F3 | GCGCAGGACACATTCTTGG | PEDV-M |
| B3 | TTGGCGACTGTGACGAAAT | ||
| FIP | CTGGGATGCAGACCTGTCGG-TCAATCCTGAAACAGACGCG | ||
| BIP | TGGAGCACCAACTGGTGTAACG-GTACGCCAGTAGCAACCTT | ||
| LB | GTGGTACATTGCTTGTAGAGGGC | ||
| PDCoV set 3 | F3 | ACCACTCGTGTTACTTGGGT | PDCoV-N |
| B3 | ACGCTCCTGAGGTCTTCC | ||
| FIP | TTGTTGGGGTTGCGTTTGGC-TAAGGGTTCGGGAGCTGAC | ||
| BIP | GCTGCTACCTCTCCGATTCCC-TCTAGCGTTGAAGGGGTCA | ||
| LB | GAGATGGCCCAGCTCAAGGT | ||
| SADS-CoV set 4 | F3 | CATTTAACCCCGAAACAGAC | SADS-CoV-M |
| B3 | ATAGTCGTGCCAGGTTTG | ||
| FIP | CTGTTGGTGCCACTGGCATA-GCCATTGCTGTCATTTCAG | ||
| BIP | AAGTGGAACACTCTTTTTCGATGG-AGTCACAAATTGCGGTAAG | ||
| LF | GGTATCGAGTAGGATCTACCAAAGA | ||
| LB | GAATTGCTACTGGTGTGCAGCC |
3.2. Evaluation of the detection performance of the microfluidic-RT-LAMP chip system
3.2.1. Specificity and sensitivity
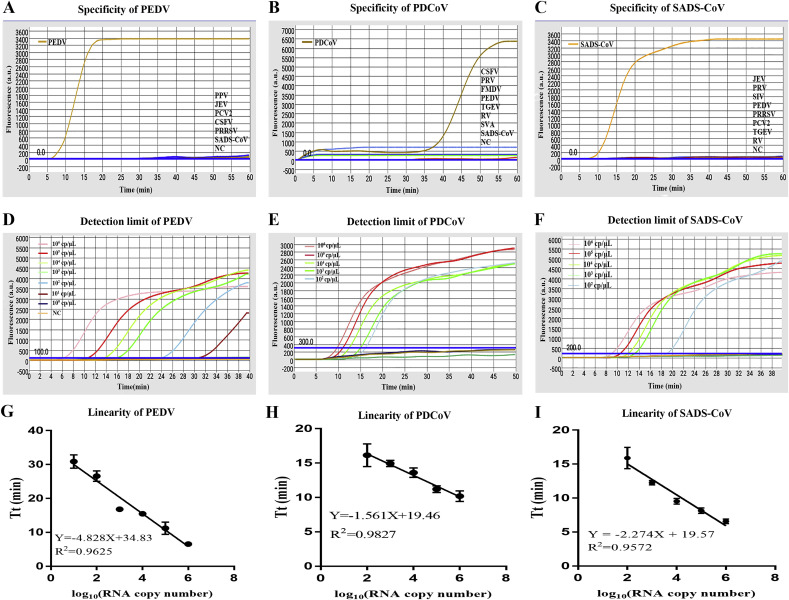
To assess the specificity of the screened LAMP primers, 11 relevant swine viral nucleic acid including that of CSFV, PPV, JEV, PCV2, PRRSV, PRV, SIV, FMDV, SVA, RV and TGEV were applied in this study. As shown in Fig. 3 (A-C) only PEDV-, SADS-CoV- and PDCoV-positive samples presented obvious amplification curves, and 11 other pathogens and the control water displayed negative reactions, which illustrated the high specificity of the established microfluidic-RT-LAMP assay. Taken together, the fluorescence curves during the amplification demonstrated that the system had no cross reaction with other common swine viruses and possessed a high degree of specificity.
Fig. 3.
Specificity and sensitivity of microfluidic-RT-LAMP chip detection system. A-C. Specificity analysis for PEDV, PDCoV and SADS-CoV; D-F. Sensitivity of the microfluidic-RT-LAMP chip system for PEDV, PDCoV and SADS-CoV; G-I. Semi-logarithmic regression between the Tt values and standards concentration by GraphPad Prism 7.0.
After evaluating the specificity of the microfluidic-RT-LAMP chip detection system, the serially diluted plasmids carrying the highly conserved genes of PEDV, PDCoV and SADS-CoV respectively were initially utilized to investigate the lowest detection limits of the established microfluidic-RT-LAMP chip system for each virus. Fig. 3D–F showed that the microfluidic-RT-LAMP chip system enabled the detection of PEDV, PDCoV and SADS-CoV within 40 min and the detection limit for each of the three viruses were maintained at 101 copies/μL, 102 copies/μL and 102 copies/μL, respectively. Those detection limits of this system were competitive for the application to POC diagnostic device [[39], [40], [41]]. The sample-to-answer time on-chip for diagnosis, including nucleic acid extraction, adding reaction solution, sealing and reaction, was approximately 1.5 h. On the other hand, the total volume of sample and reaction buffer used for one on-chip detection hole was roughly 25 μL. Undoubtedly, the new method developed here was a cost-effective and time-saving way for PEDV, PDCoV, and SADS-CoV diagnosis with POC platform when compared with existing methods [[39], [40], [41]]. In addition, the semi-logarithmic regression analysis was performed using the data from the sensitivity test results of standard samples (Fig. 3G–I). There was a significant linear relationship between the logarithm of the plasmid concentration and the Tt value for PEDV, PDCoV and SADS-CoV with the R2 values of 0.965, 0.9827 and 0.9572, respectively, which indicated the potential quantitative detection capability of this established diagnosis platform. Effective development of diagnostic platform such as microfluidic chip requires ultrahigh sensitivity and specificity to enable real-time analysis needed for clinical detection [42]. Based on above data, the established microfluidic-RT-LAMP chip detection system is competitive and competent in POC diagnosis of PEDV, PDCoV and SADS-CoV in remote areas, either for early clinical diagnosis or surveillance and outbreak investigation.
3.2.2. Stability and reproducibility analysis
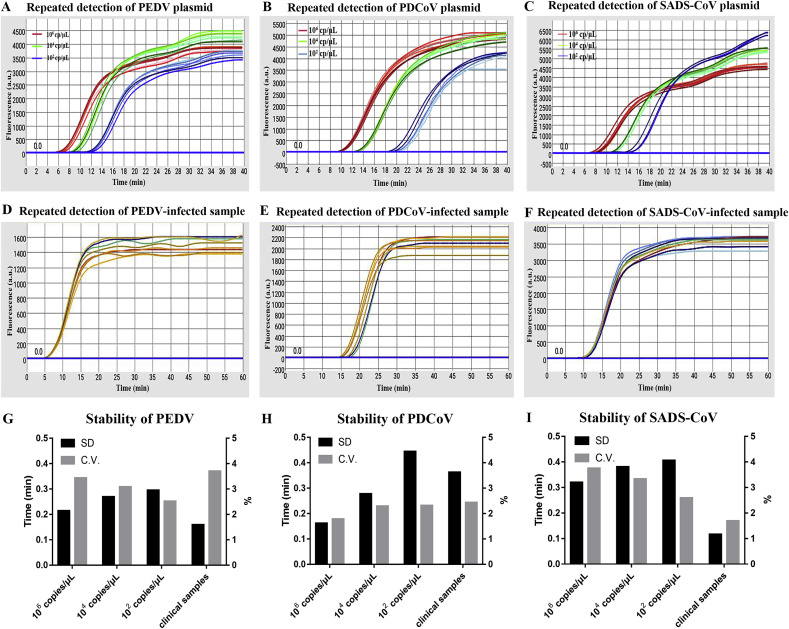
Stability is also an important indicator for a diagnostic method [43]. For this part, the high, medium and low detectable concentration standard samples were used to validate the reproducibility and stability of the established microfluidic-RT-LAMP chip detection system. We calculated the C.V. through dividing the standard deviation (SD) by the mean of different samples at the same concentration. The results of Fig. 4 A-C demonstrated the stability of our method. For the microfluidic-RT-LAMP chip detection system, the C.V. values of the standard samples of PEDV, PDCoV and SADS-CoV ranged from 2.52% to 3.43%, 1.78%–2.29% and 2.59% to 3.75% respectively with small SDs (Fig. 4G–I). All the above C.V. values were less than 5%, indicating the good reproducibility of each established microfluidic-RT-LAMP chip method. The results were further confirmed by the clinical PEDV-, PDCoV- and SADS-CoV-infected fecal samples (Fig. 4D–F), which showed ideal C.V. values of 3.71%, 2.43% and 1.69% for each coronavirus (Fig. G–I). In summary, the developed CD-like microfluidic-RT-LAMP chip system for the point-of-care diagnosis of multiple genes of PEDV, PDCoV and SADS-CoV is promising, and can be applied to the real-time qualitatively monitoring of these three coronaviruses with high specificity, sensitivity and rapidity.
Fig. 4.
Evaluation of stability and reproducibility of the microfluidic-RT-LAMP chip detection system for three swine enteric coronaviruses. A-C. Stability test of PEDV, PDCoV and SADS-CoV for 8 replicate experiments using high, medium and low (detection limits) concentration standards, respectively. D-F. Stability test of PEDV, PDCoV and SADS-CoV for 8 replicate experiments using a clinical virus-infected sample. G-I. The SD and C.V. of Tt values for each of 8 replicate experiments.
3.3. Performance of the microfluidic-RT-LAMP chip in clinical samples
The specificity, sensitivity and reproducibility test results have illustrated that the microfluidic-RT-LAMP chip detection system is stable, rapid, convenient and portable. To further verify the actual performance of the microfluidic-RT-LAMP chip detection system in clinical samples, we collected 173 swine diarrhea samples, of which 80 PEDV positive samples, 59 PDCoV positive samples, 52 SADS-CoV positive samples and 17 negative samples were detected by the microfluidic-RT-LAMP chip system and referenced qRT-PCR assays. There were 12 samples co-infected with two or three coronaviruses. No negative sample detected by the qRT-PCR assays was found to be positive by the developed microfluidic-RT-LAMP chip system. The coincidence rates (the proportion of the number of samples with the same test results in the total number of test samples) of the detection results between the qRT-PCR and the microfluidic-RT-LAMP chip system were 97.69%, 97.11% and 97.11% for PEDV, PDCoV and SADS-CoV, respectively (Table 2 ). Sensitivities of the microfluidic-RT-LAMP chip for each virus were 95.24%, 92.19% and 91.23%. Additionally, specificities of the microfluidic-RT-LAMP chip for three viruses were all 100%. The results were matched perfectly, which showed that our microfluidic-RT-LAMP chip system was effective and reliable.
Table 2.
Comparisons between the microfluidic-RT-LAMP chip system and the qRT-PCR assay for detection of three swine enteric coronaviruses. a. microfluidic-RT-LAMP chip system positive samples. b. qRT-PCR positive samples. c. microfluidic-RT-LAMP chip system negative samples. d. qRT-PCR negative samples.
| virus | No.of samples |
Performance of the microfluidic-RT-LAMP chip |
CR % | ||||
|---|---|---|---|---|---|---|---|
| Chip+a qRT-PCR + b | Chip + qRT-PCR- | Chip- qRT-PCR+ | Chip-c qRT-PCR-d | Sensitivity % | Specificity % | ||
| PEDV | 80 | 0 | 4 | 89 | 95.24 | 100 | 97.69 |
| PDCoV | 59 | 0 | 5 | 109 | 92.19 | 100 | 97.11 |
| SADS-CoV | 52 | 0 | 5 | 116 | 91.23 | 100 | 97.11 |
CR: coincidence rate. CR = (True positive + True negative)/Total∗100%.
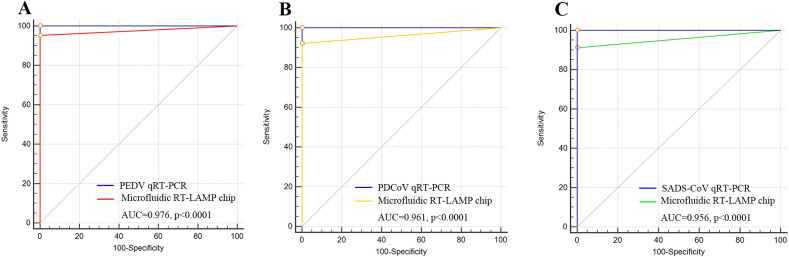
Based on the above dates, ROCs were plotted to determine the diagnostic accuracy of the microfluidic-RT-LAMP system and qRT-PCR assays. The results revealed that the AUCROC were 0.976, 0.961 and 0.956 (p < 0.0001) for PEDV, PDCoV and SADS-CoV, respectively and the corresponding Youden index were 0.9524, 0.9219 and 0.9123, which indicated the detection system we developed is an effective diagnostic tool to distinguish three kinds of coronaviruses infected and uninfected samples, with the potential to replace existing methods for POC diagnosis (Fig. 5 A–C) [[39], [40], [41]]. Simultaneously, ROC curves of the microfluidic-RT-LAMP system and qRT-PCR assays were analyzed, and the results further showed that there was no significant difference between these two kinds of methods in detecting these three coronaviruses (ZPEDV = 2.037, p < 0.0416; ZPDCoV = 2.311, p < 0.0209; ZSADS-CoV = 2.320, p < 0.0203) (Fig. 5A–C).
Fig. 5.
Diagnostic performance of the microfluidic-RT-LAMP chip system for three swine enteric coronaviruses. A-C. Receiver operating curve (ROC) analysis of the microfluidic-RT-LAMP chip detection system for the diagnosis of PEDV, PDCoV and SADS-CoV and comparison of the diagnosis accuracy between the microfluidic-RT-LAMP chip detection system and qRT-PCR assays.
4. Conclusions
Taken together, this study successfully established a rapid, simple and low-cost multiplex detection tool for PEDV, PDCoV, and SADS-CoV, which integrated a microfluidic chip, real-time RT-LAMP assay and a portable microfluidic chip fluorescence detector. The entire procedure could be completed within 1.5 h at detection limits of 1 × 101 copies/μL, 1 × 102 copies/μL and 1 × 102 copies/μL for PEDV, PDCoV and SADS-CoV, respectively. Perfect sensitivity and specificity for those three swine coronaviruses detection were also found with 173 clinical samples. This microfluidic-RT-LAMP chip system possesses the important features of a successful POC diagnostic tool, in that it is extremely portable, cost-effective and rapid. Meanwhile, it is high-throughput and easy-to-use in the field, which has given the excellent prospects of this method in swine enteric coronaviruses epidemic monitoring and control.
CRediT authorship contribution statement
Ling Zhou: Data curation, Methodology, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Yonghui Chen: Data curation, Methodology, Formal analysis, Investigation, Visualization. Xueen Fang: Data curation, Validation. Yanhong Liu: Investigation. Mengkan Du: Investigation. Xiandong Lu: Investigation. Qianniu Li: Validation. Yuan Sun: Validation. Jingyun Ma: Conceptualization, Funding acquisition, Project administration, Resources, Supervision. Tian Lan: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Science and Technology Program of Guangzhou City of China (No. 201904010433), the Research and Extension of Major Animal Epidemic Prevention and Control Technologies in the Strategic Project of Rural Revitalization of Guangdong Agricultural Department of China (Building Modern Agricultural System) (2018–2020).
References
- 1.Li Y., Wu Q., Huang L., Yuan C., Wang J., Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018;9(1):3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Vlasova A., Kenney S., Saif L. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sánchez C., Gebauer F., Suñé C., Mendez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190(1):92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pensaert M., de Bouck P. A new coronaviruslike particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Gene. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J., Wang C., Shi H., Qiu H., Liu S., Chen X., Zhang Z., Feng L. Molecular epidemiology of porcine epidemic diarrhea virus in China. Arch. Virol. 2010;155(9):1471–1476. doi: 10.1007/s00705-010-0720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun R., Cai R., Chen Y., Liang P., Chen D., Song C. Outbreak of porcine epidemic diarrhea in suckling piglets. Emerg. Infect. Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y., Dickerman A., Piñeyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States, mBio. 2013;4(5) doi: 10.1128/mBio.00737-13. e00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo P., Lau S., Lam C., Lau C., Tsang A., Lau J., Bai R., Teng J., Tsang C., Wang M. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA. Emerg. Infect. Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D., Stott C., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., Chongcharoen W. Diferent lineage of porcine deltacoronavirus in Thailand, vietnam and Lao PDR in 2015. Transbound Emerg Dis. 2017;64(1):3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madapong A., Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Srijangwad A., Tripipat T., Wegner M., Nilubol D. Complete genome sequence of porcine deltacoronavirus isolated in Thailand in 2015. Genome Announc. 2016;26(3):4. doi: 10.1128/genomeA.00408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song D., Zhang X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identifcation, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S., Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2(6) doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung K., Hu H., Saif L. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong L., Li J., Zhou Q., Xu Z., Chen L., Zhang Y., Xue C., Wen Z., Cao Y. A new bat-HKU2–like coronavirus in swine, China. Emerg. Infect. Dis. 2017;23(9) doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y., Tian X., Qin P., Wang B., Zhao P., Yang Y., Wang L., Wang D., Zhang X. Discorvey of a novel porcine enteric Alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Fan H., Lan T., Yang X., Shi W., Shi W., Zhang W., Zhu Y., Zhang Y., Xie Q., Mani S. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L., Sun Y., Lan T., Wu R., Chen J., Wu Z., Xie Q., Zhang X., Ma J. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound Emerg Dis. 2019;66(2):687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai K., Feng J., Chen G., Li D., Zhou L., Bai Y., Wu Q., Ma J. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound Emerg Dis. 2018;65(1):166–173. doi: 10.1111/tbed.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid M., Le X., Zhang H. Exponential isothermal amplification of nucleic acids and assays for proteins, cells, small molecules, and enzyme activities: an EXPAR example. Angew Chem. Int. Ed. Engl. 2018;57:11856–11866. doi: 10.1002/anie.201712217. [DOI] [PubMed] [Google Scholar]
- 22.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piepenburg O., Williams C., Stemple D., Armes N. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toley B., Covelli I., Belousov Y., Ramachandran S., Kline E., Scarr N., Vermeulen N., Mahoney W., Lutz B., Yager P. Isothermal strand displacement amplification (iSDA): a rapid and sensitive method of nucleic acid amplification for point-of-care diagnosis. Analyst. 2015;140:7540–7549. doi: 10.1039/c5an01632k. [DOI] [PubMed] [Google Scholar]
- 25.Xu G., Hu L., Zhong H., Wang H., Yusa S., Weiss T., Romaniuk P., Pickerill S., You Q. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci. Rep. 2012;2:246. doi: 10.1038/srep00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craw P., Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12(14):2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Fan P., Zhou S., Zhang L. Loop-mediated isothermal amplification (lamp): a novel rapid detection platform for pathogens. Microb. Pathog. 2017;107:54–61. doi: 10.1016/j.micpath.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Tang Y., Yu X., Chen H., Diao Y. An immunoassay-based reverse-transcription loop-mediated isothermal amplification assay for the rapid detection of avian influenza h5n1 virus viremia. Biosens. Bioelectron. 2016;86:255–261. doi: 10.1016/j.bios.2016.06.063. [DOI] [PubMed] [Google Scholar]
- 29.Hill J., Beriwal S., Chandral I., Paul V., Kapil A., Singh T., Wadowsky R., Singh V., Goyal A., Jahnukainen T. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J. Clin. Microbiol. 2008;46:2800–2804. doi: 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Wang H., Lin B., Tao Y., Zhuo K., Liao J. Loop-mediated isothermal amplification based on the mitochondrial COI region to detect Pratylenchus zeae. Eur. J. Plant Pathol. 2017;148:435–446. [Google Scholar]
- 31.Liu G., Jiang Y., Opriessnig T., Gu K., Zhang H., Yang Z. Detection and differentiation of five diarrhea related pig viruses utilizing a multiplex PCR assay. J. Virol Methods. 2019;2263:32–37. doi: 10.1016/j.jviromet.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Huang X., Chen J., Yao G., Guo Q., Wang J., Liu G. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biotechnol. 2019 Jun;103(12):4943–4952. doi: 10.1007/s00253-019-09835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Z., Wang T., Li Z., Guo X., Tian Y., Li Y., Xiao S. A novel biotinylated nanobody-based blocking ELISA for the rapid and sensitive clinical detection of porcine epidemic diarrhea virus. J. Nanobiotechnol. 2019;17(1):96. doi: 10.1186/s12951-019-0531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L., Zeng F., Huang B., Cong F., Huang R., Ma J., Guo P. Development of a conventional RT-PCR Assay for rapid detection of porcine deltacoronavirus with the same detection limit as a SYBR green-based real-time RT-PCR assay. BioMed Res. Int. 2018:5035139. doi: 10.1155/2018/5035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madou M., Zoval J., Jia G., Kido H., Kim J., Kim N. Lab on a CD. Annu. Rev. Biomed. Eng. 2006;8:601–628. doi: 10.1146/annurev.bioeng.8.061505.095758. [DOI] [PubMed] [Google Scholar]
- 36.Gorkin R., Park J., Siegrist J., Amasia M., Lee B., Park J., Kim J., Kim H., Madou M., Cho Y. Centrifugal microfluidics for biomedical applications. Lab Chip. 2010;10(14):1758–1773. doi: 10.1039/b924109d. [DOI] [PubMed] [Google Scholar]
- 37.Ma L., Zeng F., Cong F., Huang B., Zhu Y., Wu M., Xu F., Yuan W., Huang R., Guo P. Development and evaluation of a broadly reactive reverse transcription recombinase polymerase amplification assay for rapid detection of murine norovirus. BMC Vet. Res. 2018;14:399. doi: 10.1186/s12917-018-1736-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mark D., Haeberle S., Roth G., Stetten F., Zengerle R. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. Chem. Soc. Rev. 2010;39(3):1153–1182. doi: 10.1039/b820557b. [DOI] [PubMed] [Google Scholar]
- 39.Wang H., Cong F., Zeng F., Lian Y., Liu X., Luo M., Guo P., Ma J. Development of a real time reverse transcription loop-mediated isothermal amplification method (RT-LAMP) for detection of a novel swine acute diarrhea syndrome coronavirus (SADS-CoV) J. Virol Methods. 2018;260:45–48. doi: 10.1016/j.jviromet.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Zhang R., Wang J., Han Q., Liu L., Li Y., Yuan W. Real-time reverse transcription recombinase polymerase amplification assay for rapid detection of porcine epidemic diarrhea virus. J. Virol Methods. 2018;253:49–52. doi: 10.1016/j.jviromet.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Ma L., Zeng F., Huang B., Zhu Y., Wu M., Xu F., Xiao L., Huang R., Ma J., Cong F. Point-of-care diagnostic assay for rapid detection of porcine deltacoronavirus using the recombinase polymerase amplification method. Transbound Emerg Dis. 2019;66(3):1324–1331. doi: 10.1111/tbed.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beebe J., Moore S., Yu Q., Liu R., Kraft L., Jo B., Devadoss C. Microfluidic tectonics: a comprehensive construction platform for microfluidic systems. Proc. Natl. Acad. Sci. Unit. States Am. 2000;97(25):13488–13493. doi: 10.1073/pnas.250273097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye X., Li L., Li J., Wu X., Fang X., Kong J. Microfluidic-CFPA chip for the point-of-care detection of african swine fever virus with a median time to threshold in about 10 min. ACS Sens. 2019;4(11):3066–3071. doi: 10.1021/acssensors.9b01731. [DOI] [PubMed] [Google Scholar]