Abstract
Background
Diarrheagenic Escherichia coli (DEC) is a significant cause of gastroenteritis and a major public health problem. This study investigates the prevalence and the antibiotic resistance patterns of DEC that were isolated from infectious diarrhea samples of pediatric patients from central Iran.
Patients and Methods
Pediatric diarrhea samples were collected from 230 pediatric patients visiting the hospital. E. coli pathotypes were diagnosed by using conventional culture methods and PCR. Antibiotic resistance profiles, the frequency of multi-drug resistance (MDR), and the phenotypic and genotypic characteristics of extended spectrum-β-lactamase (ESBL), AmpC and integron-associated genes were analyzed.
Results
Of the 230 samples of infectious diarrhea, 91 (39.5%) produced E. coli isolates. Of these, 32 cases (35.1%) were identified as DEC by culture and PCR. The frequency of the E. coli pathotypes obtained was as follows: EAEC 11/32 (34.3%), EPEC 9/32 (28.1%), ETEC 6/32 (18.7%), EIEC 3/32 (9.3%), and EHEC 3/32 (9.3%). The antibiotic resistance rates were greater for nalidixic acid (30/32; 93.7%), ampicillin (29/32; 90.6%), and tetracycline (25/32; 78.1%) than for any of the other tested antibiotics. High levels of MDR (25/32; 78.1%) and the presence of ESBL (18/32; 56.2%) and AmpC (9/32; 28.1%) were observed in the DEC isolates. The isolates showed a higher frequency of the ESBL genes [blaTEM (18/18; 100%), blaCTX-M15 (17/18; 94.4%)], and AmpC [blaCIT (4/9; 44.4%) and blaDHA (4/9; 44.4%)] than of the other ESBL and AmpC genes.
Conclusion
Compared to the previous study, DEC appeared to be the second-most abundant agent of diarrhea in pediatric patients after Campylobacter jejuni, with frequent MDR and ESBL presence.
Keywords: diarrheagenic Escherichia coli pathotypes, pediatrics, diarrhea, antibiotic resistance, MDR, Iran
Introduction
Gastroenteritis causes 525,000 deaths per year worldwide, especially in children younger than five years old.1 It can cause serious growth retardation and malnutrition.2 Diarrheagenic Escherichia coli (DEC) is one of the most abundant agents in pediatric gastroenteritis, particularly in developing countries.3 DEC can be divided into various pathogroups: enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), entero-invasive E. coli (EIEC) and enterohemorrhagic E. coli (EHEC).4 Among these pathotypes, EAEC, EPEC and ETEC are the most significant enteric pathogens and cause 30–40% of the acute pediatric diarrhea cases in developing and developed countries.5 The emergence of multi-drug-resistant (exhibiting resistance against two or more antibiotics, MDR) E. coli strains is a public health concern and is complicating the treatment of various serious infections, particularly in pediatrics.6 Lately, worldwide diffusion of extended spectrum-β-lactamase (ESBL)-producing E. coli has been most remarkable.7 Hence, the analysis of the frequency of diarrheagenic E. coli pathotypes to the total burden of diarrheal disease and the antimicrobial resistance pattern of these pathotypes is necessary for developing useful interventions that can effectively reduce the mortality and morbidity related to diarrhea.
The number of outbreaks of diarrhea and dysentery in central Iran, especially in the summer, is very high in pediatrics,8 but the prevalence of DEC and antibiotic resistance in this region is unknown. Therefore, the present study was conducted to investigate the abundance, the phenotypic antimicrobial resistance levels and the resistance gene content of the region’s DEC by examining diarrhea samples from pediatric patients.
Patients and Methods
Sample Collection
For this cross-sectional, descriptive study, 230 samples of diarrhea were collected from children who were referred to the Amirkabir Educational-Referral Center from the beginning of May 2015 to the end of February 2016. Consent and questionnaire forms were supplied to the parents or guardians of each patient. The inclusion criteria of the study were 1- providing a signed consent form and completed questionnaire, either by the patient or the patient’s parents or caregivers, 2- providing a stool sample containing more than five white blood cells per high-power field (HPF).8 3- not having received antibiotics by the patient for at least a week before consultation at the hospital. This research obtained approval from the ethics committee of the Arak University of Medical Sciences (ARAKMU.REC. 93-176-30 and 1395.83).
Phenotypic Investigation
To identify E. coli strains and EHEC, diarrheal stool samples were cultured directly on MacConkey and Sorbitol MacConkey (SMAC) agar media (Merck, Hamburg, Germany), respectively. Next, colonies that grew on these media were identified by biochemical tests. API 20E test strips (bioMerieux, France) were used for final identification. Then, O157: H7 serological testing (SSI, Copenhagen, Denmark) of isolated colorless colonies (due to lack of sorbitol fermentation) was done. Positive controls of different E. coli pathotypes were obtained from the microbiology department of the Arak University of Medical Sciences.
Antibiotic Resistance Determination
According to the Clinical and Laboratory Standards Institute (CLSI) 2017 guidelines,9 a disk diffusion assay was performed on the isolated E. coli colonies. The antibiotic discs contained nalidixic acid (30 μg), ampicillin (10 μg), tetracycline (30 μg), cotrimoxazole (25 μg), chloramphenicol (30 μg), ceftriaxone (30 μg), cefixime (5 μg), cefotaxime (30 μg), ceftizoxime (30 μg), cefoxitin (30 μg), ceftazidime (30 μg), azithromycin (15 μg), ciprofloxacin (5 μg), gentamicin (10 μg) and imipenem (10 μg) (Mast Diagnostics, United Kingdom).
Detection of ESBL and AmpC by Phenotypic Methods
To identify ESBL-positive isolates, the samples were subjected to combination disk diffusion, and double-disk synergy testing methods, and to identify AmpC-positive isolates, disk testing and phenol boronic acid methods according to the 2017 CLSI guidelines were used.9
DNA Extraction
DNA was extracted directly from the fecal samples and the reference E. coli isolates using the QIAamp DNA stool mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s protocol. The amount and purity of the extracted DNA were measured with a NanoDrop apparatus (Thermo Fisher Scientific, Waltham. Massachusetts, United States).
Genotypic Identification
The 25 µL final volume of the PCR included 12.5 µL of master mix (1X), 2 µL of DNA template (5 ng), 1 µL each of the forward and reverse primers (10 Pm), 0.5 µL of Taq DNA polymerase (2.5 units), and 8 µL of double-distilled water (all purchased from Yekta Tajhiz Company, Iran). PCR of the uldA gene was performed to confirm E. coli at the genus level.10 PCR was performed on the pCVD432 gene for EAEC,11 eae gene for EPEC,12 elt, est genes for ETEC,13 ial gene for EIEC,4 and stx gene for EHEC.12 The ESBL genes (blaTEM, blaCTX-M-1, 2, 8, 14, 15, and blaSHV), AmpC genes (blaCMY-2, blaCIT, blaACC, blaFOX, blaMOX and blaDHA) and sul1,2 for sulfonamide resistance were identified by PCR as well.8 PCR of the qnr determinant genes qnrS, qnrA, and qnrB was performed to amplify the plasmid-mediated quinolone resistance (PMQR) targets. Mutations in the gyrA and parC genes of the quinolone-resistant E. coli isolates were identified using DNA sequencing.8 Quaternary ammonium compounds (qac) resistance genes were investigated using PCR (Table 1).8
Table 1.
The Primers Used in This Study
| References | Annealing Temperature | Amplicon Size (bp) | Sequence 5ʹ→3ʹ | Primer | Target Gene Description |
|---|---|---|---|---|---|
| 10 | 67 | 510 | 5-GCGTCTGTTGACTGGCAGGTGGTGG-3 5-GTTGCCCGCTTCGAAACCAATGCCT-3 |
uidA-F uidA-R |
E. coli |
| 11 | 57.5 | 630 | 5-CTGGCGAAAGACTGTATCAT-3 5-CAATGTATAGAAATCCGCTGTT-3 |
pCVD432-F pCVD432-R |
EAEC |
| 12 | 48 | 881 | 5-CCCGAATTCGGCACAAGCATAAGC-3 5-CCCGGATCCGTCTCGCCAGTATTCG-3 |
eae-F eae-R |
EPEC |
| 13 | 55 | 274 | 5-ACGGCGTTACTATCCTCTC-3 5-TGGTCTCGGTCAGATATGTG-3 |
elt-F elt-R |
ETEC |
| 13 | 55 | 170 | 5-TCTTTCCCCTCTTTTAGTCAGTC-3 5-CAGCACAGGCAGGATTAC-3 |
st-F st-R |
ETEC |
| 4 | 50 | 650 | 5-GGTATGATGATGATGAGTCCA-3 5- GGAGGCCAACAATTATTTCC-3 |
ial-F ial-R |
EIEC |
| 12 | 48 | 518 | 5-GAGCGAAATAATTTATATGTG-3 5-TGATGATGGCAATTCAGTAT-3 |
stx-F stx-R |
EHEC |
| 8 | 55 | 344 | 5-AAATCTGCCCGTGTCGTTGGT-3 5-GCCATACCTACGGCGATACC −3 |
gyrA-F gyrA-R |
Fluoroquinolone |
| 8 | 55 | 168 | 5-CTGAATGCCAGCGCCAAATT-3 5-GCGAACGATTTCGGATCGTC-3 |
parC-F parC-R |
|
| 8 | 60 | 656 | 5-TGGAAACCTACAATCATACATATCG-3 5-TTAGTCAGGATAAACAACAATACCC-3 |
qnrS-F qnrS-R |
|
| 8 | 60 | 593 | 5-GATAAAGTTTTTCAGCAAGAGG-3 5-ATCCAGATCGGCAAAGGTTA-3 |
qnrA-F qnrA-R |
|
| 8 | 53 | 264 | 5-GTTGGCGAAAAAATTGACAGAA-3 5-ACTCCGAATTGGTCAGATCG-3 |
qnrB-F qnrB-R |
|
| 8 | 55 | 160 | 5-CAGTGGACATAAGCCTGTTC-3 5-CCCGAGGCATAGACTGTA-3 |
Int1-F Int1-R |
Integrase1 |
| 8 | 55 | 288 | 5-TTGCGAGTATCCATAACCTG-3 5-TTACCTGCACTGGATTAAGC-3 |
Int2-F Int2-R |
Integrase2 |
| 8 | 59 | 979 | 5-GCCTCCGGCAGCGACTTTCAG-3 5-ACGGATCTGCCAAACCTGACT-3 |
Int3-F Int3-R |
Integrase3 |
| 8 | 65 | 331 | 5-TCACCGAGGACTCCTTCTTC-3 5-CAGTCCGCCTCAGCAATATC-3 |
Sul1-F Sul1-R |
Sulfonamide resistance |
| 8 | 58 | 435 | 5-CCTGTTTCGTCCGACACAGA-3 5-GAAGCGCAGCCGCAATTCAT-3 |
Sul2-F Sul2-R |
|
| 26 | 55 | 370 | 5-GCCCTACACAAATTGGGAGA-3 5-CTGCGGTACCACTGCCACAA-3 |
qac-F qac-R |
Quaternary ammonium compounds |
| 8 | 44 | 425 | 5-AAAGATGCTGAAGATCA-3 5-TTTGGTATGGCTTCATTC-3 |
TEM-F TEM-R |
β-Lactamase ESBL+ |
| 8 | 62 | 304 | 5-GCGAAAGCCAGCTGTCGGGC-3 5-GATTGGCGGCGCTGTTATCGC-3 |
SHV-F SHV-R |
|
| 8 | 52 | 670 | 5-AAGACTGGGTGTGGCATTGA-3 5-AGGCTGGGTGAAGTAAGTGA-3 |
CTX-M1-F CTX-M1-R |
|
| 8 | 60 | 552 | 5-CGACGCTACCCCTGCTATT-3 5-CCAGCGTCAGATTTTTCAGG-3 |
CTX-M2-F CTX-M2-R |
|
| 8 | 55 | 307 | 5-CGCTTTGCCATGTGCAGCACC-3 5-GCTCAGTACGATCGAGCC-3 |
CTX-M8-F CTX-M8-R |
|
| 8 | 50 | 355 | 5-TACCGCAGATAATACGCAGGTG-3 5- CAGCGTAGGTTCAGTGCGATCC-3 |
CTX-M14-F CTX-M14-R |
|
| 8 | 55 | 955 | 5-CACACGTGGAATTTAGGGACT-3 5-GCCGTCTAAGGCGATAAACA-3 |
CTX-M15-F CTX-M15-R |
|
| 8 | 58 | 758 | 5′-GCACTTAGCCACCTATACGGCAG-3′ 5′-GCTTTTCAAGAATGCGCCAGG-3′ |
CMY-2-F CMY-2-R |
|
| 8 | 64 | 190 | 5-AACATGGGGTATCAGGGAGATG-3 5-CAAAGCGCGTAACCGGATTGG-3 |
Fox-F Fox-R |
β-Lactamase AmpC+ |
| 8 | 64 | 520 | 5-GCTGCTCAAGGAGCACAGGAT-3 5-CACATTGACATAGGTGTGGTGC-3 |
Mox-F Mox-R |
|
| 8 | 64 | 405 | 5-AACTTTCACAGGTGTGCTGGGT-3 5-CCGTACGCATACTGGCTTTGC-3 |
DHA-F DHA-R |
|
| 8 | 64 | 346 | 5-AACAGCCTCAGCAGCCGGTTA-3 5-TTCGCCGCAATCATCCCTAGC-3 |
ACC-F ACC-R |
|
| 8 | 64 | 462 | 5-TGGCCAGAACTGACAGGCAAA-3 5-TTTCTCCTGAACGTGGCTGGC-3 |
CIT-F CIT-R |
Abbreviations: EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; EIEC, entero-invasive E. coli; EHEC, enterohemorrhagic E. coli.
Integron Detection
To investigate the presence of class 1, 2 and 3 integrons, PCR was performed as previously described (Table 1).8
Results and Discussion
Of the 230 analyzed samples, 91 (39.5%) produced E. coli isolates and 32 (35.1%) were identified as DEC, based on a combination of culture and PCR. All the culture-positive samples were confirmed as positive by PCR. The female-to-male DEC infection ratio was 1.1:1 (Table 2).
Table 2.
Total Number, Sex, and Age Characteristics of the Patients with E. coli Pathotypes
| E. coli Pathotypes | DEC | EACE | EPEC | ETEC | EIEC | Stx Negative EHEC O157:H7 | |
|---|---|---|---|---|---|---|---|
| LT | ST | ||||||
| Total number (%) | 32/91 (35.1%) | 11/32 (34.3%) | 9/32 (28.1%) | 6/32 (18.7%) | 3/32 (9.3%) | 3/32 (9.3%) | |
| 4/6 (66.6%) | 2/6 (33.3%) | ||||||
| Male | 15/32 (46.8%) | 4/11 (36.3%) | 5/9 (55.5%) | 4/6 (66.6%) | 1/3 (33.3%) | 1/3 (33.3%) | |
| 3/4 (75%) | 1/2 (50%) | ||||||
| Female | 17/32 (53.1%) | 7/11 (63.6%) | 4/9 (44.4%) | 2/6(33.3%) | 2/3 (66.6%) | 2/3 (66.6%) | |
| 1/4 (25%) | 1/2 (50%) | ||||||
| Average age | 4 years and 6 months | 3 years and 1 month | 2 years and 9 months | 4 years and 6 months | 5 years and 8 months | 4 years and 1 month | |
| 3 years and 8 months | 6 years | ||||||
| Youngest patient’s age | 5 months | 5 months | 8 months | 9 months | 3 years and 5 months | 1 year | |
| 9 months | 3 years | ||||||
| Oldest patient’s age | 10 years | 10 years | 10 years | 9 years | 9 years | 6 years | |
| 9 years | 9 years | ||||||
Abbreviation: DEC, diarrheagenic Escherichia coli.
Considering a study which it was done in the past, we showed that DEC (n=32, 13.9%) was second only to Campylobacter jejuni (n=45, 19.5%; p = 0.003) as the most abundant agent of diarrhea in pediatric patients, whereas in Shiraz (Iran) and India, DEC was the most common agent found.14–16 Pediatricians should therefore request standard microbiological methods for determination of the pathogens involved and for designing subsequent therapy.
Phenotypic and Genotypic Investigation
The isolates showed a greater frequency of the EAEC (11/32; 34.3%) and EPEC (9/32; 28.1%) pathotypes than of the other E. coli pathotypes. (Table 2). The PCR results were consistently in accordance with those obtained from culture. Table 3 shows the abundance of DEC and E. coli pathotypes in the present study and the pathotypes reported in previous studies. The differences in the frequencies of DEC and E. coli pathotypes may be related to a variety of factors, including differences in climate and other environmental conditions, the level of economic development, individual hygiene practices, lack of/improper medical health care, poor quality medical care, and contamination of food or of food manufacturing or processing facilities.17
Table 3.
Comparison of the Frequency, Clinical Symptoms, and Antibiotic Resistance of DEC and E. coli Pathotypes in This Study and in Other Studies
| E. coli Pathotypes | Shiraz (Iran) | India | China (Throughout the Country) | Tehran (Iran) | Sudan | Iran’s North and Northwest Provinces | China (Southeastern) | |
|---|---|---|---|---|---|---|---|---|
| DEC | 43.6% | 30.7% | 5% | 58.9% | 48% | 27% | 14.1% | |
| EAEC | 23.6% | 6.9% | 1.6% | 16.6% | 43% | 0% | 62.5% | |
| EPEC | 10.9% | 21.5% | 1.6% | 12.6% | 29% | 10.5% | 15% | |
| ETEC | 3.6% | 10.7% | 1% | 10.8% | 18% | 5.8% | 17.9% | |
| EIEC | 5.5% | – | 0.5% | – | 9% | 0% | 0.3% | |
| EHEC | 0% | 4.6% | 0.3% | 18.9% | – | 3.5% | 4% | |
| The most abundant E. coli pathotype | EAEC 84.6% | EPEC 21.5% | EAEC and EPEC 1.6% | EHEC 18.9% | EAEC 43% | EPEC 10.5% | EAEC 62.5% | |
| Clinical symptoms in patients with DEC | Mucus in the stool | – | – | 65% | 51.5% | – | – | – |
| Abdominal pain | – | – | 3.4% | – | – | – | – | |
| Vomiting | – | – | 5.1% | 52.6% | – | – | – | |
| Antibiotic resistance in patients with DEC | Nalidixic acid | – | – | – | – | 34% | – | – |
| Ampicillin | 93.8% | – | – | – | 47% | – | 91.8% | |
| Tetracycline | – | – | – | – | 24% | – | 57% | |
| Cotrimoxazole | 77.1% | – | – | – | – | – | 52.3% | |
| Cefotaxime | 66.7% | – | – | – | – | – | 35.7% | |
| Ceftriaxone | 66.7% | – | – | – | – | – | – | |
| ESBL in DEC isolates | 66.7% | – | – | – | – | – | 34.5% | |
| The most abundant ESBL genes | – | – | – | – | – | – | blaCTX-M 94.2% | |
| References | 16 | 14 | 27 | 11 | 28 | 29 | 30 | |
Note: Not reviewed.
The patients showed a greater frequency of mucus in the stool (28/32; 87.5%), abdominal pain (24/32; 75%), and vomiting (19/32; 59.3%) than of other clinical symptoms (Table 4). Table 3 shows the numerous clinical symptoms identified in this study and in other published papers. Our analysis showed differences in the clinical symptoms between various bacterial infections, but many overlaps were also evident. Hence, a laboratory-based identification of DEC is still required for etiological verification of this diarrheal disease. Nevertheless, pediatricians should seriously consider EHEC as an etiological agent of diarrhea when blood is found in stool specimens.
Table 4.
Frequency of Clinical Symptoms in Pediatric Patients with E. coli Pathotypes
| E. coli Pathotypes | Mucus in the Stool | Abdominal Pain | Vomiting | Fever | Blood in the Stool | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DEC | 28/32 (87.5%) | 24/32 (75%) | 19/32 (59.3%) | 17/32 (53.1%) | 10/32 (31.2%) | ||||||
| EAEC | 10/11 (90.9%) | 9/11 (81.8%) | 8/11 (72.7%) | 7/11 (63.6%) | 4/11 (36.3%) | ||||||
| EPEC | 9/9 (100%) | 4/9 (44.4%) | 5/9 (55.5%) | 4/9 (44.4%) | 1/9 (11.1%) | ||||||
| ETEC | LT | 5/6 (83.3%) | 3/4 (75%) | 5/6 (83.3%) | 3/4 (75%) | 2/6 (33.3%) | 1/4 (25%) | 2/6 (33.3%) | 1/4 (25%) | 0% | 0% |
| ST | 2/2 (100%) | 2/2 (100%) | 1/2 (50%) | 1/2 (50%) | 0% | ||||||
| EIEC | 3/3 (100%) | 3/3 (100%) | 3/3 (100%) | 3/3 (100%) | 2/3 (66.6%) | ||||||
| Stx negative EHEC O157:H7 | 1/3 (33.3%) | 3/3 (100%) | 1/3 (33.3%) | 1/3 (33.3%) | 3/3 (100%) | ||||||
Phenotypic and Genotypic Antibiotic Resistance Determination
The CLSI 2017 guidelines indicated greater antibiotic resistance rates for nalidixic acid (30/32; 93.7%), ampicillin (29/32; 90.6%), and tetracycline (25/32; 78.1%) than for any of the other antibiotics. All DEC isolates were susceptible to gentamicin and imipenem. DEC is a common agent in developing countries and is spread by contaminated water and food; therefore, its frequency and antimicrobial resistance are public health concerns.17 Full descriptions of the antibiotic resistance of DEC and E. coli pathotypes, from this study and others, are provided in Table 3. These results strongly suggest that nalidixic acid, ampicillin, tetracycline, and cotrimoxazole can no longer be empirically prescribed for the treatment of severe diarrhea and dysentery in central Iran. Since 1960, cotrimoxazole has been one of the World Health Organization’s essential medicines, as it is widely available and easily affordable and shows a broad spectrum of activity against an extensive range of infections, including diarrhea.
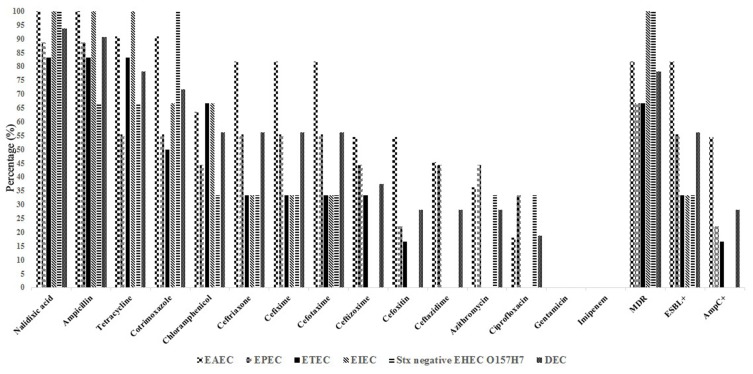
High levels of MDR (25/32; 78.1%), ESBL (18/32; 56.2%), and AmpC (9/32; 28.1%) gene expression were observed for the DEC isolates (Table 5 and Figure 1). The most extensive MDR strains for our collection of DEC showed combined resistance to nalidixic acid, ampicillin, tetracycline, and third-generation cephalosporins. The frequency of ESBL positivity was higher in the EAEC pathotype (81.8%) than in the other ESBL-positive E. coli pathotypes.
Table 5.
Phenotypic Antibiotic Resistance Rates in E. coli Pathotypes
| Antibiotic | DEC n:32 | EAEC n:11 | EPEC n:9 | ETEC n:6 | EIEC n:3 | Stx Negative EHEC O157:H7 n:3 |
|---|---|---|---|---|---|---|
| Nalidixic acid | 30 (93.7%) | 11 (100%) | 8 (88.8%) | 5 (83.3%) | 3 (100%) | 3 (100%) |
| Ampicillin | 29 (90.6%) | 11 (100%) | 8 (88.8%) | 5 (83.3%) | 3 (100%) | 2 (66.6%) |
| Tetracycline | 25 (78.1%) | 10 (90.9%) | 5 (55.5%) | 5 (83.3%) | 3 (100%) | 2 (66.6%) |
| Cotrimoxazole | 23 (71.8%) | 10 (90.9%) | 5 (55.5%) | 3 (50%) | 2 (66.6%) | 3 (100%) |
| Chloramphenicol | 18 (56.2%) | 7 (63.6%) | 4 (44.4%) | 4(66.6%) | 2 (66.6%) | 1 (33.3%) |
| Ceftriaxone | 18 (56.2%) | 9 (81.8%) | 5 (55.5%) | 2 (33.3%) | 1 (33.3%) | 1 (33.3%) |
| Cefixime | 18 (56.2%) | 9 (81.8%) | 5 (55.5%) | 2 (33.3%) | 1 (33.3%) | 1 (33.3%) |
| Cefotaxime | 18(56.2%) | 9 (81.8%) | 5 (55.5%) | 2 (33.3%) | 1 (33.3%) | 1 (33.3%) |
| Ceftizoxime | 12 (37.5%) | 6 (54.5%) | 4 (44.4%) | 2 (33.3%) | 0 (0%) | 0 (0%) |
| Cefoxitin | 9 (28.1%) | 6 (54.5%) | 2 (22.2%) | 1 (16.6%) | 0 (0%) | 0 (0%) |
| Ceftazidime | 9 (28.1%) | 5 (45.4%) | 4 (44.4%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Azithromycin | 9 (28.1%) | 4 (36.3%) | 4 (44.4%) | 0 (0%) | 0 (0%) | 1 (33.3%) |
| Ciprofloxacin | 6 (18.7%) | 2 (18.1%) | 3 (33.3%) | 0 (0%) | 0 (0%) | 1 (33.3%) |
| Gentamicin | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Imipenem | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| MDR | 25 (78.1%) | 9 (81.8%) | 6 (66.6%) | 4 (66.6%) | 3 (100%) | 3 (100%) |
| ESBL+ | 18 (56.2%) | 9 (81.8%) | 5 (55.5%) | 2 (33.3%) | 1 (33.3%) | 1 (33.3%) |
| AmpC+ | 9 (28.1%) | 6 (54.5%) | 2 (22.2%) | 1 (16.6%) | 0 (0%) | 0 (0%) |
Abbreviations: MDR, multi-drug resistance; ESBL, extended spectrum-β-lactamase.
Figure 1.
Phenotypic antibiotic resistance rates in E. coli pathotypes.
Abbreviations: MDR, Multi-drug resistance; ESBL, Extended spectrum-β-lactamase.
MDR DEC has quickly spread worldwide and represents a serious menace to the proper management of diarrhea and dysentery in developing countries.18 The frequencies of MDR DEC in the present study and in others are summarized in Table 3. The high rate of MDR-resistant isolates in pediatric patients with diarrhea can lead to more frequent treatment failures. In addition, resistant isolates demand the prescription of broad-spectrum antibiotics for the empiric treatment of infections, adding tremendous costs to treatment of these infections.19
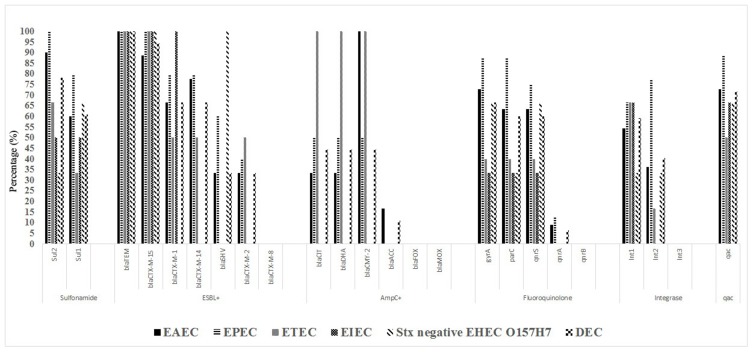
The isolates showed a higher frequency of the ESBL genes blaTEM (18/18; 100%), blaCTX-M15 (17/18; 94.4%), and AmpC [blaCIT (4/9; 44.4%) and blaDHA (4/9; 44.4%)] than of the other ESBL and AmpC genes (Table 6 and Figure 2). Table 3 shows the frequency of ESBL and its encoding genes in DEC and the other E. coli pathotype isolates. The differences in the frequencies of MDR, ESBL, and related genes across various regions may reflect different antibiotic usage patterns and different DEC community compositions.20
Table 6.
Frequency of Antibiotic Resistance Genes Among E. coli Pathotypes
| Resistance | Target Gene | DEC | EAEC | EPEC | ETEC | EIEC | Stx Negative EHEC O157:H7 |
|---|---|---|---|---|---|---|---|
| Sulfonamide | Sul2 | 18/23 (78.2%) | 9/10 (90%) | 5/5 (100%) | 2/3 (66.6%) | 1/2 (50%) | 1/3 (33.3%) |
| Sul1 | 14/23(60.8%) | 6/10 (60%) | 4/5 (80%) | 1/3 (33.3%) | 1/2 (50%) | 2/3 (66.6%) | |
| ESBL+ | blaTEM | 18/18 (100%) | 9/9 (100%) | 5/5 (100%) | 2/2 (100%) | 1/1 (100%) | 1/1 (100%) |
| blaCTX-M-15 | 17/18 (94.4%) | 8/9 (88.8%) | 5/5 (100%) | 2/2 (100%) | 1/1 (100%) | 1/1 (100%) | |
| blaCTX-M-1 | 12/18 (66.6%) | 6/9 (66.6%) | 4/5 (80%) | 1/2 (50%) | 1/1 (100%) | 0% | |
| blaCTX-M-14 | 12/18 (66.6%) | 7/9 (77.7%) | 4/5 (80%) | 1/2 (50%) | 0% | 0% | |
| blaSHV | 6/18 (33.3%) | 3/9 (33.3%) | 3/5 (60%) | 0% | 0% | 1/1 (100%) | |
| blaCTX-M-2 | 6/18 (33.3%) | 3/9 (33.3%) | 2/5 (40%) | 1/2 (50%) | 0% | 0% | |
| blaCTX-M-8 | 0% | 0% | 0% | 0% | 0% | 0% | |
| blaCIT | 4/9 (44.4%) | 2/6 (33.3%) | 1/2 (50%) | 1/1 (100%) | – | – | |
| blaDHA | 4/9 (44.4%) | 2/6 (33.3%) | 1/2 (50%) | 1/1 (100%) | – | – | |
| blaCMY-2 | 3/9 (44.4%) | 1/6 (100%) | 1/2 (100%) | 1/1 (100%) | – | – | |
| blaACC | 1/9 (11.1%) | 1/6 (16.6%) | 0% | 0% | – | – | |
| blaFOX | 0% | 0% | 0% | 0% | – | – | |
| blaMOX | 0% | 0% | 0% | 0% | – | – | |
| Fluoroquinolone | gyrA | 20/30 (66.6%) | 8/11 (72.7%) | 7/8 (87.5%) | 2/5 (40%) | 1/3 (33.3%) | 2/3 (66.6%) |
| parC | 18/30 (60%) | 7/11 (63.6%) | 7/8 (87.5%) | 2/5 (40%) | 1/3 (33.3%) | 1/3 (33.3%) | |
| qnrS | 18/30 (60%) | 7/11 (63.6%) | 6/8 (75%) | 2/5 (40%) | 1/3 (33.3%) | 2/3 (66.6%) | |
| qnrA | 2/30 (6.6%) | 1/11 (9%) | 1/8 (12.5%) | 0% | 0% | 0% | |
| qnrB | 0% | 0% | 0% | 0% | 0% | 0% | |
| Integrase | Int1 | 19/32 (59.3%) | 6/11 (54.5%) | 6/9 (66.6%) | 4/6 (66.6%) | 2/3 (66.6%) | 1/3 (33.3%) |
| Int2 | 13/32 (40.6%) | 4/11 (36.3%) | 7/9 (77.7%) | 1/6 (16.6%) | 0% | 1/3 (33.3%) | |
| Int3 | 0% | 0% | 0% | 0% | 0% | 0% | |
| Quaternary Ammonium Compounds | qac | 23/32 (71.8%) | 8/11 (72.7%) | 8/9 (88.8%) | 3/6 (50%) | 2/3 (66.6%) | 2/3 (66.6%) |
Figure 2.
Frequency of antibiotic resistance genes among E. coli pathotypes.
Among the PMQR determinants, qnrS, and qnrA were positive in 18/30 (60%) and 2/30 (6.6%) in nalidixic acid-resistant DEC strains, respectively and no case of qnrB was found. 20/30 (66.6%) isolates carrying PMQR contain similar mutations in gyrA at amino acid 83 (replacement of serine with leucine) and 18/30 (60%) parC at amino acid 80 (replacement of serine with isoleucine; GenBank accession no. HM068910). Although quinolones/fluoroquinolones are intended to be appropriate drugs against resistant isolates, the enhancement in antimicrobial resistance is a burden in controlling infections caused by DEC.21 In India, qnrS, qnrB, and qnrA were found at 46.6%, 40%, and 0% in the DEC strains, respectively.22 In Tehran (Iran), qnr was found at 72% in the DEC strains.23 These studies indicate that diffusion of qnr resistance genes among these DEC isolates is variable.
Class 1 and 2 integrons were observed in a total of 19/32 (59.3%) and 13/32 (40.6%) DEC strains: were observed. No Class 3 integrons were found (Table 6 and Figure 2). Of the three categories of integrons pertinent to antimicrobial resistance, the class I integron is the one most frequently obtained from Gram-negative bacteria.24 The prevalence of integrons in the enterobacteriaceae family has been shown to vary and has played a significant role in the spread of drug-resistance.25 In India Int1 had a prevalence of 41.6%, and no cases of Int2 or Int3 were found from DEC isolates.22 The class 1 integron is overcoming in outspreading the MDR genes among these DEC isolates despite the advent of class 2 and 3 integrons. In this study, 16/25 (64%) and 36% of MDR isolates of DEC contained Int1 and Int2 genes, respectively. In Iran (Southwest) MDR in intestinal pathogenic bacteria such as DEC is known to be related with presence.20
Conclusion
According to our pervious study, DEC is second only to Campylobacter jejuni in frequency when compared with other enteropathogens in pediatric patients with diarrhea from central Iran, and the abundance is greater for EAEC than for any other pathotype.15 Thus, pediatricians and, even more importantly, experts in laboratory diagnostics should pay special attention to the identification of these E. coli pathotypes. Pediatricians should keep these resistant bacteria in mind when treating pediatric patients with infectious diarrhea, and empiric antibiotic therapy should be adapted appropriately. Necessary measures must be taken to control the drug resistance of these bacteria.
Acknowledgments
The authors gratefully acknowledge the educational assistance of Arak University of Medical Sciences due to its financial contributions to and support of this study.
Ethics Approval and Consent to Participate
This study received ethical approval from the Arak University of Medical Sciences. Informed consent was obtained from a parent and/or guardian for participants under 16 years old. There was no access to any information that enabled authors to identify individual patient.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
AvB is an employee of bioMerieux, a company designing, developing and selling infectious disease tests. The company had no influence on the design and execution, either of the study or in the writing of the manuscript. The authors report no other conflicts of interest in this work.
References
- 1.Liu J, Platts-Mills JA, Juma J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet. 2016;388(10051):1291–1301. doi: 10.1016/S0140-6736(16)31529-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Troeger C, Colombara DV, Rao PC, et al. Global disability-adjusted life-year estimates of long-term health burden and undernutrition attributable to diarrhoeal diseases in children younger than 5 years. Lancet Glob Health. 2018;6(3):e255–e269. doi: 10.1016/S2214-109X(18)30045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canizalez-Roman A, Flores-Villaseñor HM, Gonzalez-Nuñez E, et al. Surveillance of diarrheagenic Escherichia coli strains isolated from diarrhea cases from children, adults and elderly at Northwest of Mexico. Front Microbiol. 2016;7:1924. doi: 10.3389/fmicb.2016.01924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee GY, Jang HI, Hwang IG, Rhee MS. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int J Food Microbiol. 2009;134(3):196–200. doi: 10.1016/j.ijfoodmicro.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 5.Ifeanyi CIC, Ikeneche NF, Bassey BE, Al-Gallas N, Aissa RB, Boudabous A. Diarrheagenic Escherichia coli pathotypes isolated from children with diarrhea in the Federal Capital Territory Abuja, Nigeria. J Infect Dev Ctries. 2015;9(02):165–174. doi: 10.3855/jidc.5528 [DOI] [PubMed] [Google Scholar]
- 6.Mansour Amin MS, Javaherizadeh H, Motamedifar M, et al. Antibiotic resistance pattern and molecular characterization of extended-spectrum β-lactamase producing enteroaggregative Escherichia coli isolates in children from southwest Iran. Infect Drug Resist. 2018;11:1097. doi: 10.2147/IDR.S167271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2018;61:185–188. doi: 10.1016/j.meegid.2018.04.005 [DOI] [PubMed] [Google Scholar]
- 8.Abbasi E, Abtahi H, van Belkum A, Ghaznavi-Rad E. Multidrug-resistant Shigella infection in pediatric patients with diarrhea from central Iran. Infect Drug Resist. 2019;12:1535. doi: 10.2147/IDR.S203654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel J, Weinstein M, Eliopoulos G, Jenkins S, Lewis J, Limbago B. M100 Performance standards for antimicrobial susceptibility testing. CLSI; 2017:240. [Google Scholar]
- 10.Mohammadzadeh M, Goudarzi H, Dabiri H, Fallah F. Characterization of enteropathogenic Escherichia coli associated with diarrhea among Iranian infants. Arch Pediatr Infect Dis. 2017;5(1). [Google Scholar]
- 11.Jafari F, Garcia-Gil L, Salmanzadeh-Ahrabi S, et al. Diagnosis and prevalence of enteropathogenic bacteria in children less than 5 years of age with acute diarrhea in Tehran children’s hospitals. J Infect. 2009;58(1):21–27. doi: 10.1016/j.jinf.2008.10.013 [DOI] [PubMed] [Google Scholar]
- 12.Ndlovu T, Le Roux M, Khan W, Khan S, van Overbeek LS. Co-detection of virulent Escherichia coli genes in surface water sources. PLoS One. 2015;10(2):e0116808. doi: 10.1371/journal.pone.0116808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia X, Meng J, McDermott PF, et al. Presence and characterization of Shiga toxin-producing Escherichia coli and other potentially diarrheagenic E. coli strains in retail meats. Appl Environ Microbiol. 2010;76(6):1709–1717. doi: 10.1128/AEM.01968-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrivastava AK, Kumar S, Mohakud NK, Suar M, Sahu PS. Multiple etiologies of infectious diarrhea and concurrent infections in a pediatric outpatient-based screening study in Odisha, India. Gut Pathog. 2017;9(1):16. doi: 10.1186/s13099-017-0166-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasi E, Belkum A, Ghaznavi-Rad E. Quinolone and macrolide-resistant campylobacter jejuni in pediatric gastroenteritis patients from Central Iran. Microb Drug Resist. 2019;25(7):1080–1086. doi: 10.1089/mdr.2018.0455 [DOI] [PubMed] [Google Scholar]
- 16.Aminshahidi M, Arastehfar A, Pouladfar G, Arman E, Fani F. Diarrheagenic Escherichia coli and Shigella with high rate of extended-spectrum Beta-lactamase production: two predominant etiological agents of acute diarrhea in Shiraz, Iran. Microb Drug Resist. 2017;23(8):1037–1044. doi: 10.1089/mdr.2017.0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes TA, Elias WP, Scaletsky IC, et al. Diarrheagenic escherichia coli. Braz J Microbiol. 2016;47:3–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeighami H, Haghi F, Hajiahmadi F, Kashefiyeh M, Memariani M. Multi-drug-resistant enterotoxigenic and enterohemorrhagic Escherichia coli isolated from children with diarrhea. J Chemother. 2015;27(3):152–155. doi: 10.1179/1973947813Y.0000000161 [DOI] [PubMed] [Google Scholar]
- 19.Van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin. 2016;30(2):377–390. doi: 10.1016/j.idc.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kargar M, Mohammadalipour Z, Doosti A, Lorzadeh S, Japoni-Nejad A. High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in southwest of Iran. Osong Public Health Res Perspect. 2014;5(4):193–198. doi: 10.1016/j.phrp.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namboodiri SS, Opintan JA, Lijek RS, Newman MJ, Okeke IN. Quinolone resistance in Escherichia coli from Accra, Ghana. BMC Microbiol. 2011;11(1):44. doi: 10.1186/1471-2180-11-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natarajan M, Kumar D, Mandal J, Biswal N, Stephen S. A study of virulence and antimicrobial resistance pattern in diarrhoeagenic Escherichia coli isolated from diarrhoeal stool specimens from children and adults in a tertiary hospital, Puducherry, India. J Health Popul Nutr. 2018;37(1):17. doi: 10.1186/s41043-018-0147-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heidary M, Momtaz H, Madani M. Characterization of diarrheagenic antimicrobial resistant Escherichia coli isolated from pediatric patients in Tehran, Iran. Iran Red Crescent Med J. 2014;16(4). doi: 10.5812/ircmj.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang HY, Jeong YS, Oh JY, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005;55(5):639–644. doi: 10.1093/jac/dki076 [DOI] [PubMed] [Google Scholar]
- 25.Deng Y, Bao X, Ji L, et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14(1):45. doi: 10.1186/s12941-015-0100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo W, Shan K, Xu B, Li J. Determining the resistance of carbapenem-resistant Klebsiella pneumoniae to common disinfectants and elucidating the underlying resistance mechanisms. Pathog Glob Health. 2015;109(4):184–192. doi: 10.1179/2047773215Y.0000000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Jing H, Lai S, et al. Etiology of diarrhea among children under the age five in China: results from a five-year surveillance. J Infect. 2015;71(1):19–27. doi: 10.1016/j.jinf.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeed A, Abd H, Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum, Sudan. J Med Microbiol. 2015;64(4):432. doi: 10.1099/jmm.0.000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miri ST, Dashti A, Mostaan S, Kazemi F, Bouzari S. Identification of different Escherichia coli pathotypes in north and north-west provinces of Iran. Iran J Microbiol. 2017;9(1):33. [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Chen X, Zheng S, et al. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China. Clin Microbiol Infect. 2014;20(1):52–58. doi: 10.1111/1469-0691.12188 [DOI] [PubMed] [Google Scholar]