Abstract
The only enzyme in the glycosphingolipid (GSL) metabolic pathway, which produces glucosylceramide (GlcCer) de novo is UDP-glucose ceramide glucosyltransferase (UGCG). UGCG is linked to pro-cancerous processes such as multidrug resistance development and increased proliferation in several cancer types. Previously, we showed an UGCG-dependent glutamine metabolism adaption to nutrient-poor environment of breast cancer cells. This adaption includes reinforced oxidative stress response and fueling the tricarboxylic acid (TCA) cycle by increased glutamine oxidation. In the current study, we investigated glycolytic and oxidative metabolic phenotypes following UGCG overexpression (OE). UGCG overexpressing MCF-7 cells underwent a metabolic shift from quiescent/aerobic to energetic metabolism by increasing both glycolysis and oxidative glucose metabolism. The energetic metabolic phenotype was not associated with increased mitochondrial mass, however, markers of mitochondrial turnover were increased. UGCG OE altered sphingolipid composition of the endoplasmic reticulum (ER)/mitochondria fractions that may contribute to increased mitochondrial turnover and increased cell metabolism. Our data indicate that GSL are closely connected to cell energy metabolism and this finding might contribute to development of novel therapeutic strategies for cancer treatment.
Subject terms: Breast cancer, Glycobiology, Cancer metabolism
Introduction
An essential hallmark of neoplastic diseases is persistent and uncontrolled cell proliferation. According adjustments of cellular energy metabolism provide, amongst others, sufficient macromolecule levels and energy in the form of adenosine triphosphate (ATP) to fuel cell growth and division. To understand the underlying molecular mechanisms of these metabolic changes is the first step to develop new therapeutic strategies for cancerous diseases. Glycosphingolipids (GSL) are not only important membrane components, but also act as signaling molecules in physiological and pathophysiological processes such as apoptosis and proliferation (reviewed in1,2). Numerous studies show specific expression of various GLS in particular cancers (reviewed in2) such as ganglioside GD2 in breast cancer3. Glycosylated sphingolipids cluster in the plasma membrane leading to the formation of GLS enriched microdomains (GEMs). These dynamic aggregations of sphingolipids, cholesterol and proteins are functional clusters and provide signaling platforms for membrane proteins, which are regulated by the lipid composition of the GEM (reviewed in4). Lipid microdomains are also present in the membranes of subcellular organelles modulating cytoplasmic pathways such as apoptosis (reviewed in5). Previous studies revealed that UDP-glucose ceramide glucosyltransferase (UGCG) overexpression (OE) leads to alterations of GEM composition in breast cancer cells resulting in signaling pathway activation and subsequently altered gene expression6. UGCG is a Golgi apparatus-residing enzyme that transfers an UDP-glucose molecule to ceramide to produce glucosylceramide (GlcCer), which is the precursor for all complex GSL. UGCG OE was reported in various cancers7 and is related to poor prognosis for breast cancer patients8 (reviewed in9). Otto Warburg was the first, who described aberrant characteristics of cancer cell energy metabolism as compared to non-tumor cells10,11. Specifically reprogramming of glucose metabolism to increased glycolysis, despite sufficient oxygen supply, and subsequent increased glucose consumption were observed in tumor tissues (reviewed in12). In the last years the attention was also drawn to mitochondria. Impairment of mitochondrial respiration was thought to be the reason for increased aerobic respiration of cancer cells and cancer development, but several studies showed that this is not the case for all cancer types (reviewed in13). Furthermore, it is now established that mitochondrial respiration defects are not generally the cause of reinforced aerobic glycolysis. Rather specific tumors, which are mostly glycolytic, retain a high mitochondrial respiration capacity (reviewed in13). Mitochondria are not only biosynthetic centers, for example by producing energy in form of ATP, but also are crucial signaling hubs. The organelles use various substrates from the cytoplasm to drive for example the tricarboxylic acid (TCA) cycle, mitochondrial membrane potential, fatty acid oxidation as well as de novo lipid synthesis (reviewed in13). Reactive oxygen species (ROS), which are mostly generated as a biproduct of the electron transport chain, are pro-tumorigenic and elevated levels are associated with cancer (reviewed in14). But ROS also act as signaling molecules for example by hypoxia inducible factor-1 (HIF-1) activation, which influences cellular proliferation15. Furthermore, mitochondria are important apoptosis regulators via the B-cell lymphoma protein (Bcl-2) family and associated proteins16 and maintain calcium homeostasis17. While most mitochondrial proteins are encoded by nuclear genes, mitochondria possess a small DNA genome (mtDNA) that encodes proteins essential for respiration, transfer RNAs and ribosomal RNAs. Mitochondrial morphology is regulated by various cellular pathways like mitogen activated protein kinases (MAPK), phosphoinositid-3-Kinase-Akt (PI3K-Akt) and myelocytomatosis (MYC) (reviewed in18). They form a network of long interconnected tubules and continually undergo fission and fusion. Mitochondria share nutrients, mtDNA and electron transport chain components by fusion and they divide to be distributed to daughter cells during mitosis or to be able to migrate to regions of higher energy demand (reviewed in18). Fission additionally facilitates mitophagy (reviewed in18). Mitochondria are tightly associated with membrane structures of the endoplasmic reticulum (ER). It was shown that these contact sites are functionally linked to diverse physiologic processes such as ATP production, apoptosis and mitochondrial dynamics (reviewed in5). Several studies have proven that alterations of mitochondrial biogenesis, dynamics and degradation are linked to diverse pathologies including cancer progression. Novel diagnostic and therapeutic approaches are already targeting mitochondrial redox homeostasis, TCA cycle, oxidative phosphorylation (OXPHOS) proteins or mitochondrial dynamics (reviewed in13). One example is dynamin related protein1 (DRP1), whose inhibition is currently under investigation. DRP1 is essential for mitochondrial fission and its blocking leads to reduced growth of glioblastoma cancer stem cells19 in vitro and lung adenocarcinoma cells in vivo20. In addition, DRP1 blockage inhibits tumor sphere formation of breast cancer and melanoma cells21. Since we discovered that OE of UGCG, a key enzyme of GSL metabolism, leads to increased cellular proliferation of breast cancer cells6, we were interested in the molecular mechanisms which link GSL to cellular energy metabolism. In previous studies, we were able to show that glutamine is used for reinforced oxidative stress response via glutathione production and fuels the TCA cycle to sustain the proliferative advantage of UGCG overexpressing breast cancer cells22.
In the current study we show that UGCG OE increases substrate oxidation and OXPHOS in breast cancer cells. OE of UGCG transforms cells from a quiescent/aerobic state to an energetic state, which is not ascribable to increased mitochondrial mass. Alterations of the sphingolipid composition of the ER/mitochondria fractions seem to influence OXPHOS protein activity leading to increased OXPHOS, which is accompanied by an accelerated mitochondrial turnover. Our data indicate that GSL are closely connected to cell energy metabolism, which is to our knowledge a novel finding. This finding might contribute to development of novel therapeutic strategies for cancer treatment.
Material and Methods
Cell culture
Human MCF-7 breast cancer cell line was obtained from the Health Protection Agency (European Collection of Cell Cultures EACC, Salisbury, UK). Cells were cultured as described previously22 and stably transfected cells were selected by supplementing 200 µg/ml G418 (Thermo Fisher Scientific, Waltham, USA).
Generation of stable UGCG expressing cells
The UGCG expression plasmid (pCMV6-ENTRY vector, OriGene Technologies Inc., Rockville, USA) (MCF-7/UGCG OE) and the empty control plasmid (MCF-7/empty) were stably transfected into MCF-7 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, USA), transfected cells were selected over five weeks using G418 as previously described6.
Substrate competition assay
Tracer measurements were performed as described in23. Briefly, 1.5 ×104 cells/24 well were seeded in cell culture media (a day prior to the assay) and then incubated in Krebs Ringer Phosphate (KRP) nutrient buffer containing either D-[3-3H] glucose or D-[14C (U)] glucose. Substrate oxidation was measured capturing evolved 14CO2. For glycolysis measurements, D-[3-3H] glucose was separated from tritiated [3H]2O by diffusion. To quantify tracer oxidation, media was acidified and 14CO2 trapped via reaction with 0.1 ml KOH before liquid scintillation spectrometry. To quantify tracer incorporation into cellular lipids, a chloroform–methanol (2:1 vol./vol.) extraction was performed and fractions assayed by scintillation spectrometry.
Measurement of mitochondrial respiration and glycolysis
The Seahorse XFe Analyzer (Agilent Technologies, Santa Clara, USA) was used to simultaneously measure the oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) in real-time. Cells were seeded and treated as described in24. Instead of carbonyl cyanide m-chlorophenylhydrazone (CCCP), cells were treated with 10 µM N5,N6-bis(2-fluorophenyl)-[1,2,5]oxadiazolo[3,4-b]pyrazine-5,6-diamine (BAM15) (Cayman Chemical, Ann Arbor, USA).
Quantification of mitochondrial ROS
0.5 ×104 cells/96-well (black) were seeded and incubated for 24 h at 37 °C. Subsequently, cells were incubated with 5 μM Red Mitochondrial Superoxide Indicator for live-cell imaging (MitoSOX) (Thermo Fisher Scientific, Waltham, USA) for 30 min at 37 °C. Following a three-time washing step with PBS, fluorescence was detected on a Fluostar plate reader with excitation at 510 nm and emission at 590 nm. The values were background corrected (only medium).
Comparison of mitochondrial mass using nonyl acridine orange (NAO)
Cells were seeded in 6 cm dishes and treated with 100 nM NAO the following day. After 15 minutes of incubation, cells were harvested by trypsin and pelletized. After one washing step in in PBS, mitochondrial staining of 100,000 cells per sample was analyzed via FITC channel with the BD FACS Canto II flow cytometer and the BD FACSDiva software (BD Biosciences, Franklin Lakes, USA). Data were evaluated using FlowJo software (FlowJo LLC, Ashland, USA) and related to MCF-7/empty control cells.
Protein concentration determination by Western blot analysis
Western Blot analysis was applied to investigate the relative levels of the five mitochondrial OXPHOS complexes and mitochondrial fission and fusion enzymes of MCF-7/empty and UGCG overexpressing cells. Cells were harvested and the pellet was resuspended in PhosphoSafe buffer (EMD Chemicals Inc. Billerica, USA), 2 mM DTT (AppliChem GmbH, Darmstadt, Germany), 1 x Roche Complete (Roche, Mannheim, Germany), pH 7.4 supplemented with 1% 100 X Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, Darmstadt, Germany). After sonification, the lysate was centrifuged (14,000 x g, 10 min, 4 °C). The protein concentration in the supernatant was determined via the Bradford method. 40–75 µg total protein extract (heated only at 50 °C for 5 minutes because of the heat sensitivity of the Complex IV unit) were separated by 12 or 15% SDS-PAGE and electro-blotted onto a nitrocellulose membrane (Amersham Protran, GE Healthcare Life Sciences, Freiburg, Germany). Ponceau staining (0.5% in 1% acetic acid) was applied to verify protein transfer. After 90 minutes incubation in 5% milk powder diluted in PBS with 0.1% Tween 20 (PBST) for OXPHOS proteins and 1:1 dilution of PBST and Intercept (PBS) Blocking Buffer for fission and fusion enzymes (LI-COR Biosciences, Bad Homburg, Germany), the membrane was incubated with the primary antibody (Total OXPHOS Rodent WB Antibody Cocktail (ab110413, Abcam, Cambridge, UK) (1:250 in 1% milk powder in PBST, two days, 4 °C) or antibodies from the Mitochondrial Dynamics Antibody Sampler Kit (#48799 Cell Signaling Technology, Cambridge, UK) (1:1000 (MFN1, MFN2, OPA1, MFF, DRP1 and TOM20) or 1:200 (Phospho-DRP1 (Ser616)) in 1:1 dilution of PBST and Intercept (PBS) Blocking Buffer (LI-COR Biosciences, Bad Homburg, Germany). The IRDye680 conjugated secondary antibody (LI-COR Biosciences, Bad Homburg, Germany), was used for all proteins except phospho-DRP1 (Ser616) (1:10,000 in blocking solution, one hour at RT). Fluorescence emission and densitometric analysis was performed using the Odyssey Infrared Scanner (LI-COR Biosciences, Bad Homburg, Germany) and the Image Studio Lite software (LI-COR Biosciences, Bad Homburg, Germany). The protein concentration was related to Ponceau staining intensity or heat shock protein 90 (HSP90) as a housekeeping protein (1:1000; 30 minutes incubation (BD Biosciences, Franklin Lakes, New Jersey, USA), secondary antibody IRDye 800 conjugated (LI-COR Biosciences, Bad Homburg, Germany), 60 minutes incubation). For phospho-DRP1 (Ser616) protein detection, the anti-rabbit IgG, HRP-linked secondary antibody (#7074 Cell Signaling Technology, Cambridge, UK) and the Anti-Mouse IgG (whole molecule)–Peroxidase antibody (produced in rabbit, #A9044, Sigma-Aldrich, St. Louis, USA) for HSP90 detection was applied. Protein concentration was detected via enhanced chemiluminescence method using Pierce ECL Western Blotting Substrate (#32106, Thermo Fisher Scientific, Waltham, USA).
Analysis of mitochondrial DNA copy numbers per cell
The NovaQUANT Human Mitochondrial to Nuclear DNA Ratio Kit (Merck KGaA, Darmstadt, Germany) and SYBR select Master Mix (Thermo Fisher Scientific, Waltham, USA) were used to compare the nuclear to mitochondrial DNA ratio. The qRT-PCR based kit contains primer pairs targeting two mitochondrial (ND1 and ND6) and two nuclear genes (BECN1 and NEB). By calculating ratios of the Ct values of BECN1/ND1 and NEB/ND6, the mtDNA copy number per cell was determined. DNA was isolated using KAPA Express Extract Kit (Kapa Biosystems, Wilmington, USA) and 2 ng DNA per reaction well were applied.
Quantitative real-time PCR (qRT-PCR)
Quantitative real-time PCR (qRT-PCR) was performed as described previously22. Briefly, the RNeasy Mini Kit (QIAGEN, Hilden, Germany) was used to isolate total RNA. 300 ng RNA were applied to synthesize cDNA using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, USA). Gene-specific PCR products were quantified utilizing 5 X QPCR Mix EvaGreen (ROX) (Bio&SELL, Feucht, Germany) on a QuantStudio 5 Real-time PCR System (Thermo Fisher Scientific, Waltham, USA). Relative mRNA expression was calculated according to the Δct method. Relative values were normalized to 60 S ribosomal protein L37a (RPL37A) expression level housekeeping gene. All primers, as listed in Table 1, were purchased from Eurofins (Luxembourg, Luxembourg).
Table 1.
Primer for qRT-PCR.
| Gene | Forward primer 5′ → 3′ | Reverse primer 5′ → 3′ | Amplicon (bp) |
|---|---|---|---|
| RPL37A | attgaaatcagccagcacgc | aggaaccacagtgccagatcc | 94 |
| MFN1 | atgacctggtgttagtagacagt | agacatcagcatctaggcaaaac | 90 |
| MFN2 | cacatggagcgttgtaccag | ttgagcacctccttagcagac | 104 |
| PARK2 | gtgtttgtcaggttcaactcca | gaaaatcacacgcaactggtc | 129 |
| FIS1 | gatgacatccgtaaaggcatcg | agaagacgtaatcccgctgtt | 82 |
| PINK | cccaagcaactagcccctc | ggcagcacatcagggtagtc | 107 |
| OGDH | tgccagcatattggggtgg | ggaactcctcaaacctggtgg | 160 |
| PDHA1 | tggtagcatcccgtaattttgc | attcggcgtacagtctgcatc | 151 |
| PDHB | aagaggcgctttcactggac | actaaccttgtatgccccatca | 153 |
| GOT1 | atttcttagcgcgttggtaca | acacagcattgtgattctccc | 90 |
| GOT2 | aagagtggccggtttgtcac | agaaagacatctcggctgaact | 107 |
| LDHA | atggcaactctaaaggatcagc | ccaaccccaacaactgtaatct | 86 |
| GOLPH3 | aggaagccgttcttgacaaatg | ggcatgagccaggtaaatgag | 83 |
| OPA1 | tcaagaaaaacttgatgctttca | gcagagctgattatgagtacgatt | 78 |
Measurement of mitochondrial membrane potential
To determine mitochondrial membrane depolarization of MCF-7/UGCG OE and MCF-7/empty cells, the cationic dye JC-1 that exhibits potential-dependent accumulation in mitochondria, was applied. Cells were seeded in 6 cm dishes and the next day, at 80% confluency, cells were stimulated with 2 µg/ml JC-1 for 20 minutes. Subsequently, cells were washed with PBS, harvested by trypsin and pelletized. The pellet was resuspended in 250 µl PBS and 100,000 cells per sample were measured using the BD FACS Canto II flow cytometer (monomer form: 514/529 nm; J-aggregate form: 585/590 nm) and the BD FACSDiva software (BD Biosciences, Franklin Lakes, USA). Data were evaluated using FlowJo software (FlowJo LLC, Ashland, USA).
Sphingolipid level analysis of ER/mitochondria fractions
To analyze the lipid composition of the ER/mitochondria fractions, mitochondria from 1.5 ×107 freshly harvested cells were isolated using the Qproteome Mitochondria Isolation Kit (Cat. no. 37612, QIAGEN, Hilden, Germany). All steps were conducted according to the manufacturer’s protocol not including the high-purity preparations. The fraction purity was confirmed via Western Blot analysis. The sphingolipid concentrations of the isolated ER/mitochondria fractions were determined by liquid chromatography tandem mass spectrometry (LC-MS/MS) as described previously6. Levels are related to the protein concentrations of the cytosolic fractions and normalized to MCF-7/empty control cells.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 7 software. Data are presented as mean ± standard error of the mean (SEM). Significant differences in means between two groups were assessed by unpaired t-test with Welch’s correction and by two-way ANOVA with a Bonferroni multi-comparison post-test for more than two groups.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Results
UGCG overexpression leads to a metabolic shift from an aerobic/quiescent to an energetic cell type
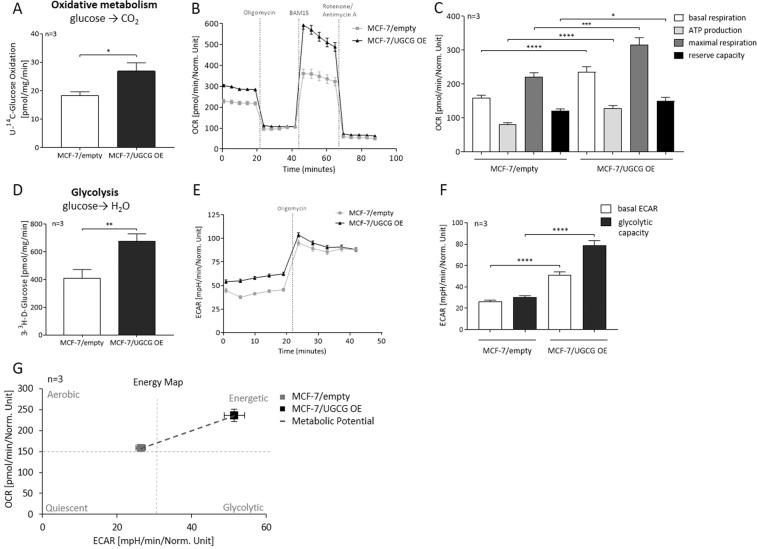
In previous studies we showed increased glutamine uptake and reinforced glutamine oxidation following UGCG overexpression (OE) in MCF-7 cells22. In this study we performed a substrate competition assay with [U-14C]-labelled glucose to track the metabolism of glucose-derived carbons in MCF-7/UGCG OE and control cells. UGCG OE led to an increased level of captured 14CO2, which demonstrates that UGCG OE increases glucose oxidation (Fig. 1A). The basal mitochondrial respiration, which is represented by oxygen consumption rate (OCR), was significantly increased in MCF-7/UGCG OE cells as compared to control cells (Fig. 1B,C). Likewise, ATP production (basal respiration − respiration after oligomycin injection) was significantly increased and the maximal respiration rate (respiration after N5,N6-bis(2-fluorophenyl)-[1,2,5]oxadiazolo[3,4-b]pyrazine-5,6-diamine (BAM15) injection − respiration after antimycin a/rotenone injection) was significantly increased in MCF-7/UGCG OE cells as compared to control cells (Fig. 1C). The maximal respiration indicates the ability of the cellular respiration to respond to an increased energy demand. Additionally, the reserve capacity (respiration after BAM15 injection − basal respiration) was significantly increased in MCF-7/UGCG OE cells as compared to control cells (Fig. 1C). The addition of 3-3H-D-glucose to the media and subsequent measurement of evolved 3H2O shows increased glycolysis following UGCG OE (Fig. 1D). In line with these data, the basal extracellular acidification rate (ECAR) (determined by the net production and extrusion of protons into the medium during lactate production from glucose) was significantly increased in UGCG overexpressing cells (Fig. 1E,F). Moreover, the glycolytic capacity (deduced from ECAR after oligomycin treatment − basal ECAR) of MCF-7/UGCG OE cells was significantly increased as compared to control cells (Fig. 1F).
Figure 1.
UGCG overexpression enhances glycolysis and cellular respiration. (A) Substrate oxidation was measured by U-14C-glucose tracing experiments. The evolved 14CO2 was determined. Data are presented as mean of n = 3 ± standard error of the mean (SEM). (B) Representative graph of the oxygen consumption rate (OCR) determined by Seahorse XFe analyzer. (C) ATP production (basal respiration − respiration after oligomycin injection), maximal respiration rate (respiration after N5,N6-bis(2-fluorophenyl)-[1,2,5]oxadiazolo[3,4-b]pyrazine-5,6-diamine (BAM15) injection − respiration after antimycin a/rotenone injection), reserve capacity (respiration after BAM15 injection − basal respiration). Data are presented as mean of n = 3 ± SEM. (D) Anaerobic glycolysis was determined by using 3-3H-D-glucose and measuring the released labelled H2O. Data are presented as mean of n = 3 ± SEM. (E) Representative graph of the extracellular acidification rate (ECAR) quantified by Seahorse XFe analyzer. (F) Glycolytic capacity (deduced from ECAR after oligomycin treatment − basal ECAR). Data are presented as mean of n = 3 ± SEM. (G) Energy map of MCF-7/UGCG OE and control cells. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
In summary, following UGCG OE, MCF-7 cells transform from an aerobic/quiescence state to an energetic metabolic state (shift of metabolic potential) showed by increased glycolysis and oxidative phosphorylation (Fig. 1G).
UGCG overexpression leads to increased superoxide levels and does not impact mitochondrial mass
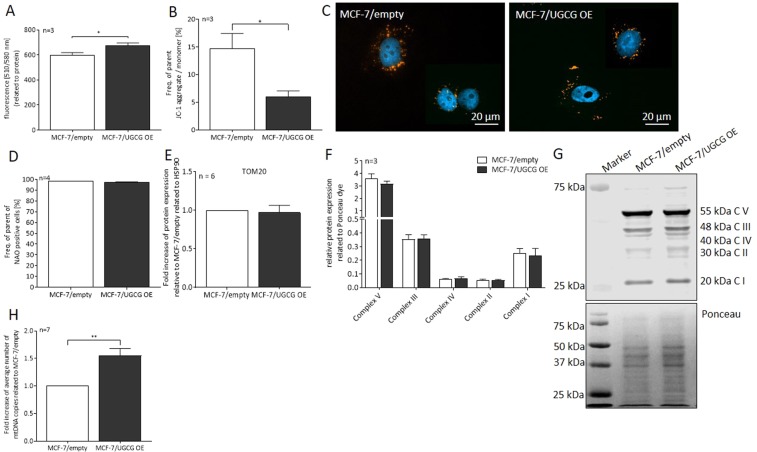
UGCG OE resulted in increased mitochondrial superoxide levels detectable as increased fluorescence of MitoSOX red dye (Fig. 2A). In addition, the mitochondrial membrane potential sensor 5,5,6,6’-tetrachloro-1,1’,3,3’ tetraethylbenzimidazoyl-carbocyanine iodide (JC-1) was detected by flow cytometry. We measured a significant decrease of JC-1 aggregate to monomer ratio, which implies an increase in mitochondrial depolarization (Fig. 2B). A representative image of mitochondrial JC-1 staining is shown in Fig. 2C. To exclude the possibility that the metabolic effects are mitochondrial mass dependent, we analyzed mitochondrial mass by several parameters in MCF-7/UGCG OE and control cells. Since the MitoTracker green reacts with ROS, we used the fluorescent mitochondrial dye nonyl acridine orange (NAO) for mitochondrial mass analysis25. We labelled the cells with NAO and analyzed vital mitochondria by flow cytometry. No significant differences between UGCG overexpressing and control cells could be detected (Fig. 2D). The protein level of the receptor translocase of the outer mitochondrial membrane 20 (TOM20), which is a part of the TOM complex that enables the import of proteins through the outer mitochondrial membrane, was not changed upon UGCG OE (Fig. 2E and supplemental data 1). The OXPHOS protein content was not significantly altered in UGCG overexpressing cells as compared to control cells (Fig. 2F,G and supplemental data 2). The results are confirmed by UGCG knockdown (Kd) experiments. MCF-7/UGCG Kd cells exhibit no significant altered total OXPHOS protein content, whereas significantly reduced complex V protein (ATP synthase) concentration could be detected (supplemental data 3). On the contrary, we detected a significantly increased mtDNA copy number in MCF-7/UGCG OE cells in comparison to control cells (Fig. 2H).
Figure 2.
Influence of UGCG overexpression on mitochondrial ROS and mass. (A) Mitochondrial reactive oxygen species (ROS) levels were quantified using the fluorogenic reagent MitoSOX. Data are presented as a mean of n = 3 ± SEM. (B) Mitochondrial membrane potential was analyzed by flow cytometry utilizing 5,5,6,6’-tetrachloro-1,1’,3,3’ tetraethylbenzimidazoyl-carbocyanine iodide (JC-1). Data are presented as a mean of n = 3 ± SEM. (C) Representative images of n = 3 of JC-1 staining by immunocytochemistry. From each of the biological samples at least 5 images were generated. (D) Mitochondrial mass determined by flow cytometry using nonyl acridine orange (NAO). Data are presented as a mean of n = 4 ± SEM. (E) Analysis of translocase of the outer mitochondrial membrane 20 (TOM20) protein concentration by Western blot analysis. Protein expression is related to the housekeeper Hsp90 and control cells. Data are presented as a mean of n = 6 ± SEM. (F) Densitometrical analysis of oxidative phosphorylation (OXPHOS) complexes I-V protein concentrations by Western blot analysis. Protein expression is related to Ponceau dye. Data are presented as a mean of n = 3 ± SEM. (G) Representative image of the OXPHOS protein Western Blot analysis (upper part) and Ponceau staining (lower part). (H) The mitochondrial DNA (mtDNA) copy number was determined using the NovaQUANT Human Mitochondrial to Nuclear DNA Ratio Kit. Data are related to MCF-7/empty and presented as a mean of n = 7 ± SEM. *p ≤ 0.05, **p ≤ 0.01.
Accordingly, our effects following UGCG OE are not ascribable to increased mitochondrial mass but might be due to increased OXPHOS protein activity.
Mitochondrial turnover is accelerated following UGCG overexpression
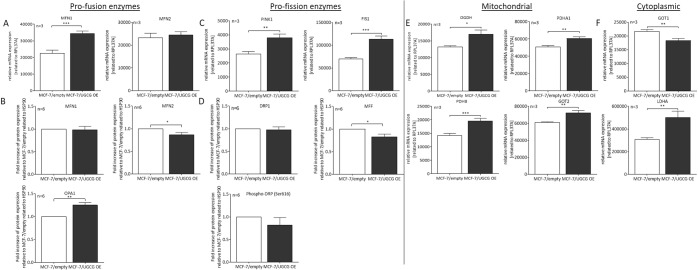
Next, we analyzed the expression of enzymes, which are involved in the process of mitochondrial fusion and fission by qRT-PCR and Western Blot analysis. We detected a significant increase of the mRNA level of mitofusin 1 (MFN1) in MCF-7/UGCG OE cells, whereas mitofusin 2 (MFN2) mRNA expression was unchanged (Fig. 3A). Contradictory, MFN1 protein level was not changed following UGCG OE (Fig. 3B and supplemental data 4), whereas MFN2 protein content was significantly decreased (Fig. 3B and supplemental data 5). Protein concentration of optic atrophy type 1 (OPA1) was significantly increased in MCF-7/UGCG OE cells as compared to control cells (Fig. 3B and supplemental data 6) and the results are confirmed on mRNA level (supplemental data 7). The mRNA level of the pro-fission enzymes PTEN-induced kinase 1 (PINK1), which accumulates in dysfunctional mitochondria, and mitochondrial fission protein 1 (FIS1) were significantly increased following UGCG OE (Fig. 3C), whereas no significant difference on Parkin 2 (PARK2) mRNA level could be detected (supplemental data 7). In addition, we determined total dynamin related protein 1 (DRP1) and total mitochondrial fission factor (MFF) and phospho-DRP1 (Ser616) protein level by Western blot analysis. No difference on total DRP1 (supplemental data 8) and phopho-DRP1 (Ser616) (supplemental data 9) protein level following UGCG OE could be detected (Fig. 3D). Total MFF protein levels are significantly reduced following UGCG OE (Fig. 3D and supplemental data 10). Phospho-MFF (Ser146) could not be detected (data not shown). Following Kd of UGCG in MCF-7 cells no significant differences on MFN1, PINK1, FIS1 and OPA1 mRNA expression levels are detectable as compared to control cells (supplemental data 11), which confirms UGCG-dependent increased mRNA expression of several proteins involved in the process of mitochondrial fusion and fission. In previous studies, UGCG OE led to an increase in glutamine metabolism in the TCA cycle22. To verify this data, we determined the mRNA levels of several mitochondrial enzymes involved in pyruvate processing and TCA cycle. The expression of the mitochondrial redox sensor alpha-ketoglutarate dehydrogenase (OGDH) is significantly increased in MCF-7/UGCG OE cells compared to MCF-7/empty control cells (Fig. 3E). OGDH catalyzes the conversion of alpha-ketoglutarate into succinyl CoA, which is the first step of the TCA cycle starting from glutamine. The pyruvate dehydrogenase E1 component subunit alpha (PDHA1) and -beta (PDHB) are units of the pyruvate dehydrogenase complex, which converts pyruvate to acetyl-CoA in the mitochondrial matrix. The mRNA level of both subunits is significantly increased in MCF-7/UGCG OE cells compared to MCF-7/empty control cells (Fig. 3E). The mitochondrial isoform 2 of the glutamate oxaloacetate transaminase (GOT2) is also significantly increased following UGCG OE (Fig. 3E) matching the increased aspartate metabolization from glutamate, which was shown by 13C5-glutamine labelling assay22. There is a significant decrease of the cytosolic GOT isoform 1 (GOT1) mRNA level in MCF-7/UGCG OE cells compared to MCF-7/empty cells (Fig. 3F). Furthermore, a significant increase of the cytosolic lactate dehydrogenase subunit A (LDHA) mRNA level following UGCG OE was measured (Fig. 3F). LDHA catalyzes the conversion of pyruvate to lactate. The increase is in line with the elevated ECAR in MCF-7/UGCG OE cells (Fig. 1E,F).
Figure 3.
UGCG overexpression impact on mitochondrial dynamics and mitochondrial and cytoplasmic enzymes. (A) The mRNA expression of the pro-fission enzymes mitofusin 1 (MFN1) and mitofusin 2 (MFN2) was determined by qRT-PCR analysis. The expression is related to the housekeeping gene (RPL37A). Data are presented as a mean of n = 3 ± SEM. (B) Protein concentration of MFN1, MFN2 and optic atrophy type 1 (OPA1) determined by Western blot analysis. Data are related to the housekeeping protein HSP90 and presented relative to MCF-7/empty control cells as a mean of n = 6 ± SEM. (C) mRNA levels of the pro-fission enzymes PTEN-induced kinase 1 (PINK1) and mitochondrial fission protein 1 (FIS1) related to RPL37A. Data are presented as a mean of n = 3 ± SEM. (D) Protein concentration determination of total dynamin related protein1 (DRP1), total mitochondrial fission factor (MFF) and phospho-DRP1 (Ser616) by Western blot analysis. Data are related to the housekeeping protein HSP90 and presented relative to MCF-7/empty control cells as a mean of n = 6 ± SEM. (E) mRNA expression analysis of alpha-ketoglutarate dehydrogenase (OGDH), pyruvate dehydrogenase E1 component subunit alpha (PDHA1), and beta (PDHB) and glutamate oxaloacetate transaminase 2 (GOT2) by qRT-PCR. The expression is related to RPL37A. Data are presented as a mean of n = 3 ± SEM. (F) mRNA expression analysis of glutamate oxaloacetate transaminase 1 (GOT1) and lactate dehydrogenase subunit A (LDHA) by qRT-PCR. The expression is related to RPL37A. Data are presented as a mean of n = 3 ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
UGCG leads to alterations of sphingolipid composition in the ER/mitochondria fraction
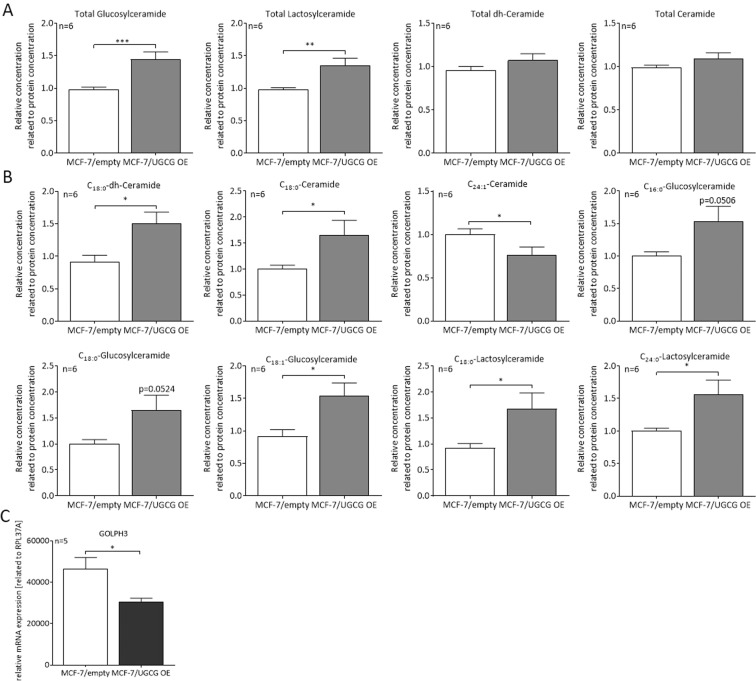
To explain the observed changes in mitochondria of MCF-7/UGCG OE cells, we compared the sphingolipid composition in endoplasmic reticulum (ER)/mitochondria fractions of MCF-7/UGCG OE and control cells. Since the ER is the place of de novo ceramide synthesis and ER and mitochondria are connected via specific ER-mitochondria contact sites, we did not isolate single ER and mitochondria fractions. Rather, we isolated ER/mitochondria fraction and the purity of the fraction was analyzed by Western blot analysis. The Western blot analysis results show high purity by protein disulfate-isomerase (PDI) (ER marker) and OXPHOS protein detection (supplemental data 12). Using liquid chromatography tandem mass spectrometry (LC-MS/MS), the sphingolipid levels of the ER/mitochondria fractions were quantified. A significant increase of total GlcCer (1.44-fold) and total lactosylceramide (LacCer) levels (1.35-fold) in MCF-7/UGCG OE cells as compared to control cells could be detected (Fig. 4A). The total dihydro (dh)-ceramide and total ceramide levels are unchanged following UGCG OE (Fig. 4A). In detail, the C18:0-dh-Cer, C18:0-Cer, C16:0-, C18:0-, C18:1-GlcCer and C24:0-LacCer levels are significantly increased in MCF-7/UGCG OE cells as compared to control cells (Fig. 4B). The C18:0-LacCer level is increased by tendency (p = 0.0512) in MCF-7/UGCG OE cells as compared to control cells (Fig. 4B). C24:1-Cer is the only sphingolipid, which is decreased in MCF-7/UGCG OE as compared to control cells (0.76-fold). All other analyzed sphingolipid concentrations are not significantly changed following UGCG upregulation (supplemental data 13). Our results are confirmed by data from MCF-7/UGCG Kd cells, which exhibit significantly lower total GlcCer, LacCer and ceramide levels, whereas total dh-ceramide levels are increased as compared to control cells (supplemental data 14). In addition, we determined the mRNA expression of the oncogene Golgi phosphoprotein 3 (GOLPH3), which localizes in the Golgi apparatus and is involved in vesicle exit for trafficking to the plasma membrane. The mRNA level of GOLPH3 is decreased in MCF-7/UGCG OE cells as compared to control cells (Fig. 4C).
Figure 4.
UGCG influences the lipid composition of ER/mitochondria fractions. (A) Total glucosylceramide, lactosylceramide, dihydro- (dh-) ceramide and ceramide levels of isolated ER/mitochondria fractions determined via LC-MS/MS. Data are presented as a mean of n = 6 ± SEM. (B) Levels of specific sphingolipids species following UGCG OE. Data are presented as a mean of n = 6 ± SEM. (C) Golgi phosphoprotein 3 (GOLPH3) mRNA level related to RPL37A determined by qRT-PCR. Data are presented as a mean of n = 5 ± SEM. *p ≤ 0.05, ***p ≤ 0.001.
Discussion
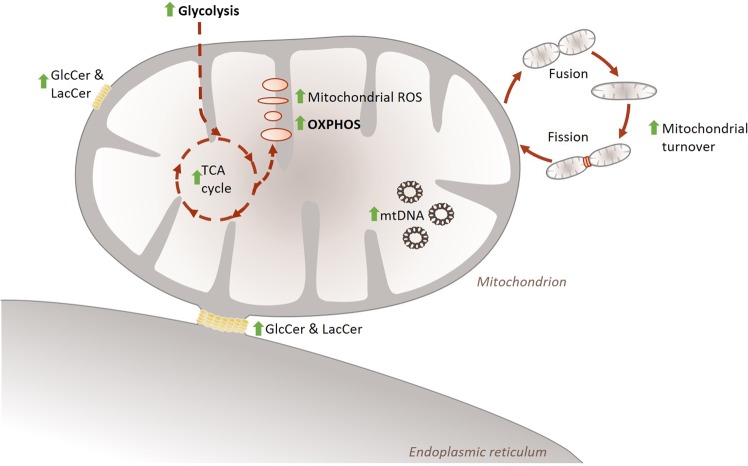
With this study we were able to show that UGCG overexpression (OE) mediates a metabolic transformation from a quiescent/aerobic to energetic breast cancer cell type indicated by increased glycolysis and OXPHOS. This is accompanied by changes in sphingolipid content in the ER/mitochondria fraction and increased mitochondrial turnover despite unchanged mitochondrial mass (Fig. 5).
Figure 5.
Schematic overview of the observed effects of UGCG overexpression on breast cancer cell metabolism. UGCG OE leads to increased glycolysis, fueled TCA cycle and increased OXPHOS. This leads to increased mitochondrial ROS level, which is accompanied by increased mitochondrial turnover. The effects are not ascribable to increased mitochondrial mass, but changes sphingolipid composition of ER/mitochondria fractions, which presumably contain ER-mitochondria contact sites. TCA = tricarboxylic acid, ROS = reactive oxygen species, OXPHOS = oxidative phosphorylation, GlcCer = glucosylceramide, LacCer = lactosylceramide, mtDNA = mitochondrial DNA, green arrow = increased in MCF-7/UGCG OE cells compared to control cells.
UGCG OE mediates increased glycolysis in breast cancer cells. Glycolysis provides not only energy, but also macro molecules to generate biomass to maintain cell proliferation. Increased glycolytic flux does not automatically mean impairment of OXPHOS (reviewed in26). Indeed, we measured increased superoxide (presumably evolved by increased OXPHOS) and decreased mitochondrial membrane potential following UGCG OE, but we could clearly show that the oxidative metabolism is not impaired, but rather increased in UGCG overexpressing cells (Fig. 5). This shows that the increased glycolysis does not need to restore mitochondrial dysfunction and that this contributes to increased proliferation of MCF-7/UGCG OE cells6. Kao et al. also observed increased basal respiration and maximal respiratory capacity accompanied by increased UGCG expression in drug resistant HL-60 cells27. By using sphingolipid enzyme inhibitors the authors showed sphingolipid level alteration-mediated changes of mitochondrial function27. We confirmed our previous results, which showed increased flux through the TCA cycle following UGCG OE22 by increased OGDH (catalyzes a rate-limiting step in the TCA cycle) and GOT2 mRNA level and decreased GOT1 mRNA level. OGDH is a mitochondrial redox sensor, which can undergo S-glutathionylation following an increase in H2O2 levels28. In addition, the pyruvate dehydrogenase mRNA is more expressed following UGCG OE, which supports fuel the TCA cycle. However, elevated 14CO2 level from the uniformly labeled [U14C]-labelled glucose could also, at least partly, result from an increase in flux through the pentose phosphate pathway. We could already show an increase in NADPH concentration following UGCG OE in MCF-7 cells22.
Since OXPHOS and TOM20 protein concentration and NAO-positive cell number is unchanged following UGCG OE, we assume that the effects on glycolysis and oxidative metabolism are not ascribable to increased mitochondrial mass. Interestingly, UGCG Kd in MCF-7 cells leads to reduced ATP synthase protein concentration indicating that the effect leading to increased ATP synthesis in MCF-7/UGCG OE cells is UGCG-dependent. However, MCF-7/UGCG OE cells exhibit a significantly higher mtDNA copy number than MCF-7/empty control cells. mtDNA exhibits high mutation rates compared to genomic DNA potentially due to persistent exposure to mutagenic ROS resulting from respiration (reviewed in29). Since MCF-7/UGCG OE cells exhibit increased mitochondrial ROS, it is likely that this is the cause for increased mtDNA amount. Hori et al. showed that redistribution of the electron transport chain during the fusion process leads to increased ROS level, which resulted in increased mtDNA copy number30. Increased mtDNA content provides sufficient mtDNA copies for persistently running of fusion and fission processes and prevents production of mitochondria without mtDNA (reviewed in31). The phenomenon of altered mtDNA content is present in various human malignancies (reviewed in32). mtDNA copy increase was shown in various cancer types such as colorectal carcinoma, prostate and ovarian cancer, while in for example hepatocellular carcinoma the mtDNA copy number is decreased (reviewed in32). In contrast to our data, Fan et al. showed a decrease in mtDNA copy number in breast cancer patients33. On the other hand, an increased mtDNA content was revealed in drug resistant human laryngeal cancer cells34.
We checked the expression of several enzymes involved in the process of mitochondrial fusion and fission to investigate whether or not one of the processes is favored following UGCG OE. Surprisingly, UGCG OE does not lead to increased fusion or fission but might be leading to increased mitochondrial turnover. This is shown by increased respectively decreased counteracting enzyme mRNA or protein expression and data are confirmed by results, which show no significant alterations on mRNA level of proteins involved in mitochondrial fusion and fission processes following UGCG Kd in MCF-7 cells as compared to control cells. PINK1 assesses the internal state of mitochondria by being quickly disposed when the inner membrane potential of the mitochondria is high. Unhealthy mitochondria fail to import and to degrade PINK1 leading to accumulation of the kinase on the cytosolic site, which represents mitochondrial dysfunction. Cytosolic site located PINK1 recruits Parkin leading to ubiquitination of outer mitochondrial membrane proteins (reviewed in35). Since PINK1 mRNA expression is increased following UGCG OE, it is possible that PINK1 synthesis is increased due to the decreased mitochondrial membrane potential we detected. Parkin mRNA expression is unaltered, but the protein could still be recruited by PINK1 to tag these mitochondria leading to increased fission rate. If this would be the case, we would have expected low MFN1 and OPA1 (pro-fusion proteins) production, because both proteins are blocked by the PINK1/Parkin signaling pathway. But we measured increased OPA1 protein concentration, whereas MFN2 is reduced on protein level. Pallanck discusses the model of PINK1 accumulation in damaged mitochondria leading to MFN2 phosphorylation, subsequent Parkin recruitment and mitochondrial degradation (reviewed in36). MFN2 is also considered anti-tumorigenic and expressed at low levels in various human cancers, including MCF-7 breast cancer cell line37. Following UGCG OE, MFN2 protein levels are even more decreased, which would indicate less mitochondrial damage and degradation respectively. This is also shown by increased oxidative metabolism in MCF-7/UGCG OE cells. That MFN1 and MFN2 exhibit functional differences is also shown by higher GTPase activity of MFN1 as compared to MFN2 and higher affinity of MFN2 for GTP as compared to MFN138. Increases of MFN1 and OPA1 were related to an increase in OCR and ATP levels in human normal dermal fibroblasts during aging39. Furthermore, FIS1 (pro-fission protein) mRNA expression is increased, whereas total DRP1 and phospho-DRP1 (Ser616) protein levels are unchanged. Phosphorylation of DRP1 at Ser616 usually promotes mitochondrial fission40. In summary, we cannot state whether or not the process of fusion or fission in mitochondria is increasingly executed. Presumably, increased superoxide levels following UGCG OE lead to reduced mitochondrial membrane potential and results in increased mitochondrial turnover (Fig. 5). Increased respiration, superoxide level and fusion-fission rate (increased OPA1 and DRP1 production) are also shown in K562 (chronic myelogenous leukemia) cells by Ruiz et al. The authors assume that increased mitochondrial turnover can lead to smaller, condensed and “boosted” mitochondria resulting in increased cell proliferation41.
We confirmed our previous results from the study in 20186, which showed an accumulation of the ER marker PDI in fraction 3, 5, 6, 7 and 9 (sucrose density centrifugation). These fractions also exhibit increased GlcCer level as compared to control cells6. Hence, together with the results of this study, we could show that UGCG OE leads to accumulation of GlcCer in specific regions of the plasma membrane and to GlcCer and LacCer accumulation in the ER/mitochondria fraction (Fig. 5) The observed UGCG mediated effects on mitochondrial enzyme activity (OXPHOS) might be triggered by changed lipid composition of the ER/mitochondria fraction, which contains ER-mitochondria contact sites. Intriguingly, UGCG Kd in MCF-7 cells abolishes the effect of UGCG OE on lipid composition in ER/mitochondria fractions and leads to reduced ATP synthase protein concentration in mitochondria. We assume that this finding leads to the effect of reduced cell proliferation of MCF-7/UGCG Kd cells, we showed in our study in 20186. In 2010, Ciarlo et al. showed that upon UGCG inhibition with [D]-PDMP, the recruitment of fission molecules to the mitochondria was impaired and consequently fission was reduced in lymphoid cells. Furthermore, apoptosis was decreased42. These findings likewise indicate an influence of the UGCG on mitochondrial raft-like structures, but also indicate a cell-type specific influence of UGCG on cellular energy metabolism. Since we already showed UGCG OE-dependent alterations of the GEM composition of the plasma membrane of MCF-7 cells6, it is likely that alterations of intracellular membranes are occurring as well. The functionally and structurally very different inner and outer mitochondrial membranes highly influence the function of the organelles (reviewed in43). Furthermore, it was shown that ER-mitochondria contact sites, which are important for mitochondrial dynamics and morphology, are influenced by lipid raft-like structures (reviewed in5). In addition, these contact sites coordinate the licensing of mtDNA replication, which is increased in MCF-7/UGCG OE cells with division to distribute newly replicated nucleosides to daughter mitochondria44. Even though the sphingolipid concentration in the mitochondrial membranes is low, its influence is huge. Accumulation of ceramide leads to the formation of large channels in the mitochondrial outer membrane45 leading to pro-apoptotic protein release during the induction phase of apoptosis. It is known that mitochondria exhibit an independent ceramide pool, whereas it is not clear from where they are delivered (reviewed in46). However, the finding that the total ceramide level in the ER/mitochondria fractions of the more viable MCF-7/UGCG OE cells is not changed as compared to control cells, indicates that mitophagy is not induced (autophagy induction could be excluded22). Interestingly, specifically C18:0-Cer concentration is increased in ER/mitochondria fractions of MCF-7/UGCG OE cells. We already showed that the C18:0-Cer synthetizing ceramide synthase 1 (CerS1), whose trafficking from ER to mitochondria mediates cellular stress and cell death47, is not expressed in MCF-7 cells48. CerS4 is also able to produce C18:0-Cer, but its mRNA expression is reduced following UGCG OE9. Accordingly, the increased C18:0-Cer concentration could be related to increased dihydroceramide desaturase (DES) activity. Contradictory, we also measured increased C18:0-dh-Cer level. C18:0-dh-Cer is a substrate for DES and we assumed that the concentration is would be decreased, which is contradictory. To our knowledge, the gangliosides GM1 and GD3 are the only complex GSL, which are related to mitochondrial lipid raft-like structures so far (reviewed in5,49). The interaction of GD3 with mitochondrial lipid raft-like structures leads to changes in the mitochondrial membrane potential presumably via enhancing ROS formation of complex III of the mitochondrial electron transport chain and subsequent opening of mitochondrial permeability transition pore and cytochrome c-dependent activation of caspase 3 (reviewed in5). Since GM1 and GD3 are downstream of GlcCer, complex GSL might also be responsible for the observed changes following UGCG OE. However, it is unclear how GSL synthesized in the Golgi apparatus reach the ER and/or mitochondria. Annunziata et al. propose redistribution of GM1 in the ER membrane by contact between ER membranes and the plasma membrane (reviewed in49). Noteworthy, b-series derived gangliosides such as GD3 and GD2 exhibit tumor-enhancing function and a-series derived gangliosides such as GM1, GM2 and GM3 often exhibit tumor-suppressing function (reviewed in2).
GOLPH3 is a membrane protein localized in the trans-Golgi and in vesicles budding from the trans-Golgi. Amongst others, GOLPH3 specifically binds phosphatidylinositol-4-phosphate, which promotes vesicle exit for trafficking to the plasma membrane50. The oncogene GOLPH3 was found to be involved in the progression of various solid tumors, amongst others breast tumors51, and related to poor clinical outcome in many cancers. In contrast to these findings, the GOLPH3 gene expression is reduced in MCF-7/UGCG OE cells. A GOLPH3 knockdown causes scission of the interconnecting Golgi ribbon tubules which leads to the division of the individual Golgi cisternae stacks. Consequently, the vesicular transport deriving from these stacks is decreased (reviewed in52). A GOLPH3 decrease-mediated reduction of the vesicular transport in MCF-7/UGCG OE cells might be an explanation for the GEM restricted (not in the whole plasma membrane) GSL level increase which was described by us in 20186. Wang et al. revealed that GOLPH3 modulates the sensitivity towards the chemotherapeutic 5-fluoruracil (5-FU). More precisely, an increased GOLPH3 expression correlates with an advantageous prognosis in patients treated with 5-FU and predicts a higher sensitivity towards 5-FU in colorectal cancer cells53. Since the GOLPH3 expression is reduced in MCF-7/UGCG OE cells, which are less sensitive towards chemotherapeutic treatment6, the findings of Wang et al. are in line with our results.
Our data indicate that UGCG is closely connected to the breast cancer cell energy metabolism. This connection allows the cells to execute increased glycolysis and OXPHOS, which maintains sufficient supply with energy and building blocks needed for augmented cell proliferation. Molecular trigger might be changes in the sphingolipid composition of ER/mitochondria fraction leading to increased OXPHOS protein activity. This is accompanied by accelerated mitochondrial turnover (Fig. 5).
Since we could show that UGCG leads to glutamine dependency in MCF-7 cells, which results in increased glycolysis, OXPHOS (current study) and cell proliferation22, pharmacological inhibition of both glutamine metabolism and GlcCer synthesis via UGCG could be beneficial for treatment of breast cancer patients. This would be a novel therapeutic strategy for breast cancer treatment. Several inhibitors of glutamine metabolism such as benzylserine and L-γ-glutamyl-p-nitroanilide (GPNA, glutamine transporter inhibitor) or CB-839 (glutaminase I inhibitor) are available but are either low in efficacy or toxic (reviewed in54). Potential side effects of GPNA, CB-839 or others could be reduced by lowering the dose and drug efficacy could be increased by co-treatment with UGCG inhibitors, because we assume that both inhibitors would work synergistically in the process of inhibiting breast cancer cell proliferation.
Supplementary information
Acknowledgements
We would like to thank Martin Wegner (Dr. Manuel Kaulich, Institute of Biochemistry II, University Clinic Frankfurt am Main) for generating MCF-7/UGCG Kd and MCF-7/NHT (non-human target) control cell lines. This work was funded by the Deutsche Forschungsgemeinschaft (WE 5825/1-1 and WE 5825/2-1), the August Scheidel-Stiftung and the Heinrich und Fritz Riese-Stiftung. In addition, funding by the Johanna Quandt-Jubiläumsfond and SFB 1039 is gratefully acknowledged.
Author contributions
N.S. performed most of the experiments and prepared the figures. L.G. and E.M.O. conducted cell culturing and S.J.A. established the U-14C-glucose tracer assay. S.T., D.T., R.G. and N.F. were involved in the LC-MS/MS analysis. K.L.H., F.L.B. and G.G. supported the study by discussing the results and by revising the manuscript. M.S.W. designed the study and wrote the main part of the mansucript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-65182-y.
References
- 1.Seo AY, et al. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. Journal of cell science. 2010;123(Pt 15):2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furukawa K, et al. New era of research on cancer-associated glycosphingolipids. Cancer science. 2019;110(5):1544–1551. doi: 10.1111/cas.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cazet A, et al. The ganglioside G(D2) induces the constitutive activation of c-Met in MDA-MB-231 breast cancer cells expressing the G(D3) synthase. Glycobiology. 2012;22(6):806–816. doi: 10.1093/glycob/cws049. [DOI] [PubMed] [Google Scholar]
- 4.Hakomori SI. Cell adhesion/recognition and signal transduction through glycosphingolipid microdomain. Glycoconjugate journal. 2000;17(3 -4):143–151. doi: 10.1023/a:1026524820177. [DOI] [PubMed] [Google Scholar]
- 5.Garofalo T, et al. Role of mitochondrial raft-like microdomains in the regulation of cell apoptosis. Apoptosis: an international journal on programmed cell death. 2015;20(5):621–634. doi: 10.1007/s10495-015-1100-x. [DOI] [PubMed] [Google Scholar]
- 6.Wegner M-S, et al. UDP-glucose ceramide glucosyltransferase activates AKT, promoted proliferation, and doxorubicin resistance in breast cancer cells. Cellular and molecular life sciences: CMLS. 2018 doi: 10.1007/s00018-018-2799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gouazé V, et al. Overexpression of glucosylceramide synthase and P-glycoprotein in cancer cells selected for resistance to natural product chemotherapy. Molecular Cancer Therapeutics. 2004;3(5):633. [PubMed] [Google Scholar]
- 8.Liu Y-Y, et al. Glucosylceramide synthase, a factor in modulating drug resistance, is overexpressed in metastatic breast carcinoma. International journal of oncology. 2011;39(2):425–431. doi: 10.3892/ijo.2011.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegner M-S, et al. The UDP-glucose ceramide glycosyltransferase (UGCG) and the link to multidrug resistance protein 1 (MDR1) BMC cancer. 2018;18(1):153. doi: 10.1186/s12885-018-4084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg O. On the origin of cancer cells. Science (New York, N.Y.) 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 11.Warburg O. On respiratory impairment in cancer cells. Science (New York, N.Y.) 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Zong W-X, Rabinowitz JD, White E. Mitochondria and Cancer. Molecular Cell. 2016;61(5):667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Seminars in cell & developmental biology. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Reyes I, et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular Cell. 2016;61(2):199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinou J-C, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Developmental cell. 2011;21(1):92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bravo-Sagua R, et al. Calcium Transport and Signaling in Mitochondria. Comprehensive. Physiology. 2017;7(2):623–634. doi: 10.1002/cphy.c160013. [DOI] [PubMed] [Google Scholar]
- 18.Trotta AP, Chipuk JE. Mitochondrial dynamics as regulators of cancer biology. Cellular and molecular life sciences: CMLS. 2017;74(11):1999–2017. doi: 10.1007/s00018-016-2451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Q, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nature Neuroscience. 2015;18:501 EP. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehman J, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2012;26(5):2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiris-Pagès M, et al. Mitochondrial fission as a driver of stemness in tumor cells: mDIVI1 inhibits mitochondrial function, cell migration and cancer stem cell (CSC) signalling. Oncotarget. 2018;9(17):13254–13275. doi: 10.18632/oncotarget.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schömel N, et al. UGCG influences glutamine metabolism of breast cancer cells. Scientific reports. 2019;9(1):15665. doi: 10.1038/s41598-019-52169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne FL, et al. Metabolic vulnerabilities in endometrial cancer. Cancer research. 2014;74(20):5832–5845. doi: 10.1158/0008-5472.CAN-14-0254. [DOI] [PubMed] [Google Scholar]
- 24.Wegner M-S, et al. GPER1 influences cellular homeostasis and cytostatic drug resistance via influencing long chain ceramide synthesis in breast cancer cells. The international journal of biochemistry & cell biology. 2019;112:95–106. doi: 10.1016/j.biocel.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Doherty E, Perl A. Measurement of Mitochondrial Mass by Flow Cytometry during Oxidative Stress. Reactive oxygen species (Apex, N.C.) 2017;4(10):275–283. doi: 10.20455/ros.2017.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L, et al. The Glycolytic Switch in Tumors: How Many Players Are Involved? Journal of Cancer. 2017;8(17):3430–3440. doi: 10.7150/jca.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao L-P, et al. Chemotherapy selection pressure alters sphingolipid composition and mitochondrial bioenergetics in resistant HL-60 cells. Journal of lipid research. 2019;60(9):1590–1602. doi: 10.1194/jlr.RA119000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mailloux RJ, Craig Ayre D, Christian SL. Induction of mitochondrial reactive oxygen species production by GSH mediated S-glutathionylation of 2-oxoglutarate dehydrogenase. Redox biology. 2016;8:285–297. doi: 10.1016/j.redox.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinovkina LA. Mechanisms of Mitochondrial DNA Repair in Mammals. Biochemistry. Biokhimiia. 2018;83(3):233–249. doi: 10.1134/S0006297918030045. [DOI] [PubMed] [Google Scholar]
- 30.Hori A, Yoshida M, Ling F. Mitochondrial fusion increases the mitochondrial DNA copy number in budding yeast. Genes to cells: devoted to molecular & cellular mechanisms. 2011;16(5):527–544. doi: 10.1111/j.1365-2443.2011.01504.x. [DOI] [PubMed] [Google Scholar]
- 31.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sciences. 2011;89(3):65–71. doi: 10.1016/j.lfs.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Fan AX-C, et al. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. Journal of cancer research and clinical oncology. 2009;135(8):983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizumachi T, et al. Increased mitochondrial DNA induces acquired docetaxel resistance in head and neck cancer cells. Oncogene. 2008;27(6):831–838. doi: 10.1038/sj.onc.1210681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narendra, D., Walker, J.E. & Youle, R. Mitochondrial Quality Control Mediated by PINK1 and Parkin: Links to Parkinsonism. Cold Spring Harbor Perspectives in Biology 4(11). 10.1101/cshperspect.a011338 (2012) [DOI] [PMC free article] [PubMed]
- 36.Pallanck L. Mitophagy: mitofusin recruits a mitochondrial killer. Current biology: CB. 2013;23(13):R570–2. doi: 10.1016/j.cub.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 37.Ma L, et al. Corrigendum Estrogen receptor β inhibits estradiol-induced proliferation and migration of MCF-7 cells through regulation of mitofusin 2. International journal of oncology. 2016;49(5):2187. doi: 10.3892/ijo.2016.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. Journal of cell science. 2004;117(Pt 26):6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 39.Son JM, et al. Mitofusin 1 and optic atrophy 1 shift metabolism to mitochondrial respiration during aging. Aging cell. 2017;16(5):1136–1145. doi: 10.1111/acel.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko A-R, et al. The Differential DRP1 Phosphorylation and Mitochondrial Dynamics in the Regional Specific Astroglial Death Induced by Status Epilepticus. Frontiers in cellular neuroscience. 2016;10:124. doi: 10.3389/fncel.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz LM, et al. Non-cytotoxic copper overload boosts mitochondrial energy metabolism to modulate cell proliferation and differentiation in the human erythroleukemic cell line K562. Mitochondrion. 2016;29:18–30. doi: 10.1016/j.mito.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Ciarlo L, et al. Association of fission proteins with mitochondrial raft-like domains. Cell death and differentiation. 2010;17(6):1047–1058. doi: 10.1038/cdd.2009.208. [DOI] [PubMed] [Google Scholar]
- 43.Praharaj PP, et al. Intricate role of mitochondrial lipid in mitophagy and mitochondrial apoptosis: its implication in cancer therapeutics. Cellular and molecular life sciences: CMLS. 2019;76(9):1641–1652. doi: 10.1007/s00018-018-2990-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis SC, Uchiyama LF, Nunnari J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science (New York, N.Y.) 2016;353(6296):aaf5549. doi: 10.1126/science.aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siskind LJ, Kolesnick RN, Colombini M. Ceramide forms channels in mitochondrial outer membranes at physiologically relevant concentrations. Mitochondrion. 2006;6(3):118–125. doi: 10.1016/j.mito.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernández-Corbacho MJ, et al. Sphingolipids in mitochondria. Biochimica et biophysica acta. Molecular and cell biology of lipids. 2017;1862(1):56–68. doi: 10.1016/j.bbalip.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oleinik N, et al. Mitochondrial protein import is regulated by p17/PERMIT to mediate lipid metabolism and cellular stress. Science advances. 2019;5(9):eaax1978. doi: 10.1126/sciadv.aax1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegner M-S, et al. Ceramide synthases CerS4 and CerS5 are upregulated by 17β-estradiol and GPER1 via AP-1 in human breast cancer cells. Biochemical pharmacology. 2014;92(4):577–589. doi: 10.1016/j.bcp.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 49.Annunziata I, Sano R, d’Azzo A. Mitochondria-associated ER membranes (MAMs) and lysosomal storage diseases. Cell death & disease. 2018;9(3):328. doi: 10.1038/s41419-017-0025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuna RS, Field SJ. GOLPH3: a Golgi phosphatidylinositol(4)phosphate effector that directs vesicle trafficking and drives cancer. Journal of lipid research. 2019;60(2):269–275. doi: 10.1194/jlr.R088328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Z, et al. Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clinical cancer research: an official journal of the American Association for. Cancer Research. 2012;18(15):4059–4069. doi: 10.1158/1078-0432.CCR-11-3156. [DOI] [PubMed] [Google Scholar]
- 52.Bergeron JJM, et al. Proteomics Identifies Golgi phosphoprotein 3 (GOLPH3) with A Link Between Golgi Structure, Cancer, DNA Damage and Protection from Cell Death. Molecular & cellular proteomics: MCP. 2017;16(12):2048–2054. doi: 10.1074/mcp.MR117.000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, et al. GOLPH3 predicts survival of colorectal cancer patients treated with 5-fluorouracil-based adjuvant chemotherapy. Journal of translational medicine. 2014;12:15. doi: 10.1186/1479-5876-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi Y-K, Park K-G. Targeting Glutamine Metabolism for Cancer Treatment. Biomolecules & therapeutics. 2018;26(1):19–28. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.