Abstract
Papillary salivary gland neoplasms are rare tumors usually arising in the minor salivary glands of the oral cavity. Their classification has been historically confusing due to overlapping histologic features, but molecular analysis may clarify these entities. Sialadenoma papilliferum (SP) is a peculiar member of this group that demonstrates both an endophytic ductal and an exophytic squamous component. SP closely resembles syringocystadenoma papilliferum of the skin, a tumor which has recently been shown to harbor BRAF V600E or HRAS mutations. We sought to perform histologic and immunophenotypic analysis of a group of SP, along with BRAF and HRAS mutational analysis. We collected 13 SP cases from 7 females and 6 males ranging from 2 to 91 years (mean 62.8). Five exophytic ductal papillomas were also analyzed as controls. Histological analysis was performed along with immunohistochemistry for CK7, p63, and SOX10. BRAF VE1 immunohistochemistry was done in all tumors, and BRAF V600E and HRAS Sanger sequencing was successfully performed in all but two cases. Histologic analysis revealed that SP consisted not only of classic SP (9 of 13 cases) but also an oncocytic variant (4 of 13 cases) characterized by a glandular component that uniformly exhibited abundant granular cytoplasm and prominent nucleoli. By immunohistochemistry, all SP demonstrated luminal CK7 and basal p63 expression, but SOX10 was expressed only in conventional SP (9 of 9 cases). BRAF VE1 immunohistochemistry was positive in 9 of 9 conventional SP but 0 of 4 oncocytic SP; staining was present in both the exophytic and endophytic components. BRAF V600E mutational status was confirmed by Sanger sequencing in 11 cases (7 conventional and 4 oncocytic). The exophytic ductal papillomas were negative for BRAF mutations, and all tumors tested were negative for HRAS mutations. In summary, we demonstrated that SP consists of two variants: (1) conventional SP which is SOX10-positive and harbors BRAF V600E mutations similar to syringocystadenoma papilliferum of the skin; and (2) an oncocytic variant which is SOX10-negative and negative for BRAF mutations. We also demonstrated that both the endophytic glandular component and exophytic squamous components of conventional SP harbor BRAF V600E mutations and are therefore neoplastic.
Keywords: Sialadenoma papilliferum, Exophytic ductal papilloma, BRAF, Salivary gland
Introduction
Papillary salivary gland tumors comprise a diverse collection of rare benign neoplasms with a tendency to occur in the minor salivary glands of the oral cavity. This group includes sialadenoma papilliferum (SP), inverted ductal papilloma, intraductal papilloma, exophytic ductal papilloma, and papillary cystadenoma [1, 2]. The classification of these tumors has been historically confusing due in part to their rarity but also due to their overlapping histologic features.
Sialadenoma papilliferum (SP) is a particularly intriguing member of the papillary salivary gland neoplasm family. SP was initially described by Abrams and Finck in 1969 who noted the histologic similarity to cutaneous syringocystadenoma papilliferum of sweat gland origin [3]. SP consists of both an exophytic squamous papillary surface component along with an inverted glandular papillary proliferation [1–3]. The nature of this tumor has been debated, particularly in regards to whether the squamous component is reactive or actually part of the neoplasm. It has been theorized that SP derives from the excretory duct or its reserve cells that give rise to both cellular components [1, 2, 4].
Salivary gland classification is undergoing a revolution with the recognition that many neoplasms harbor characteristic genetic alterations. For example, several salivary gland tumors are now known to harbor tumor-defining translocations (e.g., MAML2 for mucoepidermoid carcinoma and EWSR1 for clear cell carcinoma) [5–7] or mutations (e.g., HRAS in epithelial-myoepithelial carcinoma and CTNNB1 in basal cell adenoma) [8–10]. Knowledge of these alterations has greatly refined salivary gland tumor classification and provided helpful diagnostic tools for challenging cases. These molecular tools may help refine the diagnostic criteria for papillary salivary gland neoplasms. Indeed, Agaimy et al. recently described AKT1 mutations in a novel member of the papillary salivary gland tumor group known as intraductal papillary mucinous neoplasm [11].
Given that syringocystadenoma papilliferum was recently found to harbor BRAF V600E or HRAS mutations [12, 13], we sought to determine whether its salivary gland counterpart SP is also defined by similar underlying genetic alterations. In addition, we sought to perform a thorough characterization of the SPs by histology and immunohistochemistry to assess for any correlations with the molecular results.
Materials and Methods
Case Selection
The archives of Department of Pathology and Oral Pathology Laboratory at the National Taiwan University Hospital (NTUH) in Taiwan, the University of British Columbia (UBC) in Canada, and Texas A&M University College of Dentistry (TAM) in the United States were searched over a period from 1994 to 2018, and a total of 18 cases originally diagnosed as SP from the minor salivary gland were identified. Hematoxylin and Eosin-stained slides were reviewed by two pathologists (J.Y.C. and M.S.H.). Using the diagnostic method proposed by Fowler et al. and Ellis et al. [1, 2] 13 cases were classified as SPs (characterized by the presence of both surface papillary structures and underlying proliferation of ductal structures) and 5 were re-classified as exophytic ductal papillomas. Although re-classified, the exophytic ductal papillomas were included for analysis as controls. All 18 cases had available paraffin archive tissue blocks for immunohistochemistry and 16 cases had enough tissue for BRAF and HRAS mutation analyses. This study was approved by the Research Ethics Committee of NTUH in Taiwan, UBC in Canada, and TAM in the United States.
Immunohistochemistry (IHC)
Tissue sections (4 μm) were dewaxed and rehydrated. Immunohistochemistry was performed using Ventana BenchMark XT autostainer (Ventana, Tucson, AZ, USA). This automated process included deparaffinization by EZ prep (Ventana) and a CC1-based antigen retrieval using Cell Condition 1 solution [CC1; Tris–EDTA buffer (pH 8.0)] (Ventana) for 64 min. The slides were incubated with anti-human CK7 (SP52, ready-to-use, Ventana), CK13 (Clone KS-1A3, 1:250, Diagnostic BioSystems, Pleasanton, CA, USA), p63 (4A4, ready-to-use, Ventana), or SOX10 (EP268, ready-to-use, Bio SB, Santa Barbara, CA, USA) for 32 min. BRAF V600E mutant-specific immunohistochemistry was performed using anti-BRAF V600E (VE1, Ventana) antibody on Ventana BenchMark GX autostainer (Ventana) with the same retrieval technique for 64 min, pre-peroxidase inhibition and primary antibody incubation for 28 min according the manufacturer instructions. A BRAF V600E-mutated papillary thyroid carcinoma proven by Sanger sequencing was used as the positive control for BRAF V600E mutant-specific immunohistochemistry. BRAF V600E staining intensity was recorded as 0 (negative), 1 + (weak cytoplasmic staining), 2 + (moderate cytoplasmic staining), and 3 + (strong cytoplasmic staining). Labeling was detected with the Optiview DAB Detection Kit (Ventana) and counterstained with hematoxylin.
BRAF and HRAS Mutational Analyses
BRAF and HRAS mutations were detected by PCR and Sanger sequencing. DNA was extracted from unstained slides of the formalin-fixed, paraffin embedded tissue and extracted using a DNeasy tissue kit (Qiagen, Hilden, Germany). PCR was performed using a HotStarTaq Master Mix kit (Qiagen) with the following primers:
5ʹ-TCATAATGCTTGCTCTGATAGGA-3ʹ(BRAF-Exon15-F),
5ʹ-GGCCAAAAATTTAATCAGTGGA-3ʹ(BRAF-Exon15-R),
5ʹ-CAGGAGACCCTGTAGGAGG-3ʹ(HRAS-Exon2-F),
5ʹ-TCGTCCACAAAATGGTTCTG-3ʹ(HRAS-Exon2-R),
5ʹ-GTCCTCCTGCAGGATTCCTA-3ʹ(HRAS-Exon3-F), and
5ʹ-CGGGGTTCACCTGTACT-3ʹ (HRAS-Exon3-R).
Successfully amplified products (BRAF exon15: 224 bp; HRAS exon2: 139 bp; HRAS exon3: 179 bp) were purified, and sequenced. All sequencing reactions were conducted in both forward and reverse directions by DNA sequencing services using a Big Dye Terminator kit (Applied Biosystems, Foster City, CA, USA) and an ABI Prism 3700 DNA Analyzer (Applied Biosystems). Specimens with mutations were repeated to confirm.
Results
Clinical Findings
The clinical, immunohistochemical, and molecular features of the 13 SP and 5 EP are summarized in Table 1.
Table 1.
Clinical, immunohistochemical, and molecular findings in 13 sialadenoma papilliferum (SP) as well as 5 exophytic ductal papillomas (EP)
| Case | Sex | Age | Location | Size (mm) | Diagnosis | Immunohistochemistry | Mutation analysis | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRAF (V600E) | SOX10 | CK7 | p63 (basal) | BRAF | HRAS | ||||||
| 1 | M | 65 | Hard palate | 3.0 | Classic SP | 2 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 2 | M | 77 | Hard palate | 1.5 | Classic SP | 1 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 3 | F | 83 | Soft palate | 3.0 | Classic SP | 2 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 4 | M | 52 | Hard palate | 5.2 | Classic SP | 1 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 5 | M | 2 | Buccal mucosa | 4.0 | Classic SP | 1 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 6 | F | 91 | Hard palate | 4.5 | Classic SP | 1 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 7 | F | 58 | Tongue | 6.0 | Classic SP | 1 + , diffuse | + | + | + | BRAF V600E + | Wild type |
| 8 | F | 77 | Hard palate | 2.0 | Classic SP | 1 + , diffuse | + | + | + | N/A | N/A |
| 9 | F | 36 | Hard palate | 2.0 | Classic SP | 1 + , diffuse | + | + | + | N/A | N/A |
| 10 | M | 61 | Buccal mucosa | 2.0 | Oncocytic SP | − | − | + | + | Wild type | Wild type |
| 11 | F | 73 | Hard palate | 3.0 | Oncocytic SP | − | − | + | + | Wild type | Wild type |
| 12 | F | 77 | Hard palate | 4.0 | Oncocytic SP | − | − | + | + | Wild type | Wild type |
| 13 | M | 64 | Hard palate | 2.5 | Oncocytic SP | − | − | + | + | Wild type | Wild type |
| 14 | F | 76 | Hard palate | 3.0 | Exophytic ductal papilloma | − | − | + | + | Wild type | Wild type |
| 15 | M | 77 | Hard palate | 3.2 | Exophytic ductal papilloma | − | − | + | + | Wild type | Wild type |
| 16 | F | 87 | Buccal mucosa | 2.0 | Exophytic ductal papilloma | − | − | + | + | Wild type | Wild type |
| 17 | F | 88 | Buccal mucosa | 6.5 | Exophytic ductal papilloma | − | − | + | + | Wild type | Wild type |
| 18 | F | 73 | Upper lip | 5.0 | Exophytic ductal papilloma | − | − | + | + | Wild type | Wild type |
Measured from glass slide
N/A not available
The SPs occurred in 7 females and 6 males ranging from 2 to 91 years in age (mean 62.8 years; median 65 years). They arose from the hard palate (9 of 13), buccal mucosa (2 of 13), soft palate (1 of 13), and tongue (1 of 13). The size ranged from 1.5 to 6 mm (mean 3.3 mm).
The exophytic ductal papillomas occurred in 4 women and 1 man ranging from 73 to 88 years (mean 80.2 years; median 77 years). They arose from the hard palate (2 of 5), buccal mucosa (2 of 5) and upper lip (1 of 5) and measured 2 to 6.5 mm (mean 3.9 mm) in the largest dimension.
Histopathologic Findings
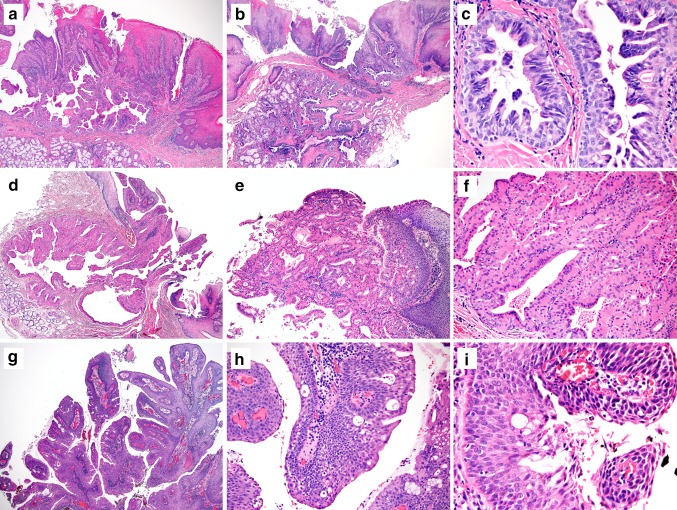
Our diagnostic criteria for SP and exophytic ductal papilloma were based on the definitions described by Fowler et al. and Ellis et al. [2, 3]: SP is a salivary gland neoplasm with an exophytic proliferation of papillary stratified squamous epithelium and a contiguously endophytic ductal proliferation underneath. All of our 13 SP cases had both exophytic and endophytic components (See examples in Fig. 1a, b). The endophytic component typically showed multiple dilated, irregularly branched ductal spaces lined by bi-layered to multilayered epithelial cells with intraluminal projections, giving the luminal spaces a fissure-like or stellate-like appearance (Fig. 1a–c). These ductal structures merged with the overlying squamous epithelium, and in some cases, the ductal cells were directly exposed on the surface of the mucosa. A plasma cell-rich inflammatory infiltrate was usually present in SP.
Fig. 1.
Examples of classic sialadenoma papilliferum (a–c), oncocytic sialadenoma papilliferum (d–f) and exophytic ductal papilloma (g–i). a Classic sialadenoma papilliferum showing a papillary squamous surface and a contiguously endophytic ductal proliferation. b The endophytic ductal proliferation sometimes extends deeply into underlying minor salivary glands. c The ductal cells are composed of columnar to cuboidal cells and arranged in bilayered to multilayered structures. d Oncocytic variant of sialadenoma papilliferum has a similar arrangement with both exophytic papillary surface and an endophytic ductal component. e The oncocytic ductal cells merge with overlying stratified squamous epithelium and form papillary structures. f Oncocytic variant of sialadenoma papilliferum comprised of oncocytic cells with round nuclei, abundant eosinophilic cytoplasm, and bland-looking nuclei. g Exophytic ductal papilloma has only exophytic papillary projections and lacks the endophytic ductal hyperplasia. h and i Exophytic ductal papilloma with mixed stratified squamous epithelium and columnar epithelium with variable numbers of mucous/goblet cells admixed (h & e staining; original magnification, a, b, d, g × 40; c,i, × 400; e: × 100; f, h: × 200)
Nine SPs exhibited classic histologic features. Eight of nine classic SPs had irregularly dilated or branched ducts lined by cuboidal or columnar epithelial cells. One classic SP had relatively smooth lumen and lack of prominent branching. These ductal cells were variably bilayered or multilayered, and occasionally morphologically similar to papillary hyperplasia of the breast (Fig. 1c).
In addition to the nine classic SPs, we also found four oncocytic variants of SP. The oncocytic SP were structurally identical to classic SP, but were unique because the ductal component was entirely lined by oncocytic cells with abundant eosinophilic cytoplasm and round hyperchromatic nuclei, with no foci of conventional-appearing ductal epithelium (Fig. 1d–f).
Both classic and oncocytic SPs were readily differentiated from the 5 cases of exophytic ductal papilloma. The exophytic ductal papillomas only had exophytic papillary projections composed of stratified squamous epithelium and/or columnar epithelium with some admixed mucocytes (Fig. 1g–i). No submucosal ductal proliferation was present in these cases.
Immunohistochemical Findings
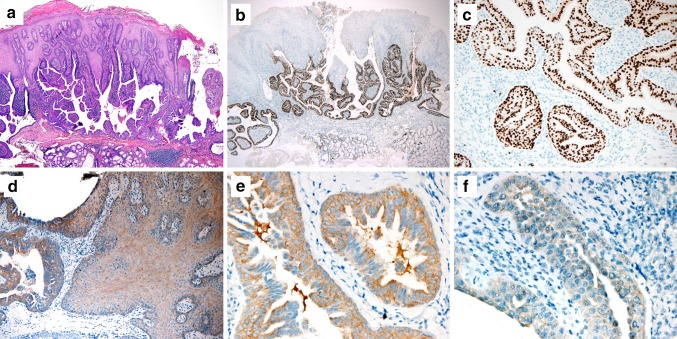
Classic SP, oncocytic SP, and exophytic ductal papilloma demonstrated similar patterns of immunostains for CK13, CK7, and p63: CK13 and CK7 highlighted the surface squamous epithelial and proliferative ductal areas, respectively, while p63 was diffusely positive in the squamous tumor epithelium and positive in the basal layer of the ductal component. On the other hand, SOX10 was diffusely and strongly positive in the ductal component of 9 of 9 classic SPs (Fig. 2a–c) but was completely negative in all 4 oncocytic SPs and all 5 exophytic ductal papillomas (Fig. 3a, b, d, e).
Fig. 2.
Examples of SOX10 and BRAF V600E Immunohistochemistry staining of classic sialadenoma papilliferum. a–c Classic sialadenoma papilliferum showing b, c positive staining for SOX10 in the endophytic ductal component and d–f BRAF V600E (VE1) in both the glandular (left) and squamous (right) tumor components and the ductal cells of classic sialadenoma papilliferum showing e moderate or f weak staining. (A: h & e staining; b, c: SOX10; d–f: BRAF V600E; original magnifications, a, b, × 40; c, d, × 200; e, f × 400)
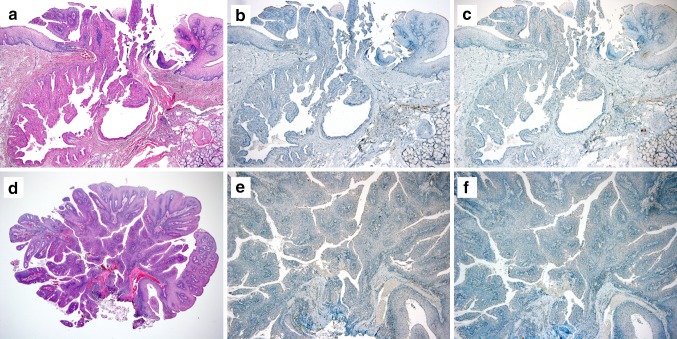
Fig. 3.
Examples of SOX10 and BRAF V600E Immunohistochemistry staining of (a–c) oncocytic sialadenoma papilliferum and (d–f) exophytic ductal papilloma. a–c) Oncocytic sialadenoma papilliferum and d–f exophytic ductal papilloma was b, e) consistently negative for SOX10 and (c, f) BRAF V600E (VE1). (a, d: h & e staining; b, e: SOX10; c, f: BRAF V600E; original magnifications, a–c and e, f, × 40; d, × 20)
Molecular Analysis
BRAF V600E (VE1) immunohistochemistry was positive in 9 of 9 classic SP. The staining was cytoplasmic in distribution, and its intensity was weak (1 +) in 7/9 cases and moderate (2 +) in 2/9 cases. (example in Fig. 2d–f). For all nine classic SPs, BRAF VE1 immunoexpression present in both the ductal and squamous tumor elements, although the intensity of staining was stronger in the ductal component. BRAF VE1 immunohistochemistry was uniformly negative in all oncocytic SP and all exophytic ductal papillomas. (Figure 3c, f).
Sixteen cases (7 classic SP, 4 oncocytic SP, and 5 exophytic ductal papillomas) had sufficient tissue available to be tested for BRAF mutation using Sanger sequencing. The BRAF c.1799 T > A mutation resulting in a p. Val600Glu substitution was confirmed to be present in 7 of 7 classic SP cases, while the 4 oncocytic SPs and 5 exophytic ductal papillomas tested were BRAF wild type. Finally, all 16 cases tested for HRAS mutations (codons 12 and 13) were negative.
Discussion
Over the past several years, an expanding list of salivary gland neoplasms has been shown to harbor characteristic genetic alterations. For example, clear cell carcinoma (EWSR1-ATF1), [5] mucoepidermoid carcinoma (CRTC1-MAML2 or CRTC3-MAML2), [6, 7] adenoid cystic carcinoma (MYB-NFIB or MYBL1-NFIB), [14, 15] secretory carcinoma (ETV6-NTRK3), [16] and polymorphous adenocarcinoma, cribriform variant (PRKD1-3 partnered with various genes) [17] are now known to harbor tumor-specific gene fusions, while basal cell adenoma/adenocarcinoma (CTNNB1), [10] classic polymorphous adenocarcinoma (PRKD1), [18] epithelial-myoepithelial carcinoma (HRAS) [8, 9] and the newly described intraductal papillary mucinous neoplasm (AKT1) [11] have characteristic mutations. The presence of these recurring tumor-specific genetic alterations has refined salivary gland tumor diagnoses by supplying benchmarks for tumor classification, facilitating an updated appreciation for complete phenotypic spectra within salivary gland tumor types. In this molecular era of salivary gland tumor classification, a reappraisal of sialadenoma papilliferum (SP) is warranted.
A careful histologic re-review of 18 cases diagnosed as SP, utilizing strict histologic criteria described by Fowler et al. and Ellis et al. [1, 2] revealed that a significant proportion of these tumors (5 of 18, 28%) actually did not meet criteria for SP because they lacked any endophytic tumor growth. These cases were more appropriately classified as exophytic ductal papillomas histologically. Moreover, the true SPs could be further refined into two groups: classic SP (9 of 13, 69%) and oncocytic SP (4 of 13, 31%).
We demonstrated that classic SP consistently harbors BRAF V600E mutations but not HRAS mutations. This is not altogether unexpected, as the cutaneous tumor for which SP was named—syringocystadenoma papilliferum – has also recently been shown to harbor BRAF V600E mutations in 52% of cases [12, 13]. This finding lends genetic support to the notion that SP and syringocystadenoma papilliferum are indeed analogous tumors. Interestingly, a significant minority (26%) of syringocystadenomas papilliferum are BRAF wild type but instead harbor HRAS G13R mutations, an alteration we did not find in our limited series of SP. Moreover, BRAF VE1 immunostaining was seen not only in the proliferative ductal elements but also the overlying squamous epithelium in classic SP. This finding seemingly confirms that notion that both components are neoplastic and driven by the BRAF V600E mutation. Finally, we demonstrated that SOX10 is robustly positive in the ductal component of classic SP. SOX10 is a transcription factor essential for the development of acinar and intercalated duct cells of the salivary gland [17]. SOX10 has been reported to be expressed in a variety of salivary gland tumors derived from acinar, intercalated duct, or myoepithelial cells (e.g., pleomorphic adenoma, polymorphous adenocarcinoma, adenoid cystic carcinoma, acinic cell carcinomas), but virtually no SOX10 staining is seen in salivary gland tumors derived from striated or excretory ducts, such as oncocytoma, mucoepidermoid carcinoma, and salivary duct carcinoma [19, 20]. The strong expression of SOX10 in classic SP suggests that these ductal cells may derive from the intercalated duct cells rather than large excretory ducts near or at the junction with the overlying stratified squamous epithelium.
While oncocytic features have been previously described in SP, they had been regarded simply as metaplastic change [1]. Our study demonstrated that there is an oncocytic form of SP that is morphologically, immunophenotypically, and molecularly distinct from classic SP suggesting differences in cell origin and pathogenesis. First, the oncocytic variants of SP had a ductal tumor component that was purely oncocytic with abundant eosinophilic cytoplasm, round vesicular nuclei, and prominent nucleoli. These oncocytic cells were multilayered or bilayered with a morphology resembling Warthin tumor. In other words, there was no conventional basophilic ductal cells to suggest that the oncocytic features were a focal metaplastic alteration. Second, the oncocytic ducts in these SPs were consistently negative for SOX10. The lack of SOX10 expression in ductal cells of oncocytic SP (or exophytic ductal papilloma) suggests the ductal component in these tumors may derive from the large excretory ducts. Finally, all oncocytic SPs were negative for BRAF V600E mutations by both immunohistochemistry and Sanger sequencing, implying different underlying genetic mechanisms. With these key differences between classic SP and oncocytic SP in mind, it is reasonable to question whether they should be regarded as entirely different tumors. However, given the similarities in anatomic distribution (oral cavity), growth pattern (exophytic squamous and inverted glandular tumor components), and immunohistochemistry (squamous elements positive for CK13, glandular elements positive for CK7 with a basal layer of p63-positive cells) we believe it is reasonable to continue classifying both tumors as forms of SP morphologically while knowing that they are distinct molecularly.
In summary, this study used histologic, immunophenotypic, and molecular analysis to shed light on SP, a rare salivary gland tumor. We found that SP actually consists of two distinct subtypes: (1) classic SP which is strongly SOX10-positive and genetically analogous to syringocystadenoma papilliferum with consistent BRAF V600E mutations; and (2) oncocytic SP which is SOX10-negative and is BRAF wild type. These findings further refine salivary gland tumor classification and suggest that SOX10 immunohistochemistry and BRAF analysis (either by immunohistochemistry or molecular testing) may be useful diagnostic adjuncts when confronted with a challenging intraoral salivary gland neoplasm.
Compliance with Ethical Standards
Conflicts of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Min-Shu Hsieh and Justin A. Bishop authors contributed equally to this work.
References
- 1.Fowler CB, Damm DD. Sialadenoma papilliferum: analysis of seven new cases and review of the literature. Head Neck Pathol. 2018;12:193–201. doi: 10.1007/s12105-017-0852-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis GL, Auclair PL. Sialadenoma papilliferum. AFIP atlas of tumor pathology: tumors of the salivary glands. Washington, DC: ARP Press; 2008. pp. 148–151. [Google Scholar]
- 3.Abrams AM, Finck FM. Sialadenoma papilliferum. A previously unreported salivary gland tumor. Cancer. 1969;24:1057–1063. doi: 10.1002/1097-0142(196911)24:5<1057::AID-CNCR2820240529>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Maiorano E, Favia G, Ricco R. Sialadenoma papilliferum: an immunohistochemical study of five cases. J Oral Pathol Med. 1996;25:336–342. doi: 10.1111/j.1600-0714.1996.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 5.Antonescu CR, Katabi N, Zhang L, et al. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer. 2011;50:559–570. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 6.Behboudi A, Enlund F, Winnes M, et al. Molecular classification of mucoepidermoid carcinomas-prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45:470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]
- 7.Seethala RR, Dacic S, Cieply K, et al. A reappraisal of the MECT1/MAML2 translocation in salivary mucoepidermoid carcinomas. Am J Surg Pathol. 2010;34:1106–1121. doi: 10.1097/PAS.0b013e3181de3021. [DOI] [PubMed] [Google Scholar]
- 8.El Hallani S, Udager AM, Bell D, et al. Epithelial-myoepithelial carcinoma: frequent morphologic and molecular evidence of preexisting pleomorphic adenoma, common HRAS mutations in PLAG1-intact and HMGA2-intact cases, and occasional TP53, FBXW7, and SMARCB1 alterations in high-grade cases. Am J Surg Pathol. 2018;42:18–27. doi: 10.1097/PAS.0000000000000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiosea SI, Miller M, Seethala RR. HRAS mutations in epithelial-myoepithelial carcinoma. Head Neck Pathol. 2014;8:146–150. doi: 10.1007/s12105-013-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato M, Yamamoto H, Hatanaka Y, et al. Wnt/beta-catenin signal alteration and its diagnostic utility in basal cell adenoma and histologically similar tumors of the salivary gland. Pathol Res Pract. 2018;214:586–592. doi: 10.1016/j.prp.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Agaimy A, Mueller SK, Bumm K, et al. Intraductal papillary mucinous neoplasms of minor salivary glands With AKT1 p.Glu17Lys mutation. Am J Surg Pathol. 2018;42:1076–1082. doi: 10.1097/PAS.0000000000001080. [DOI] [PubMed] [Google Scholar]
- 12.Konstantinova AM, Kyrpychova L, Nemcova J, et al. Syringocystadenoma papilliferum of the anogenital area and buttocks: a report of 16 cases, including human papillomavirus analysis and HRAS and BRAF V600 mutation studies. Am J Dermatopathol. 2019;41:281–285. doi: 10.1097/DAD.0000000000001285. [DOI] [PubMed] [Google Scholar]
- 13.Levinsohn JL, Sugarman JL, Bilguvar K, et al. Somatic V600E BRAF mutation in linear and sporadic syringocystadenoma papilliferum. J Invest Dermatol. 2015;135:2536–2538. doi: 10.1038/jid.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persson M, Andren Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitani Y, Liu B, Rao PH, et al. Novel MYBL1 gene rearrangements with recurrent MYBL1-NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin Cancer Res. 2016;22:725–733. doi: 10.1158/1078-0432.CCR-15-2867-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skalova A, Vanecek T, Sima R, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 17.Weinreb I, Zhang L, Tirunagari LM, et al. Novel PRKD gene rearrangements and variant fusions in cribriform adenocarcinoma of salivary gland origin. Genes Chromosomes Cancer. 2014;53:845–856. doi: 10.1002/gcc.22195. [DOI] [PubMed] [Google Scholar]
- 18.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating {PRKD}1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. doi: 10.1038/ng.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh MS, Lee YH, Chang YL. SOX10-positive salivary gland tumors: a growing list, including mammary analogue secretory carcinoma of the salivary gland, sialoblastoma, low-grade salivary duct carcinoma, basal cell adenoma/adenocarcinoma, and a subgroup of mucoepidermoid carcinoma. Hum Pathol. 2016;56:134–142. doi: 10.1016/j.humpath.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Ohtomo R, Mori T, Shibata S, et al. SOX10 is a novel marker of acinus and intercalated duct differentiation in salivary gland tumors: a clue to the histogenesis for tumor diagnosis. Mod Pathol. 2013;26:1041–1050. doi: 10.1038/modpathol.2013.54. [DOI] [PubMed] [Google Scholar]