Abstract
Next-Generation Sequencing (NGS) is being utilized with increasing frequency in the characterization of salivary gland tumours. The potential scenarios which may be encountered by using this technique in routine practice will be outlined in further text by drawing from our own clinical experience. These include oncocytic mucoepidermoid carcinomas with unusual variant morphology (and negative MAML2 fluorescent in-situ hybridization results), a diagnosis of ameloblastoma changed to adenoid cystic carcinoma (due to MYBL1 fusion presence), a salivary duct carcinoma with an ETV6-NTRK3 fusion (otherwise seen in secretory carcinomas) and novel fusion partners such as EWSR1-BEND2 (otherwise seen in pancreatic neuroendocrine carcinomas). As NGS continues to develop and more widespread clinical implementation increases, we must be cognisant of the need for proper interpretation and in some cases verification using a secondary technique, the limitations of this technique, and the ethical dilemmas one faces when encountering a novel fusion.
Keywords: Next-generation sequencing, Salivary gland neoplasms, Fusions, In-situ hybridization, Fluorescence
Introduction
The pathologic classification of salivary gland tumours has seen unparalleled growth in recent time. There are currently over 30 distinct salivary gland tumour types in the latest World Health Organization (WHO) classification system [1]; a significant progress from the mere 10–12 tumour types in the original 1953 Armed Forces Institute (AFI) classification system [2]. We have learned over time that a large proportion of salivary gland tumours are fusion-driven by tumour-specific rearrangements. This knowledge has thus placed a plethora of ancillary diagnostic tools at our disposal; immunohistochemistry (IHC), fluorescent in-situ hybridization (FISH), and in recent years, Next-Generation Sequencing (NGS) technologies [3–5]. It therefore should not come as a surprise that we have been able to refine the classification and diagnosis of salivary gland tumours; an otherwise famously histomorphologically diverse, overlapping, and diagnostically challenging group of tumours. Nonetheless, it was not until the arrival of NGS as a research and ancillary diagnostic method that the rapid 'explosion' of novel discoveries occurred. Characterization of novel salivary gland tumor types and fusion partners is occurring with ever-increasing frequency, while the 'waste-basket' category of adenocarcinoma NOS narrows [3, 6–8]. Perhaps even more significantly, new insight into tumour pathobiology and clinically actionable mutations such as NTRK are coming to light [3, 8–12]. In this review, our institutional experience with NGS—and decisions related to how these results are incorporated into the reporting of challenging and equivocal cases of salivary gland tumours—will be discussed.
Discussion
The role of Next-Generation Sequencing (NGS) as a critical ancillary diagnostic tool in solid tumours was originally affirmed in the domain of sarcoma pathology; where molecular testing of tumours for diagnostic accuracy and appropriate clinical management is now considered standard of care [13]. Briefly, the basic principle of NGS is massive parallel sequencing of nucleic acids i.e., specific DNA regions of interest either through whole exome, transcriptome or targeted panel sequencing. New types of RNA-based NGS, such as RNA-Seq have become more and more utilized in the detection of fusion transcripts, a common molecular alteration seen in lymphomas, sarcomas and salivary gland tumours [5, 6, 14, 15]. The advantages of NGS and specifically RNA-Seq, compared to other methodologies, such as FISH, include broader mutation coverage, the interrogation of anywhere between 100 to over 1000 potential fusions simultaneously, and possible identification of novel fusion partners [5, 16]. From a practical standpoint, targeted fusion panels allow for the routine use of formalin-fixed paraffin-embedded (FFPE) tissue, allowing for easier integration of this method into the workflow of a surgical pathology laboratory. However, despite the aforementioned benefits, the disadvantages are that RNA-Seq remains an expensive and time-consuming tool [5, 11, 16], thereby rendering it a challenge to implement in most laboratories.
At present, there are multiple commercially available fusion assays on the market. Many of these are broad and offer coverage of mutations which are encountered in lymphomas, sarcomas and epithelial neoplasms, such as salivary gland carcinomas. The fusion gene panel which has been used for sarcoma pathology at Mount Sinai Hospital in Toronto overlaps significantly with known salivary gland fusions and we have leveraged this platform in the routine testing of challenging salivary gland tumours in recent years. As NGS technology continues to advance, the cost is expected to drop over time, making it inevitable that the application of such tools will become more common-place in the future. However, the challenges related to the independent confirmation of the findings generated by NGS will remain, as will the question of reporting guidelines, particularly in the context of novel, unexpected findings. Moreover, this raises the question of how and when to report unexpected, but potentially targetable mutations which have been seen previously in other tumour types. We will discuss our approach to reporting these types of cases throughout further text. However, it should be mentioned that standardized reporting guidelines for molecular findings in tumours have been proposed in the “Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer” by Li et al. [17].
In further text we will aim to review the application of NGS and specifically RNA-Seq in the context of salivary gland tumours along with other ancillary diagnostic tools. As in sarcoma pathology, several possible scenarios may be encountered. To illustrate these, we will draw from our own experience with “real-life” examples. For reference, our cases were analyzed via the TruSight RNA Fusion Panel targeting 507 known fusion-related genes, using the Illumina MiSeq system. This is one of the many platforms available for this purpose. As a brief background, NGS has been applied for salivary gland tumours at our institution over the last 3 years. We would like to clarify for the purpose of this text that the ‘routine’ use of this test (as outlined in the text title) is not synonymous with ‘reflexive’ testing of all salivary gland tumours. In fact, here we do not seek to establish absolute and definitive guidelines when NGS ought to be employed but rather general guidelines that have been devised based on our personal experience. Speaking in broad terms, NGS has been employed in the testing of low to intermediate grade salivary gland tumours with unusual morphology or when the question of malignancy arose in a potentially benign appearing neoplasm (i.e. when definitive malignancy could not be ruled out by H&E and IHC). In this sense, the use of NGS is best determined on a case-by-case basis by the signing pathologist.
In most cases, the expected fusion is identified and correlates with a compatible H&E impression in the differential diagnosis. In situations such as this, confirmation of the initial H&E impression by an ancillary technique such as RT-PCR, or FISH is not warranted as the finding supports a diagnosis that would have been made anyway. This is more difficult in cases where the diagnosis is less certain, or when variant fusions or novel fusions are encountered. It has been our practice to report fusions only when they can be reasonably reconciled with the diagnosis. For any novel fusion identified that is not actionable and has no known diagnostic gene for the differential diagnosis in question, the result given is “no diagnostic and/or actionable fusion gene(s) is identified” until such a time that the result can be independently confirmed (e.g., FISH, RT-PCR). This statement is in keeping with our previously outlined institutional policy regarding not reporting fusions identified on this diagnostic panel unless they are clinically actionable or diagnostic. It must be noted that these policies are institution specific and that practices may differ. However, in keeping with Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer [17], these novel fusions would be best categorized and reported as ‘Variants of Unknown Clinical Significance; Tier III.” However, when an established driving gene typically seen in a fusion is encountered but with a novel partner, and it matches the expected diagnosis, the primary gene is reported without the novel gene partner unless secondary validation can be performed for the new partner. An example of this is a recent “spindle cell rhabdomyosarcoma” of the base of tongue with a CITED2 gene fusion. As the potential novel partner was not independently confirmed, it was not identified in the report, however the presence of a rearrangement of the CITED2 gene was reported as it is a known event in RMS and in keeping with the diagnosis [18]. These types of changes would therefore be categorized as “Variants of Potential Clinical Significance; Tier II”.
The following discussion will look at different categories of fusions encountered in our practice, how we think they should be reported, and how this impacted on the diagnosis for the case. Please note that some of the cases used as examples in this manuscript have been parts of clinical trials or research projects, however we felt that they were optimal examples which would allow us to illustrate our point to the reader of how we would deal with them if they were real time diagnostic cases. They are also illustrative of the cases that we see in our practice.
Expected Diagnosis, but with Unusual Morphology or Negative Fish Testing Results
Generally speaking, most diagnoses of salivary gland tumours are rendered or favoured almost purely on the basis of histomorphology. Despite the relatively vast panel of immunohistochemical stains used in an attempt to help diagnose salivary gland tumours, the majority of these actually do not provide definitive answers, and therefore are not particularly helpful. In the case of unusual morphology for a suspected entity, FISH testing is often employed as a reasonable next step and usually to confirm the initial working diagnosis by demonstrating the presence of a tumour-specific fusion. When FISH testing fails to prove the presence of a fusion in a case with a high 'pre-test' probability, NGS has shown itself to be an indispensable adjunctive tool [5, 15]. To illustrate this point, we will use mucoepidermoid carcinomas (MEC) with variant histomorphology as an example. The most common and perhaps, well known malignant salivary gland neoplasm, mucoepidermoid carcinomas are conventionally defined as being composed of «mucinous, intermediate and squamoid/epidermoid tumour cells forming cystic and solid patterns» [1]. These tumours are characterized by CRTC1-MAML2 fusions (most commonly), CRTC3-MAML3 [7], or rarely the EWSR1-POU5F1 fusion [19], the latter of which has not be reproduced since its original description. Variant forms such as oncocytic, Warthin-like and sclerosing MECs have been described in the literature and have demonstrated the prevalence of this fusion which is comparable to that of conventional MECs [20, 21]. One such case was recently encountered at our institution and will be discussed herein.
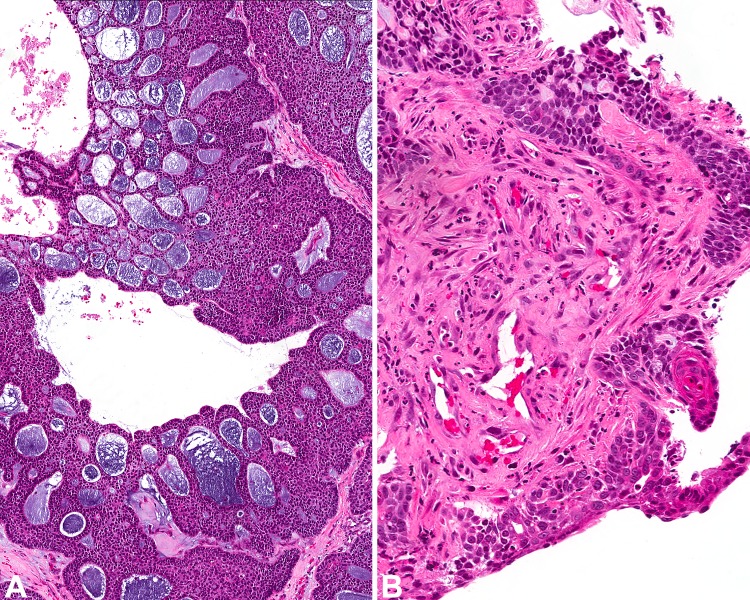
A parotid mass in a 54-year-old woman consisted of a low-grade infiltrative salivary gland carcinoma which was composed of solid lobules and ductal structures of large cells with eosinophilic, granular cytoplasm and microcystic spaces filled with eosinophilic secretions (Fig. 1). No appreciable mucous cells were noted on Hematoxylin and Eosin sections (H&E). Immunohistochemistry (IHC) showed that the tumour was positive for CK5 and p40 and possibly focal intracellular mucin on a mucicarmine special stain. The differential diagnosis included an oncocytic mucoepidermoid carcinoma, which was favoured on the basis of histomorphologic findings and IHC results, or an oncocytic carcinoma. Oncocytic mucoepidermoid carcinomas are an interesting variant in that they tend to be composed of almost a purely oncocytic cell population [20]; hence it is not surprising why oncocytic carcinoma is a common differential diagnosis. Other differential diagnoses for this tumour type include oncocytic cystadenoma, Warthin's tumour and acinic cell carcinoma [20]. Subtle vacuolated cells suggestive of mucous cells and strong p63 staining may support the diagnosis of MEC however neither are definitive and are subject to sampling error and interobserver variability. Nonetheless, this particular case was referred for further MAML2 gene testing by FISH which yielded fusion-negative results. Although between 75–93% of low to intermediate-grade MECs are MAML2 rearranged [7, 20, 22, 23], only between 46–56% of high-grade MECs are fusion positive [7, 22]. This means that a negative result does not rule out the diagnosis. Whole-exome sequencing has even demonstrated that some of these fusion-negative tumours occur via an alternate pathway with p53 mutations [24]. Alternatively and perhaps more commonly, what we have conventionally considered as being «high-grade MEC» might in fact represent other, misdiagnosed tumour types [7]. It is only recently that rare bona fide cases of MEC with high-grade transformation have been suggested in the literature; in one particular case report, conventional areas of MEC demonstrated the presence of a MAML2 rearrangement by FISH while the predominant transformed cells demonstrated multiple split signals which were interpreted as rearrangement and polyploidy [25]. However, polysomy has been demonstrated in a small number of high-grade MEC in other studies as well [22]. The tumour in our case, notably was low-grade and showed a surprising FISH-negative result. Subsequent NGS confirmed the presence of a CRTC1-MAML2 fusion. Although FISH is otherwise a robust ancillary technique which is generally used as the default gold-standard test for the detection of rearrangements in many institutions it is not without flaws. The high sensitivity and specificity of FISH can be expected in cases where canonical breakpoints are present. FISH performed on cancers with non-canonical breakpoints may be false negative [16]. Technical issues such as probe failure can also account for false negatives or interobserver variability in signal interpretation. Nonetheless cases where the histomorphology is suspicious for a certain entity and the FISH yields unexpected negative results for the fusion merit additional confirmatory testing via NGS.
Fig. 1.
Oncocytic mucoepidermoid carcinoma with CRTC1-MAML2 fusion: Despite a negative MAML2 FISH result and relative lack of mucin, this tumour with predominantly solid nests of eosinophilic cells with low-grade nuclear features and focal ductular structures, had the appearance and staining pattern of an oncocytic mucoepidermoid carcinoma and proven as such by NGS with a CRTC1-MAML2 fusion
Equivocal Histomorphologic Diagnosis, Diagnosis Determined or Altered by Fusion Results
Of all the possible scenarios outlined in this paper, this is one which carries the most immediate importance and where NGS is most valuable. By demonstrating the presence of a fusion the final diagnosis can be changed with significant clinical implications. In our instance, a nasal cavity tumour biopsy was initially diagnosed as a recurrent ameloblastoma. This was based on an unconfirmed history of ameloblastoma many years prior in another country. This pathology could not be reviewed, but was supported by a biopsy showing a basaloid tumour with focal squamous differentiation, low-grade morphology and a combination of trabecular, cystic and focal cribriform growth. The final tumour resection consisted of prominent cystic structures lined by nests of basaloid cells with extensive cribriforming and peripheral nuclear palisading (Fig. 2a). The central portions of the cyst demonstrated the presence of mucoid secretions rather than necrosis. Other areas of the tumour demonstrated more solid 'cord-like' and trabecular growth. There was evidence of invasive growth as well. The cell population was homogenous, consisting of monomorphous basaloid cells with peripheral palisading. Cell nuclei were small, hyperchromatic and with inconspicuous nucleoli and no evidence of overt cytologic atypia. No stellate reticulum or inverse polarity was noted, however. The tumour did appear biphasic in nature with focal duct formation. Focal areas of the tumour demonstrated the presence of squamous pearls (Fig. 2b). There was background stromal hyalinization and fibrosis. Adenoid cystic carcinoma, adenocarcinoma NOS, and basal cell adenocarcinoma entered the differential diagnosis, along with an adenoid variant of ameloblastoma. A previous BRAF V600E immunohistochemical stain was negative which is noted in a proportion of ameloblastomas. Because the final resection showed many features unusual for an ameloblastoma, the tumour was sent for further testing via the RNA-Seq fusion panel. A MYBL1-NFIB fusion was detected, confirming the diagnosis of an adenoid cystic carcinoma.
Fig. 2.
Adenoid cystic carcinoma with squamous differentiation and variant MYBL1-NFIB fusion: a Cystic spaces lined by basaloid cells with small hyperchromatic nuclei and peripheral palisading showing cribriform growth. b Focal squamous pearls are noted in an otherwise completely basaloid appearing neoplasm on the initial biopsy. This tumour originally called ameloblastoma in the nasal cavity was diagnosed as adenoid cystic carcinoma on the basis of an MYBL1-NFIB fusion by NGS
There were multiple unusual features in this tumour, including focal squamous metaplasia, large cysts and trabecular architecture. Adenoid cystic carcinomas are typically basaloid neoplasms with small, hyperchromatic cells and generally do not have squamous differentiation or prominent cysts (despite the name “adenoid cystic carcinoma”). Prior studies referencing elements of possible squamoid differentiation in adenoid cystic carcinomas are sparse, having either considered two simultaneous primaries, or have raised the differential diagnosis of a basaloid squamous cell carcinoma [26, 27]. Certainly, in the context of this biopsy with the prior clinical history of an ameloblastoma in the same location, the detection of the MYBL1-NFIB fusion was considered sufficient evidence that the initial diagnosis was incorrect despite some unusual morphologic features. This molecular result had significant clinical implications and hence reiterates the importance of NGS in situations like this where the diagnosis was somewhat in question but with a reasonably limited differential diagnosis. Importantly however, a negative NGS result would not have had significant impact on the diagnosis or proven the original diagnosis of ameloblastoma correct.
A similar recent case of a palate biopsy showed a tumour consisting of relatively well-circumscribed nests of basaloid cells with a predominantly trabecular and canalicular-like architecture (Fig. 3). The tumour was composed of uniform basaloid cells with enlarged elongated and round nuclei with more open chromatin and occasional nucleoli. The background stroma was fibrotic and hyalinized. No obvious infiltrative growth was noted, however the biopsy lacked a stromal interface for the most part. Although adenoid cystic carcinoma was in the differential diagnosis, the favoured diagnosis was canalicular adenoma or polymorphous adenocarcinoma. Initially, immunohistochemistry was performed and led to doubt of these diagnoses due to the absence of S100 staining and diffuse p63 and p40 staining (unusual for both canalicular adenoma and polymorphous adenocarcinoma). The case was sent for NGS where an MYBL1-NFIB was similarly detected and despite the unusual morphology the finding was considered diagnostic of adenoid cystic carcinoma in a setting of a basaloid tumour with the right immunoprofile.
Fig. 3.
Adenoid cystic carcinoma with canalicular architecture and variant MYBL1-NFIB fusion: Well-demarcated islands formed by small basaloid cells with a combination of patterns including trabecular and canalicular growth. This palatal tumour was also diagnosed as adenoid cystic carcinoma on the basis of an MYBL1-NFIB fusion by NGS
Adenoid cystic carcinomas are paradoxically considered to be 'low-grade' neoplasms, yet with aggressive behavior, perineural invasion, the tendency to keep recurring and ultimately with a fatal outcome [1]. The disease-specific survival rates in these tumour types decrease dramatically from 89% at the 5-year mark, to half this amount, 40% at the 15-year mark [28]. No standardized effective systemic therapy has been defined thus far. These tumours are generally characterized by a MYB-NFIB fusion [1, 3, 11], however cases with the variant MYBL1-NFIB fusion are well known. Both the cases described above would have been missed by MYB FISH testing alone. Molecular characterization of these tumours via NGS has attempted to identify additional events which might be a clue to potential targetable mutations [11]. Thierauf et al., performed prospective genotyping of ACCs via NGS which demonstrated that a subset of tumours, up to 65% demonstrated NOTCH1 aberrations, MYB-NFIB fusions and MYBL1-NFIB fusions. These subsets were noted to have a more aggressive course with shorter progression-free survival [11]. Importantly, in this cohort, 75% of non-resectable patients had potentially actionable mutations related to the NOTCH1 aberrations, BRCA1 mutations and alterations in the PI3K-AKT-mTOR pathways with several ongoing trials [11]. NOTCH1 aberrations in particular define patients with aggressive disease and propensity for metastases but also a potential response to NOTCH-1 inhibitors [11]. Besides serving as a confirmation of the diagnosis, the role of NGS in these tumours is expected to become more and more relevant in the detection of clinically and prognostically relevant genetic alterations. This further emphasizes the need to eventually move towards broader platforms than the currently used fusion-only NGS platform.
Positive Unexpected Result Which Does not Change the Diagnosis but may be Clinically Relevant
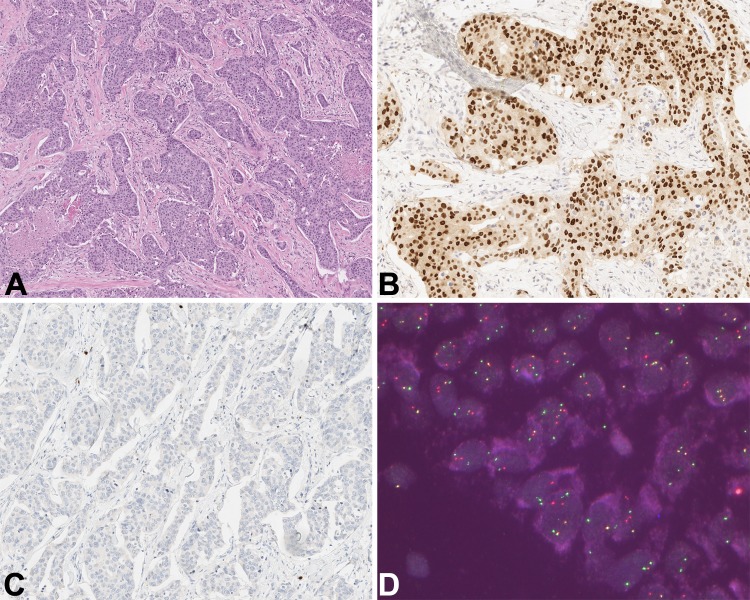
As alluded to in the context of adenoid cystic carcinomas previously, the role of NGS in the detection of clinically relevant mutations is critical. Not only can it show new insight into tumour pathobiology; it can also identify potential clinically actionable mutations. A salivary gland tumour was reviewed at our institution and was composed of eosinophilic expansile nests of high-grade apocrine-type cells with central comedonecrosis and extensive perineural and lymphovascular invasion (Fig. 4a). The tumour showed extensive growth, positive margins and numerous cervical lymph node metastases. Immunohistochemical stains showed that the tumour was positive for BRST-2, and androgen receptor (Fig. 4b), while it was negative for S100 (Fig. 4c). A diagnosis of salivary duct carcinoma was rendered on the basis of histomorphology and immunohistochemistry. As the patient happened to be enrolled in a clinical trial employing NGS to detect actionable mutations, the tumour was discovered to have an ETV6-NTRK3 fusion, which was subsequently confirmed with FISH for ETV6 (Fig. 4d). ETV6-NTRK3 fusions have been defined for some time now in secretory carcinomas of the salivary gland; first described by Skalova et al., in 2010 [29]. These tumours are defined as generally being low-grade with a lobulated growth pattern and composed of solid, microcystic, and papillary-cystic spaces. The microcystic spaces are occupied by eosinophilic luminal secretions. Tumour cells have low-grade nuclei with bubbly eosinophilic and granular cytoplasm. Immunohistochemically they are uniformly S100 and mammaglobin positive. Cases of secretory carcinomas with high-grade transformation are well known and may benefit from anti-NTRK therapies, however, they represent a minority of cases.
Fig. 4.
Salivary duct carcinoma with ETV6-NTRK3 fusion: a Infiltrative nests of apocrine-appearing cells with high-grade cytologic atypia and extensive comedo-necrosis. b Diffuse AR and BRST-2 were seen (AR shown here). c The case showed no evidence of secretory carcinoma or low-grade morphology and was S100 negative (shown here). d. FISH showed an ETV6 rearrangement (shown here) confirming the initial incidental finding of ETV6-NTRK3 by NGS
NTRK fusions in general are known to be the oncogenic drivers in multiple types of adult and pediatric tumours such as infantile fibrosarcomas and lung tumours. This led to the development of FDA-approved small-molecule inhibitors such as Larotrectinib with a documented 75% overall response rate in all solid tumours harbouring the NTRK alteration, irrespective of tumour type [12]. The appropriate detection of NTRK is therefore of paramount importance in these cases. Detection methods have included DNA and RNA-based assays, and as of late, Pan-Trk immunhistochemical staining [15, 30]. These methods have been compared against one another in respect to their utility. In a large multi-institutional study by Solomon et al., DNA-based NGS of NTRK-positive solid tumours has demonstrated an overall sensitivity of 81.1%. In fact, the greatest proportion of false-negative cases harboured NTRK2 and NTRK3 fusions. Not surprisingly, it performed worse in salivary gland tumours (characterized by ETV6-NTRK3) compared to other solid tumour types [15]. The sensitivity of DNA-based NGS is variable as it depends on tumour purity and the extent and depth of fusion NTRK1-3 breakpoints which are covered [16]. DNA-based NGS therefore may miss a breakpoint if it is not covered [16]. RNA-based sequencing on the other hand is advantageous in that it can provide ample coverage as introns are spliced out and therefore the assay for the particular coverage has less technical requirements than DNA-based assays [15]. Ultimately, RNA-based NGS directly confirms evidence of transcription [16]. However, the downside of RNA-based NGS is that RNA as a nucleic acid is easily degraded and labile in nature, making it susceptible to easy loss when extracted from FFPE. With this, it requires adequate storage and preservation of tissue and results can only be reliably interpreted if the nucleic acid quality is deemed appropriate. Although both DNA and RNA-based NGS are excellent tools for mutation detection, they are time-consuming (requiring anywhere from 2–4 weeks to receive results) and expensive, requiring appropriate validation [5, 16].
The quest for a less expensive alternative test resulted in the development of Pan-Trk immunohistochemistry for the detection of NTRK3 with variable results [15, 30]. While some studies concluded that pan-Trk is a highly specific marker for the detection of NTRK3 fusions in secretory carcinomas, other studies found that it has performed worse in salivary gland carcinomas compared to other solid tumour types [15, 16, 30]. Briefly, although the role of NTRK immunohistochemical staining is without a doubt a promising screening tool for identifying patients which may be eligible for NTRK inhibitors, it still requires additional confirmatory testing. However, the discussion regarding the application of IHC in this setting is an extensive topic meriting a separate publication.
Generally speaking, the interpretation of immunohistochemistry and NGS techniques must be done in conjunction with the histomorphologic findings as is illustrated in our case of salivary duct carcinoma with the ETV6-NTRK3 fusion confirmed via NGS and verified with ETV6 FISH. Our case both from a histomorphologic and immunohistochemical perspective was in keeping with a salivary duct carcinoma. It was re-reviewed to search for areas of more classic secretory carcinoma or any finding that would suggest a high-grade transformation event. The entire tumour consisted only of typical salivary duct carcinoma. No clinicopathologic incentive existed to test for secretory carcinoma. However, the result was incidentally detected during the clinical trial which the patient happened to be a part of. The actionable nature of this fusion warrants reporting of the finding since it is a known fusion and was validated by FISH (albeit the actionable gene, NTRK3 was not secondarily verified). At present we have no knowledge of the implications of this fusion in salivary duct carcinoma neither from a pathobiologic nor from a prognostic perspective. We have seen this result in salivary duct carcinoma on more than one occasion, however the incidence is not known in this tumour type at this time. A literature search was conducted and only one recent study demonstrated the presence of this fusion in a single case of salivary duct carcinoma [31]; all other reported cases of ETV6-NTRK3 have been diagnosed as secretory carcinomas [12, 30]. In theory, even a small incidence of 1–2% in salivary duct carcinoma would be far more relevant than in secretory carcinoma, owing to the poor prognosis for this tumour type. Salivary duct carcinomas are otherwise known to be characterized as having alterations in PI3K, p53 and HRAS [10]. Reporting this ETV6-NTRK3 finding may offer the patient additional therapeutic options, however it does not warrant a change in diagnosis on its own without a pathologic interpretation that supports it. This stresses the importance of not interpreting results of NGS testing in a vacuum; especially when advancing technology allows for not infrequent 'incidental' discoveries.
Positive Unexpected Result Which Does Not Definitively Alter the Diagnosis and has no Perceived Clinical Implications
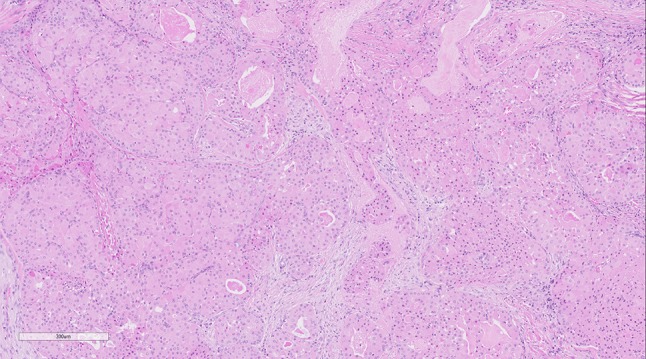
To further support the notion that NGS results should not be reported in a vacuum, the following scenario identifies a positive unexpected result which does not definitively alter the final diagnosis and does not have significant treatment implications. A middle-age male patient underwent a partial maxillectomy for a salivary gland neoplasm described as being composed of infiltrative neoplastic glands with bland nuclei, no mitotic activity or necrosis and showing focal cribriform architecture in a myxoid/desmoplastic background. The tumour showed no squamous differentiation or intracellular mucin (Fig. 5). The tumour was positive for keratins only and negative for myoepithelial markers and p63. The final diagnosis was an adenocarcinoma NOS, which showed focal bone invasion (pT4,N0). The patient has since been lost to follow up. Subsequent NGS testing was performed, for research purposes, years later and demonstrated the presence of a CRTC1-MAML2 fusion which is otherwise seen in mucoepidermoid carcinoma. However, as the morphologic features do not fit with this diagnosis definitively (no morphologic squamous differentiation or p63 positivity, and no mucin by histology or mucicarmine), the final diagnosis would not be altered in retrospect necessarily. The fusion was not reported clinically as it was a research case, however if it were a diagnostic example the fusion could be reported as being suggestive of mucoepidermoid carcinoma, or alternatively could be viewed as of uncertain significance given the lack of morphologic evidence to support the diagnosis. This would be a matter of opinion and in any event would not alter the therapy or prognosis substantively.
Fig. 5.
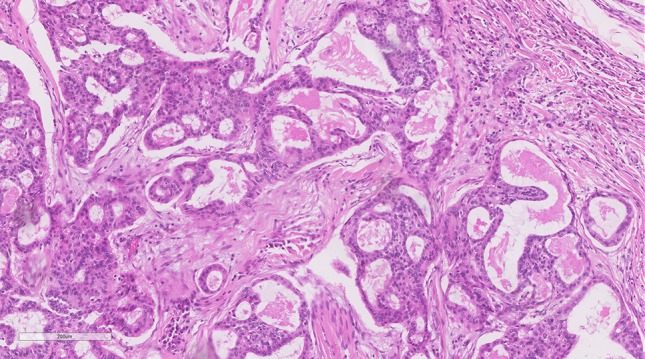
Adenocarcinoma NOS with CRTC1-MAML2 fusion: Infiltrative glands with low to intermediate-grade cytologic features and lacking any morphologic, histochemical or immunohistochemical evidence of the squamous or mucinous features typical of mucoepidermoid carcinoma. However, the case was eventually shown to be CRTC1-MAML2 positive by NGS. While it may not be appropriate to change the diagnosis it is likely this tumour is related to, or is in fact a variant morphology in mucoepidermoid carcinoma
Novel Fusion/Novel Fusion Partner
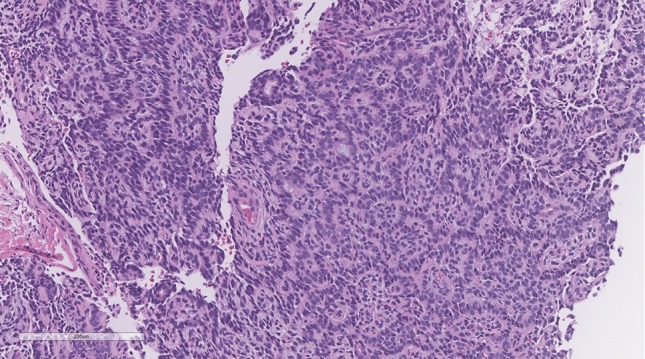
Unlike the aforementioned example of the significance of reporting NTRK fusions with known therapeutic options, one of the most compelling questions which comes with routine utilization of NGS is what to do when one detects novel fusions or fusion partners. This is commonplace in soft tissue tumours, as in the example mentioned earlier of a CITED2 rearranged spindle cell rhabdomyosarcoma. While obviously interesting from a research perspective, the question of reporting these results without secondary verification is important and not as clear cut. In one such example from our archives, a young patient was diagnosed with a high-grade adenocarcinoma NOS of the tongue base. The tumour had a polymorphous appearance composed of atypical cells arranged in cribriform structures, dyscohesive sheets and individually infiltrative cells (Fig. 6). The cells showed markedly atypical nuclei with eosinophilic and focally vacuolated cytoplasm, some of which showed a signet ring-like appearance. Mitotic activity was high. Focal intracellular mucin was noted and confirmed by the mucicarmine stain. All keratins were positive in the cribriform cells and lost in the discohesive and singly-infiltrative cells. There was focal p63 staining. The case was reported as a high-grade adenocarcinoma NOS (favoured to be arising from the minor salivary glands) and the differential diagnosis of a high-grade mucoepidermoid carcinoma and an unusual variant of adenosquamous carcinoma was raised. Subsequent NGS testing for research purposes demonstrated the presence of an EWSR1-BEND2 fusion gene. To our knowledge, EWSR1-BEND2 fusions have not been described in salivary gland neoplasia to date. This specific fusion has been rarely demonstrated in other tumour types, such as pancreatic neuroendocrine tumours [32], but these have no resemblance to this tumour. Conversely, EWSR1 in and of itself is a well-known fusion partner in several tumours, including hyalinizing clear cell carcinomas of the salivary glands and myoepithelial tumours of soft tissue (the tumour in our example did not bear histomorphologic resemblance to either of these). In the setting of a fusion with one known gene, such as EWSR1, which does not currently fit with the morphology whatsoever, the decision of whether or not to report a finding of uncertain significance is a matter of personal judgement. This was previously mentioned in the situation with the CITED2 fusion where only the primary gene was reported. Although following the reporting guidelines, one might argue that reporting it as a “Variant of Unknown Clinical Significance; Tier III”, we feel that this may actually serve to misdiagnose the patient with either a myoepithelial tumour of soft tissue or hyalinizing clear cell carcinoma (neither of which fit with the morphology in the case). Moreover, with no known clinical and therapeutic implications, reporting it would not follow our institutional policy for reporting actionable or diagnostic mutations only. A situation where this may change would be if an EWSR1 FISH rearrangement was already reported and not believed to be diagnostic. NGS would then be able to show a novel fusion that may be used to “rule out” hyalinizing clear cell carcinoma, but should be reported and interpreted with great caution. It is precisely for this reason that NGS is best used in conjunction with other ancillary techniques as independent confirmatory testing. This may be achieved through FISH, RT-PCR or Sanger sequencing which may identify the fusion end-product [5, 16].
Fig. 6.
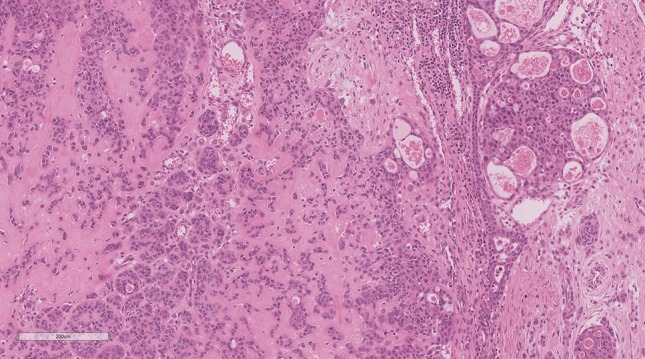
Adenocarcinoma NOS with a novel EWSR1-BEND2 fusion: Cribriform structures and individually infiltrative cells with high-grade nuclei and a signet-ring/lobular breast-like appearance in the dyscohesive cells. This tumour shows an unrecognizable morphology and was shown to have a novel EWSR1-BEND2 fusion by NGS. This type of finding may suggest a novel “cribriform and lobular adenocarcinoma” entity, but it is not appropriate to report clinically as it has no diagnostic or therapeutic implications at this time
The most difficult scenario is the detection of an entirely novel fusion, with two previously unreported genes and thus no knowledge of the clinical and pathologic implications. Generally speaking these fusions are not reported in our institution when found on this diagnostic fusion panel. Differentiating these from novel disease-defining events can be challenging and so novel, unvalidated, fusions are not reported when there is no perceived clinical implication. Reporting entirely novel fusions in the absence of known literature, and/or independent supporting data (e.g., FISH, RT-PCR) is a controversial issue. Our current policy is that novel findings are reported only if they are known to be clinically actionable or if one of the genes in the fusion sufficiently matches the morphologic interpretation such that it is considered a variant fusion for a known entity. These results cannot always be verified independently by a secondary technique, however we believe this is preferable when possible, particularly if a novel actionable fusion will alter the therapy.
These different scenarios ultimately drive home the point that NGS, as any other laboratory diagnostic test, requires proper independent confirmation. This is particularly of importance in cases of novel fusion partners [5]. The new findings must be independently confirmed via an already established diagnostic test such as FISH or RT-PCR [15]. This is also illustrated in our EWSR1-BEND2 fusion case which in our opinion is likely a novel “cribriform and lobular adenocarcinoma” for which additional cases must be sought out to better characterize.
Expanding the Range of Morphology and New Entities
One of the key reasons why NGS has been beneficial in the development of salivary gland pathology comes with the recognition of novel tumour types or novel variants [8]. By independently confirming tumour-type specific translocations via either FISH or NGS, common tumours with unusual morphology were able to be defined as simply variants; as opposed to different entities all together [21]. NGS has allowed us to prove that certain morphologies represent specific entities, such as “Microsecretory Adenocarcinoma” [6], or prove novel variants of existing tumours, such as “Mucoacinar Carcinoma”, a CRTC1-MAML2 positive mucoepidermoid carcinoma with serous acinar differentiation.
As mentioned in the opening paragraph of this paper, adenocarcinoma NOS has for a long time represented a 'waste-basket' term for tumours with ductal or glandular differentiation which could not be classified into any other diagnostic category [1]. Accounting for, not insignificantly, between 10–15% of all salivary gland carcinomas [1] this has been a 'frustrating' category one would revert to either in a limited biopsy setting or for lack of any other diagnostic options. It was not until the arrival of ancillary techniques and especially NGS, that a number of tumours from this category were re-categorized to more specific entities. Microsecretory adenocarcinomas were initially identified as being distinctive low-grade tumour types with an infiltrative growth pattern consisting of small infiltrative microcysts and cords in a fibromyxoid stroma. Tumour cells were described as being intercalated duct-like with clear to eosinophilic cytoplasm and prominent basophilic intraluminal secretions. Features of high-grade malignancy such as an elevated mitotic count and tumour-type necrosis were notably absent. All tumours were diffusely positive for S100 and p63, while negative for SMA, calponin, mammoglobin and p40 [6]. Testing by RNA-Seq by Bishop et al., all tumours with this appearance were found to harbour a novel MEF2C-SS18 fusion [6]. This fusion had not previously been seen in any solid tumour types or adenocarcinomas NOS in general. Importantly, it was the fact that the tumour was already recognized morphologically and that the fusion was seen in multiple cases and validated by RT-PCR that this could move to being a reportable fusion. A novel fusion such as this would not have been reported incidentally if this were not the case, which is the argument against reporting EWSR1-BEND2 clinically since it was an unrecognized tumour and the fusion is a “one-off” event so far. Cases such as this will likely further narrow the number of diagnosed adenocarcinoma NOS cases, which will in the future be reserved solely for the most challenging, rare cases.
Conclusion
NGS has undoubtedly contributed to the sophistication of salivary gland tumour pathology and will likely continue to reveal new discoveries. However, it must be stressed, that, despite the outlined benefits of NGS, care must be taken not to intrepret results in a vacuum. It is ultimately the combination of histology, ancillary immunohistochemical and molecular results which must be evaluated as a whole before arriving at a final diagnosis. This emphasizes the need for clear communication in complex scenarios between the pathologist and treating clinician.
To summarize our view and experience with NGS technologies in salivary gland tumours; the need for proper independent confirmation, appropriate result interpretation and clear reporting of findings are becoming the ‘pillars’ of standard of care in this era of molecular pathology.
Funding
No funding obtained.
Compliance with Ethical Standards
Conflict of interest
No conflict of interest to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization classification of tumours: pathology and genetics of head and neck tumours. 4th Edition [press release]. Lyon2017.
- 2.Foote FW, Jr, Frazell EL. Tumors of the major salivary glands. Cancer. 1953;6(6):1065–1133. doi: 10.1002/1097-0142(195311)6:6<1065::aid-cncr2820060602>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Skalova A, Stenman G, Simpson RHW, Hellquist H, Slouka D, Svoboda T, et al. The role of molecular testing in the differential diagnosis of salivary gland carcinomas. Am J Surg Pathol. 2018;42(2):e11–e27. doi: 10.1097/PAS.0000000000000980. [DOI] [PubMed] [Google Scholar]
- 4.Griffith CC, Schmitt AC, Little JL, Magliocca KR. New developments in salivary gland pathology: clinically useful ancillary testing and new potentially targetable molecular alterations. Arch Pathol Lab Med. 2017;141(3):381–395. doi: 10.5858/arpa.2016-0259-SA. [DOI] [PubMed] [Google Scholar]
- 5.Dickson BC, Swanson D. Targeted RNA sequencing: A routine ancillary technique in the diagnosis of bone and soft tissue neoplasms. Genes Chromosomes Cancer. 2019;58(2):75–87. doi: 10.1002/gcc.22690. [DOI] [PubMed] [Google Scholar]
- 6.Bishop JA, Weinreb I, Swanson D, Westra WH, Qureshi HS, Sciubba J, et al. Microsecretory adenocarcinoma: a novel salivary gland tumor characterized by a recurrent MEF2C-SS18 fusion. Am J Surg Pathol. 2019;43(8):1023–1032. doi: 10.1097/PAS.0000000000001273. [DOI] [PubMed] [Google Scholar]
- 7.Cipriani NA, Lusardi JJ, McElherne J, Pearson AT, Olivas AD, Fitzpatrick C, et al. Mucoepidermoid Carcinoma: A Comparison Of Histologic grading systems and relationship to MAML2 rearrangement and prognosis. Am J Surg Pathol. 2019;43(7):885–897. doi: 10.1097/PAS.0000000000001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonescu CR, Katabi N, Zhang L, Sung YS, Seethala RR, Jordan RC, et al. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes Chromosomes Cancer. 2011;50(7):559–570. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- 9.Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santana T, Pavel A, Martinek P, Steiner P, Grossmann P, Baneckova M, et al. Biomarker Immunoprofile and molecular characteristics in salivary duct carcinoma: Clinicopathologic and prognostic implications. Hum Pathol. 2019. [DOI] [PubMed]
- 11.Thierauf J, Ramamurthy N, Jo VY, Robinson H, Frazier RP, Gonzalez J, et al. Clinically integrated molecular diagnostics in adenoid cystic carcinoma. Oncologist. 2019. [DOI] [PMC free article] [PubMed]
- 12.Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Italiano A, Di Mauro I, Rapp J, Pierron G, Auger N, Alberti L, et al. Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): a prospective, multicentre, observational study. Lancet Oncol. 2016;17(4):532–538. doi: 10.1016/S1470-2045(15)00583-5. [DOI] [PubMed] [Google Scholar]
- 14.Dogan S. Novel rearrangements in salivary gland tumors detected by an RNA-based targeted next-generation sequencing assay. Lab Investig. 2019;99:10. [Google Scholar]
- 15.Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2019. [DOI] [PMC free article] [PubMed]
- 16.Solomon JP, Hechtman JF. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. 2019;79(13):3163–3168. doi: 10.1158/0008-5472.CAN-19-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, american society of clinical oncology, and college of american pathologists. J Mol Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alaggio R, Zhang L, Sung YS, Huang SC, Chen CL, Bisogno G, et al. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: identification of novel and recurrent VGLL2-related fusions in infantile cases. Am J Surg Pathol. 2016;40(2):224–235. doi: 10.1097/PAS.0000000000000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moller E, Stenman G, Mandahl N, Hamberg H, Molne L, van den Oord JJ, et al. POU5F1, encoding a key regulator of stem cell pluripotency, is fused to EWSR1 in hidradenoma of the skin and mucoepidermoid carcinoma of the salivary glands. J Pathol. 2008;215(1):78–86. doi: 10.1002/path.2327. [DOI] [PubMed] [Google Scholar]
- 20.Garcia JJ, Hunt JL, Weinreb I, McHugh JB, Barnes EL, Cieply K, et al. Fluorescence in situ hybridization for detection of MAML2 rearrangements in oncocytic mucoepidermoid carcinomas: utility as a diagnostic test. Hum Pathol. 2011;42(12):2001–2009. doi: 10.1016/j.humpath.2011.02.028. [DOI] [PubMed] [Google Scholar]
- 21.Bishop JA, Cowan ML, Shum CH, Westra WH. MAML2 rearrangements in variant forms of mucoepidermoid carcinoma: ancillary diagnostic testing for the ciliated and warthin-like variants. Am J Surg Pathol. 2018;42(1):130–136. doi: 10.1097/PAS.0000000000000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seethala RR, Dacic S, Cieply K, Kelly LM, Nikiforova MN. A reappraisal of the MECT1/MAML2 translocation in salivary mucoepidermoid carcinomas. Am J Surg Pathol. 2010;34(8):1106–1121. doi: 10.1097/PAS.0b013e3181de3021. [DOI] [PubMed] [Google Scholar]
- 23.Luk PP, Wykes J, Selinger CI, Ekmejian R, Tay J, Gao K, et al. Diagnostic and prognostic utility of Mastermind-like 2 (MAML2) gene rearrangement detection by fluorescent in situ hybridization (FISH) in mucoepidermoid carcinoma of the salivary glands. Oral Surg. 2016;121(5):530–541. doi: 10.1016/j.oooo.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Kang H, Tan M, Bishop JA, Jones S, Sausen M, Ha PK, et al. Whole-exome sequencing of salivary gland mucoepidermoid carcinoma. Clin Cancer Res. 2017;23(1):283–288. doi: 10.1158/1078-0432.CCR-16-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Roh JL, Choi YJ, Choi J, Cho KJ. High grade transformation in mucoepidermoid carcinoma of the minor salivary gland with polyploidy of the rearranged MAML2 gene. Head Neck Pathol. 2019. [DOI] [PMC free article] [PubMed]
- 26.Doumas S, Barrett AW, Carrillo M, Tighe JV. Synchronous metastatic adenoid cystic and squamous cell carcinoma of the cervical lymph nodes 31 years after ablation of the primary palatal tumour. Br J Oral Maxillofac Surg. 2013;51(7):e190–e191. doi: 10.1016/j.bjoms.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 27.van der Wal JE, Snow GB, Karim AB, van der Waal I. Adenoid cystic carcinoma of the palate with squamous metaplasia or basaloid-squamous carcinoma? Report of a case. J Oral Pathol Med. 1994;23(10):461–464. doi: 10.1111/j.1600-0714.1994.tb00445.x. [DOI] [PubMed] [Google Scholar]
- 28.Coca-Pelaz A, Rodrigo JP, Bradley PJ, Vander Poorten V, Triantafyllou A, Hunt JL, et al. Adenoid cystic carcinoma of the head and neck—An update. Oral Oncol. 2015;51(7):652–661. doi: 10.1016/j.oraloncology.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34(5):599–608. doi: 10.1097/PAS.0b013e3181d9efcc. [DOI] [PubMed] [Google Scholar]
- 30.Xu B, Haroon Al Rasheed MR, Antonescu CR, Alex D, Frosina D, Ghossein R, et al. Pan-Trk immunohistochemistry is a sensitive and specific ancillary tool in diagnosing secretory carcinoma of salivary gland and detecting ETV6-NTRK3 fusion. Histopathology. 2019 doi: 10.1111/his.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalin MG, Desrichard A, Katabi N, Makarov V, Walsh LA, Lee KW, et al. comprehensive molecular characterization of salivary duct carcinoma reveals actionable targets and similarity to apocrine breast cancer. Clin Cancer Res. 2016;22(18):4623–4633. doi: 10.1158/1078-0432.CCR-16-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543(7643):65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]