Summary
A change in Presenilin (PS)/γ-secretase activity is linked to essential biological events as well as to the progression of many diseases. However, not much is known about how PS/γ-secretase activity is spatiotemporally regulated in cells. One of the limitations is lack of tools to directly monitor dynamic behavior of the PS/γ-secretase in intact/live cells. Here we present successful development and validation of the Förster resonance energy transfer (FRET)-based biosensors that enable quantitative monitoring of endogenous PS/γ-secretase activity in live cells longitudinally on a cell-by-cell basis. Using these FRET biosensors, we uncovered that PS/γ-secretase activity is heterogeneously regulated among live neurons.
Subject Areas: Biological Sciences, Cell Biology, Methodology in Biological Sciences, Neuroscience
Graphical Abstract
Highlights
-
•
C99 R-G biosensor reports endogenous PS/γ-secretase activity in live cells
-
•
Modified donor/acceptor and linker length result in higher sensitivity C99 Y-T sensor(s)
-
•
APP C99 in the C99 Y-T biosensor can be replaced with Notch1 N100
-
•
PS/γ-secretase activity is heterogeneously regulated among live neurons
Biological Sciences; Cell Biology; Methodology in Biological Sciences; Neuroscience
Introduction
Presenilin (PS)/γ-secretase is a membrane-embedded aspartic protease responsible for the proteolytic processing of a wide variety of membrane-associated proteins that include the amyloid precursor protein (APP) and Notch1 (De Strooper et al., 1998, De Strooper et al., 1999, Wolfe et al., 1999). The proteolytic cleavage of APP by BACE1/β-secretase releases the APP extracellular domain and generates the membrane-bound APP C99 fragment that is an immediate substrate of PS/γ-secretase. The first step cleavage of APP C99 by PS/γ-secretase, which is known as epsilon-cleavage, generates longer Aβ48 or Aβ49 peptides and the APP intracellular domain (AICD). Longer Aβ peptides are subsequently processed by PS/γ-secretase (so-called gamma cleavages) to produce shorter Aβ37- Aβ43 peptides (Qi-Takahara et al., 2005). The accumulation in the brain of Aβ peptides, Aβ42 in particular, is one of the hallmarks of Alzheimer's disease (AD). In the case of Notch1, the N-terminally truncated Notch1 by a furin-like protease and ADAMs/α-secretase is further processed by PS/γ-secretase. This PS/γ-secretase-mediated Notch1 processing generates Notch intracellular domain (NICD), resulting in transcriptional changes in the nucleus, which is known as Notch signaling (Schroeter et al., 1998).
PS/γ-secretase is widely expressed throughout the body and plays a pivotal role in essential biological events during development, e.g., neurogenesis and skeletal formation (Shen et al., 1997, Wong et al., 1997). It has been also suggested that changes in PS/γ-secretase activity are linked to the pathogenesis of numerous diseases that are related to brain, skin, immune system, etc. (reviewed in Jurisch-Yaksi et al., 2013). Yet, little is known about how PS/γ-secretase activity is spatiotemporally regulated, as there are no tools currently available to “visualize” PS/γ-secretase activity in living cells. A few PS/γ-secretase activity assays, including the cell-free in vitro activity assay (Kakuda et al., 2006) or the cell-based reporter assay (Cao and Sudhof, 2001, Hansson et al., 2006, Ilagan et al., 2011) have been developed; however, shortcomings in these assays do not permit investigation of the dynamics of endogenous PS/γ-secretase activity on a cell-by-cell basis and over time. Here we report successful development, optimization, and validation of the Förster resonance energy transfer (FRET)-based biosensors for monitoring the activity of endogenous PS/γ-secretase in intact and/or live cells. These biosensors allow direct visualization of PS/γ-secretase within an individual neuron and demonstrate that PS/γ-secretase activity is differently regulated in various neurons over time.
Results
Development of the C99 R-G Biosensor
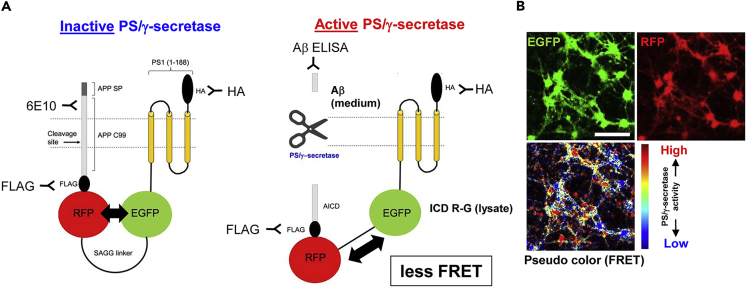
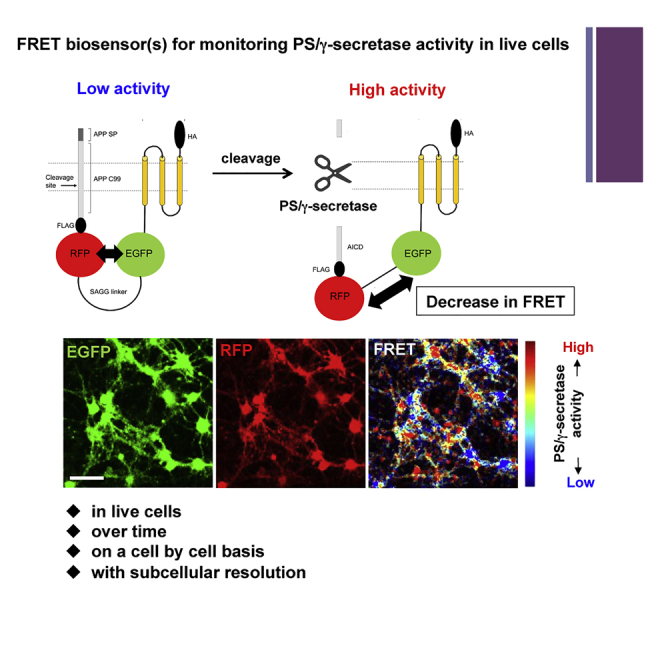
PS/γ-secretase is responsible for the processing of a wide variety of membrane associated proteins and thus potentially influences numerous cellular pathways. However, there are no tools or assays currently available to monitor dynamic changes in the activity of PS/γ-secretase in live cells over time on a cell-by-cell basis and ultimately to help identify molecular regulators of its activity. This study aims to develop a biosensor that would enable exploring the dynamic nature of PS/γ-secretase. The C99 RFP-EGFP (C99 R-G) biosensor is an ideal molecular probe for the FRET-based assay, since in addition to an immediate PS/γ-secretase substrate, APP C99, it contains two fluorescent proteins, EGFP (donor) and RFP (acceptor) expressed in a 1:1 ratio (Figure 1A left and 1B). The C terminus of human APP C99 is tagged with RFP and EGFP is connected to the RFP with 20 amino acids (a.a.) SAGG-repeat linker (Komatsu et al., 2011). To increase FRET detection capability, the EGFP is stabilized near the membrane by fusion to the N-terminal portion of PS1. This anchor domain contains 1–188 a.a. spanning only the N terminus and first three transmembrane domains of PS1. Of note, this fragment does not contain any known binding sites for co-factors (nicastrin, Aph1, and Pen2) (Kaether et al., 2004, Watanabe et al., 2005) or the catalytic core (Wolfe et al., 1999) of PS/γ-secretase and thus does not possess functional PS/γ-secretase activity. For selective biochemical detection of the C99 R-G processing, FLAG and HA tags are inserted at the C terminus of APP C99 and after the PS1 a.a. 188, respectively. The cleavage of APP C99 within the C99 R-G biosensor by endogenous PS/γ-secretase releases Aβ peptides and APP intracellular domain (ICD) R-G (Figure 1A right). This results in a change in the proximity and/or orientation between the EGFP and the RFP, which we record by ratiometric spectral FRET analysis (Uemura et al., 2009, Maesako et al., 2017) as a reduction in the FRET efficiency. The value of the FRET efficiency can be color coded and mapped over the entire image of a cell. Therefore, the measurement of FRET efficiency permits the “visualization” of PS/γ-secretase-mediated APP C99 processing within a cell (Figure 1B).
Figure 1.
Development of the C99 R-G Biosensor
(A) Schematic representation of the C99 R-G FRET biosensor. Endogenous PS/γ-secretase cleaves the APP C99, which results in the production of Aβ peptides and the APP intracellular domain (ICD) R-G, and causes a low FRET efficiency between the EGFP and the RFP.
(B) The AAV-mediated expression of C99 R-G probe in mouse cortex primary neurons verified by confocal microscopy, and the pseudo-colored image corresponding to FRET efficiency measured by spectral FRET analysis. Scale bar, 100 μm.
The C99 R-G Biosensor Is Cleaved by PS/γ-Secretase
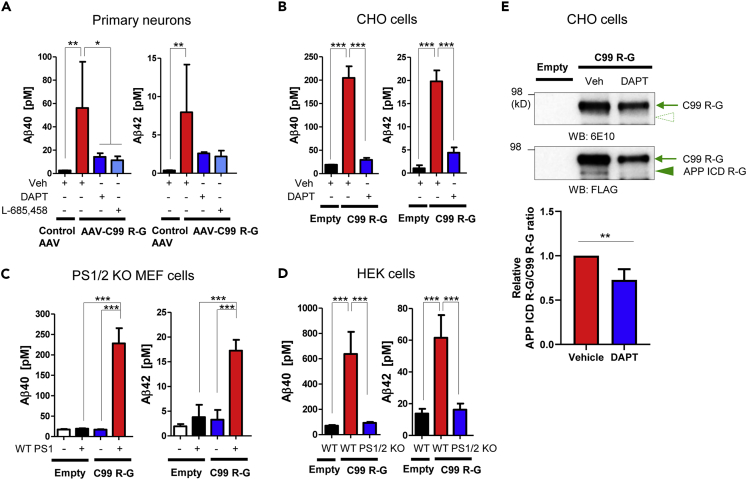
Cell fractionation revealed that the C99 R-G probe is integrated into the membrane (Figure S1A). Furthermore, the cell surface expression of C99 R-G was verified by a biotinylation assay confirming that the C99 R-G biosensor is trafficked through the secretory pathway (Figure S1B). To further ensure that the C99 R-G biosensor is successfully cleaved by endogenous PS/γ-secretase, we measured the level of Aβ in the conditioned medium using human Aβ40 and Aβ42 ELISA. The expression of C99 R-G in primary neurons as well as in CHO cells allowed for a clear detection of human Aβ40 and Aβ42 in the conditioned medium at approximately 10:1 ratio of the Aβ40 to Aβ42. The Aβ generation was inhibited by the treatment with PS/γ-secretase inhibitor(s), DAPT or L-685,458, verifying the specificity of the detection (Figures 2A and 2B). Human Aβ40 and Aβ42 were also detected in the conditioned media of genetically manipulated MEF and HEK cells expressing C99 R-G and PS1 (Figures 2C and 2D). In addition, western blotting was performed to detect the APP intracellular domain R-G (APP ICD R-G) in the lysate of C99 R-G expressing cells. The 6E10 antibody binding to an epitope on the N terminus of C99 R-G detected a single band around 95 kD corresponding to C99 R-G in the lysates of both vehicle and DAPT-treated cell. On the other hand, the FLAG antibody detected a lower-molecular-weight band corresponding to APP ICD R-G, which was significantly diminished by the DAPT treatment (Figure 2E). To examine the stability of C99 R-G and APP ICD R-G, the cycloheximide chase assay was performed. We found that the half-life of C99 R-G and APP ICD R-G is approximately 4 h (Figure S1C), which is significantly longer than that of endogenous C99 and APP ICD (Dyrks et al., 1993, Kimberly et al., 2001). This suggests that the fusion with RFP-linker-EGFP-PS1 (1–188)-HA might stabilize the C99 and APP ICD. Overall, these results verify that the C99 R-G biosensor is cleaved by endogenous PS/γ-secretase.
Figure 2.
The C99 R-G Biosensor Is Cleaved by PS/γ-Secretase
Detection of human Aβ40 and Aβ42 in the conditioned medium of C99 R-G expressing primary neurons (A), CHO (B), PS1/2 KO MEF + WT PS1 (C), or HEK cells (D). Aβ generation was inhibited by the treatment with PS/γ-secretase inhibitor(s) or the knockdown of PS1/2 expression. n = 4–5 biological replicates, mean ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way factorial ANOVA. (E) Western blotting using Novex 6% Tris-Glycine gels reveals APP ICD R-G band (arrowhead) detected with the FLAG antibody but not with the 6E10 antibody in CHO cell lysates. The arrow indicates full-length C99 R-G. The ratio of APP ICD R-G band over C99 R-G was quantified and the relative ratio was shown (vehicle is set as 1). n = 3 biological replicates, mean ± SD, ∗∗p < 0.01, one-sample t test.
PS/γ-Secretase-Mediated Processing of the C99 R-G Biosensor Causes Decrease in FRET between the EGFP and the RFP
Next, we used the ratiometric spectral FRET assay we set up in the laboratory (Uemura et al., 2009, Maesako et al., 2017) to determine if the FRET signal between the EGFP and RFP fluorophores within the C99 R-G probe would change after endogenously expressed PS/γ-secretase cleaves the probe. In the spectral FRET assay, the EGFP (donor) within C99 R-G is selectively excited by an Argon laser at 488 nm and emitted fluorescence is simultaneously detected by eight channels within the 456- to 617-nm wavelength range (21.4 nm bandwidth). The ratio of the fluorescence emission intensity in the 596 ± 10.7-nm channel (emission peak of RFP) to the emission in 510 ± 10.7-nm channel (emission peak of EGFP) yields the 596(R)/510(G) ratio that is used as readout of the FRET efficiency. The lower 596(R)/510(G) ratio denotes lower FRET efficiency, and in the case of C99 R-G biosensor, would indicate higher PS/γ-secretase activity.
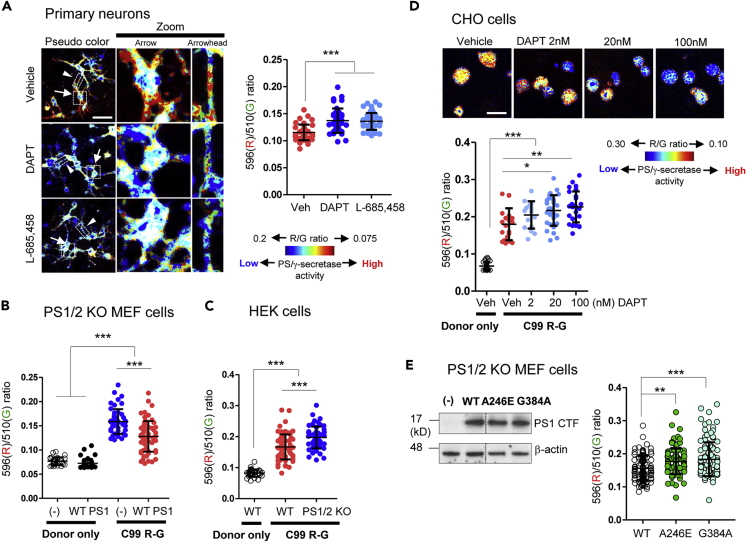
First, we verified the cleavage of the C99 R-G biosensor by PS/γ-secretase indeed results in reduced FRET efficiency. For this, mouse primary neurons expressing the C99 R-G biosensor were treated with DAPT or L-685,458 (PS/γ-secretase inhibitors, GSIs) to block the activity of endogenous PS/γ-secretase. The overall 596(R)/510(G) ratio in DAPT- or L-685,458-treated neurons was significantly higher than that in the vehicle-treated cells, indicating that high FRET efficiency is associated with low PS/γ-secretase activity (Figure 3A). The pseudo-colored image allows one to visualize regional differences in the FRET efficiency in vehicle-treated cells, revealing that PS/γ-secretase activity varies at the cell periphery versus perinuclear area or in different loci within the processes (Figure 3A). To corroborate the finding in different cell types and with a complementary genetic approach, we measured FRET efficiency in C99 R-G biosensor expressing MEF cells lacking both PS1 and PS2 (PS1/2 KO). The rescue of PS/γ-secretase activity by the stable expression of wild-type (WT) PS1 resulted in lowering of the 596(R)/510(G) ratio (Figure 3B). Similarly, the 596(R)/510(G) ratio was significantly lower in C99 R-G-expressing WT HEK cells, as compared with HEK cells in which PS1/2 expression is suppressed by the CRISPR-Cas9. These results are consistent with the higher 596(R)/510(G) ratio in cells representing reduced (lacking) PS/γ-secretase activity (Figure 3C).
Figure 3.
PS/γ-Secretase-Mediated Processing of the C99 R-G Biosensor Causes Decrease in FRET between the EGFP and the RFP
(A) The pseudo-colored images of the 596(R)/510(G) ratio in primary neurons infected with the AAV-C99 R-G. The square and rectangle in low-magnification images (left) pointed by arrowhead and arrow, respectively, correspond to the high magnification images (middle and right). The color-coded images were generated by MATLAB using 0.075–0.2 range of the 596(R)/510(G) ratios to visually amplify the difference between experimental conditions. The overall 596(R)/510(G) ratio in processes of neurons treated with DAPT or L-685,458 (1 μM,16 h) is significantly higher than that in vehicle control (n = 40–47 regions of interest [ROIs] on neuronal processes), as shown by predominantly blue pixels. Scale bar, 50 μm.
(B) The stable expression of WT PS1 in the PS1/2 KO MEF cells, i.e., re-constitution of the γ-secretase activity, results in the lower 596/510 ratio (n = 31–64 cells).
(C) The knockdown of PS1/2 in HEK cells significantly increases the 596(R)/510(G) ratio (n = 31–73 cells).
(D) Dose-dependent increase in the 596(R)/510(G) ratio in CHO cells treated with DAPT for 16 h (n = 16–30 cells). The pseudo-colored images suggest heterogeneous inhibition of γ-secretase activity. Scale bar, 20 μm.
(E) Comparable expression levels of the active PS1 in the PS1/2 KO MEF cells stably expressing WT, A246E, or G384A PS1 (top panel). The 596(R)/510(G) ratio in the PS1/2 KO MEF + A246E PS1 or G384A PS1 cells was significantly higher than that in the PS1/2 KO MEF + WT PS1 cells (n = 102–115 cells) (bottom). Mean ± SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way factorial ANOVA.
To determine the ability of the C99 R-G biosensor to report progressing changes in the PS/γ-secretase activity, as oppose to “on”/“off” activity recording, the CHO cells expressing C99 R-G probe were treated with DAPT (IC50 = 20 nM) at different concentrations. The spectral FRET imaging revealed a dose-dependent increase in the 596(R)/510(G) ratio with increasing DAPT concentrations, demonstrating that the C99 R-G sensor can report gradual changes in the PS/γ-secretase activity (Figure 3D). To ensure that differences in the 596(R)/510(G) FRET ratio do not come from a different expression level of the C99 R-G probe, we performed a correlation analysis between the 596(R)/510(G) ratio and the EGFP donor fluorescence emission intensity, which reflects the expression level of C99 R-G sensor in each cell. We found that there was no positive correlation between the 596(R)/510(G) ratio and the expression level of C99 R-G biosensor in primary neurons, CHO cells or the PS1/2 DKO MEF cells stably expressing WT PS1 (Figure S2A-C). This indicates that the difference in the FRET ratio is not dependent on the level of substrate but rather on the change in proximity and/or orientation between EGFP and RFP due to changes in the PS/γ-secretase activity.
To further validate the C99 R-G spectral FRET assay, we evaluated the relative proximity between the EGFP and RFP fluorophores in vehicle/GSIs-treated cells using a complementary FRET assay: fluorescence lifetime imaging microscopy (FLIM). FLIM monitors lifetimes of the donor fluorophore (EGFP in this case) as a measure of the proximity to an acceptor (RFP) fluorophore, with shorter lifetimes reflective of a closer proximity between the donor and acceptor (Berezovska et al., 2003). In cells expressing EGFP donor fluorophore only, the EGFP lifetime is significantly longer compared with that in the cells expressing C99 R-G that contain both donor and acceptor fluorophores (Figure S2D). The shortened EGFP lifetime in DAPT- or L-685,458-treated cells as compared with the vehicle control indicates closer EGFP-RFP proximity when PS/γ-secretase activity is inhibited and the probe is not cleaved (Figure S2D). This is consistent with the findings from the spectral FRET analysis.
Recent studies suggest that the majority of familial AD (FAD) mutations in PS1 cause a decrease or loss of PS/γ-secretase activity (Quintero-Monzon et al., 2011, Chávez-Gutiérrez et al., 2012, Sun et al., 2017). To test whether the C99 R-G probe would be processed differently by the WT and FAD mutant PS1/γ-secretases, we expressed the C99 R-G biosensor in PS1/2 dKO MEF cells stably transfected with WT, A246E, or G384A PS1. Western blotting revealed comparable level of PS1 expression between the WT and mutant PS1 cell lines (Figure 3E top). The spectral FRET analysis, however, showed that the 596(R)/510(G) ratio was significantly higher in the A246E- or G384A PS1-expressing MEF cells compared with that in the WT PS1 cells (Figure 3E bottom), reflecting reduced overall activity of the PS/γ-secretase in FAD mutants. Thus, the C99 R-G biosensor can reliably document the reduced PS/γ-secretase activity due to FAD PS1 mutations in intact cells.
Optimization of the C99 R-G Biosensor
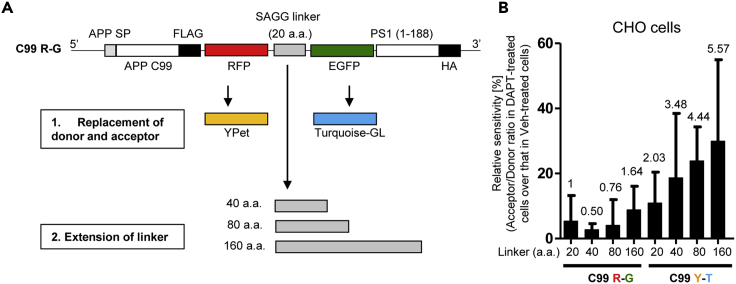
It has been previously reported that replacement of the donor/acceptor fluorescent proteins and extension of the linker length significantly improves the sensitivity of FRET biosensor monitoring kinases activity such as PKA or ERK (Komatsu et al., 2011). Thus, we next tested whether replacing the EGFP and RFP in the C99 R-G biosensor with Turquoise-GL (T) and YPet (Y) as a new donor and acceptor pair and/or extending the 20 a.a. SAGG linker to 40, 80, or 160 a.a. would enhance the sensitivity of the original C99 R-G biosensor (Figure 4A). The original C99 R-G (20 a.a.) biosensor and the optimized biosensors were expressed in CHO cells. The cells were treated with DAPT or vehicle control, and the 596(R)/510(G) or the 531(Y)/489(T) FRET ratio was measured by the spectral FRET analysis. To compare the sensitivity of the optimized FRET probes with the original C99 R-G (20 a.a.) biosensor, we calculated relative sensitivity of each biosensor. For this, the R/G or Y/T FRET ratio in cells expressing each probe and treated with DAPT was divided by the FRET ratio in the corresponding vehicle-treated cells. We found that replacement of the donor/acceptor fluorescent proteins and the extension of the linker length in C99 Y-T probe significantly increased the sensitivity of the original C99 R-G biosensor (Figure 4B). The cytotoxicity assay showed no significant toxicity owing to biosensor expression in CHO cells (Figure S3A). Of note, we found much lower transfection efficiency for the C99 Y-T (160 a.a.) construct compared with the other biosensors (data not shown). The C99 Y-T (80 a.a.) biosensor permits one to identify “the exceptional cells” in vehicle-treated primary neurons that show extremely high or low PS/γ-secretase activity (Figure S3B). This previously lacking capability to distinguish individual WT neurons with active and inactive PS/γ-secretase would help to understand the consequences of changes in PS/γ-secretase activity over time.
Figure 4.
Optimization of the C99 R-G Biosensor
(A) Schematic representation of the sequence of C99 R-G and C99 Y-T FRET biosensors. The EGFP (donor) and RFP (acceptor) in the C99 R-G biosensor were replaced with Turquoise-GL and YPet, respectively. The 20-a.a. SAGG linker in the C99 R-G or C99 Y-T biosensor was extended to 40, 80, or 160 a.a.
(B) CHO cells expressing different biosensors were treated with 1 μM DAPT or vehicle control for 16 h. The 596(R)/510(G) ratio or 531(Y)/489(T) ratio in cells treated with DAPT was divided by that in corresponding vehicle-treated cells, and the value was multiplied by 100 to calculate the relative sensitivity of each biosensor (%) (n = 76–149 cells). The fold-change compared with the original C99 R-G (20 a.a.) biosensor (set as 1) is shown. Mean ± SD.
To determine whether fusion of the donor/acceptor florescent proteins and the PS1 (1–188) affects the trafficking of C99 and thus cleavage by PS/γ-secretase, we evaluated the processing efficiency between the cells expressing C99 FLAG and the C99 Y-T (80 a.a.) probe. For this we compared the ratio of the cleavage product (Aβ40 in the medium) with the level of the substrate in respective cell lysate. We found that there was no significant difference in the product/substrate ratio between the C99 FLAG- and the C99 Y-T (80 a.a.)-expressing CHO cells (Figure S3C). Thus, we conclude that fusion of the donor/acceptor florescent proteins and the PS1 (1–188) does not significantly affect the cleavage efficiency of C99 by PS/γ-secretase.
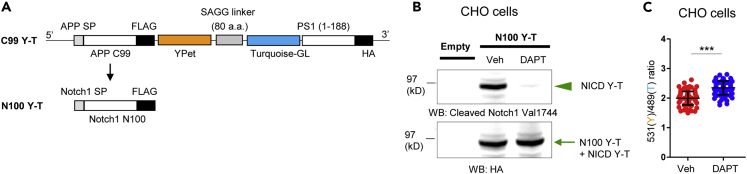
Development and Validation of the Notch1-Based N100 Y-T Biosensor
To further verify the assay is not “APP C99-specific,” we have generated a Notch1-substrate-based PS/γ-secretase activity biosensor, N100 Y-T, by replacing the APP C99 sequence in the C99 Y-T (80 a.a.) biosensor with the N- and C-terminally truncated Notch1 (N100), which is known to be an immediate substrate of PS/γ-secretase (Esler et al., 2002) (Figure 5A). In western blotting, the HA antibody reveals both N100 Y-T and NICD Y-T bands and thus shows a mixture of these two bands in vehicle-treated lysates, and mainly N100 Y-T in DAPT condition, whereas the Notch intracellular domain (NICD) antibody selectively detects only NICD Y-T band present in the lysate of vehicle treated CHO cells, but not DAPT, verifying that the N100 Y-T biosensor is processed by endogenous PS/γ-secretase (Figure 5B). Spectral FRET analysis reveals that DAPT significantly increases the 531(Y)/489(T) ratio compared with vehicle control in N100 Y-T expressing cells, showing that the cleavage of N100 Y-T biosensor by endogenous PS/γ-secretase reduces FRET efficiency between the Turquoise-GL donor and YPet acceptor (Figure 5C). Thus, similarly to the APP C99-based biosensors, the Notch1 N100 Y-T probe is successfully cleaved by and therefore could be used to report activity of the endogenous PS/γ-secretase.
Figure 5.
Development and Validation of the Notch1-based N100 Y-T Biosensor
(A) Schematic representation of the sequence of N100 Y-T biosensor. The APP C99 in the C99 Y-T (80 a.a.) biosensor was replaced with Notch1 N100.
(B) Combined full-length N100 Y-T and NICD Y-T bands are detected with the HA antibody in Western blotting using NuPAGE 4%–12% Bis-Tris Protein gels. The NICD-specific cleaved Notch1 Val1744 antibody reveals cleaved NICD Y-T band only, which is diminished by the treatment with DAPT.
(C) The overall 531(Y)/489(T) ratio in DAPT-treated CHO cells expressing the N100 Y-T biosensor is significantly higher than that in vehicle control (n = 78–81 cells). Mean ± SD, ∗∗∗p < 0.001, Student's t test.
Heterogeneous Regulation of PS/γ-Secretase Activity among Different Neurons
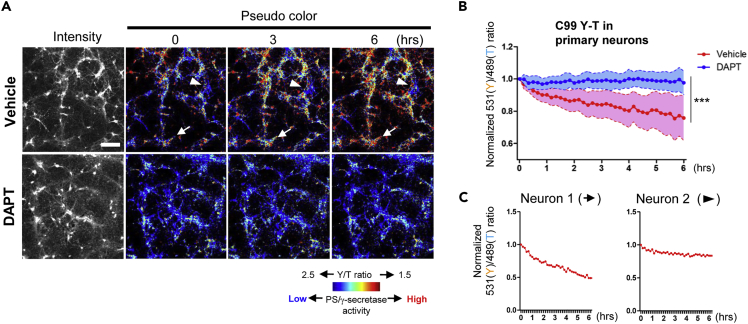
Finally, to examine if the C99 Y-T biosensor could monitor dynamic longitudinal changes in PS/γ-secretase activity within a single cell, and thus to elucidate whether PS/γ-secretase activity is temporally and/or differentially regulated among neurons, the 531(Y)/489(T) ratio was longitudinally monitored for 6 h in live neurons treated with DAPT or vehicle control in the presence of cycloheximide. As expected, there was no change in the 531(Y)/489(T) ratio over time in neuronal cultures where the PS/γ-secretase activity was inhibited by DAPT (Figures 6A and 6B). However, we observed a gradual decrease in the overall 531(Y)/489(T) ratio in the vehicle control-treated neurons (Figures 6A and 6B), indicating that the C99 Y-T biosensor is being cleaved by PS/γ-secretase over time. More importantly, cell-by-cell analysis of the 531(Y)/489(T) ratio permitted dissociation of neuronal populations in which PS/γ-secretase would be active or inactive (Figure 6C), highlighting the heterogeneous regulation in PS/γ-secretase-mediated C99 Y-T biosensor processing among different neurons. Altogether, these results provide strong evidence that cleavage of the C99/N100 donor-acceptor biosensor(s) by endogenous PS/γ-secretase results in reduced FRET efficiency between the donor (EGFP/Turquoise-GL) and the acceptor (RFP/YPet), and thus they could be used as reliable reporters of the activity of endogenous PS/γ-secretase in its native environment in intact/live cells. Furthermore, the FRET-based biosensors we developed allowed us to uncover the dynamic and heterogeneous nature of the PS/γ-secretase in different neurons.
Figure 6.
Heterogeneous Regulation of PS/γ-Secretase Activity among Different Neurons
(A) The pseudo-colored images of longitudinal (for 6 h) imaging of the 531(Y)/489(T) ratio in live primary neurons infected with AAV-C99 Y-T.
(B) Time-dependent decrease in the 531(Y)/489(T) ratio observed in vehicle-treated neurons is abolished by the treatment with DAPT (n = 39 cells). Mean ± SD, ∗∗∗p < 0.001, Repeated Measures ANOVA, Scale bar, 100 μm.
(C) Cell-by-cell analysis reveals neurons with active (Neuron 1 highlighted by an arrow in Figure 6A) or less active PS/γ-secretase (Neuron 2 by an arrowhead).
Discussion
PS knockout mice revealed that PS/γ-secretase is required for normal neurogenesis, neuronal survival, and skeletal and vasculature formation in embryos, indicating that PS/γ-secretase plays a significant role during development (Shen et al., 1997, Wong et al., 1997). Approximately 100 different substrates of PS/γ-secretase have been identified (Haapasalo and Kovacs, 2011), stressing that changes in the activity of PS/γ-secretase would potentially affect numerous cell pathways. Indeed, conditional knockout of the PS1/2, Nicastrin, or Aph1 in the excitatory neurons of adult mouse brain reveals age-dependent neuronal loss (Saura et al., 2004; Tabuchi et al., 2009; Acx et al., 2017), supporting a crucial role of PS/γ-secretase in neuronal survival. Furthermore, over 300 missense mutations that cause early onset of AD have been identified on the genes encoding PS1 and PS2 (http://www.alzforum.org/mutations), emphasizing the importance of PS/γ-secretase in the pathogenesis of AD. Together, these highlight the significant roles of PS/γ-secretase in neuronal survival and neurodegeneration.
Several assays for monitoring PS/γ-secretase activity have been previously developed, such as the cell-free in vitro PS/γ-secretase activity assay (Kakuda et al., 2006), the cell-based reporter assays (Cao and Sudhof, 2001, Hansson et al., 2006, Ilagan et al., 2011), or the C99 GFP-based fluorescence membrane retention assay (Florean et al., 2008). The major advantage of the cell-free in vitro activity assay, which is widely used in many studies (including ours), is its high sensitivity; however, it allows neither a cell-by-cell-based analysis nor longitudinal monitoring of the PS/γ-secretase activity. Moreover, it is unclear if the activity of PS/γ-secretase in vitro truly reflects that in live/intact cells. On the other hand, the cell-based reporter assay (Gal4-dependent Luciferase expression, split GFP, etc.) and the fluorescence membrane retention assay could be potentially influenced by stability of the intracellular domain. Additionally, these assays have been verified in a limited number of cell types and thus may not allow monitoring PS/γ-secretase activity in neurons. To overcome these shortcomings, we present FRET-based biosensors based on APP-C99 and Notch1-N100 substrates that enable reporting of endogenous PS/γ-secretase activity over time on a cell-by-cell basis and within the physiological environment of the cell.
A broad range of evidence from studies in C. elegans, flies, and mice has implicated that FAD PS1/2 mutations result in the loss of proteolytic activity of PS/γ-secretase (Levitan et al., 1996, Seidner et al., 2006, Saito et al., 2011, Xia et al., 2015). Partial loss of PS/γ-secretase activity results in multiple biological events that include relative increase in the membrane-bound C-terminal fragment and decrease in the intracellular domain of PS/γ-secretase substrates, as well as relative increase in the Aβ42 over Aβ40 peptides. Although over 20 years have passed from the first discovery of missense mutations on the genes encoding PS1/PS2 (Sherrington et al., 1995, Levy-Lahad et al., 1995), it is still unclear what is the exact molecular mechanism by which PS missense mutations result in FAD. It is worth to note that patients carrying FAD-linked PS1 mutations also express one normal copy of PS1 and two normal copies of PS2 alleles, which could partially compensate for the shortcomings of the diseased allele. On the other hand, recent studies have shown that FAD-linked mutant PS1/γ-secretase interacts with the WT PS1/γ-secretase and decreases overall PS/γ-secretase activity, demonstrating the dominant negative effect of FAD-linked mutant over WT PS1/γ-secretase (Heilig et al., 2013, Zhou et al., 2017). Our novel FRET-based biosensors could help to elucidate whether overall PS/γ-secretase activity is decreased or is not changed in the “FAD (i.e., mutant PS1 is co-expressed with WT PS1 and PS2)” intact/live neurons compared with WT neurons. Furthermore, the Aβ49/48 generated by PS/γ-secretase-mediated epsilon-cleavage of APP C99 is sequentially processed by consecutive gamma-cleavages to yield the predominant Aβ species, Aβ40/42/43 (Kakuda et al., 2006). Since the APP-C99-based biosensors report more specifically the first epsilon-cleavage by PS/γ-secretase, identification of the cells that have increased epsilon cleavage could also indicate, although indirectly, the cells in which Aβ production occurs at different rates.
Disappointingly, the AD clinical trials testing the efficacy of γ-secretase inhibitors failed owing to adverse events that include cognitive worsening (Doody et al., 2013). On the other hand, γ-secretase modulators, GSMs, have been developed and their efficacy is being assessed in clinical trials. The major advantage of the GSMs is that they spare the epsilon-cleavage of APP, as well as other substrates such as Notch1, but shift the gamma-cleavages of APP toward generation of the shorter Aβ38/37. The novel biosensors we developed would not only help to test directly whether FAD mutations diminish the Notch1/APP epsilon cleavage by an endogenous PS/γ-secretase enzyme in its normal physiological environment but also, importantly, may have preclinical therapeutic application by enabling to screen for PS/γ-secretase modulators that spare the ICD-generating γ-secretase-dependent cleavage of the substrates, as well as for PS/γ-secretase activators that could restore the decreased overall PS/γ-secretase activity due to, for example, FAD mutations.
PS/γ-secretase also plays a significant role in the pathogenesis of other diseases beyond AD (reviewed in Jurisch-Yaksi et al., 2013). For example, one of the adverse events triggered by γ-secretase inhibitors in the AD trial was skin cancer (Doody et al., 2013). Moreover, mutations on the genes of γ-secretase components such as PS1, Pen2, or Nicastrin that cause γ-secretase haploinsufficiency are reported to cause familial acne inversa (Wang et al., 2010) and/or Dowing-Degos disease associated with acne inversa (Ralser et al., 2017). It has never been clarified how the inhibition of PS/γ-secretase activity causes these skin diseases. The FRET biosensors we developed could provide important spatiotemporal information about the role of PS/γ-secretase in skin biology. Since the APP C99 part in the C99 R-G/Y-T biosensors could be easily replaced by other PS/γ-secretase substrates to test their cleavability and thus their role in various cellular pathways (Figure 5), the “γ-Substrate” donor-acceptor biosensor could be applied more broadly to investigate the roles of PS/γ-secretase in multiple cellular events or diseases.
In summary, we have developed, validated, and optimized APP C99- and Notch1 N100-based FRET biosensors for monitoring PS/γ-secretase activity in intact/live cells. Using these biosensors, we showed uneven PS/γ-secretase activity in neuronal processes as well as in different compartments of an individual neurite, suggesting that the PS/γ-secretase-mediated processing of substrates is differentially regulated within a single neuron. Moreover, the longitudinal imaging in live neurons revealed that overall PS/γ-secretase activity is heterogeneously regulated among live neurons. These findings suggest the spatiotemporally heterogeneous nature of the PS/γ-secretase activity. The FRET biosensors developed in this study would not only be useful tools to identify the molecular regulators of PS/γ-secretase activity but provide previously missing capability to distinguish cell populations with active and inactive PS/γ-secretase activity in WT neurons. Such capability would allow direct examination of the mechanistic link between change in PS/γ-secretase activity and its biological consequences such as neuronal vulnerability in WT neurons in future studies.
Limitations of the Study
The common issues related to fluorescent imaging such as potential photobleaching due to high laser power excitation could affect the readout, and thus should be controlled for. It should also be noted that the change in FRET efficiency while using the FRET biosensors could reflect changes not only in PS/γ-secretase activity but also in the amount of active PS/γ-secretase complex. Therefore, further refinement of the assay and mechanistic studies are required in the follow-up studies.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Masato Maesako (mmaesako@mgh.harvard.edu).
Materials Availability
Plasmids generated in this study will be made available on reasonable requests with a completed Materials Transfer Agreement.
Data and Code Availability
The published article includes all datasets generated during the study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We are grateful to Dr. Simon Dujardin (Massachusetts General Hospital) for helpful discussion during the design of biosensor, to Dr. Steven Hou (Massachusetts General Hospital) for support with data analysis, to Dr. Maria Calvo-Rodriguez (Massachusetts General Hospital) and Ms. Maya Modi (Massachusetts General Hospital) for performing unpublished experiments, and to Ms. Miwei Hu (Massachusetts General Hospital) for the preparation of primary neuron. We also appreciate Dr. Michiyuki Matsuda and Dr. Kenta Terai (Kyoto University, Kyoto, Japan) for sharing the AKAR3EV plasmid as well as technical support for the optimization of biosensor and Dr. Dennis Selkoe (Brigham and Women's Hospital, Harvard Medical School, Boston MA) and Dr. Bart De Strooper (University of Leuven, Leuven, Belgium) for sharing the PS1/2 knockout HEK cells and MEF cells, respectively. This work was supported by the National Institutes of Health grants AG 44486 (O.B.) and AG 15379 (O.B.). M.M. was supported by the research fellowships from the Japan Society for the Promotion of Science (JSPS), the Kyoto University Foundation, and the BrightFocus Foundation A2019056F.
Author Contributions
M.M. designed the study, performed the experiments, and drafted the manuscript; N.M.S., A.A., P.F., and L.C.A. performed the experiments; and O.B. designed the study, discussed the data, and proofread the manuscript.
Declaration of Interests
The authors have no competing interests.
Published: Jun 26, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101139.
Contributor Information
Masato Maesako, Email: mmaesako@mgh.harvard.edu.
Oksana Berezovska, Email: oberezovska@mgh.harvard.edu.
Supplemental Information
References
- Acx H., Serneels L., Radaelli E., Muyldermans S., Vincke C., Pepermans E., Müller U., Chávez-Gutiérrez L., De Strooper B. Inactivation of γ-secretases leads to accumulation of substrates and non-Alzheimer neurodegeneration. EMBO Mol. Med. 2017;9:1088–1099. doi: 10.15252/emmm.201707561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovska O., Ramdya P., Skoch J., Wolfe M.S., Bacskai B.J., Hyman B.T. Amyloid precursor protein associates with a nicastrin-dependent docking site on the presenilin 1-gamma-secretase complex in cells demonstrated by fluorescence lifetime imaging. J. Neurosci. 2003;23:4560–4566. doi: 10.1523/JNEUROSCI.23-11-04560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Sudhof T.C. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Chávez-Gutiérrez L., Bammens L., Benilova I., Vandersteen A., Benurwar M., Borgers M., Lismont S., Zhou L., Van Cleynenbreugel S., Esselmann H. The mechanism of γ-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B., Saftig P., Craessaerts K., Vanderstichele H., Guhde G., Annaert W., Von Figura K., Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J.S., Schroeter E.H., Schrijvers V., Wolfe M.S., Ray W.J. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Doody R.S., Raman R., Farlow M., Iwatsubo T., Vellas B., Joffe S., Kieburtz K., He F., Sun X., Thomas R.G. A phase 3 trial of semagacestat for treatment of Alzheimer's disease. N. Engl. J. Med. 2013;369:341–350. doi: 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- Dyrks T., Dyrks E., Mönning U., Urmoneit B., Turner J., Beyreuther K. Generation of beta A4 from the amyloid protein precursor and fragments thereof. FEBS Lett. 1993;335:89–93. doi: 10.1016/0014-5793(93)80446-2. [DOI] [PubMed] [Google Scholar]
- Esler W.P., Kimberly W.T., Ostaszewski B.L., Ye W., Diehl T.S., Selkoe D.J., Wolfe M.S. Activity-dependent isolation of the presenilin- gamma -secretase complex reveals nicastrin and a gamma substrate. Proc. Natl. Acad. Sci. U S A. 2002;99:2720–2725. doi: 10.1073/pnas.052436599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florean C., Zampese E., Zanese M., Brunello L., Ichas F., De Giorgi F., Pizzo P. High content analysis of γ-secretase activity reveals variable dominance of presenilin mutations linked to familial Alzheimer's disease. Biochim. Biophys. Acta. 2008;1783:1551–1560. doi: 10.1016/j.bbamcr.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Haapasalo A., Kovacs D.M. The many substrates of presenilin/γ-secretase. J. Alzheimers Dis. 2011;25:3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson E.M., Teixeira A.I., Gustafsson M.V., Dohda T., Chapman G., Meletis K., Muhr J., Lendahl U. Recording Notch signaling in real time. Dev. Neurosci. 2006;28:118–127. doi: 10.1159/000090758. [DOI] [PubMed] [Google Scholar]
- Heilig E.A., Gutti U., Tai T., Shen J., Kelleher R.J., 3rd Trans-dominant negative effects of pathogenic PSEN1 mutations on γ-secretase activity and Aβ production. J. Neurosci. 2013;33:11606–11617. doi: 10.1523/JNEUROSCI.0954-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan M.X., Lim S., Fulbright M., Piwnica-Worms D., Kopan R. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci. Signal. 2011;4:rs7. doi: 10.1126/scisignal.2001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurisch-Yaksi N., Sannerud R., Annaert W. A fast growing spectrum of biological functions of γ-secretase in development and disease. Biochim. Biophys. Acta. 2013;1828:2815–2827. doi: 10.1016/j.bbamem.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Kakuda N., Funamoto S., Yagishita S., Takami M., Osawa S., Dohmae N., Ihara Y. Equimolar production of amyloid beta-protein and amyloid precursor protein intracellular domain from beta-carboxyl-terminal fragment by gamma-secretase. J. Biol. Chem. 2006;281:14776–14786. doi: 10.1074/jbc.M513453200. [DOI] [PubMed] [Google Scholar]
- Kaether C., Capell A., Edbauer D., Winkler E., Novak B., Steiner H., Haass C. The presenilin C-terminus is required for ER-retention, nicastrin-binding and gamma-secretase activity. EMBO J. 2004;23:4738–4748. doi: 10.1038/sj.emboj.7600478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly W.T., Zheng J.B., Guénette S.Y., Selkoe D.J. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J. Biol. Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- Komatsu N., Aoki K., Yamada M., Yukinaga H., Fujita Y., Kamioka Y., Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D., Doyle T.G., Brousseau D., Lee M.K., Thinakaran G., Slunt H.H., Sisodia S.S., Greenwald I. Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E., Wasco W., Poorkaj P., Romano D.M., Oshima J., Pettingell W.H., Yu C.E., Jondro P.D., Schmidt S.D., Wang K. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- Maesako M., Horlacher J., Zoltowska K.M., Kastanenka K.V., Kara E., Svirsky S., Keller L.J., Li X., Hyman B.T., Bacskai B.J. Pathogenic PS1 phosphorylation at Ser367. Elife. 2017;6 doi: 10.7554/eLife.19720. e19720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi-Takahara Y., Morishima-Kawashima M., Tanimura Y., Dolios G., Hirotani N., Horikoshi Y., Kametani F., Maeda M., Saido T.C., Wang R. Longer forms of amyloid beta protein: implications for the mechanism of intramembrane cleavage by gamma-secretase. J. Neurosci. 2005;25:436–445. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Monzon O., Martin M.M., Fernandez M.A., Cappello C.A., Krzysiak A.J., Osenkowski P., Wolfe M.S. Dissociation between the processivity and total activity of γ-secretase: implications for the mechanism of Alzheimer's disease-causing presenilin mutations. Biochemistry. 2011;25:9023–9035. doi: 10.1021/bi2007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser D.J., Basmanav F.B., Tafazzoli A., Wititsuwannakul J., Delker S., Danda S., Thiele H., Wolf S., Busch M., Pulimood S.A. Mutations in γ-secretase subunit-encoding PSENEN underlie Dowling-Degos disease associated with acne inversa. J. Clin. Invest. 2017;127:1485–1490. doi: 10.1172/JCI90667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Suemoto T., Brouwers N., Sleegers K., Funamoto S., Mihira N., Matsuba Y., Yamada K., Nilsson P., Takano J. Potent amyloidogenicity and pathogenicity of Aβ43. Nat. Neurosci. 2011;14:1023–1032. doi: 10.1038/nn.2858. [DOI] [PubMed] [Google Scholar]
- Saura C.A., Choi S.Y., Beglopoulos V., Malkani S., Zhang D., Shankaranarayana Rao B.S., Chattarji S., Kelleher R.J., 3rd, Kandel E.R., Duff K. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger J.A., Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Shen J., Bronson R.T., Chen D.F., Xia W., Selkoe D.J., Tonegawa S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- Seidner G.A., Ye Y., Faraday M.M., Alvord W.G., Fortini M.E. Modeling clinically heterogeneous presenilin mutations with transgenic Drosophila. Curr. Biol. 2006;16:1026–1033. doi: 10.1016/j.cub.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Sherrington R., Rogaev E.I., Liang Y., Rogaeva E.A., Levesque G., Ikeda M., Chi H., Lin C., Li G., Holman K. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Sun L., Zhou R., Yang G., Shi Y. Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Abeta42 and Abeta40 peptides by gamma-secretase. Proc. Natl. Acad. Sci. U S A. 2017;114:E476–E485. doi: 10.1073/pnas.1618657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura K., Lill C.M., Li X., Peters J.A., Ivanov A., Fan Z., DeStrooper B., Bacskai B.J., Hyman B.T., Berezovska O. Allosteric modulation of PS1/gamma-secretase conformation correlates with amyloid beta(42/40) ratio. PLoS One. 2009;4:e7893. doi: 10.1371/journal.pone.0007893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K., Chen G., Südhof T.C., Shen J. Conditional forebrain inactivation of nicastrin causes progressive memory impairment and age-related neurodegeneration. J. Neurosci. 2009;29:7290–7301. doi: 10.1523/JNEUROSCI.1320-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yang W., Wen W., Sun J., Su B., Liu B., Ma D., Lv D., Wen Y., Qu T. Gamma-secretase gene mutations in familial acne inversa. Science. 2010;330:1065. doi: 10.1126/science.1196284. [DOI] [PubMed] [Google Scholar]
- Wong P.C., Zheng H., Chen H., Becher M.W., Sirinathsinghji D.J., Trumbauer M.E., Chen H.Y., Price D.L., Van der Ploeg L.H., Sisodia S.S. Presenilin 1 is required for Notch1 and DII1 expression in the paraxial mesoderm. Nature. 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Tomita T., Sato C., Kitamura T., Morohashi Y., Iwatsubo T. Pen-2 is incorporated into the gamma-secretase complex through binding to transmembrane domain 4 of presenilin 1. J. Biol. Chem. 2005;280:41967–41975. doi: 10.1074/jbc.M509066200. [DOI] [PubMed] [Google Scholar]
- Wolfe M.S., Xia W., Ostaszewski B.L., Diehl T.S., Kimberly W.T., Selkoe D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- Xia D., Watanabe H., Wu B., Lee S.H., Li Y., Tsvetkov E., Bolshakov V.Y., Shen J., Kelleher R.J., 3rd Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer's disease. Neuron. 2015;85:967–981. doi: 10.1016/j.neuron.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Yang G., Shi Y. Dominant negative effect of the loss-of-function γ-secretase mutants on the wild-type enzyme through heterooligomerization. Proc. Natl. Acad. Sci. U S A. 2017;114:12731–12736. doi: 10.1073/pnas.1713605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during the study.