Abstract
Advancements in cell-free synthetic biology are enabling innovations in sustainable biomanufacturing, that may ultimately shift the global manufacturing paradigm toward localized and ecologically harmonized production processes. Cell-free synthetic biology strategies have been developed for the bioproduction of fine chemicals, biofuels and biological materials. Cell-free workflows typically utilize combinations of purified enzymes, cell extracts for biotransformation or cell-free protein synthesis reactions, to assemble and characterize biosynthetic pathways. Importantly, cell-free reactions can combine the advantages of chemical engineering with metabolic engineering, through the direct addition of co-factors, substrates and chemicals –including those that are cytotoxic. Cell-free synthetic biology is also amenable to automatable design cycles through which an array of biological materials and their underpinning biosynthetic pathways can be tested and optimized in parallel. Whilst challenges still remain, recent convergences between the materials sciences and these advancements in cell-free synthetic biology enable new frontiers for materials research.
Keywords: cell-free synthetic biology, biological materials, biomaterials, biomimetics, metabolic engineering
Introduction
We live in a material world (Callén Moreu and López Gómez, 2019). There are more than five trillion plastic pieces in the world’s oceans (Eriksen et al., 2014), an estimated 15 billion trees are cut down each year (Crowther et al., 2015) and global natural fiber production is estimated to have already exceeded 30 million tons per annum (Townsend and Sette, 2016). Clearly, the mass production, processing and disposal of an array of materials has significantly shaped the global ecosystem. Whilst we continue to approach irreversible climate change (Lenton et al., 2019), it is of increasing importance that positive advances in the materials sciences are balanced against any negative environmental consequences that relate to material consumption. Arguably, these challenges warrant significant shifts in the manufacturing paradigm, from global mass production to local, sustainable and personalized manufacturing strategies (Hu, 2013; Stock and Seliger, 2016; Kleer and Piller, 2019). Likewise, instead of the chemical industries, the next generation of materials may come through developments in synthetic biology, sustainable biotechnology and the burgeoning bioeconomy (Le Feuvre and Scrutton, 2018; Philp, 2018; Freemont, 2019; French, 2019).
The field of synthetic biology has emerged over the last twenty years into a highly dynamic community of interdisciplinary researchers and societal stakeholders, that are working toward the responsible development and implementation of cutting-edge biotechnologies (Cameron et al., 2014; El Karoui et al., 2019; Lai et al., 2019). Synthetic biology combines knowledge across numerous scientific disciplines including molecular biology, biochemistry, biophysics and the social sciences, whilst also integrating them within an engineering design framework (Anderson et al., 2019; Trump et al., 2019). Ultimately, the synthetic biology approach is geared toward the rational engineering or repurposing of biological parts, devices and systems into solutions that can help address societal challenges (Ausländer et al., 2017). Synthetic biologists envision a broad application space for the field and anticipate positive societal impacts in medical technologies (Pardee, 2018; Hicks et al., 2020) (e.g., biosensors and therapeutics), food security and sustainable energy (Russo et al., 2019; Roell and Zurbriggen, 2020) (e.g., biofuels and synthetic photosynthesis), bioremediation (Dvořák et al., 2017; Wan et al., 2019) (e.g., pollution monitoring and sequestration), education (Kelwick et al., 2015a; Huang et al., 2018; Dy et al., 2019) (e.g., iGEM competition) and biomanufacturing (Le Feuvre and Scrutton, 2018; Choi K.R. et al., 2019; Gilbert and Ellis, 2019; Roberts et al., 2019) (e.g., fine chemicals and materials production).
However, biological systems are highly complex and initial attempts at engineering biological systems to fulfill specific application goals, are often only partially successful. To help overcome these challenges, synthetic biology employs the concept of the design-cycle, through which biotechnologies are iteratively designed, built and tested (Arpino et al., 2013; Church et al., 2014; Kelwick et al., 2014). Learning how to improve a biological system may require multiple attempts, which could be made easier by more rapid and systematic workflows. One potential solution is to utilize cell-free synthetic biology for rapid prototyping (Moore et al., 2017b; Kelwick et al., 2018). Typically, cell-free reactions make use of isolated cellular components and machinery (e.g., ribosomes and recombinant proteins), rather than live whole-cells. Whilst, some cell-free reaction components may be cell-derived (e.g., cell extracts), once prepared, cell-free workflows can be completed within hours (Chappell et al., 2013). In contrast, typical whole-cell experiments may involve several days or weeks of delays that are associated with plasmid cloning, transformation and cell growth (Sun et al., 2014). Furthermore, cell-free reactions are accessible and can combine the advantages of chemical engineering with metabolic engineering, through the direct addition of enzyme co-factors, substrates and chemicals – including those that are cytotoxic (Dudley et al., 2015; Karim and Jewett, 2016; Kay and Jewett, 2020). These advantages are increasingly being exploited for cell-free applications including biopart prototyping, cell-free metabolic engineering, medical or environmental biosensors and on-demand therapeutics production (Kightlinger et al., 2019; Silverman et al., 2019). Based upon an expanding repertoire of examples in the literature, we envision that biological materials and bio-functionalized smart materials are the next frontier for cell-free synthetic biology (Table 1). To this end, this review will introduce key concepts and recent developments in cell-free synthetic biology, with a focus on examples relevant to the materials sciences. Examples will be given of industrially and societally important biological materials that have been generated using cell-free synthetic biology. Cell-free synthetic biology can also be utilized to bio-functionalize materials, which may further enable the emergence of new types of smart materials. This review will also explore future trends and challenges in cell-free synthetic biology and speculate on their potential impact on biological materials of the future.
TABLE 1.
Cell-free strategies for biological material biomanufacturing or material bio-functionalization.
| Cell-free reaction format | Material | Application | References |
| Recombinant enzymes | Polyhydroxyalkanoates (PHAs) | Biopolymer production | Thomson et al., 2009; Han et al., 2011; Tomizawa et al., 2012; Opgenorth et al., 2016 |
| Lactic acid | Platform material for polymer production | Kopp et al., 2019 | |
| Cell extract biotransformation | Bio-cellulose | Bio-cellulose production | Ullah et al., 2015 |
| Chitin | Chitin synthesis | Jaworski et al., 1963 | |
| Poly-3-hydroxybutyrate [P(3HB)] | Optimizing PHAs biopolymer production | Kelwick et al., 2018 | |
| Gold nanoparticles (AuNPs) | Medical and industrial | Chauhan et al., 2011; Krishnan et al., 2016 | |
| Silver nanoparticles (AgNPs) | Nanobiotechnology, therapeutic development | Costa Silva et al., 2017 | |
| Cell-free protein synthesis | Bacteriophages | De novo synthesis and phage engineering | Garamella et al., 2016; Rustad et al., 2018 |
| Chitin | Chitinase expression | Endoh et al., 2006 | |
| Clay microgels | Protein production | Jiao et al., 2018 | |
| DNA hydrogels/Protein-producing gels (P-gel) | Protein production | Park et al., 2009a; Ruiz et al., 2012 | |
| Elastin-like polypeptides (ELPs) | Biopolymer with non-canonical amino acids | Martin et al., 2018 | |
| Extracellular vesicles (EVs) | Therapeutics/EV biogenesis research | Shurtleff et al., 2016; García-Manrique et al., 2018 | |
| Freeze-dried pellets | In vitro diagnostics or therapeutic production | Pardee et al., 2016b; Salehi et al., 2016, 2017 | |
| Liposomes and nanodiscs | Membrane protein production, drug discovery or protocell production | Garamella et al., 2016; Rues et al., 2016; Shinoda et al., 2016; Contreras-Llano and Tan, 2018; Gessesse et al., 2018; Dubuc et al., 2019; Shelby et al., 2019 | |
| Microfluidic devices (various) | Antibody development and protein microarrays | Kilb et al., 2014; Georgi et al., 2016; Contreras-Llano and Tan, 2018 | |
| Microparticles/nanoparticles | On-demand functional biomaterials/therapeutics | Lim et al., 2009; Benítez-Mateos et al., 2018 | |
| Paper | In vitro diagnostics | Pardee et al., 2014, 2016a; Duyen et al., 2017; Gräwe et al., 2019; Thavarajah et al., 2020 | |
| PEG hydrogels | Education | Huang et al., 2018 | |
| Poly-3-hydroxybutyrate (P(3HB)) | Polyhydroxyalkanoates (PHAs) biosynthetic operon prototyping | Kelwick et al., 2018 | |
| Protein biologics | Cancer therapeutics, protein therapeutics | Zawada et al., 2011; Sullivan et al., 2016; Salehi et al., 2017; Kightlinger et al., 2019 | |
| Silk fibroin | Silk fibroin production | Greene et al., 1975; Lizardi et al., 1979 |
Cell-Free Synthetic Biology Reaction Formats and Strategies
Cell-free synthetic biology is a broad term that encompasses many different in vitro biotechnologies. Broadly, the term cell-free synthetic biology refers to different methods and technologies for engineering or using biological processes outside of a cell. For example, cell-free protein synthesis reactions enable the production of proteins within biochemical reactions. Thus, cell-free reactions typically make use of isolated cellular components (e.g., recombinant proteins) and/or cell extracts, rather than live whole-cells. In the context of this review four commonly used cell-free reaction formats will be discussed (Figure 1). We describe these cell-free reaction formats as either (i) recombinant enzyme-based, (ii) protein synthesis using recombinant elements (PURE)-based cell-free protein synthesis, (iii) wildtype and/or engineered cell extract biotransformation or (iv) cell extract-based cell-free protein synthesis.
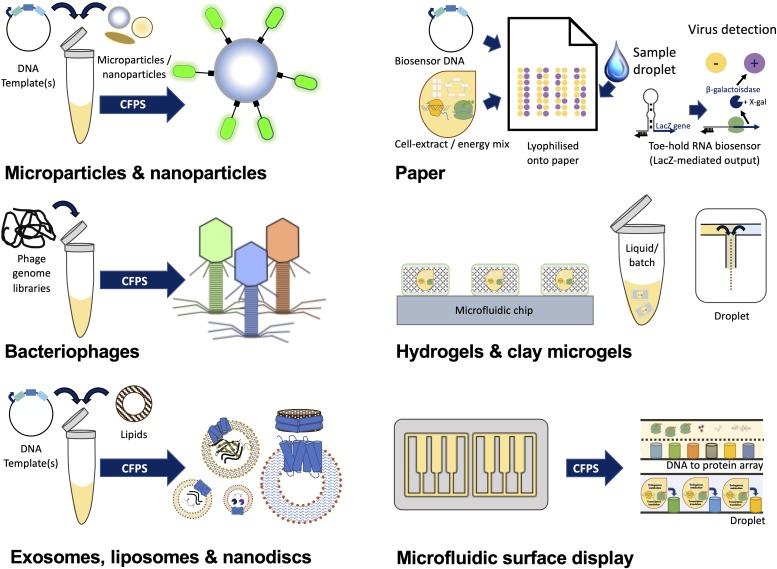
FIGURE 1.
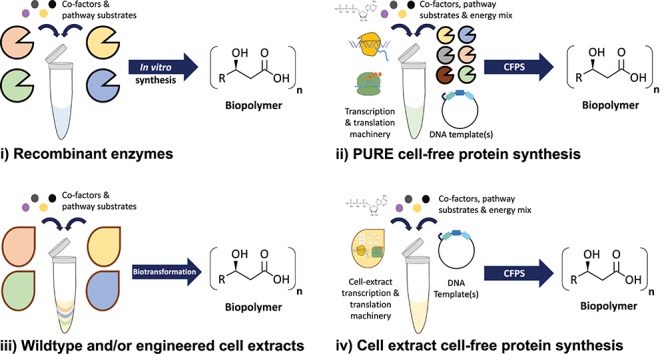
Cell-free synthetic biology reaction formats and strategies. (i) Recombinant enzymes can be mixed together along with enzyme co-factors and substrates to form biosynthetic pathways. (ii) The PURE cell-free protein synthesis system utilizes reconstituted Escherichia coli transcription and translation machinery, DNA templates, purified enzymes and other factors. (iii) Cell extracts from lysed wildtype or engineered cells can be mixed together along with enzyme co-factors and substrates to form biosynthetic pathways. (iv) Cell extract-based cell-free protein synthesis reactions utilize the transcription and translation machinery within cell lysates, along with exogenously added energy mix components (e.g., amino acids) and DNA templates for in vitro protein production.
Recombinant enzyme-based reaction formats utilize purified enzymes, along with any required co-factors and pathway substrates, to produce fine chemicals, polymer monomers or other molecules of interest. The PURE-based cell-free protein synthesis format reconstitutes the transcription and translation machinery from Escherichia coli using purified histidine (His)-tagged proteins (Shimizu et al., 2001, 2005). In this reaction format, the exact components are known, including the co-factors, substrates and energy mixes. Since PURE reaction components are known they can be standardized and rationally optimized. However, PURE cell-free reactions typically produce lower protein yields than cell-free protein synthesis reactions that use E. coli extracts (Shimizu et al., 2005). The third cell-free reaction format uses cell extracts from lysed wildtype and/or engineered cells, which can be mixed together along with relevant required enzyme co-factors and substrates to form multicomponent biosynthetic pathways. Finally, the last format, cell extract-based cell-free protein synthesis (CFPS), uses the transcription and translation machinery from lysed cells, along with added co-factors and energy mixes to produce proteins in vitro. Cellular extract-based cell-free reactions use the host cells native transcription and translation machinery as well as other metabolic components, including energy providing enzymes such as those involved in glycolysis and the Krebs cycle, which are released when the cells are lysed either mechanically (French pressure cell press), by sonication or osmotically (Gregorio et al., 2019). These reactions were originally developed as experimental tools to enable the fundamental understanding of aspects of cellular biochemistry, molecular biology and in vitro production of various proteins of interest (Gagoski et al., 2016). A range of different host cells have been used to develop these reactions, including bacteria such as Bacillus subtilis (Kelwick et al., 2016), Streptomyces venezuelae (Moore et al., 2017a; Li et al., 2018) and E. coli (Sun et al., 2013) as well as insect (Ezure et al., 2006), wheat germ (Harbers, 2014), yeast (Hodgman and Jewett, 2013; Aw and Polizzi, 2019), protozoans such as Leishmania tarentolae (Mureev et al., 2009; Kovtun et al., 2010, 2011) and mammalian cells (Weber et al., 1975; Martin et al., 2017).
It is important to note that these different cell-free reaction formats are not mutually exclusive and can be combined together. Recombinant enzymes or small molecule substrates can also be added into cell-free protein synthesis reactions to complete biosynthetic pathways, or to use exogenous chemistries within the reaction. It is this flexibility that we envision being particularly useful in terms of exploring how cell-free reactions could create novel types of biological materials or bio-functionalized smart materials. In the following sections we discuss exemplars where cell-free synthetic reactions have been utilized to prototype, manufacture or bio-functionalize, biological materials (Table 1).
Cell-Free Strategies for Sustainable Materials Biomanufacturing
Living cells and organisms have evolved highly complex enzymes and metabolic processes that generate extremely diverse biochemistries. Exploring these natural biochemistries may lead to important foundational advances in our understanding of natural product synthesis. Foundational discoveries in functional genomics, cellular metabolism and natural product synthesis are also important, because they might inspire novel biosynthetic pathway designs for biological materials production. In synthetic biology, cell-free metabolic engineering (CF-ME) approaches can reconstitute entire biosynthetic pathways using either cell extracts from diverse species, engineered cells and/or cell-free synthesized recombinant enzymes (Karim and Jewett, 2018; Martin et al., 2018; Yim et al., 2019; Bowie et al., 2020) (Figure 1). Also, cell-free protein synthesis and cell extract biotransformation reactions can be combined to create more complex cell-free reactions (Karim and Jewett, 2018; Kelwick et al., 2018). Another important advantage in using cell-free approaches is that pathway reaction bottlenecks can be identified, through the direct addition of the required recombinant enzymes, enzyme co-factors or chemical substrates needed for each stage of a biosynthetic pathway (Dudley et al., 2015). Increasingly sophisticated combinatorial CF-ME strategies, together with high-throughput automation, deep data omics and design of experiments (DoE) approaches to cell-free reaction optimization have been deployed (Caschera et al., 2018; Jiang et al., 2018; Dopp et al., 2019). These advancements have considerably improved the feasibility of refactoring and optimizing fine chemical or natural product biosynthetic pathways within short timeframes (Dudley et al., 2015; Korman et al., 2017; Moore et al., 2017a; Wilding et al., 2018).
Cell-free synthetic biology approaches have also been directed toward the de novo bioproduction of biological materials, including biopolymers or their monomers, cellulosic materials and nanoparticles (Table 1). However, the maximum cell-free bioproduction yields or reaction efficiencies of several reported materials were generally low or unspecified. Examples of cell-free produced materials and their reported maximum production yields and reaction efficiencies include bio-cellulose (3.726 ± 0.05 g/L; 57.68%) (Ullah et al., 2015), chitin (yields not stated) (Jaworski et al., 1963; Endoh et al., 2006), lactic acid (6.6 ± 0.1 mM; 47.4 ± 3.9%) (Kopp et al., 2019), gold nanoparticles (yields not stated) (Chauhan et al., 2011; Krishnan et al., 2016), (R)-3-hydroxybutyrate-CoA (32.87 ± 6.58 μM) (Kelwick et al., 2018), silver nanoparticles (yields not specified) (Costa Silva et al., 2017) and silk fibroin (yields not specified) (Greene et al., 1975; Lizardi et al., 1979). Poor cell-free production yields and efficiencies can be due to a variety of factors including rapid depletion of reaction energy mix components (e.g., ATP, amino acids), the formation of inhibitory waste products (e.g., inorganic phosphates) or unwanted side reactions that divert reaction fluxes away from desirable pathways (Caschera and Noireaux, 2014). Because of these limitations, cell-free synthetic biology may not be an ideal production method for some biological materials. Nevertheless, whilst actual cell-free material production yields can be relatively low, these approaches are still beneficial for prototyping different biosynthetic pathways, substrates or reaction conditions to boost both in vitro and whole-cell production yields. An exemplar is the use of cell-free assays to characterize polyhydroxyalkanoates (PHAs) biosynthetic pathways from phaCAB operons that also enhanced in vivo PHAs production (Kelwick et al., 2018). Furthermore, the same study also demonstrated that the cell-extract biotransformation of whey permeate into 3-hydroxybutyrate (3HB), could be simultaneously coupled with the cell-free protein synthesis of a potential Acetyl-CoA recycling enzyme (Kelwick et al., 2018). Thus, highlighting that combinatorial cell-free reaction formats can be a useful strategy for bioplastic pathway prototyping and optimization.
Interestingly, in some cases, cell-free bioproduction may actually be a more desirable manufacturing route. For instance, several in vitro gold or silver nanoparticle production studies reported desirable nanoparticle characteristics (e.g., size/zeta potential) and/or easier purification protocols within cell-free bioproduction reactions than whole-cell production methods (Krishnan et al., 2016; Costa Silva et al., 2017). Cell-free bioproduction can also be carried out at industrially relevant scales, as illustrated by Sutro biopharma who have developed a highly scalable good manufacturing practices (GMP)-compliant cell-free protein synthesis platform, for producing therapeutic proteins within 100 L bioreactor reaction volumes (Zawada et al., 2011). For cell-free materials production, a highly efficient synthetic biochemistry module was developed to convert glucose into bio-based chemicals, including the PHA bioplastic monomer polyhydroxybutyrate (PHB) (Opgenorth et al., 2016). To achieve this, purified recombinant enzymes were used to reconstitute core elements of the pentose, bifido, glycolysis and PHB pathways (Opgenorth et al., 2016). Cell-free PHB production yields (40 g/L) and efficiencies (90%) were impressive and are promisingly close to industrially attractive scales (Opgenorth et al., 2016). These improvements in PHB production are also welcome since PHB, as well as other PHAs biopolymers, are biodegradable and can potentially be used as ‘drop-in’ replacements for oil-derived plastics (e.g., food packaging) or as biomaterials for tissue engineering (Choi S.Y. et al., 2019; Tarrahi et al., 2020). PHAs are also an interesting example because of their industrial importance and the diversity of cell-free strategies that have been applied to PHAs research (Table 1). Building upon these examples, CFME approaches could be used to explore a greater diversity of PHAs biopolymers given that PHA biopolymers can be composed of a variety of different wildtype and/or synthetic monomers (∼160 different monomers exist) to create complex co-polymers, with an array of material characteristics (Choi S.Y. et al., 2019). Future cell-free synthetic biology explorations of PHAs are likely to unlock novel PHAs biopolymers with unique characteristics (Chen and Hajnal, 2015) and therefore, accelerate bioplastic materials development.
Microbial (in vivo) PHAs production has been commercially manufactured at industrial scales over the last several decades. Unfortunately, the commercial impact of PHA-based bioplastics has been historically prohibited by their higher production costs than oil-derived plastics (Chen et al., 2020). However, more efficient PHAs production processes have been devised through the rational design of phaCAB biosynthetic pathways (Hiroe et al., 2012; Kelwick et al., 2015b; Li et al., 2016; Tao et al., 2017; Zhang X. et al., 2019), key metabolite recycling processes (e.g., Acetyl-CoA) (Matsumoto et al., 2013; Beckers et al., 2016), alternative microbial production hosts (e.g., Halomonas sp., Tan et al., 2011) and the use of industrially sourced, low-cost feedstocks (e.g., whey permeate) (Wong and Lee, 1998; Ahn et al., 2000; Kim, 2000; Nikel et al., 2006; Cui et al., 2016; Nielsen et al., 2017). Interestingly, several of these microbial PHAs production strategies are also compatible with cell-free synthetic biology reactions. In particular, using locally sourced, low-cost feedstocks (e.g., whey permeate) may help to make cell-extract based PHAs production more economically viable (Kelwick et al., 2018). A similar approach has already been piloted for cell-free lactic acid production from spent coffee grounds (Kopp et al., 2019) and could become a generalized strategy for sustainable cell-free materials bioproduction (Figure 2). We would argue that combining cell-free extracts with local feedstocks enables immediate access to highly diverse cellular biochemistries and low-cost substrates (e.g., waste feedstocks), that could potentially be used for the sustainable biomanufacturing of a diverse array of biological materials (Yan and Fong, 2015; Le Feuvre and Scrutton, 2018). Furthermore, just as lyophilized cell-free reactions enable the on-demand production of biotherapeutics (Pardee et al., 2016b), we likewise envision that cell-free reactions might one day lead to rapid and distributed, on-demand biological materials production or bio-functionalization.
FIGURE 2.
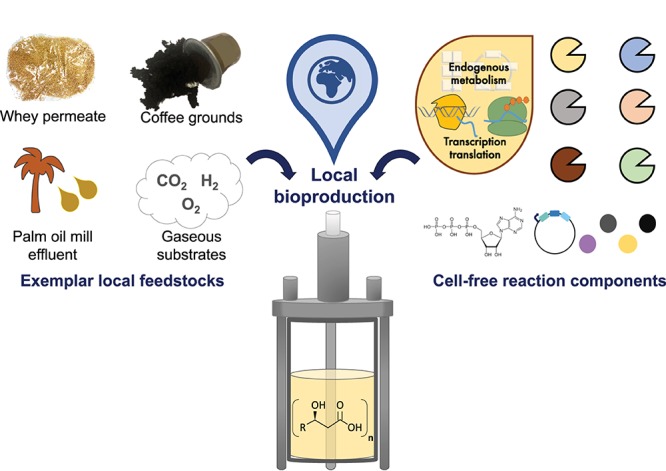
Sustainable cell-free biomanufacturing of biological materials. Schematic depicts the local, on-demand cell-free mediated, biomanufacturing of biological materials. Local feedstocks can potentially be utilized as replacements for expensive reaction energy mix components, or to provide the enzymatic co-factors and biosynthetic pathway substrates that are required to produce biological materials of interest.
Cell-Free Material bio-Functionalization and Biomimetics
Cell-free systems are still being applied to understand the foundational principles and molecular mechanisms of transcription and translation (Hecht et al., 2017; Borkowski et al., 2018). Unlocking these principles can also potentially identify novel strategies to enhance in vitro protein production, which continues to be a core application for cell-free synthetic biology (Gagoski et al., 2016; Khambhati et al., 2019). Simplified or improved cell-free methodologies (Sun et al., 2013; Kwon and Jewett, 2015; Krinsky et al., 2016; Katsura et al., 2017; Lavickova and Maerkl, 2019), coupled with lower-cost and rationally optimized cell-free reaction energy mixes (Cai et al., 2015), has enabled the implementation of in vitro therapeutic protein production at industrial scales (Ogonah et al., 2017; Garenne and Noireaux, 2019). Interestingly, the integration of cell-free reactions within materials can also enhance cell-free protein synthesis yields. For instance, the Luo group has developed the protein-producing gel (P-gel) platform, which integrates protein-producing cell-free reactions within enzyme-catalyzed, 3D DNA hydrogel matrices (Park et al., 2009a). P-gels have been used to produce a panel of model proteins including green fluorescent protein (GFP), chloramphenicol acetyltransferase (CAT) and Renilla reniformis luciferase protein (Park et al., 2009a, b). Optimized P-gels reportedly generated between 1.87 and 5 mg/ml–1 of functional luciferase protein, which was significantly higher (>90-fold) than comparative, standard liquid-format cell-free reactions (Park et al., 2009a, b). However, the authors compared P-gels against linear DNA templates within standard, liquid-format cell-free reactions. In comparison to plasmid DNA, linear DNA fragments can be relatively unstable in cell-free reactions and this instability can result in relatively low cell-free protein production yields (Chappell et al., 2013; Sun et al., 2014). Nevertheless, the P-gel platform is potentially a flexible and versatile platform for cell-free protein production. Furthermore, P-gel droplets can also be rapidly fabricated using microfluidics – thus making scale-up more feasible (Ruiz et al., 2012). In a separate study, a microfluidic fabrication strategy has been developed to embed cell-free protein synthesis reactions within clay microgels that could produce >1 mg/ml of GFP protein (Jiao et al., 2018). Importantly, P-gels and clay microgels illustrate that the cell-free functionalization of materials can lead to novel approaches for protein biomanufacturing.
Combining cell-free synthetic biology with materials can also be important for other applications. For example, by combining the cell-free protein synthesis of membrane proteins with liposomes or nanodiscs can potentially facilitate the production and stable integration of membrane proteins within lipid bilayers – thus enabling structure-function studies or drug discovery applications (Shinoda et al., 2016; Gessesse et al., 2018; Shelby et al., 2019). In a separate study, the Schekman group recreated aspects of exosome biogenesis in vitro, by using cell-free reactions to examine exosome membrane protein topology and exosome-associated miRNA sorting (Shurtleff et al., 2016). These cell-free approaches also enable directed membrane functionalization - potentially leading to rationally engineered protocells and designer exosomes. Combining cell-free synthetic biology with materials has led to the bottom-up engineering of biomimetics, including synthetic protocells (Shin and Noireaux, 2012; Huang et al., 2013; Garamella et al., 2016; Dubuc et al., 2019; Yue et al., 2019), entire phages (Rustad et al., 2018) and more recently, a partially self-replicating in vitro translation system was reported that functions by activating a 116 kb genome and is an important step toward a living, synthetic cell (Libicher et al., 2020).
Cell-Free Synthetic Biology Enabled Smart Materials
Cell-free synthetic biology reactions also enable smart materials and biosensor applications. Cell-free synthetic biology reactions can be programmed with plasmid-encoded gene circuits, lyophilized and embedded within paper (Pardee et al., 2014, 2016a; Smith and Berkheimer, 2014; Pardee, 2018), as well as potentially other materials such P-gels or clay microgels. Cell-free paper-based biosensors can be activated, post-lyophilization, using water or liquid samples and have been shown to maintain activity even after several months of storage at room temperature (Pardee et al., 2014; Smith and Berkheimer, 2014). Once activated, these paper-based cell-free reactions enable in vitro biosensor applications where these cell-free reactions are programmed to generate detectable signal outputs in response to the presence of relevant molecules (e.g., Mercury) or disease biomarkers (Lee and Kim, 2019; Silverman et al., 2019). Beneficially, these cell-free smart materials also provide greater flexibilities in terms of their usage beyond the laboratory and outside in the field (Pardee et al., 2016a, b). The complexity of synthetic biology genetic circuits and the strategies used to devise them has increased significantly in recent years (Nielsen et al., 2016; Grunberg and Del Vecchio, 2020). This has resulted in the development of an array of cell-free compatible genetic circuits that incorporate a variety of different regulatory elements that are applicable to different biosensing applications. For example, cell-free compatible gene networks can be transcription-based such that the presence of a small molecule induces reporter gene expression, through binding to, and activation or repression of, a transcriptional regulator. These types of transcriptional gene circuits have been used to develop cell-free biosensors for detecting heavy metals (Gräwe et al., 2019; Gupta et al., 2019), a date-rape drug (Gräwe et al., 2019), metabolites (Voyvodic et al., 2019) and quorum sensing molecules from Pseudomonas aeruginosa-infected respiratory samples (Wen et al., 2017). Small molecules can also regulate transcription by binding to endogenous or engineered RNA aptamer-regions within 5′ untranslated (UTR) mRNA regions – termed riboswitches. Small molecule binding to the riboswitch facilitates the emergence of stable RNA structures that permit continued reporter gene transcription. Small molecules can regulate translation via a different mechanism, whereby binding of the small molecule to the riboswitch influences the mRNA structure such that it occludes ribosome binding and downstream translation (Nahvi et al., 2002). Utilizing these principles a cell-free biosensor was developed that exploits a riboswitch to detect environmental fluoride (Thavarajah et al., 2020). Cell-free biosensors can also utilize toeholds to detect RNAs (e.g., viral RNA). Essentially, toehold aptamers function in a similar way to riboswitches except that the presence of a complementary RNA is responsible for a conformational change in the mRNA that enables ribosome access and reporter protein translation (Figure 3). Engineered cell-free toehold biosensors can also differentiate between different Ebola (Pardee et al., 2014) and Zika (Pardee et al., 2016a) strains. Conceivably, RNA aptamer-based approaches could be adapted to detect Coronaviruses such as SARS-CoV and 2019 n-CoV (Ahn et al., 2009). Indeed, the 2020 COVID-19 pandemic might be a catalyst for the development of cell-free viral biosensors and the accompanying clinical studies that will be required for comparative testing against existing technologies (e.g., quantitative reverse transcription polymerase chain reaction, RT-qPCR) (Liu et al., 2020). Post-translational cell-free biosensors have also been developed to detect glucose. In this example, cell-free protein synthesis was used to produce fusion protein pairs that elicit changes in Fluorescence Resonance Energy Transfer (FRET) signals in response to bound glucose (Pardee et al., 2014). Furthermore, purified fusion proteins can also enable other types of smart material applications. For instance, Wagner et al. (2019) developed information-processing materials that function by using protease-based signal-amplifying cascades, that integrate both proteolytic activities and ligand-receptor sensing within its logic circuits. As an exemplar, the same authors reported on the development of smart materials, inspired by synthetic biology logic circuits, that can detect novobiocin antibiotics (Wagner et al., 2019). These, as well as many other transcriptional and translational regulatory mechanisms can be combined into highly complex cell-free executable circuit designs (Jeong et al., 2019). Therefore, it is conceivable that these studies might inspire future efforts to embed cell-free executable logic circuits within a broad array of synthetic biology-based smart materials.
FIGURE 3.
Cell-free synthetic biology-based material functionalization. Schematic depicts examples of materials that have been bio-functionalized using cell-free protein synthesis (CFPS) reactions.
Automated Design-Cycles for Cell-Free Biological Materials
An important future trend in synthetic biology is the maturation of rational, engineering-led strategies for designing and implementing complex biological systems. Essentially, the field of synthetic biology envisions a future where model-guided, forward-engineering strategies will be routinely used to iterate design-build-test-learn cycles toward the final biotechnology production and application (Kelwick et al., 2014; Bultelle et al., 2016; Misirli et al., 2019). However, the scale of experiments needed to realize this vision may require continued advancements in synthetic biology machine-learning strategies, coupled with automation equipment (e.g., liquid handling robots) (Bultelle et al., 2016; Rajakumar et al., 2019). Indeed, automation capabilities can greatly expand the scale, sophistication and scope of synthetic biology design cycles (Kelwick et al., 2014; Carbonell et al., 2019; El Karoui et al., 2019; Hillson et al., 2019; Jessop-Fabre and Sonnenschein, 2019). Co-ordinated efforts toward improving inter-laboratory data reproducibility, standardizing experimental metrology and an emphasis on industrially scalable biotechnology are also driving the adoption of automation in synthetic biology workflows (Kelly et al., 2009; Beal et al., 2016, 2018a,b; de Lorenzo and Schmidt, 2018; Carbonell et al., 2019; Exley et al., 2019). In a broader sense, automation is also becoming more accessible through reductions in gene synthesis costs (Carlson, 2014), advancements in automated DNA assembly protocols (Kanigowska et al., 2016; Rajakumar et al., 2019; Storch et al., 2019; Walsh et al., 2019), rapid mass spectrometry of complex biological samples (Gowers et al., 2019; Miguez et al., 2019; O’Kane et al., 2019) and through the emergence of academic biofoundries (Chambers et al., 2016; Hillson et al., 2019).
Cell-free prototyping strategies can be readily integrated into design-cycles, including those applications that are intended to be functional in living cells (in vivo). Several studies have described comparability between DNA regulatory elements and genetic circuits tested in vitro (cell-free) and in vivo (whole-cells) across several model (e.g., E. coli, Bacillus subtilis) (Chappell et al., 2013; Sun et al., 2014; Kelwick et al., 2016) and non-model (e.g., Bacillus megaterium, Vibrio natriegens) (Failmezger et al., 2018; Moore et al., 2018) organisms. Thus, cell-free rapid prototyping strategies can also be applied to speed up the development of in vivo (whole-cell) applications. Cell-free workflows are also amenable to automation. Indeed, several studies have successfully utilized acoustic liquid handling robots to rapidly setup large-scale, low-volume (≤10 μl) prototyping cell-free reactions in a 384 well plate format (Moore et al., 2018; Kopniczky et al., 2020). Microfluidic (Swank et al., 2019), droplet array (Zhang Y. et al., 2019) or multiplex (Yim et al., 2019) strategies have also been used to enable high-throughput cell-free experiments. These approaches enable the testing of large numbers of regulatory elements or enzymes, which could potentially inform material biosynthetic pathway optimization. Conceivably, cell-free optimized material biosynthetic pathways could also be implemented within in vivo production strategies (e.g., microbial cell factories). These data sets also enable accompanying quantitative modeling that can potentially predict unknown model parameters, such as transcription factor binding affinities or cell-free energy utilization (Bujara and Panke, 2012; Moore et al., 2018). These data can potentially be entered into biopart data repositories (Ham et al., 2012; Bultelle et al., 2016; McLaughlin et al., 2018) for use in machine learning-enhanced design of experiments (DoE) approaches, to speed up materials development (Liu et al., 2017; Exley et al., 2019; Madsen et al., 2019). High-throughput cell-free experiments can also tease apart where cell-free reactions are fundamentally different to native intracellular environments. Molecular crowding, metabolic fluxes, co-factor regeneration rates and other biophysiochemical characteristics may all differ between cell-free reactions and in vivo (whole-cell) contexts. Also, cell extract processing breaks down cellular organelles and other intracellular compartments, thus perturbing native biomolecule localization. Yet, despite these potential limitations complex genetic circuits (e.g., oscillators), that are functional in vivo, have been prototyped using cell-free forward-engineering design cycles (Niederholtmeyer et al., 2015). This review has also explored several other examples where cell-free approaches have been used to produce or bio-functionalize materials (Table 1). It is therefore conceivable, that cell-free synthetic biology design-cycles may strengthen future efforts to accelerate the development of an array of bio-functionalized smart materials (Figure 4).
FIGURE 4.
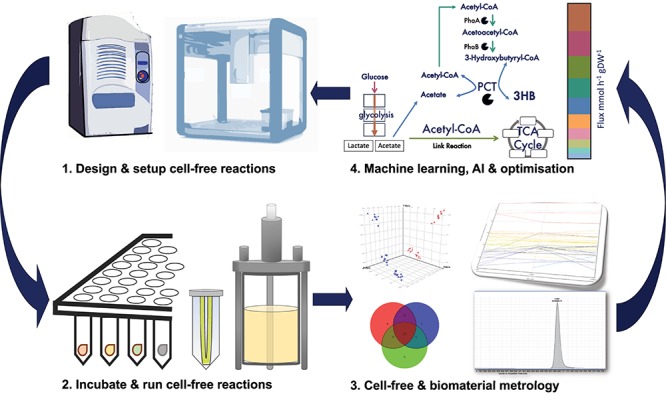
Automated cell-free design-cycles for biological materials development. Schematic depicts a cell-free design-cycle for prototyping or biomanufacturing biological materials. Firstly, cell-free reactions are designed and then setup using liquid handling robots. Secondly, these cell-free reactions are incubated in an array of reaction formats including batch or continuous feed, at a range of scales (μl to L). Thirdly, cell-free reactions and biological materials are assayed and characterized. Finally, these data inform cell-free reaction models (e.g., metabolic flux models) or material quality control benchmarks that ultimately inform the next iteration of biosynthetic pathways and cell-free reaction parameters to be tested. Sophisticated design-cycle workflows may also utilize machine learning and design-of-experiment (DOE) approaches, to rationally iterate design cycles toward the desired biological material.
Summary and Outlook
In an era of extreme climate events there is an increasing global consensus to collectively implement more sustainable and carbon-neutral policies (Gills and Morgan, 2019). However, there continues to be an insatiable global demand for materials and the natural resources that are used to manufacture them. These activities have, however, negatively impacted the global ecosystem through for example deforestation, industrial pollution and biodiversity destruction. To partially address some of these challenges, there is increasing support for more locally based bioeconomies underpinned by sustainable practices. Cell-free biomanufacturing could have an important role in the emerging bioeconomy by enabling biological materials to be produced locally, on-demand and more sustainably from waste feedstocks (e.g., whey permeate). Furthermore, centrally produced cell-free reaction components can be lyophilized and then distributed to different geographical locations (Pardee et al., 2016b). Once distributed, these cell-free reactions could potentially make use of highly customized DNA-encoded biosynthetic operons to produce personalized materials that suit local needs. Conceivably, these cell-free produced biological materials could be fabricated using synthetic biochemistries [e.g., unnatural amino acids (Martin et al., 2018; Des Soye et al., 2019; Gao et al., 2019) and xeno nucleic acids (Glasscock et al., 2016; Hu et al., 2019)], mixed cell extracts from diverse bacterial species (Yim et al., 2019), or de novo biological components [e.g., engineered ribosomes (Caschera et al., 2018; d’Aquino et al., 2020) and rationally designed proteins (Huang et al., 2016; Rolf et al., 2019)]. These synthetic components might confer cell-free produced materials with unique physical characteristics or other attributes that are not typically associated with natural fibers. Likewise, we envision that future cell-free materials will have integrated smart-features including biosensing capabilities, the ability to change material properties in response to specific stimuli or self-healing capabilities when damaged. Such capabilities may require further advancements in cell-free genetic circuits and the methods used to embed them within different materials.
A panoply of recent examples, including those discussed in this review, are indicative that biological materials are the next frontier for cell-free synthetic biology – but challenges remain. A deeper understanding of the compositions and biochemical activities of processed cell extracts and cell-free reactions are needed (Foshag et al., 2018) to mitigate cell-free extract batch variability and increase protein synthesis yields. Compounding these challenges are the need for improved cell-free metrology and cell-free biomanufacturing quality control standards, which are especially important for industrial applications (de Lorenzo and Schmidt, 2018). Equally, the acceptability of cell-free produced biological materials may need to be considered through stakeholder engagement, consumer awareness activities and through, where necessary, the establishment of specialist recycling/waste management infrastructure. The scalability of cell free reactions to industrial manufacturing is also extremely challenging. Despite these challenges, Genomatica, Greenlight Biosciences, Sutro Biopharma, Tierra Biosciences and other companies have already successfully developed scalable cell-free synthetic biology platforms that have at least pilot tested materials production (e.g., Genomatica’s cell-free platform produces polymer chemicals including 1,4-butanediol). In addition, the synthetic biology companies Bolt Threads, Spiber, Colorifix, and Ginkgo Bioworks are also working toward a future sustainable fashion industry. Bolt Threads produce spinnable recombinant spider silk in yeast and are also developing scalable biomanufacturing processes for mushroom-based leather. Spiber are also producing recombinant silk, as well as other synthetic protein materials. Both Bolt Threads and Spiber have successfully manufactured pilot batches of clothing products for a future sustainable fashion industry. Colorifix have developed a biological textile dye process that is designed to be less polluting than traditional chemical methods. Meanwhile, Ginkgo Bioworks have made significant investments into their organism-engineering foundry to support an array of manufacturing applications – including the production of materials. Ginkgo Bioworks also occasionally uses cell-free metabolic engineering strategies within its innovation pipelines.
In summary, cell-free synthetic biology is a powerful, highly customizable and promising biotechnology that is beginning to have a positive impact on the industrialization of sustainable materials production. Importantly, synthetic biology companies continue to champion interdisciplinary collaborations with designers and materials scientists as part of the development process. We envision that a continuation of these trends will result in a new frontier for sustainable cell-free materials production and the growing bioeconomy.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank colleagues in the Section of Structural and Synthetic Biology, WISER project colleagues and Dr. Amélie Heliot.
Footnotes
Funding. RK was supported by a BBSRC-funded Royal Society of Edinburgh Enterprise Fellowship. AW was supported by the UK Government’s Global Challenges Research Fund (GCRF) through the EPSRC grant (EP/P028519/1) as part of the WISER project. We acknowledge the support of Imperial Confidence in Concept (MRC/EPSRC), Imperial College London EPSRC Impact Acceleration Account (EP/R511547/1), EPSRC grant (EP/L011573/1), BBSRC grant (BB/L027852/1), and the Imperial College London CRUK Development Fund.
References
- Ahn D.-G., Jeon I.-J., Kim J. D., Song M.-S., Han S.-R., Lee S.-W., et al. (2009). RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst 134:1896. 10.1039/b906788d [DOI] [PubMed] [Google Scholar]
- Ahn W. S., Park S. J., Lee S. Y. (2000). Production of Poly(3-hydroxybutyrate) by fed-batch culture of recombinant Escherichia coli with a highly concentrated whey solution. Appl. Environ. Microbiol. 66 3624–3627. 10.1128/aem.66.8.3624-3627.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. A., Jones R. D., Arkin A. P., Weiss R. (2019). Principles of synthetic biology: a MOOC for an emerging field. Synth. Biol. 4:ysz010 10.1093/synbio/ysz010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpino J. A. J., Hancock E. J., Anderson J., Barahona M., Stan G.-B. V., Papachristodoulou A., et al. (2013). Tuning the dials of synthetic biology. Microbiology 159 1236–1253. 10.1099/mic.0.067975-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausländer S., Ausländer D., Fussenegger M. (2017). synthetic biology-the synthesis of biology. Angew. Chemie Int. Ed. 56 6396–6419. 10.1002/anie.201609229 [DOI] [PubMed] [Google Scholar]
- Aw R., Polizzi K. M. (2019). Biosensor-assisted engineering of a high-yield Pichia pastoris cell-free protein synthesis platform. Biotechnol. Bioeng. 116 656–666. 10.1002/bit.26901 [DOI] [PubMed] [Google Scholar]
- Beal J., Haddock-Angelli T., Baldwin G., Gershater M., Dwijayanti A., Storch M., et al. (2018a). Quantification of bacterial fluorescence using independent calibrants. PLoS One 13:e0199432. 10.1371/journal.pone.0199432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal J., Haddock-Angelli T., Farny N., Rettberg R. (2018b). Time to get serious about measurement in synthetic biology. Trends Biotechnol. 36 869–871. 10.1016/j.tibtech.2018.05.003 [DOI] [PubMed] [Google Scholar]
- Beal J., Haddock-Angelli T., Gershater M., de Mora K., Lizarazo M., Hollenhorst J., et al. (2016). Reproducibility of fluorescent expression from engineered biological constructs in E. coli. PLoS One 11:e0150182. 10.1371/journal.pone.0150182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers V., Poblete-Castro I., Tomasch J., Wittmann C. (2016). Integrated analysis of gene expression and metabolic fluxes in PHA-producing Pseudomonas putida grown on glycerol. Microb. Cell Fact. 15:73. 10.1186/s12934-016-0470-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez-Mateos A. I., Llarena I., Sánchez-Iglesias A., López-Gallego F. (2018). Expanding one-pot cell-free protein synthesis and immobilization for on-demand manufacturing of biomaterials. ACS Synth. Biol. 7 875–884. 10.1021/acssynbio.7b00383 [DOI] [PubMed] [Google Scholar]
- Borkowski O., Bricio C., Murgiano M., Rothschild-Mancinelli B., Stan G.-B., Ellis T. (2018). Cell-free prediction of protein expression costs for growing cells. Nat. Commun. 9:1457. 10.1038/s41467-018-03970-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U., Sherkhanov S., Korman T. P., Valliere M. A., Opgenorth P. H., Liu H. (2020). Synthetic biochemistry: the bio-inspired cell-free approach to commodity chemical production. Trends Biotechnol. 10.1016/j.tibtech.2019.12.024 [DOI] [PubMed] [Google Scholar]
- Bujara M., Panke S. (2012). In silico assessment of cell-free systems. Biotechnol. Bioeng. 109 2620–2629. 10.1002/bit.24534 [DOI] [PubMed] [Google Scholar]
- Bultelle M., de Murieta I. S., Kitney R. (2016). “Introducing SynBIS - the synthetic biology information system,” in Proceedings of the IET/SynbiCITE Engineering Biology Conference, Michael Faraday House. [Google Scholar]
- Cai Q., Hanson J. A., Steiner A. R., Tran C., Masikat M. R., Chen R., et al. (2015). A simplified and robust protocol for immunoglobulin expression in E scherichia coli cell-free protein synthesis systems. Biotechnol. Prog. 31 823–831. 10.1002/btpr.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callén Moreu B., López Gómez D. (2019). Intimate with your junk! A waste management experiment for a material world. Sociol. Rev. 67 318–339. 10.1177/0038026119830318 [DOI] [Google Scholar]
- Cameron D. E., Bashor C. J., Collins J. J. (2014). A brief history of synthetic biology. Nat. Rev. Microbiol. 12 381–390. 10.1038/nrmicro3239 [DOI] [PubMed] [Google Scholar]
- Carbonell P., Radivojevic T., García Martín H. (2019). Opportunities at the intersection of synthetic biology, machine learning, and automation. ACS Synth. Biol. 8 1474–1477. 10.1021/acssynbio.8b00540 [DOI] [PubMed] [Google Scholar]
- Carlson R. (2014). Time for New DNA Sequencing And Synthesis Cost Curves. Available online at: https://synbiobeta.com/time-new-dna-synthesis-sequencing-cost-curves-rob-carlson/ (accessed February 1, 2020). [Google Scholar]
- Caschera F., Karim A. S., Gazzola G., D’Aquino A. E., Packard N. H., Jewett M. C. (2018). High-throughput optimization cycle of a cell-free ribosome assembly and protein synthesis system. ACS Synth. Biol. 7 2841–2853. 10.1021/acssynbio.8b00276 [DOI] [PubMed] [Google Scholar]
- Caschera F., Noireaux V. (2014). Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription–translation system. Biochimie 99 162–168. 10.1016/j.biochi.2013.11.025 [DOI] [PubMed] [Google Scholar]
- Chambers S., Kitney R., Freemont P. (2016). The foundry: the DNA synthesis and construction foundry at imperial college. Biochem. Soc. Trans. 44 687–688. 10.1042/BST20160007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J., Jensen K., Freemont P. S. (2013). Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 41 3471–3481. 10.1093/nar/gkt052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A., Zubair S., Tufail S., Sherwani A., Sajid M., Raman S. C., et al. (2011). Fungus-mediated biological synthesis of gold nanoparticles: potential in detection of liver cancer. Int. J. Nanomedicine 2305:195. 10.2147/IJN.S23195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.-Q., Chen X.-Y., Wu F.-Q., Chen J.-C. (2020). Polyhydroxyalkanoates (PHA) toward cost competitiveness and functionality. Adv. Ind. Eng. Polym. Res. 3 1–7. 10.1016/j.aiepr.2019.11.001 [DOI] [Google Scholar]
- Chen G.-Q., Hajnal I. (2015). The ‘PHAome.’. Trends Biotechnol. 33 559–564. 10.1016/j.tibtech.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Choi K. R., Jang W. D., Yang D., Cho J. S., Park D., Lee S. Y. (2019). Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37 817–837. 10.1016/j.tibtech.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Choi S. Y., Rhie M. N., Kim H. T., Joo J. C., Cho I. J., Son J., et al. (2019). Metabolic engineering for the synthesis of polyesters: a 100-year journey from polyhydroxyalkanoates to non-natural microbial polyesters. Metab. Eng. 58 47–81. 10.1016/j.ymben.2019.05.009 [DOI] [PubMed] [Google Scholar]
- Church G. M., Elowitz M. B., Smolke C. D., Voigt C. A., Weiss R. (2014). Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 15 289–294. 10.1038/nrm3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Llano L. E., Tan C. (2018). High-throughput screening of biomolecules using cell-free gene expression systems. Synth. Biol. 3 1–13. 10.1093/synbio/ysy012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Silva L. P., Pinto Oliveira J., Keijok W. J., da Silva A. R., Aguiar A. R., Guimarães M. C. C., et al. (2017). Extracellular biosynthesis of silver nanoparticles using the cell-free filtrate of nematophagous fungus duddingtonia flagrans. Int. J. Nanomedicine Volume 12 6373–6381. 10.2147/IJN.S137703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther T. W., Glick H. B., Covey K. R., Bettigole C., Maynard D. S., Thomas S. M., et al. (2015). Mapping tree density at a global scale. Nature 525 201–205. 10.1038/nature14967 [DOI] [PubMed] [Google Scholar]
- Cui Y.-W., Zhang H.-Y., Lu P.-F., Peng Y.-Z. (2016). Effects of carbon sources on the enrichment of halophilic polyhydroxyalkanoate-storing mixed microbial culture in an aerobic dynamic feeding process. Sci. Rep. 6:30766. 10.1038/srep30766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Aquino A. E., Azim T., Aleksashin N. A., Hockenberry A. J., Krüger A., Jewett M. C. (2020). Mutational characterization and mapping of the 70S ribosome active site. Nucleic Acids Res. 48 2777–2789. 10.1093/nar/gkaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V., Schmidt M. (2018). Biological standards for the knowledge-based bioeconomy: what is at stake. N. Biotechnol. 40 170–180. 10.1016/j.nbt.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Des Soye B. J., Gerbasi V. R., Thomas P. M., Kelleher N. L., Jewett M. C. (2019). A highly productive, one-pot cell-free protein synthesis platform based on genomically recoded Escherichia coli. Cell Chem. Biol. 26 1743.e–1754.e. 10.1016/j.chembiol.2019.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopp J. L., Jo Y. R., Reuel N. F. (2019). Methods to reduce variability in E. Coli-based cell-free protein expression experiments. Synth. Syst. Biotechnol. 4 204–211. 10.1016/j.synbio.2019.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuc E., Pieters P. A., van der Linden A. J., van Hest J. C., Huck W. T., de Greef T. F. (2019). Cell-free microcompartmentalised transcription–translation for the prototyping of synthetic communication networks. Curr. Opin. Biotechnol. 58 72–80. 10.1016/j.copbio.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley Q. M., Karim A. S., Jewett M. C. (2015). Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J. 10 69–82. 10.1002/biot.201400330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyen T. T. M., Matsuura H., Ujiie K., Muraoka M., Harada K., Hirata K. (2017). Paper-based colorimetric biosensor for antibiotics inhibiting bacterial protein synthesis. J. Biosci. Bioeng. 123 96–100. 10.1016/j.jbiosc.2016.07.015 [DOI] [PubMed] [Google Scholar]
- Dvořák P., Nikel P. I., Damborský J., de Lorenzo V. (2017). Bioremediation 3. 0: Engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 35 845–866. 10.1016/j.biotechadv.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Dy A. J., Aurand E. R., Friedman D. C. (2019). YouTube resources for synthetic biology education. Synth. Biol. 4:ysz022 10.1093/synbio/ysz022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Karoui M., Hoyos-Flight M., Fletcher L. (2019). Future trends in synthetic biology—a report. Front. Bioeng. Biotechnol. 7:175. 10.3389/fbioe.2019.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh T., Kanai T., Sato Y. T., Liu D. V., Yoshikawa K., Atomi H., et al. (2006). Cell-free protein synthesis at high temperatures using the lysate of a hyperthermophile. J. Biotechnol. 126 186–195. 10.1016/j.jbiotec.2006.04.010 [DOI] [PubMed] [Google Scholar]
- Eriksen M., Lebreton L. C. M., Carson H. S., Thiel M., Moore C. J., Borerro J. C., et al. (2014). plastic pollution in the world’s oceans: more than 5 Trillion plastic pieces weighing over 250,000 tons afloat at Sea. PLoS One 9:111913. 10.1371/journal.pone.0111913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley K., Reynolds C. R., Suckling L., Chee S. M., Tsipa A., Freemont P. S., et al. (2019). Utilising datasheets for the informed automated design and build of a synthetic metabolic pathway. J. Biol. Eng. 13:8. 10.1186/s13036-019-0141-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezure T., Suzuki T., Higashide S., Shintani E., Endo K., Kobayashi S.-I., et al. (2006). Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol. Prog. 22 1570–1577. 10.1021/bp060110v [DOI] [PubMed] [Google Scholar]
- Failmezger J., Scholz S., Blombach B., Siemann-Herzberg M. (2018). Cell-free protein synthesis from fast-growing vibrio natriegens. Front. Microbiol. 9:1146 10.3389/fmicb.2018.01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foshag D., Henrich E., Hiller E., Schäfer M., Kerger C., Burger-Kentischer A., et al. (2018). The E. coli S30 lysate proteome: a prototype for cell-free protein production. N. Biotechnol. 40 245–260. 10.1016/j.nbt.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Freemont P. S. (2019). Synthetic biology industry: data-driven design is creating new opportunities in biotechnology. Emerg. Top. Life Sci. 3 651–657. 10.1042/ETLS20190040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French K. E. (2019). Harnessing synthetic biology for sustainable development. Nat. Sustain. 2 250–252. 10.1038/s41893-019-0270-x [DOI] [Google Scholar]
- Gagoski D., Polinkovsky M. E., Mureev S., Kunert A., Johnston W., Gambin Y., et al. (2016). Performance benchmarking of four cell-free protein expression systems. Biotechnol. Bioeng. 113 292–300. 10.1002/bit.25814 [DOI] [PubMed] [Google Scholar]
- Gao W., Cho E., Liu Y., Lu Y. (2019). Advances and challenges in cell-free incorporation of unnatural amino acids into proteins. Front. Pharmacol. 10:611 10.3389/fphar.2019.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garamella J., Marshall R., Rustad M., Noireaux V. (2016). The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol. 5 344–355. 10.1021/acssynbio.5b00296 [DOI] [PubMed] [Google Scholar]
- García-Manrique P., Matos M., Gutiérrez G., Pazos C., Blanco-López M. C. (2018). Therapeutic biomaterials based on extracellular vesicles: classification of bio-engineering and mimetic preparation routes. J. Extracell. Vesicles 7:1422676. 10.1080/20013078.2017.1422676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garenne D., Noireaux V. (2019). Cell-free transcription–translation: engineering biology from the nanometer to the millimeter scale. Curr. Opin. Biotechnol. 58 19–27. 10.1016/j.copbio.2018.10.007 [DOI] [PubMed] [Google Scholar]
- Georgi V., Georgi L., Blechert M., Bergmeister M., Zwanzig M., Wüstenhagen D. A., et al. (2016). On-chip automation of cell-free protein synthesis: new opportunities due to a novel reaction mode. Lab. Chip. 16 269–281. 10.1039/C5LC00700C [DOI] [PubMed] [Google Scholar]
- Gessesse B., Nagaike T., Nagata K., Shimizu Y., Ueda T. (2018). G-protein coupled receptor protein synthesis on a lipid bilayer using a reconstituted cell-free protein synthesis system. Life 8:54. 10.3390/life8040054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Ellis T. (2019). Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 8 1–15. 10.1021/acssynbio.8b00423 [DOI] [PubMed] [Google Scholar]
- Gills B., Morgan J. (2019). Global climate emergency: after COP24, climate science, urgency, and the threat to humanity. Globalizations 1–18. 10.1080/14747731.2019.1669915 [DOI] [Google Scholar]
- Glasscock C. J., Lucks J. B., DeLisa M. P. (2016). Engineered protein machines: emergent tools for synthetic biology. Cell Chem. Biol. 23 45–56. 10.1016/j.chembiol.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Gowers G.-O. F., Cameron S. J. S., Perdones-Montero A., Bell D., Chee S. M., Kern M., et al. (2019). Off-colony screening of biosynthetic libraries by rapid laser-enabled mass spectrometry. ACS Synth. Biol. 8 2566–2575. 10.1021/acssynbio.9b00243 [DOI] [PubMed] [Google Scholar]
- Gräwe A., Dreyer A., Vornholt T., Barteczko U., Buchholz L., Drews G., et al. (2019). A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PLoS One 14:e0210940. 10.1371/journal.pone.0210940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene R. A., Morgan M., Shatkin A. J., Gage L. P. (1975). Translation of silk fibroin messenger RNA in an Ehrlich ascites cell-free extract. J. Biol. Chem. 250 5114–5121. [PubMed] [Google Scholar]
- Gregorio N. E., Levine M. Z., Oza J. P. (2019). A User’s guide to cell-free protein synthesis. Methods Protoc. 2:24. 10.3390/mps2010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg T. W., Del Vecchio D. (2020). Modular analysis and design of biological circuits. Curr. Opin. Biotechnol. 63 41–47. 10.1016/j.copbio.2019.11.015 [DOI] [PubMed] [Google Scholar]
- Gupta S., Sarkar S., Katranidis A., Bhattacharya J. (2019). Development of a cell-free optical biosensor for detection of a broad range of mercury contaminants in water: a plasmid DNA-Based approach. ACS Omega 4 9480–9487. 10.1021/acsomega.9b00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham T. S., Dmytriv Z., Plahar H., Chen J., Hillson N. J., Keasling J. D. (2012). Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res. 40:e141. 10.1093/nar/gks531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Satoh Y., Satoh T., Matsumoto K., Kakuchi T., Taguchi S., et al. (2011). Chemo-enzymatic synthesis of polyhydroxyalkanoate (PHA) incorporating 2-hydroxybutyrate by wild-type class i PHA synthase from Ralstonia eutropha. Appl. Microbiol. Biotechnol. 92 509–517. 10.1007/s00253-011-3362-8 [DOI] [PubMed] [Google Scholar]
- Harbers M. (2014). Wheat germ systems for cell-free protein expression. FEBS Lett. 588 2762–2773. 10.1016/j.febslet.2014.05.061 [DOI] [PubMed] [Google Scholar]
- Hecht A., Glasgow J., Jaschke P. R., Bawazer L. A., Munson M. S., Cochran J. R., et al. (2017). Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 45 3615–3626. 10.1093/nar/gkx070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks M., Bachmann T. T., Wang B. (2020). Synthetic biology enables programmable cell-based biosensors. Chemphyschem 21 132–144. 10.1002/cphc.201900739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillson N., Caddick M., Cai Y., Carrasco J. A., Chang M. W., Curach N. C., et al. (2019). Building a global alliance of biofoundries. Nat. Commun. 10:2040. 10.1038/s41467-019-10079-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroe A., Tsuge K., Nomura C. T., Itaya M., Tsuge T. (2012). Rearrangement of gene order in the phaCAB operon leads to effective production of ultrahigh-molecular-weight poly[(R)-3-hydroxybutyrate] in genetically engineered Escherichia coli. Appl. Environ. Microbiol. 78 3177–3184. 10.1128/AEM.07715-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman C. E., Jewett M. C. (2013). Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis. Biotechnol. Bioeng. 110 2643–2654. 10.1002/bit.24942 [DOI] [PubMed] [Google Scholar]
- Hu J., Xiao K., Jin B., Zheng X., Ji F., Bai D. (2019). Paper-based point-of-care test with xeno nucleic acid probes. Biotechnol. Bioeng. 116 2764–2777. 10.1002/bit.27106 [DOI] [PubMed] [Google Scholar]
- Hu S. J. (2013). Evolving paradigms of manufacturing: from mass production to mass customization and personalization. Procedia CIRP 7 3–8. 10.1016/j.procir.2013.05.002 [DOI] [Google Scholar]
- Huang A., Nguyen P. Q., Stark J. C., Takahashi M. K., Donghia N., Ferrante T., et al. (2018). BiobitsTM explorer: a modular synthetic biology education kit. Sci. Adv. 4 1–10. 10.1126/sciadv.aat5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S., Boyken S. E., Baker D. (2016). The coming of age of de novo protein design. Nature 537 320–327. 10.1038/nature19946 [DOI] [PubMed] [Google Scholar]
- Huang X., Li M., Green D. C., Williams D. S., Patil A. J., Mann S. (2013). Interfacial assembly of protein–polymer nano-conjugates into stimulus-responsive biomimetic protocells. Nat. Commun. 4:2239. 10.1038/ncomms3239 [DOI] [PubMed] [Google Scholar]
- Jaworski E., Wang L., Marco G. (1963). Synthesis of chitin in cell-free extracts of prodenia eridania. Nature 198 790–790. 10.1038/198790a013978870 [DOI] [Google Scholar]
- Jeong D., Klocke M., Agarwal S., Kim J., Choi S., Franco E., et al. (2019). Cell-free synthetic biology platform for engineering synthetic biological circuits and systems. Methods Protoc. 2:39. 10.3390/mps2020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop-Fabre M. M., Sonnenschein N. (2019). Improving reproducibility in synthetic biology. Front. Bioeng. Biotechnol. 7:18. 10.3389/fbioe.2019.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zhao J., Lian J., Xu Z. (2018). Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology. Synth. Syst. Biotechnol. 3 90–96. 10.1016/j.synbio.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Liu Y., Luo D., Huck W. T. S., Yang D. (2018). Microfluidic-assisted fabrication of clay microgels for cell-free protein synthesis. ACS Appl. Mater. Interfaces 10 29308–29313. 10.1021/acsami.8b09324 [DOI] [PubMed] [Google Scholar]
- Kanigowska P., Shen Y., Zheng Y., Rosser S., Cai Y. (2016). Smart DNA fabrication using sound waves. J. Lab. Autom. 21 49–56. 10.1177/2211068215593754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim A. S., Jewett M. C. (2016). A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 36 116–126. 10.1016/j.ymben.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Karim A. S., Jewett M. C. (2018). Cell-free synthetic biology for pathway prototyping. Methods Enzymol. 608 31–57. 10.1016/bs.mie.2018.04.029 [DOI] [PubMed] [Google Scholar]
- Katsura K., Matsuda T., Tomabechi Y., Yonemochi M., Hanada K., Ohsawa N., et al. (2017). A reproducible and scalable procedure for preparing bacterial extracts for cell-free protein synthesis. J. Biochem. 162 357–369. 10.1093/jb/mvx039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J. E., Jewett M. C. (2020). A cell-free system for production of 2,3-butanediol is robust to growth-toxic compounds. Metab. Eng. Commun. 10:e00114. 10.1016/j.mec.2019.e00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. R., Rubin A. J., Davis J. H., Ajo-Franklin C. M., Cumbers J., Czar M. J., et al. (2009). Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 3:4. 10.1186/1754-1611-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R., Bowater L., Yeoman K. H., Bowater R. P. (2015a). Promoting microbiology education through the iGEM synthetic biology competition. FEMS Microbiol. Lett. 362:fnv129. 10.1093/femsle/fnv129 [DOI] [PubMed] [Google Scholar]
- Kelwick R., Kopniczky M., Bower I., Chi W., Chin M. H. W., Fan S., et al. (2015b). A Forward-design approach to increase the production of poly-3-hydroxybutyrate in genetically engineered Escherichia coli. PLoS One 10:e0117202. 10.1371/journal.pone.0117202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R., MacDonald J. T., Webb A. J., Freemont P. (2014). Developments in the tools and methodologies of synthetic biology. Front. Bioeng. Biotechnol. 2:60. 10.3389/fbioe.2014.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R., Ricci L., Chee S. M., Bell D., Webb A. J., Freemont P. S. (2018). Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synth. Biol. 3:ysy016 10.1093/synbio/ysy016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R., Webb A. J., MacDonald J. T., Freemont P. S. (2016). Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng. 38 370–381. 10.1016/j.ymben.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Khambhati K., Bhattacharjee G., Gohil N., Braddick D., Kulkarni V., Singh V. (2019). Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems. Front. Bioeng. Biotechnol. 7:248. 10.3389/fbioe.2019.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kightlinger W., Duncker K. E., Ramesh A., Thames A. H., Natarajan A., Stark J. C., et al. (2019). A cell-free biosynthesis platform for modular construction of protein glycosylation pathways. Nat. Commun. 10 1–13. 10.1038/s41467-019-12024-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilb N., Burger J., Roth G. (2014). Protein microarray generation by in situ protein expression from template DNA. Eng. Life Sci. 14 352–364. 10.1002/elsc.201300052 [DOI] [Google Scholar]
- Kim B. S. (2000). Production of poly(3-hydroxybutyrate) from inexpensive substrates. Enzyme Microb. Technol. 27 774–777. 10.1016/s0141-0229(00)00299-4 [DOI] [PubMed] [Google Scholar]
- Kleer R., Piller F. T. (2019). Local manufacturing and structural shifts in competition: market dynamics of additive manufacturing. Int. J. Prod. Econ. 216 23–34. 10.1016/j.ijpe.2019.04.019 [DOI] [Google Scholar]
- Kopniczky M. B., Canavan C., McClymont D. W., Crone M. A., Suckling L., Goetzmann B., et al. (2020). Cell-free protein synthesis as a prototyping platform for mammalian synthetic biology. ACS Synth. Biol. 9 144–156. 10.1021/acssynbio.9b00437 [DOI] [PubMed] [Google Scholar]
- Kopp D., Willows R. D., Sunna A. (2019). Cell-free enzymatic conversion of spent coffee grounds into the platform chemical lactic acid. Front. Bioeng. Biotechnol. 7:389. 10.3389/fbioe.2019.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korman T. P., Opgenorth P. H., Bowie J. U. (2017). A synthetic biochemistry platform for cell free production of monoterpenes from glucose. Nat. Commun. 8:15526. 10.1038/ncomms15526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O., Mureev S., Johnston W., Alexandrov K. (2010). Towards the construction of expressed proteomes using a Leishmania tarentolae based cell-free expression system. PLoS One 5:e14388. 10.1371/journal.pone.0014388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O., Mureev S., Jung W., Kubala M. H., Johnston W., Alexandrov K. (2011). Leishmania cell-free protein expression system. Methods 55 58–64. 10.1016/j.ymeth.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Krinsky N., Kaduri M., Shainsky-Roitman J., Goldfeder M., Ivanir E., Benhar I., et al. (2016). A simple and rapid method for preparing a cell-free bacterial lysate for protein synthesis. PLoS One 11:e0165137. 10.1371/journal.pone.0165137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S., Narayan S., Chadha A. (2016). Whole resting cells vs. cell free extracts of Candida parapsilosis ATCC 7330 for the synthesis of gold nanoparticles. AMB Express 6:92. 10.1186/s13568-016-0268-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.-C., Jewett M. C. (2015). High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 5:8663. 10.1038/srep08663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H.-E., Canavan C., Cameron L., Moore S., Danchenko M., Kuiken T., et al. (2019). Synthetic biology and the united nations. Trends Biotechnol. 37 1146–1151. 10.1016/j.tibtech.2019.05.011 [DOI] [PubMed] [Google Scholar]
- Lavickova B., Maerkl S. J. (2019). A simple, robust, and low-cost method to produce the pure cell-free system. ACS Synth. Biol. 8 455–462. 10.1021/acssynbio.8b00427 [DOI] [PubMed] [Google Scholar]
- Le Feuvre R. A., Scrutton N. S. (2018). A living foundry for synthetic biological materials: a synthetic biology roadmap to new advanced materials. Synth. Syst. Biotechnol. 3 105–112. 10.1016/j.synbio.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-H., Kim D.-M. (2019). In vitro use of cellular synthetic machinery for biosensing applications. Front. Pharmacol. 10:1166 10.3389/fphar.2019.01166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton T. M., Rockström J., Gaffney O., Rahmstorf S., Richardson K., Steffen W., et al. (2019). Climate tipping points — too risky to bet against. Nature 575 592–595. 10.1038/d41586-019-03595-0 [DOI] [PubMed] [Google Scholar]
- Li J., Wang H., Jewett M. C. (2018). Expanding the palette of Streptomyces -based cell-free protein synthesis systems with enhanced yields. Biochem. Eng. J. 130 29–33. 10.1016/j.bej.2017.11.013 [DOI] [Google Scholar]
- Li T., Ye J., Shen R., Zong Y., Zhao X., Lou C., et al. (2016). Semirational approach for ultrahigh Poly(3-hydroxybutyrate) accumulation in Escherichia coli by combining one-step library construction and high-throughput screening. ACS Synth. Biol. 5 1308–1317. 10.1021/acssynbio.6b00083 [DOI] [PubMed] [Google Scholar]
- Libicher K., Hornberger R., Heymann M., Mutschler H. (2020). In vitro self-replication and multicistronic expression of large synthetic genomes. Nat. Commun. 11:904. 10.1038/s41467-020-14694-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. Y., Kim K.-O., Kim D.-M., Park C. B. (2009). Silica-coated alginate beads for in vitro protein synthesis via transcription/translation machinery encapsulation. J. Biotechnol. 143 183–189. 10.1016/j.jbiotec.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Liu R., Fu A., Deng Z., Li Y., Liu T. (2020). Promising methods for detection of novel coronavirus SARS-CoV-2. View 1:e4 10.1002/viw2.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhao T., Ju W., Shi S., Shi S., Shi S. (2017). Materials discovery and design using machine learning. J. Mater. 3 159–177. 10.1016/j.jmat.2017.08.002 [DOI] [Google Scholar]
- Lizardi P. M., Mahdavi V., Shields D., Candelas G. (1979). Discontinuous translation of silk fibroin in a reticulocyte cell-free system and in intact silk gland cells. Proc. Natl. Acad. Sci. 76 6211–6215. 10.1073/pnas.76.12.6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen C., Goñi Moreno A., Umesh P., Palchick Z., Roehner N., Atallah C., et al. (2019). Synthetic biology open language (SBOL) Version 2.3. J. Integr. Bioinform. 16:25. 10.1515/jib-2019-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. W., Des Soye B. J., Kwon Y.-C., Kay J., Davis R. G., Thomas P. M., et al. (2018). Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun. 9:1203. 10.1038/s41467-018-03469-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. W., Majewska N. I., Chen C. X., Albanetti T. E., Jimenez R. B. C., Schmelzer A. E., et al. (2017). Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies. ACS Synth. Biol. 6 1370–1379. 10.1021/acssynbio.7b00001 [DOI] [PubMed] [Google Scholar]
- Matsumoto K. K., Okei T., Honma I., Ooi T., Aoki H., Taguchi S. (2013). Erratum to: Efficient (R)-3-hydroxybutyrate production using acetyl CoA-regenerating pathway catalyzed by coenzyme A transferase. Appl. Microbiol. Biotechnol. 97 439–439. 10.1007/s00253-012-4563-5 [DOI] [PubMed] [Google Scholar]
- McLaughlin J. A., Myers C. J., Zundel Z., Mısırlı G., Zhang M., Ofiteru I. D., et al. (2018). SynBioHub: a standards-enabled design repository for synthetic biology. ACS Synth. Biol. 7 682–688. 10.1021/acssynbio.7b00403 [DOI] [PubMed] [Google Scholar]
- Miguez A. M., McNerney M. P., Styczynski M. P. (2019). Metabolic profiling of Escherichia coli -based cell-free expression systems for process optimization. Ind. Eng. Chem. Res. 58 22472–22482. 10.1021/acs.iecr.9b03565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misirli G., Nguyen T., McLaughlin J. A., Vaidyanathan P., Jones T. S., Densmore D., et al. (2019). A computational workflow for the automated generation of models of genetic designs. ACS Synth. Biol. 8 1548–1559. 10.1021/acssynbio.7b00459 [DOI] [PubMed] [Google Scholar]
- Moore S. J., Lai H.-E., Needham H., Polizzi K. M., Freemont P. S. (2017a). Streptomyces venezuelae TX-TL - a next generation cell-free synthetic biology tool. Biotechnol. J. 12:1600678. 10.1002/biot.201600678 [DOI] [PubMed] [Google Scholar]
- Moore S. J., MacDonald J. T., Freemont P. S. (2017b). Cell-free synthetic biology for in vitro prototype engineering. Biochem. Soc. Trans. 45 785–791. 10.1042/BST20170011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. J., MacDonald J. T., Wienecke S., Ishwarbhai A., Tsipa A., Aw R., et al. (2018). Rapid acquisition and model-based analysis of cell-free transcription–translation reactions from nonmodel bacteria. Proc. Natl. Acad. Sci. U.S.A. 115 E4340–E4349. 10.1073/pnas.1715806115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mureev S., Kovtun O., Nguyen U. T. T., Alexandrov K. (2009). Species-independent translational leaders facilitate cell-free expression. Nat. Biotechnol. 27 747–752. 10.1038/nbt.1556 [DOI] [PubMed] [Google Scholar]
- Nahvi A., Sudarsan N., Ebert M. S., Zou X., Brown K. L., Breaker R. R. (2002). Genetic control by a metabolite binding mRNA. Chem. Biol. 9 1043–1049. 10.1016/S1074-5521(02)00224-7 [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H., Sun Z. Z., Hori Y., Yeung E., Verpoorte A., Murray R. M., et al. (2015). Rapid cell-free forward engineering of novel genetic ring oscillators. eLife 4:e09771. 10.7554/eLife.09771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A. A. K., Der B. S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E. A., et al. (2016). Genetic circuit design automation. Science 352:7341. 10.1126/science.aac7341 [DOI] [PubMed] [Google Scholar]
- Nielsen C., Rahman A., Rehman A. U., Walsh M. K., Miller C. D. (2017). Food waste conversion to microbial polyhydroxyalkanoates. Microb. Biotechnol. 2:12776. 10.1111/1751-7915.12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel P. I., de Almeida A., Melillo E. C., Galvagno M. A., Pettinari M. J. (2006). New recombinant Escherichia coli strain tailored for the production of poly(3-hydroxybutyrate) from agroindustrial by-products. Appl. Environ. Microbiol. 72 3949–3954. 10.1128/AEM.00044-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogonah O. W., Polizzi K. M., Bracewell D. G. (2017). Cell free protein synthesis: a viable option for stratified medicines manufacturing? Curr. Opin. Chem. Eng. 18 77–83. 10.1016/j.coche.2017.10.003 [DOI] [Google Scholar]
- O’Kane P. T., Dudley Q. M., McMillan A. K., Jewett M. C., Mrksich M. (2019). High-throughput mapping of CoA metabolites by SAMDI-MS to optimize the cell-free biosynthesis of HMG-CoA. Sci. Adv. 5:eaaw9180. 10.1126/sciadv.aaw9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opgenorth P. H., Korman T. P., Bowie J. U. (2016). A synthetic biochemistry module for production of bio-based chemicals from glucose. Nat. Chem. Biol. 12 1–4. 10.1038/nchembio.2062 [DOI] [PubMed] [Google Scholar]
- Pardee K. (2018). Perspective: solidifying the impact of cell-free synthetic biology through lyophilization. Biochem. Eng. J. 138 91–97. 10.1016/j.bej.2018.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K., Green A. A., Ferrante T., Cameron D. E., DaleyKeyser A., Yin P., et al. (2014). Paper-based synthetic gene networks. Cell 159 940–954. 10.1016/j.cell.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K., Green A. A., Takahashi M. K., Braff D., Lambert G., Lee J. W., et al. (2016a). Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell 165 1255–1266. 10.1016/j.cell.2016.04.059 [DOI] [PubMed] [Google Scholar]
- Pardee K., Slomovic S., Nguyen P. Q., Lee J. W., Donghia N., Burrill D., et al. (2016b). Portable, on-demand biomolecular manufacturing. Cell 167 248.e–259.e. 10.1016/j.cell.2016.09.013 [DOI] [PubMed] [Google Scholar]
- Park N., Kahn J. S., Rice E. J., Hartman M. R., Funabashi H., Xu J., et al. (2009a). High-yield cell-free protein production from P-gel. Nat. Protoc. 4 1759–1770. 10.1038/nprot.2009.174 [DOI] [PubMed] [Google Scholar]
- Park N., Um S. H., Funabashi H., Xu J., Luo D. (2009b). A cell-free protein-producing gel. Nat. Mater. 8 432–437. 10.1038/nmat2419 [DOI] [PubMed] [Google Scholar]
- Philp J. (2018). The bioeconomy, the challenge of the century for policy makers. N. Biotechnol. 40 11–19. 10.1016/j.nbt.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Rajakumar P. D., Gowers G.-O. F., Suckling L., Foster A., Ellis T., Kitney R. I., et al. (2019). Rapid prototyping platform for Saccharomyces cerevisiae using computer-aided genetic design enabled by parallel software and workcell platform development. SLAS Technol. Transl. Life Sci. Innov. 24 291–297. 10.1177/2472630318798304 [DOI] [PubMed] [Google Scholar]
- Roberts A. D., Finnigan W., Wolde-Michael E., Kelly P., Blaker J. J., Hay S., et al. (2019). Synthetic biology for fibers, adhesives, and active camouflage materials in protection and aerospace. MRS Commun. 9 486–504. 10.1557/mrc.2019.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roell M.-S., Zurbriggen M. D. (2020). The impact of synthetic biology for future agriculture and nutrition. Curr. Opin. Biotechnol. 61 102–109. 10.1016/j.copbio.2019.10.004 [DOI] [PubMed] [Google Scholar]
- Rolf J., Rosenthal K., Lütz S. (2019). Application of cell-free protein synthesis for faster biocatalyst development. Catalysts 9:190 10.3390/catal9020190 [DOI] [Google Scholar]
- Rues R.-B., Henrich E., Boland C., Caffrey M., Bernhard F. (2016). Cell-free production of membrane proteins in Escherichia coli lysates for functional and structural studies. Methods Mol. Biol. 1432 1–21. 10.1007/978-1-4939-3637-3_1 [DOI] [PubMed] [Google Scholar]
- Ruiz R. C. H., Kiatwuthinon P., Kahn J. S., Roh Y. H., Luo D. (2012). Cell-free protein expression from DNA-based hydrogel (P-Gel) droplets for scale-up production. Ind. Biotechnol. 8 372–377. 10.1089/ind.2012.0024 [DOI] [Google Scholar]
- Russo D. A., Zedler J. A. Z., Jensen P. E. (2019). A force awakens: exploiting solar energy beyond photosynthesis. J. Exp. Bot. 70 1703–1710. 10.1093/jxb/erz054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad M., Eastlund A., Jardine P., Noireaux V. (2018). Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth. Biol. 3 1–7. 10.1093/synbio/ysy002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi A. S. M., Shakalli Tang M. J., Smith M. T., Hunt J. M., Law R. A., Wood D. W., et al. (2017). Cell-Free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal. Chem. 89 3395–3401. 10.1021/acs.analchem.6b04034 [DOI] [PubMed] [Google Scholar]
- Salehi A. S. M., Smith M. T., Bennett A. M., Williams J. B., Pitt W. G., Bundy B. C. (2016). Cell-free protein synthesis of a cytotoxic cancer therapeutic: onconase production and a just-add-water cell-free system. Biotechnol. J. 11 274–281. 10.1002/biot.201500237 [DOI] [PubMed] [Google Scholar]
- Shelby M. L., He W., Dang A. T., Kuhl T. L., Coleman M. A. (2019). Cell-free co-translational approaches for producing mammalian receptors: expanding the cell-free expression toolbox using nanolipoproteins. Front. Pharmacol. 10:744 10.3389/fphar.2019.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19 751–755. 10.1038/90802 [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Kanamori T., Ueda T. (2005). Protein synthesis by pure translation systems. Methods 36 299–304. 10.1016/j.ymeth.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Shin J., Noireaux V. (2012). An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 1 29–41. 10.1021/sb200016s [DOI] [PubMed] [Google Scholar]
- Shinoda T., Shinya N., Ito K., Ishizuka-Katsura Y., Ohsawa N., Terada T., et al. (2016). Cell-free methods to produce structurally intact mammalian membrane proteins. Sci. Rep. 6 1–15. 10.1038/srep30442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff M. J., Temoche-Diaz M. M., Karfilis K. V., Ri S., Schekman R. (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5 1–23. 10.7554/eLife.19276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A. D., Karim A. S., Jewett M. C. (2019). Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21 151–170. 10.1038/s41576-019-0186-3 [DOI] [PubMed] [Google Scholar]
- Smith M. T., Berkheimer S. D. (2014). Lyophilized Escherichia coli -based cell-free systems for robust, high-density, long-term storage. Biotechniques 56:158. 10.2144/000114158 [DOI] [PubMed] [Google Scholar]
- Stock T., Seliger G. (2016). Opportunities of sustainable manufacturing in industry 4.0. Proc. CIRP 40 536–541. 10.1016/j.procir.2016.01.129 [DOI] [Google Scholar]
- Storch M., Haines M. C., Baldwin G. S. (2019). DNA-BOT: a low-cost, automated DNA assembly platform for synthetic biology. bioRxiv [Preprint], 10.1101/832139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C. J., Pendleton E. D., Sasmor H. H., Hicks W. L., Farnum J. B., Muto M., et al. (2016). A cell-free expression and purification process for rapid production of protein biologics. Biotechnol. J. 11 238–248. 10.1002/biot.201500214 [DOI] [PubMed] [Google Scholar]
- Sun Z. Z., Hayes C. A., Shin J., Caschera F., Murray R. M., Noireaux V. (2013). Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp. e50762. 10.3791/50762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. Z., Yeung E., Hayes C. A., Noireaux V., Murray R. M. (2014). Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli Based TX-TL cell-free system. ACS Synth. Biol. 3 387–397. 10.1021/sb400131a [DOI] [PubMed] [Google Scholar]
- Swank Z., Laohakunakorn N., Maerkl S. J. (2019). Cell-free gene-regulatory network engineering with synthetic transcription factors. Proc. Natl. Acad. Sci. U.S.A. 116 5892–5901. 10.1073/pnas.1816591116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Xue Y.-S., Aibaidula G., Chen G.-Q. (2011). Unsterile and continuous production of polyhydroxybutyrate by halomonas TD01. Bioresour. Technol. 102 8130–8136. 10.1016/j.biortech.2011.05.068 [DOI] [PubMed] [Google Scholar]
- Tao W., Lv L., Chen G.-Q. (2017). Engineering halomonas species TD01 for enhanced polyhydroxyalkanoates synthesis via CRISPRi. Microb. Cell Fact. 16:48. 10.1186/s12934-017-0655-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrahi R., Fathi Z., Seydibeyoğlu M. Ö., Doustkhah E., Khataee A. (2020). Polyhydroxyalkanoates (PHA): from production to nanoarchitecture. Int. J. Biol. Macromol. 146 596–619. 10.1016/j.ijbiomac.2019.12.181 [DOI] [PubMed] [Google Scholar]
- Thavarajah W., Silverman A. D., Verosloff M. S., Kelley-Loughnane N., Jewett M. C., Lucks J. B. (2020). Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor. ACS Synth. Biol. 9 10–18. 10.1021/acssynbio.9b00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson N., Roy I., Summers D., Sivaniah E. (2009). In vitro production of polyhydroxyalkanoates: achievements and applications. J. Chem. Technol. Biotechnol. 85 760–767. 10.1002/jctb.2299 [DOI] [Google Scholar]
- Tomizawa S., Yoshioka M., Ushimaru K., Tsuge T. (2012). Preparative synthesis of Poly(R)-3-hydroxybutyrate monomer for enzymatic cell-free polymerization. Polym. J. 44 982–985. 10.1038/pj.2012.34 [DOI] [Google Scholar]
- Townsend T., Sette J. (2016). “Natural fibres and the world economy,” in Natural Fibres: Advances in Science and Technology Towards Industrial Applications, eds Fangueiro R., Rana S. (Dordrecht: Springer; ), 381–390. 10.1007/978-94-017-7515-1_30 [DOI] [Google Scholar]
- Trump B. D., Cegan J., Wells E., Poinsatte-Jones K., Rycroft T., Warner C., et al. (2019). Co-evolution of physical and social sciences in synthetic biology. Crit. Rev. Biotechnol. 39 351–365. 10.1080/07388551.2019.1566203 [DOI] [PubMed] [Google Scholar]
- Ullah M. W., Ul-Islam M., Khan S., Kim Y., Park J. K. (2015). Innovative production of bio-cellulose using a cell-free system derived from a single cell line. Carbohydr. Polym. 132 286–294. 10.1016/j.carbpol.2015.06.037 [DOI] [PubMed] [Google Scholar]
- Voyvodic P. L., Pandi A., Koch M., Conejero I., Valjent E., Courtet P., et al. (2019). Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nat. Commun. 10:1697. 10.1038/s41467-019-09722-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H. J., Engesser R., Ermes K., Geraths C., Timmer J., Weber W. (2019). Synthetic biology-inspired design of signal-amplifying materials systems. Mater. Today 22 25–34. 10.1016/j.mattod.2018.04.006 [DOI] [Google Scholar]
- Walsh D. I., Pavan M., Ortiz L., Wick S., Bobrow J., Guido N. J., et al. (2019). Standardizing automated DNA assembly: best practices, metrics, and protocols using robots. SLAS Technol. Transl. Life Sci. Innov. 24 282–290. 10.1177/2472630318825335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., Volpetti F., Petrova E., French C., Maerkl S. J., Wang B. (2019). Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 15 540–548. 10.1038/s41589-019-0244-3 [DOI] [PubMed] [Google Scholar]
- Weber L. A., Feman E. R., Baglioni C. (1975). Cell free system from HeLa cells active in initiation of protein synthesis. Biochemistry 14 5315–5321. 10.1021/bi00695a015 [DOI] [PubMed] [Google Scholar]
- Wen K. Y., Cameron L., Chappell J., Jensen K., Bell D. J., Kelwick R., et al. (2017). A Cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa -infected respiratory samples. ACS Synth. Biol. 6 2293–2301. 10.1021/acssynbio.7b00219 [DOI] [PubMed] [Google Scholar]
- Wilding K. M., Schinn S.-M., Long E. A., Bundy B. C. (2018). The emerging impact of cell-free chemical biosynthesis. Curr. Opin. Biotechnol. 53 115–121. 10.1016/j.copbio.2017.12.019 [DOI] [PubMed] [Google Scholar]
- Wong H. H., Lee S. Y. (1998). Poly-(3-hydroxybutyrate) production from whey by high-density cultivation of recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 50 30–33. 10.1007/s002530051252 [DOI] [PubMed] [Google Scholar]
- Yan Q., Fong S. S. (2015). Bacterial chitinase: nature and perspectives for sustainable bioproduction. Bioresour. Bioprocess. 2:31 10.1186/s40643-015-0057-5 [DOI] [Google Scholar]
- Yim S. S., Johns N. I., Park J., Gomes A. L., McBee R. M., Richardson M., et al. (2019). Multiplex transcriptional characterizations across diverse bacterial species using cell-free systems. Mol. Syst. Biol. 15:875. 10.15252/msb.20198875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue K., Zhu Y., Kai L. (2019). Cell-free protein synthesis: chassis toward the minimal cell. Cells 8:315. 10.3390/cells8040315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawada J. F., Yin G., Steiner A. R., Yang J., Naresh A., Roy S. M., et al. (2011). Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 108 1570–1578. 10.1002/bit.23103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lin Y., Wu Q., Wang Y., Chen G.-Q. (2019). Synthetic biology and genome-editing tools for improving pha metabolic engineering. Trends Biotechnol. 10.1016/j.tibtech.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Minagawa Y., Kizoe H., Miyazaki K., Iino R., Ueno H., et al. (2019). Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array. Sci. Adv. 5:eaav8185. 10.1126/sciadv.aav8185 [DOI] [PMC free article] [PubMed] [Google Scholar]