Abstract
Circadian clocks are intrinsic, time-tracking systems that bestow upon organisms a survival advantage. Under natural conditions, organisms are trained to follow a 24-h cycle under environmental time cues such as light to maximize their physiological efficiency. The exact timing of this rhythm is established via cell-autonomous oscillators called cellular clocks, which are controlled by transcription/translation-based negative feedback loops. Studies using cell-based systems and genetic techniques have identified the molecular mechanisms that establish and maintain cellular clocks. One such mechanism, known as post-translational modification, regulates several aspects of these cellular clock components, including their stability, subcellular localization, transcriptional activity, and interaction with other proteins and signaling pathways. In addition, these mechanisms contribute to the integration of external signals into the cellular clock machinery. Here, we describe the post-translational modifications of cellular clock regulators that regulate circadian clocks in vertebrates.
Keywords: Circadian clock, cellular clock, clock protein, post-translational modification, transcription, clock gene
1. INTRODUCTION
Circadian clocks regulate a number of physiological functions, such as sleep and metabolism, in a broad spectrum of organisms ranging from bacteria to humans [1]. They generate daily changes (circadian rhythms) in various biochemical, physiological and behavioral processes. Under natural conditions, circadian clocks are trained according to the 24-h cycle based on environmental time cues, such as light, to maximize an organism’s physiological efficiency [2]. Thus, disrupting circadian clocks can have a profound effect on organisms’ health and is linked to various diseases, including sleep disorders and metabolic syndromes [3, 4].
At the molecular level, circadian clocks can be divided into three conceptual components [1, 5]. The first is the pacemaker that is dedicated to generating and sustaining circadian rhythms by receiving and integrating signals from external time cues [6]. The second component is the input, which refers to the pathway through which external time cues are perceived and act upon the central pacemaker. The third element relates to how the circadian clock affects physiology and is achieved through the output pathways.
The pacemaker in the members of Neurospora and Drosophila and in vertebrates are transcription/translation-based negative feedback loops that rely on positive and negative oscillator elements [7]. In vertebrates, three basic helix-loop-helix Per-ARNT-Sim (PAS) domain-containing transcription factors, known as CLOCK, neuronal PAS domain protein 2 (NPAS2), and brain-muscle-ARNT-like protein (BMAL), constitute the positive elements [1, 5, 8]. CLOCK or NPAS2 heterodimerizes with BMAL to form a transcriptionally active complex that binds to E-box elements (CACGTG) present in the promoters of the members of the period (per) and cryptochrome (cry) families. Once PER and CRY proteins are translated, they form heterodimers that can then translocate into the nucleus to repress CLOCK(NPAS2)–BMAL-mediated transcription through direct protein−protein interaction. Importantly, when active, the CLOCK(NPAS2)–BMAL complex stimulates the transcription of various other clock-controlled genes. The protein products of these genes, in turn, influence functions external to the oscillatory mechanism itself and mediate the “output” function of the clock. This partly accounts for the presence of circadian rhythms in a variety of physiological processes [3, 4].
Although the relatively straightforward mechanisms of positive and negative feedback loops are necessary to establish and maintain cellular clocks, these clocks are complex and involve processes such as the post-transcriptional regulation of cellular clock components (clock proteins) [4, 9, 10]. These modifications have essential roles in appropriately regulating clock protein stability, cellular localization, transcriptional activity, and interaction with other proteins. In addition, a variety of studies revealed that the post-transcriptional modifications of clock proteins are involved in the regulation of the input and output processes of circadian clocks [11-13]. Here, we describe the roles of the post-translational modifications of clock proteins involved in controlling circadian clocks in vertebrates.
2. IMPORTANCE OF THE PHOSPHORYLATION OF CELLULAR CLOCK PROTEINS FOR REGULATING THE CLOCK’S PERIODICITY AND LIGHT RESPONSE
The functions of various mammalian cellular clock proteins, including CLOCK, BMAL1, PER1, PER2, PER3, CRY1, and CRY2, are regulated via phosphorylation by various kinases [4, 9, 10]. The first major step towards understanding the importance of phosphorylation in vertebrate circadian clock regulation was taken when the tau mutation, which causes a short-period phenotype in the Syrian hamster, was identified [14]. The tau locus encodes casein kinase I epsilon (CKIε) that phosphorylates PERs. The short-period phenotype observed in the mutant hamster was generated because of a low rate of CKI-dependent phosphorylation of PER2. Defects in the phosphorylation of cellular clock proteins have been implicated in human disorders [15, 16]. A missense mutation in the circadian clock gene Per2 is associated with familial advanced sleep phase syndrome. The corresponding mutated PER2 protein is less effectively phosphorylated than the wild-type PER2; moreover, the phosphorylation-dependent stability control of the mutated PER2 was demonstrated to have been eliminated in vitro. Additionally, polymorphism in a region of human Per3, the presumed CKIε-binding domain, may be associated with delayed sleep phase syndrome [17]. CKIε specifically interacts with and phosphorylates PER1, 2 and 3 proteins, and thus regulates each of them differently [4, 9, 10, 18, 19].
A number of pharmacological studies have used luciferase-based clock reporter cells to identify the molecules that regulate cellular clocks. These studies identified various kinases that are important for establishing or regulating cellular clocks. For example, using mammalian clock reporter cells, a small-scale screening of kinase inhibitors identified candidate molecules affecting the period-length of the mammalian cellular clock [20]. As a result, casein kinase II (CKII), PI3-kinase (PI3K), and c-Jun N-terminal kinases (JNKs) were identified as candidate regulators for the cellular clock. Another research group examined the effect of various kinase inhibitors on the periodicity of cellular clocks and found that the period-length of the clocks of mammalian cultured cells increased following treatment with p38 (SB203580), JNK (SP600125), CKI (IC261) and CDK (roscovitine) kinase inhibitor and decreased following treatment with GSK-3 (SB216763) or CaMKII (KN93) inhibitors [21]. It is important to stress that several of the already-mentioned inhibitors have off-target effects. Therefore, confirming these findings by using pharmacological approaches in other experimental settings is warranted. In fact, a number of genetic and biochemical analyses reported the involvement of these kinases in the functional regulation of clock proteins and the maintenance of circadian clocks at the molecular, cellular, and organismic levels [11, 22-27]. Below, we describe the role of JNKs in circadian clock regulation.
3. ROLES OF THE JNK-MEDIATED SIGNALING PATHWAY IN CIRCADIAN CLOCK REGULATION
JNK activity is regulated via the phosphorylation of particular tyrosine and threonine residues located in the kinase domain [28]. JNK phosphorylation is catalyzed by two dual-specificity kinases, MKK4 and MKK7, which act in a synergistic manner [29, 30]. Although it is primarily activated in response to external stress (osmolarity changes, heat shock and UV irradiation), phosphorylated JNK has been detected in unstressed cultured cells and in isolated mouse tissues, such as the brain [30-32]. Notably, previous studies have shown that JNK phosphorylation levels, and thus its kinase activity, fluctuate in a circadian manner in both the suprachiasmatic nucleus (SCN), the location of the central clock in mammals, and in cultured mammalian cells [33, 34]. These studies indicate the importance of JNK signaling in physiological processes other than cellular stress responses, including circadian clocks.
In this context, it was previously reported that the MKK7–JNK signaling pathway is an essential regulator of periodicity in the cellular clocks of mammals. The MKK7–JNK signaling pathway induces PER2 phosphorylation and stabilizes PER2 by inhibiting its ubiquitination, which has an effect opposite to that of CKIε-induced PER2 destabilization [24] (Fig. 1A). Because genetically inhibiting MKK7’s function results in the extension of the cellular clock’s periodicity in cultured cells, this phosphorylation-mediated PER2 stability control may be necessary to maintain the normal periodicity of cellular clocks. In addition, a recent study generated neuron-specific Mkk7-deleted mice, in which MKK7 was genetically inactivated in the central clocks of the SCN [32]. A behavioral analysis of these mice revealed that the neuron-specific disruption of Mkk7 resulted in longer periods of circadian behavioral rhythms and also reduced the amplitude of rhythmicity compared with wild-type mice. These findings provide evidence that the MKK7–JNK signaling pathway is involved in the regulation of the circadian pacemaker at an organismic level.
Fig. (1).
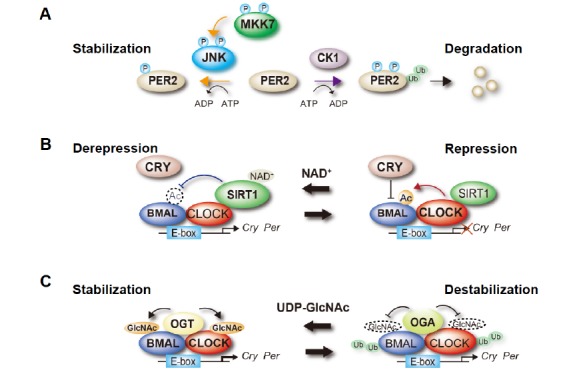
Functional regulation of clock proteins through post-translational modifications. (A) Phosphorylation-dependent control of PER2 stability. The activity of JNK is regulated via the phosphorylation of the tyrosine and threonine residues located in the kinase domain, which is catalyzed by MKK7. MKK7-mediated JNK activation induces phosphorylation of PER2 and increases its protein half-life by competing with the CKI-induced ubiquitination and the subsequent degradation of PER2. (B) Regulation of CLOCK:BMAL-mediated transcription by BMAL acetylation. CLOCK acetylates its heterodimeric partner BMAL. This CLOCK-mediated acetylation increases the interaction of the CLOCK:BMAL complex with CRY, facilitating repression of the CLOCK:BMAL complex’s activity. SIRT1 deacetylates BMAL, which cancels the CRY-dependent repression of CLOCK:BMAL-mediated transcription. (C) O-GlcNAcylation-dependent regulation of CLOCK:BMAL complex’s stability. Both CLOCK and BMAL1 are O-GlcNAcylated, which is catalyzed by OGT and reversed by OGA. This modification stabilizes both proteins by inhibiting their ubiquitination.
In addition to PER2, JNK phosphorylates BMAL1 and CLOCK in mammals. In particular, Yoshitane et al. reported that neuron-specific isoform JNK3-deficient mice have longer free-running periods of behavioral rhythms and compromised phase shifts to light [11]. In nocturnal animals, the higher the light intensity in constant light conditions, the longer the circadian period becomes (and vice versa in diurnal species). This phenomenon is known as Aschoff’s rule [35, 36]. In JNK3-deficient mice, behavioral rhythms are insensitive to intensity changes in constant light, thus deviating from Aschoff’s rule [11]. These findings provide solid evidence that JNK-mediated BMAL1 phosphorylation is an important regulatory mechanism underlying the circadian pacemaker, as well as the light input pathway of the circadian clock in vivo.
4. REGULATION OF CELLULAR CLOCKS VIA BMAL1 AND DIFFERENTIATED EMBRYOCHONDROCYTE EXPRESSED GENE 1 (DEC1) SUMOylation
SUMOylation is the covalent linking of small ubiquitin-related modifier proteins to lysine residues [37, 38]. This modification is a reversible post-translational modification that has been implicated in transcriptional regulation by a number of mechanisms. Previous studies found that BMAL1 is Sumoylated on a highly conserved lysine residue (Lys259) in cultured cells and that BMAL1 SUMOylation shows a circadian pattern in mouse liver tissue [39]. In addition, BMAL SUMOylation has been demonstrated to control BMAL protein stability and to play a critical role in the regulation of the pacemaker in the circadian clock.
Reportedly, BMAL1 SUMOylation promotes the interaction of CREB-binding protein to the CLOCK–BMAL1 complex, which resets the cellular clock in response to serum stimuli [40]. The formation of this ternary complex induces the acute activation of the CLOCK–BMAL1-mediated transcription of Per1, resetting the phase of the cellular clock. These findings clearly demonstrate that BMAL1 SUMOylation is a regulatory element for the input pathway of the circadian clock.
Differentiated embryo-chondrocyte expressed gene 1 (DEC1) is a basic mammalian helix-loop-helix protein that acts as a transcription factor [41, 42]. DEC1 inhibits CLOCK–BMAL1-mediated transcription through direct interaction with BMAL1 and/or competition for E-box elements in the promoters of cellular clock-controlled genes [43]. Moreover, DEC1 is Sumoylated on highly conserved lysine residues (Lys159 and Lys279) at its C-terminal domain [44]. SUMOylation stabilizes DEC1 by inhibiting its ubiquitination and promoting the inhibition of CLOCK–BMAL1-mediated transcription. These findings suggest that SUMOylation serves as a key regulatory element of cellular clocks by controlling multiple transcriptional factors.
5. IMPORTANCE OF THE ACETYLATION OF CELLULAR CLOCK PROTEINS FOR THE MAINTENANCE OF CLOCK PERIODICITY
Chromatin, the nucleoprotein structure into which the eukaryotic genome is organized, enables the functioning of essential biological processes, such as the regulation of transcription, DNA repair, apoptosis, and cell division [45-47]. Histone acetylation plays a pivotal role in the modulation of the chromatin structure associated with transcriptional activation [48, 49]. The activation of clock-controlled genes by the CLOCK–BMAL1 complex has been shown to be coupled to circadian changes in histone acetylation at their promoters, evidence that transcription-permissive chromatin states are dynamically established in a circadian-time-specific manner [50-52].
In mammals, the core circadian regulator, CLOCK, has intrinsic histone acetyltransferase (HAT) activity [53]. Moreover, CLOCK regulates the circadian pattern of gene expression by the virtue of its HAT activity. This finding modifies the common view of the CLOCK protein, demonstrating that it operates not only as a transcription factor but also as an enzyme. The finding of CLOCK’s HAT activity suggests that the HAT enzymatic activity also targets other non-histone proteins, which is a characteristic of other HATs. In fact, CLOCK acetylates its heterodimeric partner, BMAL1 [54, 55] (Fig. 1B). This CLOCK-mediated acetylation increases the interaction of the CLOCK–BMAL1 complex with CRY1. One study reported that BMAL1 is deacetylated by SIRT1, a nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase (HDAC) [56, 57], in a time-dependent manner [58] (Fig. 1B). Accordingly, BMAL1 acetylation is significantly rhythmic in mice liver as well as in cultured cells and SCNs. Another study demonstrated that SIRT1 also deacetylates PER2, giving SIRT1 an additional function in the transcriptional regulation of the cellular clock [59].
Notably, several pharmacological studies have confirmed the importance of the acetylation of cellular clock proteins in clock regulation. For example, it has been reported that the inhibition of SIRT1 activity by its inhibitors, such as nicotinamide and the drug splitomicin, disturbs the circadian expression of clock-controlled genes and histone H3 and BMAL1 acetylations [58]. In addition, the study using several specific SIRT1 activators demonstrated that SIRT1 activation led to the suppression of clock-controlled gene expressions and H3 acetylation at corresponding promoters in vitro and in vivo [60].
6. THE POSSIBLE ROLE OF O-LINKEDNACETYLGLUCOSAMINYLATION (O-GLCNACYLATION) OF CLOCK PROTEINS IN TRANSDUCING NUTRITIONAL SIGNALS TO THE CIRCADIAN CLOCK MACHINERY
O-GlcNAcylation is one of the most common post-translational protein modification with the high-energy compound, UDP-GlcNAc, as the direct donor [61]. Two enzymes regulate O-GlcNAcylation: O-GlcNAc transferase (OGT), which attaches UDP-GlcNAc to the serine and threonine residues of proteins through a β-glycosidic O-linkage, and O-GlcNAcase (OGA), which hydrolyzes O-GlcNAc in proteins [61, 62]. Analyses with O-GlcNAcylation inhibitors (the OGT inhibitor Alloxan) and activators (the OGA inhibitor PUGNAc) have revealed that the suppression of O-GlcNAcylation shortened the periodicity of cellular clocks, whereas its activation lengthened the periodicity of cellular clocks in mammalian cultured cells [13]. These findings provide evidence that O-GlcNAcylation is involved in cellular clock regulation.
Both CLOCK and BMAL1 are rhythmically O-GlcNAcylated, and this modification stabilizes both the proteins by inhibiting their ubiquitination [12, 63] (Fig. 1C). Consistent with these findings, OGT facilitates CLOCK–BMAL1-mediated transcription. Another study reported that PER2 is also O-GlcNAcylated at the region that regulates the human sleep phase in humans, competing with phosphorylation in this region [13].
Glucose flux via the hexosamine biosynthesis pathway leads to intracellular glycosylation by increasing the O-GlcNAcylation of proteins [61]. It is well established that O-GlcNAcylation regulates fundamental cellular processes in response to diverse nutritional cues. Because circadian clocks are coupled with metabolic oscillations through nutrient-sensing pathways [3, 6], these facts indicate that the O-GlcNAcylation of clock proteins plays a crucial role in transducing nutritional signals to the cellular clock machinery.
7. ROLE OF POLY(ADP-ribosyl)ATION OF CLOCK IN FEEDING-DEPENDENT CIRCADIAN CLOCK REGULATION
Mammals have no photoreception in peripheral tissues; therefore, the effect of light on peripheral clocks is indirect [6]. SCN, the site of the master clock in mammals, integrates photic cues from the retina and transmits this information to peripheral clocks, synchronizing them through humoral signals [2, 4]. In addition to the input of the central clock (SCN), peripheral cellular clocks respond to cellular metabolism [3, 6]. When feeding in nocturnal animals is restricted to daytime, the phase of peripheral cellular clocks differs from that of SCN. Feeding cue affects peripheral cellular clocks via the poly(ADP-ribosyl)ation of CLOCK [64]. PARP-1, an NAD(+)-dependent ADP-ribosyltransferase, has been shown to interact with and poly(ADP-ribosyl)ate CLOCK. The poly(ADP-ribosyl)ation of CLOCK leads to reduced DNA binding ability in the CLOCK–BMAL1 complex, and modulates the interaction of this complex with PER and CRY. These poly(ADP-ribosyl)ation-dependent functional regulations of the CLOCK–BMAL1 complex, in turn, influence the expression patterns of cellular clock-controlled genes. Notably, PARP-1-deficient mice show altered expression profiles in CLOCK–BMAL1-dependent gene expression, particularly in response to changes in feeding times [64]. In addition, PARP-1-deficient mice exhibit impaired food entrainment of peripheral cellular clocks. These findings support the idea that the PARP-1-mediated poly(ADP-ribosyl)ation of CLOCK plays a role in connecting feeding with the circadian clock system in mammals.
8. POSSIBLE CROSSTALK BETWEEN THE CIRCADIAN CLOCK AND CELLULAR PROCESSES THROUGH SHARED POST-TRANSLATIONAL MODIFICATIONS
Reportedly, clock proteins have important physiological roles that are not restricted to their functions as cellular clock regulators. Previous studies in mice and zebrafish reported that PER2 physically interacts with nuclear receptors such as PPARα, REV-ERBα, RORα, and PPARγ to regulate their transcriptional activities [65-67]. Particularly, the PER2-mediated regulation of PPARγ’s transcriptional activity contributes to the control of metabolism in mice [66]. It is tempting to speculate that post-translational modifications of PER2 would regulate the interactions between PER2 and nuclear receptors, contributing to metabolic controls.
Recent studies found that BMAL1 interacts with HIF-1α to regulate the expression of HIF-1α target genes [68, 69]. The BMAL1–HIF-1α complex targets glycolysis genes, suggesting its control over cellular ATP levels. Furthermore, Bmal1-/- mice show reduced life spans with the symptoms of premature aging, implicating BMAL1 in control of aging [70]. Notably, a recent study reported that cellular clocks are not involved in the process of in vitro cellular senescence, suggesting that the BMAL1-mediated control of aging is not dependent on BMAL1’s function in cellular clock control [71]. Particularly, as various studies have reported SIRT1’s roles in aging [72], it is tempting to speculate that BMAL1 acetylation, which is regulated by SIRT1 [58], is involved in the BMAL1-mediated control of aging.
Cellular responses to the UV component of solar light and/or photo-oxidative stress have been proposed to be the evolutionary origin of circadian clocks [4, 73]. In support of this idea, the alteration of a cell’s reduction–oxidation state triggers the transduction of photic signals that regulate circadian clock gene transcription [74-76]. In addition, various studies have implicated the role of core cellular clock components in the regulation of both, the cell cycle and DNA damage responses (DDRs) [4, 73]. Indeed, cellular clocks control the timing of cell proliferation by regulating the expression of key cell cycle genes in mammals and zebrafish [77, 78]. Moreover, post-translational modifications are vital for the regulation of the cell cycle and DDR. SIRT1 and casein kinase 2, already identified as being responsible for the post-translational modifications of clock proteins [22, 23, 58, 79] (Fig. 1), have also been implicated in the post-translational modifications of proteins such as p53 and E-cadherin, which are involved in the cell cycle and DDR [80, 81]. These findings support the hypothesis that circadian clock regulators may be linked to non-circadian physiologies through shared post-translational modifications.
CONCLUSION
It is now clear that the circadian-time dependent regulation of clock proteins’ phosphorylation is important for fine-tuning the period of the circadian clock. A number of kinases, such as CKIε, CK2, GSK3β and JNK, contribute to the circadian oscillation of phosphorylated clock proteins, regulating the clock proteins’ functions. Besides phosphorylation, other posttranslational modifications, such as SUMOylation, acetylation, O-GlcNAcylation and ADP-ribosylation, are essential modulators of clock proteins, regulating their transcriptional activity, subcellular localization, and protein stability. These modifications play a role in controlling the core mechanism of the circadian clock itself as well as the light signaling pathway to the circadian clock. In addition, it has been implicated that the post-translational modifications of clock proteins are import regulators of cellular physiology, apart from circadian timekeeping. The findings of new clock proteins’ post-translational modifications will reveal a yet unappreciated level of regulation within the core mechanism of the circadian clock and cellular physiology.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work was supported in part by:
• The Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research [18KT0068 (J. H.)].
• The grant from the smoking research foundation (J. H.).
• The grants from the cooperative research program of the Institute of Nature and Environmental Technology Kanazawa University [No. 19024].
• The grant from the Komatsu University.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18(3):164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okamura H. Clock genes in cell clocks: Roles, actions, and mysteries. J. Biol. Rhythms. 2004;19(5):388–399. doi: 10.1177/0748730404269169. [DOI] [PubMed] [Google Scholar]
- 3.Sahar S., Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat. Rev. Cancer. 2009;9(12):886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 4.Uchida Y., Hirayama J., Nishina H. A common origin: Signaling similarities in the regulation of the circadian clock and DNA damage responses. Biol. Pharm. Bull. 2010;33(4):535–544. doi: 10.1248/bpb.33.535. [DOI] [PubMed] [Google Scholar]
- 5.Reppert S.M., Weaver D.R. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Schibler U., Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111(7):919–922. doi: 10.1016/S0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap J.C. Molecular bases for circadian clocks. Cell. 1999;96(2):271–290. doi: 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 8.DeBruyne J.P., Weaver D.R., Reppert S.M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 2007;10(5):543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirayama J., Sassone-Corsi P. Structural and functional features of transcription factors controlling the circadian clock. Curr. Opin. Genet. Dev. 2005;15(5):548–556. doi: 10.1016/j.gde.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Gallego M., Virshup D.M. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8(2):139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 11.Yoshitane H., Honma S., Imamura K., Nakajima H., Nishide S.Y., Ono D., Kiyota H., Shinozaki N., Matsuki H., Wada N., Doi H., Hamada T., Honma K., Fukada Y. JNK regulates the photic response of the mammalian circadian clock. EMBO Rep. 2012;13(5):455–461. doi: 10.1038/embor.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y.T., Luo H., Guan W.J., Zhang H., Chen C., Wang Z., Li J.D. O-GlcNAcylation of BMAL1 regulates circadian rhythms in NIH3T3 fibroblasts. Biochem. Biophys. Res. Commun. 2013;431(3):382–387. doi: 10.1016/j.bbrc.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 13.Kaasik K., Kivimäe S., Allen J.J., Chalkley R.J., Huang Y., Baer K., Kissel H., Burlingame A.L., Shokat K.M., Ptáček L.J., Fu Y.H. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17(2):291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowrey P.L., Shimomura K., Antoch M.P., Yamazaki S., Zemenides P.D., Ralph M.R., Menaker M., Takahashi J.S. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288(5465):483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toh K.L., Jones C.R., He Y., Eide E.J., Hinz W.A., Virshup D.M., Ptácek L.J., Fu Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291(5506):1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y., Padiath Q.S., Shapiro R.E., Jones C.R., Wu S.C., Saigoh N., Saigoh K., Ptácek L.J., Fu Y.H. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434(7033):640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 17.Ebisawa T., Uchiyama M., Kajimura N., Mishima K., Kamei Y., Katoh M., Watanabe T., Sekimoto M., Shibui K., Kim K., Kudo Y., Ozeki Y., Sugishita M., Toyoshima R., Inoue Y., Yamada N., Nagase T., Ozaki N., Ohara O., Ishida N., Okawa M., Takahashi K., Yamauchi T. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2(4):342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akashi M., Tsuchiya Y., Yoshino T., Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol. Cell. Biol. 2002;22(6):1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takano A., Isojima Y., Nagai K. Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J. Biol. Chem. 2004;279(31):32578–32585. doi: 10.1074/jbc.M403433200. [DOI] [PubMed] [Google Scholar]
- 20.Yagita K., Yamanaka I., Koinuma S., Shigeyoshi Y., Uchiyama Y. Mini screening of kinase inhibitors affecting period-length of mammalian cellular circadian clock. Acta Histochem. Cytochem. 2009;42(3):89–93. doi: 10.1267/ahc.09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kon N., Sugiyama Y., Yoshitane H., Kameshita I., Fukada Y. Cell-based inhibitor screening identifies multiple protein kinases important for circadian clock oscillations. Commun. Integr. Biol. 2015;8(4): e982405. doi: 10.4161/19420889.2014.982405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier B., Wendt S., Vanselow J.T., Wallach T., Reischl S., Oehmke S., Schlosser A., Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23(6):708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamaru T., Hirayama J., Isojima Y., Nagai K., Norioka S., Takamatsu K., Sassone-Corsi P. CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 2009;16(4):446–448. doi: 10.1038/nsmb.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchida Y., Osaki T., Yamasaki T., Shimomura T., Hata S., Horikawa K., Shibata S., Todo T., Hirayama J., Nishina H. Involvement of stress kinase mitogen-activated protein kinase kinase 7 in regulation of mammalian circadian clock. J. Biol. Chem. 2012;287(11):8318–8326. doi: 10.1074/jbc.M111.308908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kon N., Yoshikawa T., Honma S., Yamagata Y., Yoshitane H., Shimizu K., Sugiyama Y., Hara C., Kameshita I., Honma K., Fukada Y. CaMKII is essential for the cellular clock and coupling between morning and evening behavioral rhythms. Genes Dev. 2014;28(10):1101–1110. doi: 10.1101/gad.237511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Top D., Harms E., Syed S., Adams E.L., Saez L. GSK-3 and CK2 kinases converge on timeless to regulate the master clock. Cell Rep. 2016;16(2):357–367. doi: 10.1016/j.celrep.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang E.E., Liu A.C., Hirota T., Miraglia L.J., Welch G., Pongsawakul P.Y., Liu X., Atwood A., Huss J.W., III, Janes J., Su A.I., Hogenesch J.B., Kay S.A. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139(1):199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada T., Stepniak E., Hui L., Leibbrandt A., Katada T., Nishina H., Wagner E.F., Penninger J.M. Antagonistic control of cell fates by JNK and p38-MAPK signaling. Cell Death Differ. 2008;15(1):89–93. doi: 10.1038/sj.cdd.4402222. [DOI] [PubMed] [Google Scholar]
- 29.Asaoka Y., Nishina H. Diverse physiological functions of MKK4 and MKK7 during early embryogenesis. J. Biochem. 2010;148(4):393–401. doi: 10.1093/jb/mvq098. [DOI] [PubMed] [Google Scholar]
- 30.Yamasaki T., Kawasaki H., Nishina H. Diverse roles of JNK and MKK pathways in the brain. J. Signal Transduct. 2012;2012:459265. doi: 10.1155/2012/459265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamasaki T., Kawasaki H., Arakawa S., Shimizu K., Shimizu S., Reiner O., Okano H., Nishina S., Azuma N., Penninger J.M., Katada T., Nishina H. Stress-activated protein kinase MKK7 regulates axon elongation in the developing cerebral cortex. J. Neurosci. 2011;31(46):16872–16883. doi: 10.1523/JNEUROSCI.1111-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamasaki T., Deki-Arima N., Kaneko A., Miyamura N., Iwatsuki M., Matsuoka M., Fujimori-Tonou N., Okamoto-Uchida Y., Hirayama J., Marth J.D., Yamanashi Y., Kawasaki H., Yamanaka K., Penninger J.M., Shibata S., Nishina H. Age-dependent motor dysfunction due to neuron-specific disruption of stress-activated protein kinase MKK7. Sci. Rep. 2017;7(1):7348. doi: 10.1038/s41598-017-07845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pizzio G.A., Hainich E.C., Ferreyra G.A., Coso O.A., Golombek D.A. Circadian and photic regulation of ERK, JNK and p38 in the hamster SCN. Neuroreport. 2003;14(11):1417–1419. doi: 10.1097/00001756-200308060-00002. [DOI] [PubMed] [Google Scholar]
- 34.Chansard M., Molyneux P., Nomura K., Harrington M.E., Fukuhara C. c-Jun N-terminal kinase inhibitor SP600125 modulates the period of mammalian circadian rhythms. Neuroscience. 2007;145(3):812–823. doi: 10.1016/j.neuroscience.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terman M., Remé C.E., Wirz-Justice A. The visual input stage of the mammalian circadian pacemaking system: II. The effect of light and drugs on retinal function. J. Biol. Rhythms. 1991;6(1):31–48. doi: 10.1177/074873049100600105. [DOI] [PubMed] [Google Scholar]
- 36.Carpenter G.A., Grossberg S. A neural theory of circadian rhythms: Aschoff’s rule in diurnal and nocturnal mammals. Am. J. Physiol. 1984;247(6 Pt 2):R1067–R1082. doi: 10.1152/ajpregu.1984.247.6.R1067. [DOI] [PubMed] [Google Scholar]
- 37.Müller S., Hoege C., Pyrowolakis G., Jentsch S. SUMO, ubiquitin’s mysterious cousin. Nat. Rev. Mol. Cell Biol. 2001;2(3):202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 38.Gill G. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 2003;13(2):108–113. doi: 10.1016/S0959-437X(03)00021-2. [DOI] [PubMed] [Google Scholar]
- 39.Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo J.J., Sassone-Corsi P. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309(5739):1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 40.Lee J., Lee Y., Lee M.J., Park E., Kang S.H., Chung C.H., Lee K.H., Kim K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell. Biol. 2008;28(19):6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen M., Kawamoto T., Yan W., Nakamasu K., Tamagami M., Koyano Y., Noshiro M., Kato Y. Molecular characterization of the novel basic helix-loop-helix protein DEC1 expressed in differentiated human embryo chondrocytes. Biochem. Biophys. Res. Commun. 1997;236(2):294–298. doi: 10.1006/bbrc.1997.6960. [DOI] [PubMed] [Google Scholar]
- 42.Fujimoto K., Shen M., Noshiro M., Matsubara K., Shingu S., Honda K., Yoshida E., Suardita K., Matsuda Y., Kato Y. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem. Biophys. Res. Commun. 2001;280(1):164–171. doi: 10.1006/bbrc.2000.4133. [DOI] [PubMed] [Google Scholar]
- 43.Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419(6909):841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 44.Hong Y., Xing X., Li S., Bi H., Yang C., Zhao F., Liu Y., Ao X., Chang A.K., Wu H. SUMOylation of DEC1 protein regulates its transcriptional activity and enhances its stability. PLoS One. 2011;6(8):e23046. doi: 10.1371/journal.pone.0023046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheung P., Allis C.D., Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103(2):263–271. doi: 10.1016/S0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 46.Peterson C.L., Laniel M.A. Histones and histone modifications. Curr. Biol. 2004;14(14):R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 49.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Ripperger J.A., Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38(3):369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 51.Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sahar S., Sassone-Corsi P. The epigenetic language of circadian clocks. Handb. Exp. Pharmacol. 2013;(217):29–44. doi: 10.1007/978-3-642-25950-0_2. [DOI] [PubMed] [Google Scholar]
- 53.Doi M., Hirayama J., Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125(3):497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 54.Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450(7172):1086–1090. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 55.Hata S., Hirayama J., Kajiho H., Nakagawa K., Hata Y., Katada T., Furutani-Seiki M., Nishina H. A novel acetylation cycle of transcription co-activator yes-associated protein that is downstream of Hippo pathway is triggered in response to SN2 alkylating agents. J. Biol. Chem. 2012;287(26):22089–22098. doi: 10.1074/jbc.M111.334714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 57.Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 58.Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L.P., Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F.W., Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 60.Bellet M.M., Nakahata Y., Boudjelal M., Watts E., Mossakowska D.E., Edwards K.A., Cervantes M., Astarita G., Loh C., Ellis J.L., Vlasuk G.P., Sassone-Corsi P. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc. Natl. Acad. Sci. USA. 2013;110(9):3333–3338. doi: 10.1073/pnas.1214266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanover J.A., Krause M.W., Love D.C. Bittersweet memories: Linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 2012;13(5):312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 62.Hart G.W., Slawson C., Ramirez-Correa G., Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M.D., Ruan H.B., Hughes M.E., Lee J.S., Singh J.P., Jones S.P., Nitabach M.N., Yang X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013;17(2):303–310. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asher G., Reinke H., Altmeyer M., Gutierrez-Arcelus M., Hottiger M.O., Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142(6):943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 65.Schmutz I., Ripperger J.A., Baeriswyl-Aebischer S., Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24(4):345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grimaldi B., Bellet M.M., Katada S., Astarita G., Hirayama J., Amin R.H., Granneman J.G., Piomelli D., Leff T., Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12(5):509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang M., Zhong Z., Zhong Y., Zhang W., Wang H. The zebrafish period2 protein positively regulates the circadian clock through mediation of retinoic acid receptor (RAR)-related orphan receptor α (Rorα). J. Biol. Chem. 2015;290(7):4367–4382. doi: 10.1074/jbc.M114.605022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peek C.B., Levine D.C., Cedernaes J., Taguchi A., Kobayashi Y., Tsai S.J., Bonar N.A., McNulty M.R., Ramsey K.M., Bass J. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 2017;25(1):86–92. doi: 10.1016/j.cmet.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Y., Tang D., Liu N., Xiong W., Huang H., Li Y., Ma Z., Zhao H., Chen P., Qi X., Zhang E.E. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 2017;25(1):73–85. doi: 10.1016/j.cmet.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 70.Kondratov R.V., Kondratova A.A., Gorbacheva V.Y., Vykhovanets O.V., Antoch M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakahata Y., Yasukawa S., Khaidizar F.D., Shimba S., Matsui T., Bessho Y. Bmal1-deficient mouse fibroblast cells do not provide premature cellular senescence in vitro. Chronobiol. Int. 2018;35(5):730–738. doi: 10.1080/07420528.2018.1430038. [DOI] [PubMed] [Google Scholar]
- 72.Ong A.L.C., Ramasamy T.S. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res. Rev. 2018;43:64–80. doi: 10.1016/j.arr.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 73.Kondratov R.V., Gorbacheva V.Y., Antoch M.P. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr. Top. Dev. Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- 74.Hirayama J., Cho S., Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: Catalase represses light-dependent clock gene expression in the zebrafish. Proc. Natl. Acad. Sci. USA. 2007;104(40):15747–15752. doi: 10.1073/pnas.0705614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osaki T., Uchida Y., Hirayama J., Nishina H. Diphenyleneiodonium chloride, an inhibitor of reduced nicotinamide adenine dinucleotide phosphate oxidase, suppresses light-dependent induction of clock and DNA repair genes in zebrafish. Biol. Pharm. Bull. 2011;34(8):1343–1347. doi: 10.1248/bpb.34.1343. [DOI] [PubMed] [Google Scholar]
- 76.Pagano C., Siauciunaite R., Idda M.L., Ruggiero G., Ceinos R.M., Pagano M., Frigato E., Bertolucci C., Foulkes N.S., Vallone D. Evolution shapes the responsiveness of the D-box enhancer element to light and reactive oxygen species in vertebrates. Sci. Rep. 2018;8(1):13180. doi: 10.1038/s41598-018-31570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 78.Dekens M.P., Santoriello C., Vallone D., Grassi G., Whitmore D., Foulkes N.S. Light regulates the cell cycle in zebrafish. Curr. Biol. 2003;13(23):2051–2057. doi: 10.1016/j.cub.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 79.Tsuchiya Y., Akashi M., Matsuda M., Goto K., Miyata Y., Node K., Nishida E. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci. Signal. 2009;2(73):ra26. doi: 10.1126/scisignal.2000305. [DOI] [PubMed] [Google Scholar]
- 80.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17(3):349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 81.Gorospe M., de Cabo R. AsSIRTing the DNA damage response. Trends Cell Biol. 2008;18(2):77–83. doi: 10.1016/j.tcb.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


