Graphical abstract
Abbreviations: AKT3, AKT serine/threonine kinase 3; CCR4-NOT, carbon catabolite repressor 4-negative on TATA; CRISPR-Cas9, clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (cas9); DGCR8, DiGeorge syndrome critical region gene 8; EGFR, epidermal growth factor receptor; GW182 protein, glycine/tryptophan repeats-containing protein with molecular weight of 182 kDa; H3K9, histone H3 lysine 9; Hsp70/90, heat shock proteins 70/90; JEV, Japanese encephalitis virus; KRAS, Kirsten rat sarcoma oncogene; PIWI, P-element-induced wimpy testis; PAZ, PIWI-argonaute-zwille; P4H, prolyl 4-hydroxylase; PAM, protospacer adjacent motif; RISCs, small RNA-induced silencing complexes; TRBP, the transactivating response (TAR) RNA-binding protein; TRIM71/LIN41, tripartite motif-containing 71, known as Lin41; WSSV, white spot syndrome virus
Keywords: Argonaute protein, Small RNA, miRNAs, piRNAs, Post-translational modification
Abstract
Argonaute proteins are highly conserved in almost all organisms. They not only involve in the biogenesis of small regulatory RNAs, but also regulate gene expression and defend against foreign pathogen invasion via small RNA-mediated gene silencing pathways. As a key player in these pathways, the abnormal expression and/or mis-modifications of Argonaute proteins lead to the disorder of small RNA biogenesis and functions, thus influencing multiply biological processes and disease development, especially cancer. In this review, we focus on the post-translational modifications and novel functions of Argonaute proteins in alternative splicing, host defense and genome editing.
Introduction
The gene regulation mediated by small nucleic acid sequences, including small interfering RNA (siRNA), microRNA (miRNA), P-element-induced wimpy testis (PIWI)-interacting RNA (piRNA) and small interfering DNA (siDNA), has demonstrated to be the fundamental principle in regulating many biological processes, including cell proliferation, differentiation and homeostasis [1], [2], [3], [4], [5]. The key protein effector element in those processes is the Argonaute protein family. Argonaute proteins are found in almost all eukaryotes, bacteria and archaea (Table 1) [6].
Table 1.
Argonaute proteins, their guides and functions.
| Argonaute proteins | Types | Host | Guide | Classify | Function | Reference |
|---|---|---|---|---|---|---|
| Ago1-4 | Ago-like family | Homo sapiens | miRNA-guided RNA | eAgos | Interference and/or cleavage target gene | [16], [41] |
| PIWI1-4 | PIWI-like family | Homo sapiens | piRNA-guided RNA | eAgos | Transposon silencing and infertility | [57], [109] |
| Ago1-10 | Ago-like family | Arabidopsis thaliana | miRNA/siRNA-guided RNA | eAgos | Interference and/or cleavage target gene | [110] |
| Ago1-27 | Ago-like family | Caenorhabditis elegans | mRNA/piRNA-guided RNA | eAgos | Interference, cleavage and silencing | [17] |
| TtAgo | Long Ago | Thermus thermophilus | siDNA-guided DNA/RNA | pAgos | Host defence, cleavage, gene-editing | [92] |
| PfAgo | Long Ago | Pyrococcus furiosus | siDNA-guided DNA | pAgos | Host defence, cleavage, interference | [72] |
| MjAgo | Long Ago | Methanocaldococcus jannaschii | siDNA-guided DNA | pAgos | Silencing, cleavage and gene-editing | [15] |
| NgAgo | Long Ago | Natronobacterium gregoryi | siDNA-guided DNA | pAgos | Silencing, interference | [87] |
| MpAgo | Long Ago | Marinitoga piezophila | siRNA-guided RNA/DNA | pAgos | Cleavage ssDNA | [111] |
| TpAgo | Long Ago | Thermotoga profunda | siRNA/miRNA-guided DNA | pAgos | Cleavage ssDNA | [111] |
| AfAgo | Long Ago | Archaeoglobus fulgidus | siRNA-guided mRNA | pAgos | Cleavage mRNA | [112] |
| AaAgo | Long Ago | Aquifex aeolicus | siDNA-guided RNA | pAgos | Cleavage, interference | [113] |
| RsAgo | Long Ago | Rhodobacter sphaeroides | siRNA-guided DNA | pAgos | Host defence and target cleavage | [5] |
Note: miRNA: microRNA; eAgos: eukaryotic Argonaute proteins; siRNA: small interfering RNA; piRNA: P-element-induced wimpy testis (PIWI)-interacting RNA; pAgos: prokaryotic Argonaute proteins; ssDNA: single-stranded DNA.
Eukaryotic Argonaute proteins (eAgos) and prokaryotic Argonaute proteins (pAgos) have very low sequence homology, but their overall architecture and functions are extremely conserved in almost all organisms [7], [8]. Both eAgos and pAgos with catalytic activity have the similar catalytic cycle, mainly including guide binding, target recognition and annealing, and target cleavage and release [9]. However, the structural differences of Argonaute proteins render them to have distinct functions. For example, almost all eAgos and most pAgos guide small RNA to regulate gene-silencing [10], [11], [12], while a small part of pAgos utilize DNA as a guide [9], [13], [14], [15]. However, the amount of Argonaute proteins in each organism is considerably different. For instance, there exist 8 different Argonaute proteins in human beings, including 4 Ago proteins and 4 PIWI proteins [16], while there are 27 Argonaute proteins in Caenorhabditis elegans [17].
Argonaute proteins constitute a main component of the small RNA-induced silencing complexes (RISCs) with small RNAs and/or additional proteins by direct or indirect binding, including Dicer, glycine/tryptophan (GW) repeats-containing 182 protein, heat shock proteins 70/90 (Hsp70/Hsp90) and so on [18], [19], [20], [21]. These complexes not only involve in small RNA biogenesis by Drosha/DiGeorge syndrome critical region gene 8 (DGCR8)-dependent or -independent pathways, but also provide anchor sites for small RNA to regulate gene expression in cytoplasm [2], [12]. In the process of target recognition, Ago proteins can bypass secondary structure and protein barriers to diffuse along the substrate and effectively scan a canonical target through loose protein-nucleic acid interactions and intersegmental transfer [22]. Meanwhile, it also was reported that Argonaute proteins were able to be localized into the nucleus, where engaged in remodeling chromatins through histone modification, DNA methylation, alternative splicing of precursor mRNA, and double-strand DNA break repair processes [3], [23], [24], [25]. In addition, Argonaute proteins are also closely associated with some human cancer [26], [27], [28]. These findings are expanding our understanding of the role of Argonaute proteins beyond gene silencing.
Evolutionary and structural characters of Argonaute proteins
According to the domain architecture, eAgos are divided into four classes: Ago-like family, PIWI-like family, WAGO family and Trypanosoma Ago family, while pAgos are grouped into three, including long pAgos, short pAgos and PIWI-RE proteins [7], [29]. Of these categories, long pAgos contain the very similar domains to eAgos’, including N-terminal (amino-terminal), PAZ (PIWI-Argonaute-Zwille), MID (Middle) and PIWI domains (Fig. 1) [7], [30]. Their overall structure is a two-lobe scaffold, with one lobe comprising the N-PAZ domains and the other consisting of the MID-PIWI domains. The bilobed configuration of Argonaute proteins is connected by Linker 1 (L1) and Linker 2 (L2), and they occur structural rearrangement when binding to small RNA molecules. Unlike long pAgos, short pAgos only consist of the MID and PIWI domains.
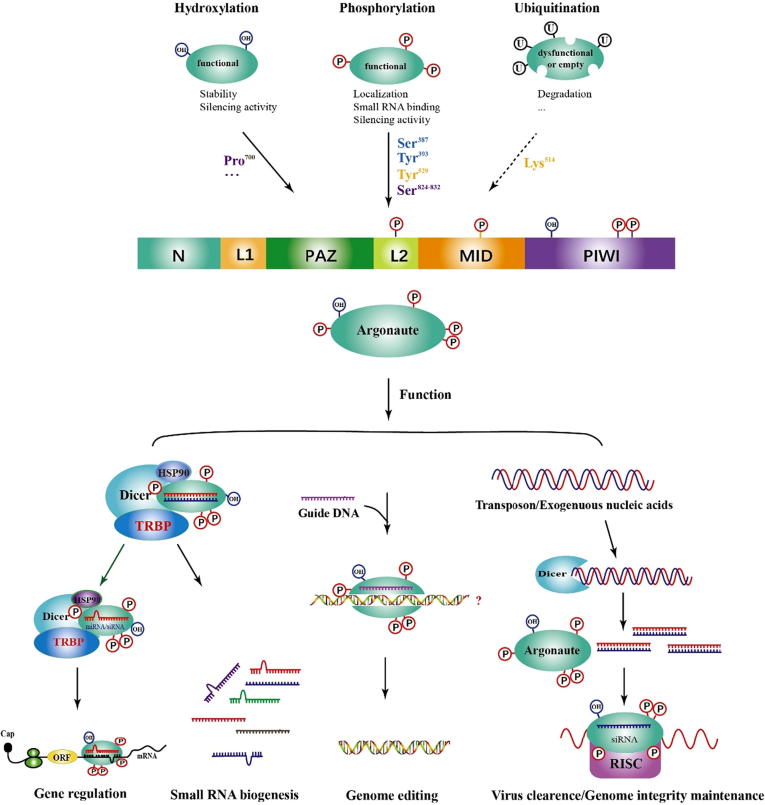
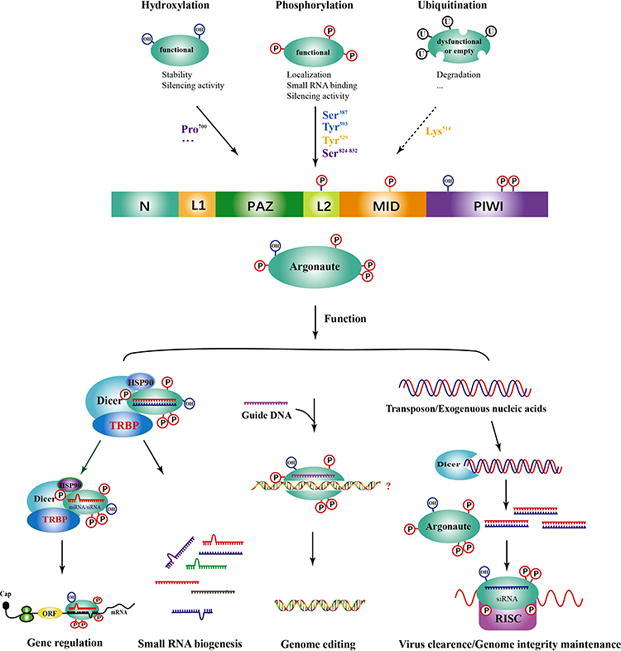
Fig. 1.
Argonaute protein architecture, post-transcriptional modifications and functions Generally, Argonaute proteins contain four domains, including N, PAZ, MID and PIWI. Of them, the N and PAZ domains are connected by Linker 1 (L1) and the rest two connected by Linker 2 (L2). At the post-transcriptional level, functional Argonaute proteins are phosphorylated and/or hydroxylated at specific residues to enhance their silencing activity, stability, binding to small RNAs and so on. However, dysfunctional or empty Argonaute proteins are normally subject to ubiquitylation at Lys514 for degradation. Together with other auxiliary molecules, modified proteins play a role in multiple biological processes, such as gene regulation and genome integrity maintenance. N, N-terminal; PAZ, PIWI-Argonaute-Zwille domain; MID, middle domain; PIWI, P-element-Induced Wimpy testis domain; L1, linker 1; L2, linker 2 and RISC, small RNA-induced silencing complex.
Structural and biochemical studies have revealed that the N-terminal domain is required for deriving duplex small RNAs unwinding and loading a guide strand to assemble mature RISCs [31]. The PAZ domain can non-specifically anchor the 3′ end overhang of small RNA or siDNA into the specific binding pocket of Argonaute proteins (Fig. 1) [32], [33], [34]. The insertion of siRNA/miRNA 3′-hydroxy or methylated piRNA 3′ ends into the PAZ domain is essential for protecting small RNA from degradation [34], [35]. To functionally coordinate with the PAZ domain, the MID domain, being located between the PAZ and PIWI domains, provides an insertion site for the 5′-phosphorylated terminal base of guide RNA through a highly conserved tyrosine in the nucleotide binding pocket [36]. The interaction of the 5′-end nucleotide binding pocket in the MID domain with the phosphorylated small RNA is more critical for guide binding and cleavage activity than the one with the 3′-hydroxyl end nucleotide binding pocket in the PAZ [37]. In some Argonaute proteins, which are structurally similar to ribonuclease H (RNase H), the PIWI domain contains a slicer active site that cleaves target RNA complementary with small RNA [38]. In the cleavage-compatible conformation, several highly conserved amino acid residues, such as DDX/DEDX (X stands for D or H), form a slicer activity center with an Mg2+ cation [15], [39]. Surprisingly, only a part of Argonaute protein members have cleavage activity. For example, only Ago2 possesses catalytic and slicing activity in mammals [40]. However, it has been recently demonstrated that human Ago3 also has cleavage activity through selecting the guide RNA with bearing 5′- and 3′-flanking sequences in the guide-target complementary region [41]. Thus, it is possible that the structural difference of Argonaute proteins may be closely related to small RNA types they bind to and also to binding manners.
Mechanisms of RISC assembly
In the small RNA-mediated gene silencing pathway, the binding process of the guide strand to Argonaute protein is linked to the biogenesis of the guide strand itself. When pre-miRNAs are exported to the cytoplasm, they are cut by Dicer, a kind of RNase III enzyme, to produce a short double-stranded RNA duplex (guide strand and passenger strand). This duplex together with Dicer is directly embedded into an Argonaute protein to recognize and select the guide strand through the thermodynamic asymmetry rule, followed by cleavage and ejection of passenger strand [42]. However, the mechanism how to eject the passenger stand is still elusive.
Together with small RNA, Argonaute proteins compose the core of RISC to repress translation or to enhance the degradation of target mRNA. Previous studies have demonstrated that Argonaute proteins divide the guide-strand RNAs into five distinct function regions: 5′ anchor into the MID domain (g1), the seed sequence of miRNAs (g2-g8), the central region (g9-g12), 3′ complementary sequence (g13–g16), and 3′ tail region into the PAZ pocket (g17-g21/more) [43], [44]. In addition to Argonaute proteins and small RNA, the assembly of RISC also requires the involvement of other key proteins, such as transactivating response RNA-binding protein (TRBP) and Hsp70/Hsp90. TRBP is a dsRNA-binding protein in human and senses the asymmetry and location of RNA to select and load the correct guide strand as R2d2 does in Drosophila melanogaster [45]. The Hsp70/Hsp90 multi-chaperone systems play a major role in protein function and structural activation, being involved in the conformation change of Argonaute proteins in an open and active status by hydrolyzing ATP that allows them to accommodate the RNA duplex [21]. Therefore, it is likely that Dicer, Hsp70/Hsp90 system, Argonaute and dsRNA-binding protein consist of a minimal RISC-loading complex.
Post-translational modification of Argonaute proteins
Like most proteins, Argonaute proteins also undergo post-translational modifications in multiple amino acid residues, mainly including phosphorylation, ubiquitylation and hydroxylation, which affect the functions of Argonaute proteins (Fig. 1).
Phosphorylation
Phosphorylation is the most basic and common mechanism for regulating protein activity and functions based on the signaling cascade effect. Several studies have reported that phosphorylated Argonaute proteins possess the ability to regulate its localization, binding to small RNA and gene silencing activity [46], [47], [48]. For example, human Ago2 protein contains three main phosphorylation regions, including Ser387 and Tyr393 in the L2 region [49], [50], Tyr529 in the MID domain [47] and a Ser824-832 cluster in the PIWI domain [48]. It is noted that hyper-phosphorylated Ago2 protein in humans does not affect miRNA binding, localization or cleavage activity, but hyper-phosphorylation is essential for Argonaute proteins to execute these functions in C. elegans [51], [52]. Although these studies are contradictory, it is clear that the phosphorylation of Argonaute proteins directly affects their localization and functions.
Ubiquitylation
In addition to phosphorylation, Argonaute proteins can also be ubiquitylated. This modification may play a more important role in the degradation of Argonaute proteins than that in their localization, turnover and regulation. In Drosophila, a RING-type E3 ubiquitin ligase can preferentially recognize and eliminate the dysfunctional and empty Argonaute proteins by selective ubiquitination at Lys514 to control the quality of Argonaute proteins and miRNA-mediated silencing [53]. It was also reported that the tripartite motif-containing 71, known as Lin41 (TRIM71/LIN41) in C. elegans and mouse acted as E3 ubiquitin ligase for the ubiquitination of Argonaute proteins [54], [55], but these findings were refuted in a subsequent study [56]. Moreover, in male sterile germ cell lines, deficient mutations in human and mouse PIWI proteins prevent their ubiquitination and degradation, and eventually lead to azoospermia via disrupting histone retention during spermiogenesis [57], [58]. Together, these studies reveal that the ubiquitin–proteasome system mainly regulates the abundance of Argonaute proteins.
Hydroxylation
The hydroxylation modification in proteins is quite essential for their biological roles, which may alter the interactions between proteins and their downstream signaling events. The modification not only occurs at proline residues, but also at lysine, aspartate and histidine residues [59]. Although studies on the hydroxylation in Argonaute proteins are scarce, it is likely to play a role in their stability and activity. In mouse and human, the hydroxylation of Ago2 at Pro700 increases its stability [60]. Meanwhile, it was also reported that hypoxia-induced hydroxylation modification in Ago2 enhanced its endonuclease activity and the association with Hsp90 [61].
The biological functions and associated diseases of Argonaute proteins
As a key player in the small RNA-mediated gene silencing network, Argonaute proteins not only regulate the biogenesis of small RNA, mRNA translation and decay, but also involve in small RNA-guided host defense against exogenous nucleic acids, genome editing and even disease development (Fig. 1).
Biogenesis of Argonaute protein-associated small RNA
It has been known that small RNA exerts the functions in the developmental processes by associating with Argonaute proteins. Of them, miRNAs regulate their target mRNA expression as a post-transcriptional regulator [62]. Generally, most miRNAs are processed from primary transcripts (pri-miRNAs) to form precursor transcripts (pre-miRNAs) in the nucleus by Drosha, which were exported to the cytoplasm via Exportin 5 and further processed into dsRNAs by Dicer. When dsRNAs are successfully loaded into Argonaute proteins in an ATP-dependent manner, the duplexes will be unwound following wedged by the N-terminal domain of Argonaute protein. Subsequently, one guide strand is selected based on the unstably paired 5′ end and thermodynamic asymmetry rule, whereas the passenger strand is cleaved and/or discarded [63], [64]. This process is referred as the canonical pathway. Additionally, there are also other biogenesis pathways of Argonaute protein-associated small RNA, termed as non-canonical pathways. Those small RNA species bypass the canonical Drosha and Dicer processing, such as piRNAs and agotrons [65], [66].
piRNAs
The biogenesis of piRNAs is realized by the loading of PIWI proteins with piRNAs via the ping-pong cycle [67]. Briefly, antisense pri-piRNAs are cleaved by the nuclease Zuc to generate pre-piRNAs with U-preference at the 5′ end. The piRNAs are loaded into PIWI proteins to form cleavage-active piRNA complexes, which recognize, bind and cleave sense pre-piRNAs under the guiding of piRNAs to generate mature sense piRNAs. In subsequent steps, the sense piRNAs are loaded into other PIWI proteins to cleave antisense pre-piRNAs in the same way. In the cycle, sense and/or antisense piRNAs with PIWI proteins silence transposon activity to maintain genome integrity and other mobile genetic elements in germinal cells by targeting the transposon RNA via a miRNA-like pairing rule [68]. Although the ping-pang cycle model provides a reasonable explanation for the generation and functions of piRNAs, the detailed mechanism still remains unclear, such as piRNA initiation and specific target selection.
Agotrons
Agotrons are transcribed from small gene introns with a size of 80–100 nt and exported to the cytoplasm possibly via Exportin 5 [2]. Afterwards, although the agotrons have a complex secondary structure, they are loaded into Argonaute proteins to regulate mRNA expression in a seed-dependent manner [69]. Agotrons are conserved in mammals and act as a miRNA-like regulator with lower off-target or as an Argonaute protein protector by preventing proteasome-mediated degradation due to the high stability after miRNA loading [70]. However, the mechanism and biological functions still need to be further explored.
Argonaute protein-mediated gene regulation
In addition to orchestrating small RNA generation, Argonaute proteins have also been reported to be involved in gene regulation by directly binding and cleaving the targeted RNA or indirectly silencing the translation of the targeted RNA together with additional silencing proteins [71], [72]. For the small RNA that is perfectly complementary with their target genes, mRNA targets will be directly cleaved if Argonaute proteins possess catalytic activity [73], but this phenomenon mostly occurs in plants [74]. In mammals, Argonaute proteins interact with several auxiliary factors to silence the targeted mRNA through the “seed sequence” complementary to miRNAs [62].
The GW protein family containing many glycine-tryptophan (GW) repeats harbors multiple binding sites for Argonaute proteins, and the affinity with Argonaute is increased upon miRNA binding [75]. Meanwhile, it was also demonstrated that a human GW182 protein recruited up to three human Argonaute proteins based on the multiple repeat binding sites in the GW motifs [76]. The GW182 protein is thought to act as a flexible structural scaffold to bridge the interactions between Argonaute proteins and downstream effectors [77]. These effectors mainly include the deadenylase complexes (such as poly A binding protein cytoplasmic [78], [79], carbon catabolite repressor 4-negative on TATA (CCR4-NOT) and poly A Nuclease 2/3 [80]) and the decapping complexes (such as decapping 1/2, maternal expression at 31B and human pluripotency-associated transcripts [81]).
In addition, Ago1 and Ago2 are also present in the nucleus in mammals, and are involved into chromatin modification and alternative splicing [23]. The process mainly implicates the methylation of histone H3 lysine 9 (H3K9) and recruitment of the spliceosome complex to regulate gene expression, but the details of this mechanism are still largely unknown. Therefore, more studies will be required to explore the model for Argonaute protein-mediated chromatin modification and alternative splicing.
Defense mechanisms of Argonaute proteins
The survival of healthy cells depends not only on the integrity and precise replication mechanism of genetic information, but also on the defense mechanisms that prevent the infiltration and invasion of foreign nucleic acids. A number of studies have demonstrated that Argonaute proteins play a crucial role in defense against viral infection and transposon insertion into the genome [5], [68], [82], [83], [84]. For instance, during HIV infection, Ago2 has been shown to bind preferentially to unspliced HIV pre-mRNA to regulate HIV-1 multiply-spliced transcripts and the production of viral particles in a Dicer-independent manner [82].
When Ago1 and 3 are double-knocked out, the susceptibility and mortality of mice to influenza A infection are enhanced [85]. Similarly, the inhibition of Ago1 using isoform-specific siRNAs resulted in greater load of white spot syndrome virus (WSSV) in Marsupenaeus japonicus shrimp [86]. It was also reported that Ago2 was involved in significant inhibition of Japanese encephalitis virus infection in Aedes aegypti (JEV) [84]. Moreover, the NgAgo-siDNA complex efficiently suppressed HBV replication by accelerating pre-viral RNA decay [87]. These studies provide compelling evidence that Argonaute proteins are closely involved in host antiviral infection.
Transposons have the ability to randomly insert or extract themselves into the genome [88]. Although transposon symbiosis may cause DNA damages, living organisms have evolved an elaborative Argonaute-dominating mechanism to reduce the mobility of transposable elements and preserve genome integrity, namely the piRNA-dependent transposon silencing mechanism [66], [68], [89].
Argonaute proteins as a novel genome-editing tool
The recently developed clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR–Cas9) complexes are the most extensively used tool for editing genomes, and can cleave genomic dsDNA sequences based on RNA–DNA hybridization [90], [91]. However, the application of CRISPR-Cas9 is greatly limited due to the high tolerance to guide-target mismatches, relatively easy-to-form RNA-directed secondary structures and the requirement of a protospacer adjacent motif (PAM).
In view of the above-mentioned various characteristics of Argonaute proteins, more and more researchers have begun to speculate whether Argonaute proteins have DNA-editing functions. At present, a growing number of studies have been demonstrated that some pAgos not only bind siDNA to mediate DNA-guided DNA silencing, but also cleave the siDNA cognate single-stranded target DNA in vitro, such as pAgos from Thermus thermophilus (TtAgo), Methanocaldococcus jannaschii (MjAgo) and Pyrococcus furiosus (PfAgo) [15], [72], [92]. However, TtAgo requires the minimum reaction temperature above 65 °C, MjAgo is most active around 85 °C and PfAgo most active between 87 °C and 99 °C. Therefore, the activity of these three pAgos is obviously limited in mammalian cells. Interestingly, the gDNA-Natronobacterium gregoryi Argonaute system (gDNA/NgAgo) was used as a genetic tool to knock out a specific gene fabp11a, eventually causing eye developmental defects in zebrafish [93]. It was also reported that the NgAgo-gDNA system was able to efficiently inhibit HBV replication through gene silencing but not DNA editing [87]. Nevertheless, a recent study showed that NgAgo accelerated homologous arm-guided gene editing rather than relying on its enzymatic activity and gDNA, with the editing efficiency of 80–100% in Pasteurella multocida and Escherichia coli. The principle where NgAgo system works is that the interaction of PIWI-like domain with recombinase A (RecA) enhances RecA-guided DNA strand exchange [94]. Recently, a great progress has been made from two Argonaute proteins from human gastrointestinal bacteria Clostridium perfringens (CpAgo) and Intestinibacter bartlettii (IbAgo), and they are demonstrated to cleave single- and double-stranded DNA sequences at 37 °C in a DNA guide-dependent manner [95]. The discovery raises exciting possibility for developing CpAgo- or IbAgo-based novel tools to accurately edit genomic DNA.
Taken together, it is becoming clear that Argonaute proteins contain nucleic acid-guided nuclease activity, but there are still no characterized Argonaute proteins that show robust genome editing ability.
Argonaute proteins in diseases
A large amount of data has demonstrated that Argonaute proteins are involved in the development of diseases through a miRNA-dependent or -independent way, such as tumorgenesis [96] and infertility [57]. Argonaute proteins, especially human Ago2, have been found to be overexpressed in most cancer, including colon cancer [28], ovarian carcinoma [97], gastric cancer [98], [99] and Gliomas [100]. The upgraduated Ago2 eventually leads to the disbalance of small RNA and facilitates oncogenic miRNAs to repress or activate their target genes by a feed-back regulatory loop in special tissues [97], [98]. Moreover, accumulating studies have reported that more than one-third of human cancers exist a Kirsten rat sarcoma oncogene (KRAS)-oncogenic mutation [101], [102], [103] and the mutant KRAS directly interacts with Ago2 to promote cellular transformation and specifically potentiate the KRAS-mediated tumorigenesis [103]. It also was found that Ago2 was post-translationally modified by some tumor factors, such as AKT serine/threonine kinase 3 (AKT3), epidermal growth factor receptor (EGFR) and prolyl 4-hydroxylase (P4H), to promote cancer cell growth, angiogenesis and metastasis [26]. However, there is an inconsistency that the expression of Ago2 is strongly reduced in melanoma and breast cancer [96], [104], [105]. This discrepancy may be due to the differences in cancer types and miRNAs expression patterns. In addition to a role of Ago2 in cancer, PIWI proteins also play a key role in tumorgenesis. A number of studies have demonstrated that PIWI proteins are not only involved in cancer cell apoptosis and proliferation, but also in cancer metastasis and invasion [106], [107], [108]. It was also reported that the mutations in human PIWI protein caused male infertility [57], [109]. However, whether Argonaute proteins are dysfunctional in these disorders still requires further exploration.
Concluding remarks
Argonaute proteins play an important role in post-translational gene silencing and the biogenesis of small RNA. Accumulating studies have revealed that Argonaute proteins also implicate several other cellular processes, such as host defense, gene regulation and genome editing. Although considerable breakthroughs have been made in the structure and functions, there are still many issues that need to be further addressed. For example, most eAgos trend to target RNA using RNA guides, while pAgos seem to target DNA based on either DNA or RNA guides. What is the basis of the preference and specificity of Argonaute proteins for their guide strands and target genes? Next, it is known that Argonaute proteins without small RNA loading are extremely unstable, and whether these loaded small RNAs protect Argonaute proteins from degradation and why. Furthermore, what are the functions and mechanism of short pAgos?
For Argonaute, another emerging field is the use in genome editing, which has rapidly grown at an unprecedented rate in the past few years. Although some pAgos have been demonstrated to selectively cleave target DNA based on the complementary guide strand, their applications are greatly restricted due to high working temperature required, such as TtAgo and PfAgo. Fortunately, CpAgo and IbAgo give us a new hope for the development of Argonaute-based genome editing tools though a long way to go in clinic applications.
In conclusion, a comprehensive understanding the functions of Argonaute proteins will help us better reveal and explore the causes of disease development. Meanwhile, it is expected to discover more Argonaute proteins with genome-editing functions in future, which will bring the gospel to the treatment of human genetic diseases.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
This work was financially supported by the National Natural Science Foundation of China (31472185 and U1703104) and the National Key Basic Research Program (973 program) of China (2015CB150300).
Author contributions
Yadong Zheng had the idea for the article. Jin’en Wu and Yang Jing performed the literature search and data analysis. Jin’en Wu drafted the work. Yadong Zheng and William C. Cho critically revised the work.
Biography
Yadong Zheng, a parasitologist, received PhD in Molecular Bioscience (2012) from the University of Nottingham, UK. Since 2012, he has been working as Principle Investigator at Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Zheng’s research interests are mainly focused on small RNAs of pathogens in animals and humans and of their hosts, including viruses and parasites, and gene silencing pathways involved: (1) analysis of expression profiling of srRNAs and characterization of their roles in the pathogens’ growth and development, and infection and immunity; (2) investigation of the biogenesis pathways of srRNAs; (3) revelation of miRNA-induced silencing gene regulation networks; (4) assessment of the possibility of miRNAs and their pathways as therapeutic targets. He is Associate Editor of Frontiers in Genetics (2018–), Frontiers in Veterinary Science (2018–) and BMC Veterinary Research (2014–), and member of Young Parasitologist Board of Parasitological Sub-association, China Zoological Society (2011–).
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Sheu-Gruttadauria J., MacRae I.J. Structural Foundations of RNA Silencing by Argonaute. J Mol Biol. 2017;429(17):2619–2639. doi: 10.1016/j.jmb.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daugaard I., Hansen T.B. Biogenesis and Function of Ago-Associated RNAs. Trends Genet. 2017;33(3):208–219. doi: 10.1016/j.tig.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14(7):447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 4.Aalto A.P., Pasquinelli A.E. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol. 2012;24(3):333–340. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol Cell. 2013;51(5):594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolia N.H., Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3(1):36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 7.Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 2014;21(9):743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olina A.V., Kulbachinskiy A.V., Aravin A.A., Esyunina D.M. Argonaute Proteins and Mechanisms of RNA Interference in Eukaryotes and Prokaryotes. Biochemistry (Mosc) 2018;83(5):483–497. doi: 10.1134/S0006297918050024. [DOI] [PubMed] [Google Scholar]
- 9.Lisitskaya L., Aravin A.A., Kulbachinskiy A. DNA interference and beyond: structure and functions of prokaryotic Argonaute proteins. Nat Commun. 2018;9(1):5165. doi: 10.1038/s41467-018-07449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doxzen K.W., Doudna J.A. DNA recognition by an RNA-guided bacterial Argonaute. PLoS ONE. 2017;12(5) doi: 10.1371/journal.pone.0177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn C.D., Joshua-Tor L. Eukaryotic Argonautes come into focus. Trends Biochem Sci. 2013;38(5):263–271. doi: 10.1016/j.tibs.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Azlan A., Dzaki N., Azzam G. Argonaute: The executor of small RNA function. J Genet Genomics. 2016;43(8):481–494. doi: 10.1016/j.jgg.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Willkomm S., Makarova K., Grohmann D. DNA-silencing by prokaryotic Argonaute proteins adds a new layer of defence against invading nucleic acids. FEMS Microbiol Rev. 2018 doi: 10.1093/femsre/fuy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei J., Sheng G., Cheung P.P., Wang S., Li Y., Gao X. Two symmetric arginine residues play distinct roles in Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc Natl Acad Sci U S A. 2019;116(3):845–853. doi: 10.1073/pnas.1817041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat Microbiol. 2017;2:17035. doi: 10.1038/nmicrobiol.2017.35. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki T., Shiohama A., Minoshima S., Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82(3):323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 17.Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127(4):747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 18.Faehnle C.R., Joshua-Tor L. Argonautes confront new small RNAs. Curr Opin Chem Biol. 2007;11(5):569–577. doi: 10.1016/j.cbpa.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D. Structural features of Argonaute-GW182 protein interactions. Proc Natl Acad Sci U S A. 2013;110(40):E3770–E3779. doi: 10.1073/pnas.1308510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi H., Tomari Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta. 2016;1859(1):71–81. doi: 10.1016/j.bbagrm.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Tsuboyama K., Tadakuma H., Tomari Y. Conformational Activation of Argonaute by Distinct yet Coordinated Actions of the Hsp70 and Hsp90 Chaperone Systems. Mol Cell. 2018;70(4):722–729. doi: 10.1016/j.molcel.2018.04.010. e4. [DOI] [PubMed] [Google Scholar]
- 22.Cui T.J., Klein M., Hegge J.W., Chandradoss S.D., van der Oost J., Depken M. Argonaute bypasses cellular obstacles without hindrance during target search. Nat Commun. 2019;10(1):4390. doi: 10.1038/s41467-019-12415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19(10):998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 24.Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149(1):101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Batsche E., Ameyar-Zazoua M. The influence of Argonaute proteins on alternative RNA splicing. Wiley Interdiscip Rev RNA. 2015;6(1):141–156. doi: 10.1002/wrna.1264. [DOI] [PubMed] [Google Scholar]
- 26.Ye Z., Jin H., Qian Q. Argonaute 2: A Novel Rising Star in Cancer Research. J Cancer. 2015;6(9):877–882. doi: 10.7150/jca.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M., Zhang L., Liu Z., Zhou J., Pan Q., Fan J. AGO1 may influence the prognosis of hepatocellular carcinoma through TGF-beta pathway. Cell Death Dis. 2018;9(3):324. doi: 10.1038/s41419-018-0338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L., Yu C., Gao H., Li Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer. 2010;10:38. doi: 10.1186/1471-2407-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegge J.W., Swarts D.C., van der Oost J. Prokaryotic Argonaute proteins: novel genome-editing tools? Nat Rev Microbiol. 2018;16(1):5–11. doi: 10.1038/nrmicro.2017.73. [DOI] [PubMed] [Google Scholar]
- 30.Parker J.S. How to slice: snapshots of Argonaute in action. Silence. 2010;1(1):3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwak P.B., Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19(2):145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- 32.Hock J., Meister G. The Argonaute protein family. Genome Biol. 2008;9(2):210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon B., Kirkpatrick J.P., Eckhardt S., Reuter M., Rocha E.A., Andrade-Navarro M.A. Recognition of 2'-O-methylated 3'-end of piRNA by the PAZ domain of a Piwi protein. Structure. 2011;19(2):172–180. doi: 10.1016/j.str.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Tian Y., Simanshu D.K., Ma J.B., Patel D.J. Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc Natl Acad Sci U S A. 2011;108(3):903–910. doi: 10.1073/pnas.1017762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J.H., Shin S.Y., Shin C. Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. 2017;45(4):1569–1583. doi: 10.1093/nar/gkx029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boland A., Huntzinger E., Schmidt S., Izaurralde E., Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci U S A. 2011;108(26):10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456(7224):921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M., Doudna J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457(7228):405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J. Structure of yeast Argonaute with guide RNA. Nature. 2012;486(7403):368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 41.Park M.S., Phan H.D., Busch F., Hinckley S.H., Brackbill J.A., Wysocki V.H. Human Argonaute3 has slicer activity. Nucleic Acids Res. 2017;45(20):11867–11877. doi: 10.1093/nar/gkx916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 43.Song G., Chen H., Sheng G., Wang Y., Lou J. Argonaute Facilitates the Lateral Diffusion of the Guide along Its Target and Prevents the Guide from Being Pushed Away by the Ribosome. Biochemistry. 2018;57(15):2179–2183. doi: 10.1021/acs.biochem.8b00213. [DOI] [PubMed] [Google Scholar]
- 44.Ma J.B., Ye K., Patel D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429(6989):318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noland C.L., Ma E., Doudna J.A. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43(1):110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng Y., Sankala H., Zhang X., Graves P.R. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413(3):429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 47.Rudel S., Wang Y., Lenobel R., Korner R., Hsiao H.H., Urlaub H. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39(6):2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Golden R.J., Chen B., Li T., Braun J., Manjunath H., Chen X. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542(7640):197–202. doi: 10.1038/nature21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bridge K.S., Shah K.M., Li Y., Foxler D.E., Wong S.C.K., Miller D.C. Argonaute Utilization for miRNA Silencing Is Determined by Phosphorylation-Dependent Recruitment of LIM-Domain-Containing Proteins. Cell Rep. 2017;20(1):173–187. doi: 10.1016/j.celrep.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajgor D., Sanderson T.M., Amici M., Collingridge G.L., Hanley J.G. NMDAR-dependent Argonaute 2 phosphorylation regulates miRNA activity and dendritic spine plasticity. EMBO J. 2018 doi: 10.15252/embj.201797943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quevillon Huberdeau M., Zeitler D.M., Hauptmann J., Bruckmann A., Fressigne L., Danner J. Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 2017;36(14):2088–2106. doi: 10.15252/embj.201696386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Orozco J., Pare J.M., Holme A.L., Chaulk S.G., Fahlman R.P., Hobman T.C. Functional analyses of phosphorylation events in human Argonaute 2. RNA. 2015;21(12):2030–2038. doi: 10.1261/rna.053207.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi H., Shoji K., Kiyokawa K., Negishi L., Tomari Y. Iruka Eliminates Dysfunctional Argonaute by Selective Ubiquitination of Its Empty State. Mol Cell. 2019;73(1):119–129. doi: 10.1016/j.molcel.2018.10.033. e5. [DOI] [PubMed] [Google Scholar]
- 54.Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E.A., Michel G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11(12):1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 55.Hammell C.M., Lubin I., Boag P.R., Blackwell T.K., Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136(5):926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loedige I., Gaidatzis D., Sack R., Meister G., Filipowicz W. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 2013;41(1):518–532. doi: 10.1093/nar/gks1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gou L.T., Kang J.Y., Dai P., Wang X., Li F., Zhao S., Zhang M., Hua M.M., Lu Y., Zhu Y., Li Z., Chen H., Wu L.G., Li D., Fu X.D., Li J., Shi H.J., Liu M.F. Ubiquitination-Deficient Mutations in Human Piwi Cause Male Infertility by Impairing Histone-to-Protamine Exchange during Spermiogenesis. Cell. 2017;169(6):1090–1104. doi: 10.1016/j.cell.2017.04.034. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao S., Gou L.T., Zhang M., Zu L.D., Hua M.M., Hua Y. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24(1):13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 59.Zurlo G., Guo J., Takada M., Wei W., Zhang Q. New Insights into Protein Hydroxylation and Its Important Role in Human Diseases. Biochim Biophys Acta. 2016;1866(2):208–220. doi: 10.1016/j.bbcan.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi H.H., Ongusaha P.P., Myllyharju J., Cheng D., Pakkanen O., Shi Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455(7211):421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu C., So J., Davis-Dusenbery B.N., Qi H.H., Bloch D.B., Shi Y. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31(23):4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czech B., Hannon G.J. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12(1):19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noland C.L., Doudna J.A. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013;19(5):639–648. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 65.Hansen T.B., Veno M.T., Jensen T.I., Schaefer A., Damgaard C.K., Kjems J. Argonaute-associated short introns are a novel class of gene regulators. Nat Commun. 2016;7:11538. doi: 10.1038/ncomms11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamashiro H., Siomi M.C. PIWI-Interacting RNA in Drosophila: Biogenesis, Transposon Regulation, and Beyond. Chem Rev. 2018;118(8):4404–4421. doi: 10.1021/acs.chemrev.7b00393. [DOI] [PubMed] [Google Scholar]
- 67.Czech B., Hannon G.J. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci. 2016;41(4):324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen E.Z., Chen H., Ozturk A.R., Tu S., Shirayama M., Tang W. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell. 2018;172(5):937–951. doi: 10.1016/j.cell.2018.02.002. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen T.B. Detecting Agotrons in Ago CLIPseq Data. Methods Mol Biol. 1823;2018:221–232. doi: 10.1007/978-1-4939-8624-8_17. [DOI] [PubMed] [Google Scholar]
- 70.Stagsted L.V., Daugaard I., Hansen T.B. The agotrons: Gene regulators or Argonaute protectors? BioEssays. 2017;39(4) doi: 10.1002/bies.201600239. [DOI] [PubMed] [Google Scholar]
- 71.Hutvagner G., Simard M.J. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 72.Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015;43(10):5120–5129. doi: 10.1093/nar/gkv415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 74.Mallory A., Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22(12):3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang K., Zhang X., Cai Z., Zhou J., Cao R., Zhao Y. A novel class of microRNA-recognition elements that function only within open reading frames. Nat Struct Mol Biol. 2018;25(11):1019–1027. doi: 10.1038/s41594-018-0136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elkayam E., Faehnle C.R., Morales M., Sun J., Li H., Joshua-Tor L. Multivalent Recruitment of Human Argonaute by GW182. Mol Cell. 2017;67(4):646–658. doi: 10.1016/j.molcel.2017.07.007. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 78.Jinek M., Fabian M.R., Coyle S.M., Sonenberg N., Doudna J.A. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17(2):238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozlov G., Safaee N., Rosenauer A., Gehring K. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J Biol Chem. 2010;285(18):13599–13606. doi: 10.1074/jbc.M109.089540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuzuoglu-Ozturk D., Huntzinger E., Schmidt S., Izaurralde E. The Caenorhabditis elegans GW182 protein AIN-1 interacts with PAB-1 and subunits of the PAN2-PAN3 and CCR4-NOT deadenylase complexes. Nucleic Acids Res. 2012;40(12):5651–5665. doi: 10.1093/nar/gks218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nishihara T., Zekri L., Braun J.E., Izaurralde E. miRISC recruits decapping factors to miRNA targets to enhance their degradation. Nucleic Acids Res. 2013;41(18):8692–8705. doi: 10.1093/nar/gkt619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eckenfelder A., Segeral E., Pinzon N., Ulveling D., Amadori C., Charpentier M. Argonaute proteins regulate HIV-1 multiply spliced RNA and viral production in a Dicer independent manner. Nucleic Acids Res. 2017;45(7):4158–4173. doi: 10.1093/nar/gkw1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seth M., Shirayama M., Tang W., Shen E.Z., Tu S., Lee H.C. The Coding Regions of Germline mRNAs Confer Sensitivity to Argonaute Regulation in C. elegans. Cell Rep. 2018;22(9):2254–2264. doi: 10.1016/j.celrep.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sasaki T., Kuwata R., Hoshino K., Isawa H., Sawabe K., Kobayashi M. Argonaute 2 Suppresses Japanese Encephalitis Virus Infection in Aedes aegypti. Jpn J Infect Dis. 2017;70(1):38–44. doi: 10.7883/yoken.JJID.2015.671. [DOI] [PubMed] [Google Scholar]
- 85.Van Stry M., Oguin T.H., 3rd, Cheloufi S., Vogel P., Watanabe M., Pillai M.R. Enhanced susceptibility of Ago1/3 double-null mice to influenza A virus infection. J Virol. 2012;86(8):4151–4157. doi: 10.1128/JVI.05303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang T., Zhang X. Contribution of the argonaute-1 isoforms to invertebrate antiviral defense. PLoS ONE. 2012;7(11):e50581. doi: 10.1371/journal.pone.0050581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu Z., Tan S., Xu L., Gao L., Zhu H., Ma C. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017;145:20–23. doi: 10.1016/j.antiviral.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 88.Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 89.Iwasaki Y.W., Murano K., Ishizu H., Shibuya A., Iyoda Y., Siomi M.C. Piwi Modulates Chromatin Accessibility by Regulating Multiple Factors Including Histone H1 to Repress Transposons. Mol Cell. 2016;63(3):408–419. doi: 10.1016/j.molcel.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 90.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507(7491):258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qi J., Dong Z., Shi Y., Wang X., Qin Y., Wang Y. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell Res. 2016;26(12):1349–1352. doi: 10.1038/cr.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu L., Xie C., Jin Z., Tu Z., Han L., Jin M. The prokaryotic Argonaute proteins enhance homology sequence-directed recombination in bacteria. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cao Y., Sun W., Wang J., Sheng G., Xiang G., Zhang T. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single-and double-stranded DNA at 37° C. Cell Discovery. 2019;5(1):38. doi: 10.1038/s41421-019-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Voller D., Linck L., Bruckmann A., Hauptmann J., Deutzmann R., Meister G. Argonaute Family Protein Expression in Normal Tissue and Cancer Entities. PLoS ONE. 2016;11(8):e0161165. doi: 10.1371/journal.pone.0161165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vaksman O., Hetland T.E., Trope C.G., Reich R., Davidson B. Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum Pathol. 2012;43(11):2062–2069. doi: 10.1016/j.humpath.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 98.Zhang J., Fan X.S., Wang C.X., Liu B., Li Q., Zhou X.J. Up-regulation of Ago2 expression in gastric carcinoma. Med Oncol. 2013;30(3):628. doi: 10.1007/s12032-013-0628-2. [DOI] [PubMed] [Google Scholar]
- 99.Gao C.L., Sun R., Li D.H., Gong F. PIWI-like protein 1 upregulation promotes gastric cancer invasion and metastasis. Onco Targets Ther. 2018;11:8783–8789. doi: 10.2147/OTT.S186827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng B., Hu P., Lu S.J., Chen J.B., Ge R.L. Increased argonaute 2 expression in gliomas and its association with tumor progression and poor prognosis. Asian Pac J Cancer Prev. 2014;15(9):4079–4083. doi: 10.7314/apjcp.2014.15.9.4079. [DOI] [PubMed] [Google Scholar]
- 101.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23(11):1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aguirre A.J., Hahn W.C. Synthetic Lethal Vulnerabilities in KRAS-Mutant Cancers. Cold Spring Harb Perspect Med. 2018;8(8) doi: 10.1101/cshperspect.a031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shankar S., Pitchiaya S., Malik R., Kothari V., Hosono Y., Yocum A.K. KRAS Engages AGO2 to Enhance Cellular Transformation. Cell Rep. 2016;14(6):1448–1461. doi: 10.1016/j.celrep.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Casey M.C., Prakash A., Holian E., McGuire A., Kalinina O., Shalaby A. Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer. 2019;19(1):712. doi: 10.1186/s12885-019-5884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bellissimo T., Tito C., Ganci F., Sacconi A., Masciarelli S., Di Martino G. Argonaute 2 drives miR-145-5p-dependent gene expression program in breast cancer cells. Cell Death Dis. 2019;10(1):17. doi: 10.1038/s41419-018-1267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xie Y., Yang Y., Ji D., Zhang D., Yao X., Zhang X. Hiwi downregulation, mediated by shRNA, reduces the proliferation and migration of human hepatocellular carcinoma cells. Mol Med Rep. 2015;11(2):1455–1461. doi: 10.3892/mmr.2014.2847. [DOI] [PubMed] [Google Scholar]
- 107.Wang X., Tong X., Gao H., Yan X., Xu X., Sun S. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45(6):2385–2392. doi: 10.3892/ijo.2014.2673. [DOI] [PubMed] [Google Scholar]
- 108.Han Y.N., Li Y., Xia S.Q., Zhang Y.Y., Zheng J.H., Li W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell Physiol Biochem. 2017;44(1):1–20. doi: 10.1159/000484541. [DOI] [PubMed] [Google Scholar]
- 109.Yan W. piRNA-independent PIWI function in spermatogenesis and male fertility. Biol Reprod. 2017;96(6):1121–1123. doi: 10.1093/biolre/iox055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13(7):350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 111.Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J. A bacterial Argonaute with noncanonical guide RNA specificity. Proc Natl Acad Sci U S A. 2016;113(15):4057–4062. doi: 10.1073/pnas.1524385113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parker J.S., Roe S.M., Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434(7033):663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19(3):405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]